Abstract
A novel PCR-restriction fragment length polymorphism analysis of the hsp65 gene was developed. The restriction patterns for Mycobacterium tuberculosis and Mycobacterium avium complex (MAC) species were designed to be highly distinct, and the overall number of restriction patterns was designed to be limited. Four hundred specimens (17 reference strains and 383 clinical isolates) were tested, of which 98 were M. tuberculosis and 132 were MAC species. The assay was virtually 100% sensitive and specific for M. tuberculosis and MAC species. Moreover, it gave highly concordant results for other mycobacterial species other than M. terrae complex species. This assay can be completed in one day and is user-friendly and robust. Therefore, it is highly suitable for large-scale use in a clinical laboratory.
In the past two decades, there has been a dramatic increase in the numbers of diseases caused by Mycobacterium tuberculosis complex and other nontuberculous mycobacteria, in particular, members of the M. avium complex (MAC) (2). This increase is driven mainly by the AIDS pandemic, with both M. tuberculosis and MAC species causing disseminated disease in AIDS patients. Therefore, there is great pressure on clinical laboratories to rapidly and accurately detect and identify clinically important mycobacteria. Conventionally, identification of mycobacteria grown in culture is achieved by standard culture and biochemical tests that are time-consuming and not always accurate (5, 6, 15). Other methods such as high-performance, gas-liquid, and thin-layer chromatographies and DNA sequence analysis of the 16S rRNA gene (rDNA) region are either too labor-intensive, difficult, or expensive for routine use (3, 9, 12, 15, 21). Rapid and simple genotypic assays for the identification of mycobacteria, such as Accuprobe (Gen-Probe Inc., San Diego, Calif.), are available commercially. However, the high costs of these assays have prohibited their large-scale use in most clinical laboratories, especially in developing areas with a high incidence of tuberculosis (14).
PCR-restriction fragment length polymorphism analysis (PRA) is simple to perform, rapid, and economical, features that make it highly attractive for routine clinical laboratories. However, assays for PRA have often been criticized as being difficult to read because of minor differences in patterns between some species that are made worse by gel-to-gel variations. As a result, time-consuming computer-assisted analysis is often required. PRA techniques have been developed for several mycobacterial genes, such as hsp65, the 16S-23S rDNA spacer, and rpoB (7, 13, 20). Of these, the one most investigated and validated is hsp65 (1, 8, 11, 17, 18, 19, 20). However, assays for that gene have been impeded by difficulties such as minor differences in band sizes between some species and the occurrence of new patterns that have not been reported previously (1, 10). In view of this, we decided to redesign an assay for PRA of the hsp65 gene using available DNA sequence data so that M. tuberculosis, MAC species, and other clinically important mycobacteria can be identified with ease. To this end, the restriction patterns of M. tuberculosis and MAC species were designed to be highly distinct and the overall number of possible restriction patterns was designed to be limited; thus, the patterns are highly recognizable.
Development of assay.
Seventy-six hsp65 sequences from 36 different mycobacterial species and subspecies were obtained from GenBank (Table 1). hsp65 sequences for different M. kansasii and M. scrofulaceum subspecies were obtained from other sources (10, 18). All MAC strains had a restriction site at nucleotide position 671. Primers specific for hsp65 were then designed so that digestion of the product with Sau96I gave a unique and highly distinctive pattern for MAC strains. Having designed the primers, 305 other restriction enzymes were screened with Genamics Expression software for suitability for use for PRA. Accordingly, CfoI was chosen as the second restriction enzyme because it gave a unique pattern for M. tuberculosis. Moreover, the CfoI patterns for M. avium, M. intracellulare, and M. scrofulaceum were different, so these species could readily be differentiated from each other. The sizes of the restriction fragments produced by Sau96I and CfoI for each species were then calculated. In all, 8 different restriction patterns were predicted for Sau96I (Table 2) and 10 were predicted for CfoI (Table 3) (fragments which were similar in size were grouped together so that a range of sizes is given, and fragments smaller than 30 bp were excluded). Accordingly, an algorithm was drawn up for PRA (Table 4).
TABLE 1.
Sources of hsp65 sequences used for construction of algorithm
| Organism | Strain |
|---|---|
| M. tuberculosis complex | M15467, S76635, U17957, AD000014, U55825, AL021932 (H37Rv), M17705 (BCG), U55833 (M. bovis), U17925 (M. bovis) |
| M. avium | AF281650, AF126033, AF126030, AF126031, AF234261, U85632, U17922, AF126032, AF281650 |
| M. intracellulare | U85637, U55828, U85638, U85638, U55830, U85636, U85635, U17944, AF126035, AF126034, U55829, U85633, U17943 |
| M. scrofulaceum | U17955 (cluster A), sequences of cluster B genotypes from Swanson et al. (18) |
| M. kansasii | U17947 (subspecies I), sequences of subspecies II, III, IV, and VI from Richter et al. (10) |
| M. gastri | U17931 |
| M. Marinum | U55831, U17949 |
| M. asiaticum | U17921 |
| M. genavense | U17932 |
| M. malmoense | U17948 |
| M. shimoidi | U17956 |
| M. gordonae | U17933, U17934, U17935, U17936, U17938, U17939 |
| M. xenopi | U17959 |
| M. neoaurum | U17950 |
| M. nonchromogenicum | U17951 |
| M. ulcerans | U34034 |
| M. habana | AF129011 |
| M. fortuitum | AF140677, AF140676 |
| M. chelonae | AF071142, AF071141, AF071130 |
| M. smegmetis | AF071138 |
| M. phlei | U17952 |
| M. agri | U17920 |
| M. fallax | U17930 |
| M. rhodesiae | U17954 |
| M. vaccae | U17958 |
| M. chitae | U17929 |
| M. senegalese | AF071137 |
| M. peregenium | AF071136, AF257467 |
| M. mucogenicum | AF071135 |
| M. brumae | AF071129 |
| M. confluentis | AF071132 |
| M. pulveris | U17953 |
| M. abscessus | AF071139, U17927, AF071128 |
| M. terrae | AF257468 |
| M. simiae | AF247570, AF247569 |
TABLE 2.
Sau961 restriction patterns
| Type | Fragment(s) size (bp) |
|---|---|
| A | 102, 81, 36–75 |
| B | 117, 102, 54–66 |
| C | 138–145, 93–102, 54 |
| D | 183, 57, 54 |
| E | 183, 75, 36 |
| F | 183, 111 |
| G | 219–240, 54–75 |
| H | 294 |
TABLE 3.
CfoI restriction patterns
| Type | Fragment size (bp) |
|---|---|
| a | 103, 83, 36 |
| b | 122, 57, 50 |
| c | 122, 83, 50 |
| d | 122, 83, 72 |
| e | 122, 89, 63 |
| f | 122, 100, 50–72 |
| g | 172–180, 53–63, 39–50 |
| h | 172–194, 83 |
| i | 172, 100 |
| j | 211, 63 |
TABLE 4.
Algorithm for PRA of the hsp65 gene
| Pattern with Sau96I | Pattern with CfoI | Species |
|---|---|---|
| A | g | M. simiae, M. habana |
| B | g | M. scrofulaceuma |
| h | M. avium | |
| i | M. intracellure | |
| C | g | M. marinum |
| h | M. mucogenicum | |
| j | M. pulveris | |
| D | h | M. genavense |
| E | b | M. kansasii type I and II, M. gastri |
| g | M. asiaticum, M. kansasii type III, M. gordonae, M. scrofulaceum type B | |
| F | b | M. kansasii type IV |
| g | M. gordonae, M. terrae, M. scrofulaceumb | |
| G | a | M. neoaurum |
| c | M. fortuitum, M. senegalense | |
| d | M. tuberculosis complex | |
| e | M. shimoidi | |
| f | M. abscessus | |
| g | M. scrofulaceum,b M. kansasii type VI, M. ulcerans, M. szulgai, M. marinum, M. flavescens, M. malmoense, M. chitae | |
| h | M. fortuitum, M. smegmatis, M. nonchromogenicum, M. phlei, M. fallax, M. perigenicum, M. brumae | |
| H | c | M. rhodesiae |
| f | M. abscessus | |
| g | M. gordonae, M. scrofulaceum,b M. xenopi, M. agri | |
| h | M. vaccae, M. aurum, M. confluentis | |
| i | M. chelonae |
Specimens.
A total of 400 isolates consisting of 17 American Type Culture Collection (ATCC) reference strains and 383 clinical isolates were used in the study (Table 5). All clinical isolates were from the Tuberculosis Reference Laboratory, Yung Fung Shee Memorial Center, Hong Kong, and were identified by standard laboratory methods (5).
TABLE 5.
Reference strains and clinical isolates used in the study
| Mycobacterium species | Reference strain(s) | No. of clinical isolates |
|---|---|---|
| M. tuberculosis complex | H37Rv (M. tuberculosis), ATCC 35720 (M. bovis) | 96 |
| MAC species | ATCC 13950 (M. intracellulare) | 131 |
| M. scrofulaceum | ATCC 19981 | 23 |
| M. kansasii | ATCC 12478 | 22 |
| M. gordonae | ATCC 14470 | 21 |
| M. terrae | 37 | |
| M. szulgai | ATCC 35799 | 2 |
| M. marinum | ATCC 927 | 2 |
| M. simiae | 1 | |
| M. flavescens | ATCC 14474 | 1 |
| M. asiaticum | ATCC 25276 | |
| M. gastri | ATCC 15754 | |
| M. lentiflavum | 4a | |
| M. fortuitum | ATCC 6841 | 30 |
| M. chelonae | ATCC 14472 | 13 |
| M. aurum | ATCC 23366 | |
| M. neoaurum | ATCC 25795 | |
| M. chitae | ATCC 19627 | |
| M. smegmatis | ATCC 19420 |
Identified by 16S rDNA sequencing (16).
Assay conditions.
A loopful of a bacterial colony was suspended in 400 μl of distilled water, and the suspension was boiled for 5 min. The suspension was then centrifuged at 13,000 rpm for 5 min, and the supernatant was used for PCR amplification. A 294-bp region of the hsp65 gene was amplified with primers HSP-1 (5′-GCCAAGAAGACCGAYGACGT) and HSP-2 (5′-GGTGATGACGCCCTCGTTGC). PCR was carried out in a final volume of 50 μl consisting of 5 μl of the DNA preparation, each primer at a concentration of 0.2 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2, and 1.25 U of Taq polymerase (all reagents were from Pharmacia-Biotech, Freiburg, Germany). The thermal profile consisted of an initial denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 30s, 62°C for 30s, and 72°C for 1 min and a final period of extension of 6 min at 72°C. The amplicon was digested with 1 U of Sau96I and CfoI (both from Boehringer Mannheim Biochemicals, Mannheim, Germany) in separate reactions. The restriction digests were carried out with 10 μl of the amplicon at 37oC for at least 1 h. The digests were then electrophoresed in 3% Metaphor agarose gel (FMC Bioproducts, Rockland, Maine) with ethidium bromide. The fragment sizes were determined visually by comparison with the DNA V marker (Boehringer Mannheim).
Selected specimens which gave discrepant results by biochemical and genotypic tests were further investigated by sequencing hsp65 and the hypervariable region of 16S rDNA (12). The hsp65 sequences of reference strains M. aurum ATCC 23366, M. flavescens ATCC 14474), and M. szulgai ATCC 35799 (GenBank accession numbers AF350414, AF350413, and AF350412, respectively) were determined; and these species were added to the algorithm accordingly.
All mycobacterial isolates were amplified by the primers without any problems. During the study, 7 of 8 predicted Sau96I restriction patterns and 8 of 10 predicted CfoI restriction patterns were seen (Fig. 1). All could readily be identified by eye, without computer assistance. Moreover, the patterns could readily be recognized in the presence of gel artifacts such as a marked “smiling effect.” Digestion of M. tuberculosis amplicons with CfoI produced a characteristic restriction pattern (pattern d). All M. tuberculosis clinical isolates and the two reference strains were correctly identified by PRA. None of the other 302 non-M. tuberculosis isolates were identified as M. tuberculosis by PRA. Digestion of MAC amplicons with Sau96I produced a characteristic restriction pattern (pattern B). Digestion with CfoI allowed differentiation between M. avium (pattern h), M. intracellulare (pattern i), and M. scrofulaceum (pattern g). Reference strain ATCC 13950 was correctly identified by PRA. Of the 131 clinical isolates, 28 were identified as M. avium and 97 were identified as M. intracellulare. 16S rDNA sequencing was carried out for 6 M. avium isolates and 13 M. intracellulare isolates, and the results concurred with those of PRA. Of the six isolates identified as non-MAC mycobacteria by PRA, one was found to be M. fortuitum, one was found to be M. simiae, and the other four could not be matched definitively to any Mycobacterium species in GenBank. Of the 268 non-MAC isolates, 2 M. scrofulaceum clinical isolates, 1 M. terrae complex clinical isolate, and 1 M. kansasii clinical isolate were identified as M. intracellulare by PRA; and the results were confirmed by 16S rDNA sequencing. All other reference strains had PRA patterns consistent with that of the algorithm (Table 5). Most of the PRA results were consistent with the biochemical results with the exception of those for isolates of the M. terrae complex, for which the PRA profiles were very heterogeneous.
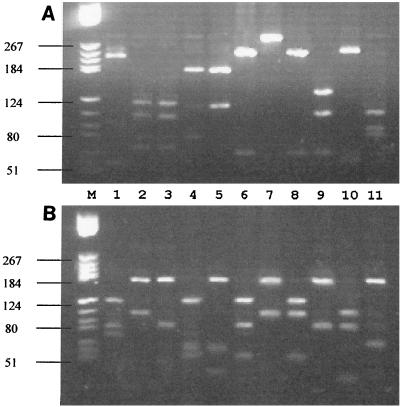
FIG. 1.
PRA profiles for different mycobacterial species. (A) Sau96I digests; (B) CfoI digests. Lanes (in the designations after the strain names, the corresponding Sau96I patterns are presented as capital letters and the CfoI patterns are presented as lowercase letters): M, DNA V marker (Boehringer Mannheim); 1, M. tuberculosis complex (Gd); 2, M. intracellulare (Bi); 3, M. avium (Bh); 4, M. kansasii (Eb); 5, M. gordonae (Fg); 6, M. fortuitum (Gc); 7, M. chelonae (Hi); 8, M. abscessus (Gf); 9, M. mucogenicum (Ch); 10, M. neoaurum (Ga); 11, M. simiae (Ag). Marker positions are indicated on the left (base pairs).
We sought to develop a molecular biology-based method for the identification of commonly isolated mycobacteria for large-scale use in a routine clinical laboratory. For this purpose, the assay should be highly validated, economical, and easy to perform, read, and interpret. Accordingly, the hsp65 gene was chosen for use in the assay because it is the best-investigated gene other than 16S rDNA for taxonomic purposes. Furthermore, it is already used by a well-established PRA (1, 20). One salient feature of our assay is the small number of bands present in different restriction patterns, which makes the results much easier to read but which results in reduced discriminatory power compared to other those of PRAs. However, unlike other PRAs for mycobacteria, this assay was never intended to be a catchall assay for all mycobacteria but was designed so that the most commonly isolated mycobacteria, in particular, M. tuberculosis and MAC species, could be identified with ease. To this end, the use of frequently cutting enzymes such as HaeIII was purposefully avoided because it would have generated too many patterns. The use of HaeIII in other PRAs was understandable because of the desire to identify as many species as possible. We believe that this approach is potentially hazardous in view of the fact that many species are rarely encountered in a routine laboratory and thus are not well investigated. The heterogeneity of our M. terrae complex isolates convinced us of the correctness of our approach.
In use, we found this assay to be virtually 100% sensitive and specific for M. tuberculosis and MAC species. Although the numbers of isolates tested are small, the initial results for mycobacteria other than those belonging to the M. terrae complex were encouraging. The heterogeneity of the M. terrae complex isolates was expected and had been reported in other studies (6, 7, 21). The PRA described here could be completed on the same day that specimens were received and is cost-effective compared to other assays (4): its cost when used on a regular basis is estimated to be US$1.50 per sample, and it requires 3.5 min of technical time per sample (for a batch size of 40 to 80 specimens). It is based on openly available DNA sequence data from a well-researched mycobacterial gene. Above all, it is user-friendly and robust, and it is therefore highly suitable for large-scale use in a routine clinical laboratory. Moreover, the cost can be further reduced by the use of CfoI only for the identification of M. tuberculosis. Since M. tuberculosis and MAC species are the most important mycobacterial pathogens and account for more than 90% of our mycobacterial isolates, early identification of these organisms is of great clinical and public health importance. We anticipate carrying out this assay with more than 10,000 specimens per year.
Acknowledgments
We thank Kent Lai, Jacky Cheng, and other colleagues at the TB Reference Laboratory, Yung Fung Shee Memorial Center, for technical assistance with this assay. We also thank Anna Ng and Viola Tung at the Public Health Laboratory, Kowloon Hospital, for carrying out the DNA sequencing, and Margaret Chan, director of health, HKSAR Government, for permission to publish the manuscript.
REFERENCES
- 1.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkinham J O. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glickman S E, Kilburn J O, Butler W R, Ramos L S. Rapid identification of mycolic acid patterns of mycobacteria by high-performance liquid chromatography using pattern recognition software and a Mycobacterium library. J Clin Microbiol. 1994;32:740–745. doi: 10.1128/jcm.32.3.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kam K M, Yip C W, Chan M Y, Mok C Y, Wong W S. IS6110 dot blot hybridization for the identification of Mycobacterium tuberculosis complex. Diagn Microbiol Infect Dis. 1999;33:13–18. doi: 10.1016/s0732-8893(98)00136-9. [DOI] [PubMed] [Google Scholar]
- 5.Kent P T, Kubica G P. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 6.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Park H J, Cho S N, Bai G H, Kim S J. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000;38:2966–2971. doi: 10.1128/jcm.38.8.2966-2971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pai S, Esen N, Pan X, Musser J M. Routine rapid Mycobacterium species assignment based on species-specific allelic variation in the 65-kilodalton heat shock protein gene (hsp65) Arch Pathol Lab Med. 1997;121:859–864. [PubMed] [Google Scholar]
- 9.Patel J B, Leonard D G B, Pan X, Musser J M, Berman R E, Nachamkin I. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000;38:246–251. doi: 10.1128/jcm.38.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter E, Niemann S, Rüsch-Gerdes S, Hoffner S. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J Clin Microbiol. 1999;37:964–970. doi: 10.1128/jcm.37.4.964-970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ringuet H, Akoua-Koffi C, Honore S, Varnerot A, Vincent V, Berche P, Gaillard J L, Pierre-Audigier C. hsp65 sequencing for identification of rapidly growing mycobacteria. J Clin Microbiol. 1999;37:852–857. doi: 10.1128/jcm.37.3.852-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogall T, Flohr T, Bottger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 13.Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt R M, Habicht M, Fischer M, Mausch H. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S–23S rRNA gene spacer and restriction endonculeases. J Clin Microbiol. 2000;38:1094–1104. doi: 10.1128/jcm.38.3.1094-1104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snider D E, Jr, Raviglione M, Kochi A. Global burden of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 3–11. [Google Scholar]
- 15.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Springer B, Wu W K, Bodmer T, Haase G, Pfyffer G E, Kroppenstedt R M, Schroder K H, Emler S, Kilburn J O, Kirschner P, Telenti A, Coyle M B, Bottger E C. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996;34:1100–1107. doi: 10.1128/jcm.34.5.1100-1107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steingrube A V, Gibson J L, Brown B A, Zhang Y, Wilson R W, Rajagopalan M, Wallace R J. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J Clin Microbiol. 1995;33:149–153. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson D S, Pan X, Musser J M. Identification and subspecific differentiation of Mycobacterium scrofulaceum by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. J Clin Microbiol. 1996;34:3151–3159. doi: 10.1128/jcm.34.12.3151-3159.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor T B, Patterson C, Hlae Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila M-L. Separation among species of Mycobacterium terrae complex by lipid analysis: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998;36:499–505. doi: 10.1128/jcm.36.2.499-505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]