Abstract
Background:
The carboxylation status of Osteocalcin (Ocn) not only influences formation and structure in bones but also has important endocrine functions affecting energy metabolism and expenditure. In this study, the role of γ-carboxylation of the glutamate residues in the structure-dynamics-function relationship in Ocn is investigated.
Methods:
Three forms of Ocn, differentially carboxylated at the Glu-17, 21 and 24 residues, along with a mutated form of Ocn carrying Glu/Ala mutations, are modeled and simulated using molecular dynamics (MD) simulation in the presence of calcium ions.
Results:
Characterization of the global conformational dynamics of Ocn, described in terms of the orientational variations within its 3-helical domain, highlights large structural variations in the non-carboxylated osteocalcin (nOcn). The bi-carboxylated Ocn (bOcn) and tri-carboxylated (tOcn) species, in contrast, display relatively rigid tertiary structures, with the dynamics of most regions strongly correlated. Radial distribution functions calculated for both bOcn and tOcn show long-range ordering of the calcium ion distribution around the carboxylated glutamate (γGlu) residues, likely playing an important role in promoting stability of these Ocns. Additionally, the same calcium ions are observed to coordinate with neighboring γGlu, better shielding their negative charges and in turn stabilizing these systems more than do the singly coordinating calcium ions observed in the case of nOcn. bOcn is also found to exhibit a more helical C-terminal structure, that has been shown to activate its cellular receptor GPRC6A, highlighting the allosteric role of Ocn carboxylation in modulating the stability and binding potential of the active C-terminal.
Conclusions:
The carboxylation status of Ocn as well and its calcium coordination appear to have a direct influence on Ocn structure and dynamics, possibly leading to the known differences in Ocn biological function.
General significance:
Modification of Ocn sequence or its carboxylation state may provide the blueprint for developing high-affinity peptides targeting its cellular receptor GPRC6A, with therapeutic potential for treatment of metabolic disorders.
1. Introduction
Osteocalcin (Ocn), an abundant single chain protein produced by osteoblasts, forms an important component of the extracellular bone matrix [1–3]. Ocn consists of 40–50 amino acid residues depending on the species, and is highly conserved among vertebrates [4]. Post-translational modification by a vitamin K-dependent carboxylase produces three γ-carboxyglutamic acid (γGlu) residues at positions 17, 21 and 24 in human Ocn, that are found to associate with Ca2+ in hydroxyapatite, thus contributing its extracellular matrix function to regulate bone formation [5,6].
In vivo studies have shown that mineral-bound Ocn is predominantly present in the tricarboxylated form (tOcn), with a small amount found circulating in the blood [7–13]. In vivo studies in mice and cell-based in vitro studies support a role of Ocn in bone formation [14]. Ocn is purported to also have important endocrine functions, affecting energy metabolism and male fertility [15–20]. Ocn released from bone has been proposed to function as a hormone through the binding to, and activation of, GPRC6A, a class-C G-protein coupled receptor in target tissues [21–24]. Ocn activation of GPRC6A directly regulates signaling pathways and metabolic processes controlling glucose and fat metabolism in multiple tissues, including liver hepatocytes [25], skeletal muscle [26,27], pancreatic β-cells [24,28–30], and testicular Leydig cells [20]. GPRC6A has also been shown to be directly activated by an Ocn-derived C-terminal hexapeptide (Ocn-6aa-C), that have been proposed to bind in the GPRC6A allosteric binding pocket [28], as well as an N-terminal 15 amino acid fragment of Ocn present in the blood with the sequence NH2-YLGASVPSPDPLEP-COOH [31]. These findings suggest that both bicarboxylated and noncarboxylated forms of Ocn (bOcn and nOcn respectively), but not tOcn, act as bone-derived multifunctional hormones [15–17,20]. Decarboxylation of tOcn has been shown to take place in vivo in bone resorption lacunae through acidification of the extracellular matrix operated by osteoclasts, a process under the control of insulin signaling in osteoblasts [16], shifting the target tissue of Ocn from the bone to extra-osseous organs.
The three forms of Ocn seem to share a similar 3-dimensional tertiary structure, but may have differing biological functions, as discussed above. Previous studies using circular dichroism (CD) and nuclear magnetic resonance (NMR) have shown that without Ca2+ bound, Ocn is predominantly a random coil, but were indicative of a folded protein in the presence of Ca2+ [32–36]. Previous computational studies also predicted both tOcn and bOcn to explore a larger conformational space in the absence of Ca2+ ions [37]. High-resolution structures from NMR and X-ray crystallography of porcine and fish tOcn, respectively, show a fold characterized by a protein core formed by three α-helical segments stabilized by a disulfide salt-bridge between two cysteines, with Ca2+ binding to the three γGlu residues [38,39]. These γGlu residues occupy positions complementary to the Ca2+ sites presented in the hydroxyapatite crystal, involved in the formation of a high-affinity mineral-protein complex in bones [5,33,36,38,40]. A recently available crystal structure of bovine nOcn shows a similar structure in the presence of calcium ions, along with highly flexible N- and C-terminal regions [41]. Although most of these data demonstrate that mature Ocn is composed of an α-helical domain in the presence of Ca2+, there is still some uncertainty regarding the helical and unstructured random coil content in different carboxylated forms of Ocn. Data on human Ocn is also still inconclusive with regard to the role of the protein, as most studies do not account for the influence of vitamin K or differentiate between the different γ-carboxylated forms. Furthermore, the carboxylation state of the active form of Ocn that interacts with the receptor GPRC6A still remains controversial and conflicting results exist regarding both the structure and function of the different forms, leading to uncertainty over their exact biological functions [3,19,42–45]. The three forms of Ocn can thus be expected to exhibit different conformational properties that are likely linked with different functions.
In the present article, molecular dynamics (MD) simulations, totaling 1.5 μs in duration, are used to sample the dynamics of different species of Ocn in solution and in the presence of Ca2+ ions, shedding light on differences in their secondary structure as well as tertiary structure arrangement of the three constituent helices. The role of Ca2+ is further characterized in this study in stabilizing multiple negative charges of γ-carboxylated glutamates present in close proximity. To delineate the role of the extra negative charges in the modified Ocn, simulations of the protein with Glu/Ala mutations have also been carried out. In order to identify the Ocn form showing the most stable/helical structure for the 6 amino acid C-terminus proposed to bind and activate GPRC6A [28], the dynamics of the flexible C-terminal region present in the different forms of Ocn is characterized in terms of fluctuations observed in the individual residues as well as the helical content of the peptide sampled during the simulation, and compared to the dynamics of the isolated Ocn-6aa-C.
2. Methods
In order to characterize the conformational dynamics of the different carboxylated forms of Ocn, models of the different forms were first constructed, and atomistic simulations carried out. The different steps involved in the modeling and the simulations are provided below.
2.1. Ocn multiple sequence alignment and homology modeling
The multiple sequence alignment, using the human Ocn protein sequence as the query sequence, was performed using HHblits [46] with an E-value of e−10 and using 4 iterations for alignment enrichment. The alignment was further filtered to remove sequences that did not cover at least 75% of the query sequence or with greater than 75% gaps. A graphical representation of the alignment was generated using Jalview [47].
To investigate the role of carboxylation on the structure and dynamics of Ocn, structural models of human Ocn were constructed. Bovine Ocn, for which the crystal structure is available (PDB: 4MZZ) [41], was used as the main template for modeling the human Ocn structure. Missing regions in the N-terminal (res 13–16) and C-terminal (res 49) of the bovine Ocn template structure were modeled using the crystal structure of porcine Ocn (PDB: 1Q8H) [38]. N-terminal residues 1–12 were not present in the generated models as the bovine/porcine Ocn template structures do not contain this flexible region. Ten main-chain models (res 13–49) with ten sidechain conformers per main-chain model were generated for the main template using MOE-2013 [48] homology modeling facility with the CHARMM27 force-field [49]. The best-scoring homology model generated by using bovine Ocn as a template was selected for further MD studies based on the predicted GB/VI scores [50] that ranks the models based on calculated Generalized Born interaction energies. The electrostatic surface for the generated model was also calculated in MOE-2013 and the secondary structure was calculated using DSSP [51] and STRIDE [52].
bOcn and tOcn were obtained from the nOcn homology model by replacing the γH atoms of the glutamate residues with a carboxylate group in MOE, generating the γ-carboxylated glutamate residues at the 17, 21 and 24 positions for the tOcn, and at the 21 and 24 positions for the bOcn. Mutated Ocn (mutOcn) was generated by replacing the 3 Glu residues with alanine. The Ocn-derived C-terminal hexapeptide, Ocn-6aa-C, consisting of 6 residues, was obtained from the nOcn homology model by deleting the rest of the protein in MOE.
2.2. MD simulations
The topology for the γGlu residue was generated using CHARMM General Force Field (CGenFF) [53]. With the γH atom being replaced by the carboxylate group, the total charge of the residue was set to (−2). The bonded, angle, dihedral, improper and non-bonded parameters for all the atom types in the residue and CMAP corrections (correction to the backbone dihedral energy) were derived from the CHARMM36 protein parameters for the Glu residue.
CHARMM-GUI [54] was used to build the simulation systems for nOcn, bOcn, tOcn, mutOcn and Ocn-6aa-C models. A disulfide bond was built between Cys-23 and Cys-29, residues known to form this bond in other species of Ocn. A rectangular water box was built using a water thickness of 10 Å around the edges of the protein. The choice of Ca2+ concentrations was based on two considerations: (i) higher than the normal Ca2+ concentration in blood (2.25–2.75 mM) [55] was taken to mimic the interactions between Ocn and the calcium-sensing receptor GPRC6A [56], as the proper interactions between Ocn and GPRC6A can be expected to take place in an increased local concentration of Ca2+ [57]; (ii) minimum concentration of Ca2+ needed to saturate all negatively-charged residue side-chains of Ocn. Here, the maximum negative charge (−12) observed in the case of tOcn was used to calculate the number of ions added to the system. The same ionic concentration of Ca2+ ions (as taken for tOcn) was used for all other systems to maintain consistency. It is to be noted that the ionic concentration of Ca2+ used in the study is higher than that observed under physiological conditions. As such, higher ionic concentrations can be used to enable interactions of ions with the protein in the limited timescales of the MD simulations that would not allow ample sampling otherwise. Cl− ions were also added accordingly to neutralize the extra positive charges in the different systems. The details of the setup for the different systems are summarized in Table 1.
Table 1.
Simulation setup for MD simulations. The details for the systems constructed for different forms of Ocn is provided. The Ca2+ concentration was calculated based on a cubic box with the side length of 52 Å in the case of the first 4 systems and 39 Å in the case of Ocn-6aa-C.
| System | Number of Atoms | Number of water atoms | Number of ions | Ionic concentration (M) | Simulation time (ns) |
|---|---|---|---|---|---|
| nOcn | 12,120 | 11,505 | 12 Ca2+; 19 Cl− |
0.14 Ca2+; 0.22 Cl− |
300 |
| bOcn | 12,152 | 11,535 | 12 Ca2+; 17 Cl− |
0.14 Ca2+; 0.20 Cl− |
300 |
| tOcn | 12,150 | 11,532 | 12 Ca2+; 16 Cl− |
0.14 Ca2+; 0.19 Cl− |
300 |
| mutOcn | 12,168 | 11,565 | 12 Ca2+; 22 Cl− |
0.14 Ca2+; 0.26 Cl− |
300 |
| Ocn-6aa-C | 5464 | 5343 | 5 Ca2+; 11 Cl− |
0.14 Ca2+; 0.31 Cl− |
300 |
The system size and cell origin were calculated with VMD-1.9.2 [58], and periodic boundary conditions were applied using these values. A switching distance of 10 Å, a cut-off value of 12 Å and a pair-list distance of 14 Å were used to calculate non-bonded interactions. A time-step of 2 fs was used in all simulations. A constant temperature of 300 K was maintained throughout the simulations using Langevin dynamics [59]. Particle Mesh Ewald (PME) [60,61] was used for calculation of the electrostatic interactions under periodic boundary conditions and the simulations were performed in the NVT ensemble.
The MD simulations were performed using NAMD [62] on the Newton high performance cluster at the University of Tennessee. All simulations used the CHARMM-36 force field parameters for the protein [49] and ions, TIP3 model for water molecules and the generated parameters for the γGlu residues. Two equilibrium steps were performed as follows. In the first step, the protein was restrained and the water molecules/ions were energy minimized for 5000 steps using the conjugate gradient method [63] and further equilibrated for 0.5 ns. In the second step, restraints on the protein were removed and the system was energy minimized for another 2500 steps (5 ps), allowing the protein to relax in the water box, and further equilibrated for 0.5 ns. For the Ocn-6aa-C system, only the second energy minimization/equilibration step was performed. Finally, 300 ns of trajectory was generated in the production run for each of the 5 systems.
2.3. Analysis
The conformational dynamics of different carboxylated forms of Ocn was characterized in terms of the local secondary structure changes as well as in the global motions in its tertiary structure. Additionally, the role of the Ca2+in modulating the conformational ensemble of the different Ocn forms was elucidated by calculation of the ionic distribution around the carboxylated residues.
2.4. Local structural changes
The local structural changes in the different forms of Ocn simulated in the study were quantified in terms of the dynamics of the individual residues, as well as changes in the secondary structure. The root-mean-square fluctuation (RMSF) is used as a measure of the dynamics of each residue with respect to an average structure calculated over the entire trajectory. This measure can highlight how different regions of the structure fluctuate from their mean structures. In the case of Ocn, RMSF of five regions in the protein were calculated independent of each other, with the average structure of each region calculated individually, using Bio3D and Matplotlib. These five regions are: N-terminal (res 13–17), helix-1:H1 (res 18–26), helix-2:H2 (res 28–36), helix-3:H3 (res 38–46) and C-terminal (res 44–49).
The DSSP implementation in Bio3D [51] was used to calculate the secondary structure assignment of each residue during the entire trajectory. This allowed for the calculation of the percentage helicity for each residue during the simulation.
2.5. Global conformational changes
The global conformational changes in different Ocn were determined through the calculation of the root-mean-square deviation (RMSD) of the structures and the orientational changes in the 3-helical tertiary structure. RMSD is a standard measure of the structural difference between coordinate sets. RMSD values for each system over the 300 ns trajectory, using Cα atoms for trajectory frame superposition, were calculated using the Bio3D [64] module in R [65], Prody v1.7 [66] and Matplotlib library in Python [67]. As the N- terminal (res13–17) and part of the C-terminal (res 47–49) remained highly disordered and mobile during the simulations, these were not included in these calculations.
The angles between the 3 helices of Ocn and the distances between their center of masses were calculated over the entire trajectory using Python scripts in Pymol [68]. The backbone atoms (C, O, N) positions of each residue in a helix were used to compute the best-fit line through a given structure by using a parametric least squares algorithm [69], giving the center and direction of a helix as vectors. The angles between two helices were calculated using the dot-product between the respective vectors. The relationship between inter-helix angles and distances was calculated by binning the values at 5°/5 Å increments to generate angle-angle and distance-distance frequency distributions. The frequency distributions were normalized with respect to the bin displaying the highest frequency.
2.6. Cross-correlation analysis
The extent to which the atomic fluctuations/displacements of different regions in a system are correlated with one another can be assessed by examining the magnitude of the pairwise cross-correlation coefficients. Bio3D was used to calculate residue-wise cross-correlations for the different Ocn systems using the Cα atoms for each residue in the full trajectory. In order to check the convergence of the cross-correlation calculated over the course of the simulation, the correlation analysis was also conducted on 50% (150 ns) and 75% of the trajectory (225 ns).
2.7. Radial distribution function
The interactions of Ca2+ with the different forms of Ocn were quantified in terms of their radial distribution functions. Radial distribution function, g(r), describes the probability of finding a particle at a distance ‘r’ from another particle, and is useful in describing the distribution of solvent particles like water and ions around the protein (solute) molecules. The radial distribution of Ca2+ ions as a function of the Ca2+ − Glu/ γGlu distances was calculated using radial distribution function implemented in Gromacs v5.0.5 [70]. The DCD trajectory files were converted to TRR format read by Gromacs using VMD v1.9.2. To calculate the radial distribution function from a simulation, the neighbors (Ca2+ ions) around each reference atom or molecule (center of mass of carboxylate group in Glu/γGlu residues) were sorted into distance bins set to a value of 0.01 Å. The number of neighbors in each bin were then averaged over the entire simulation. The function can be represented by the following formula:
where, g(r) is the radial distribution function, n(r) is the mean number of atoms in a shell of width Δr (bin size) at distance r, and ρ is the mean atom density. The function was normalized with respect to the bulk density of the ions. In the case of γGlu residue, the function values obtained for the two carboxylate groups individually in the same residue were averaged.
3. Results
MD simulations of different carboxylated forms of Ocn allowed characterization of the structural differences present between the different states, also highlighting the differential role played by the Ca2+ ions in modulating the dynamical properties of the three systems. The results presented in this study are primarily divided into three sections describing the: (1) overall Ocn sequence and structure, (2) local and global conformational changes in the different carboxylated Ocn forms, and (3) interactions of Ocn with Ca2+ ions.
3.1. Sequential features of Ocn
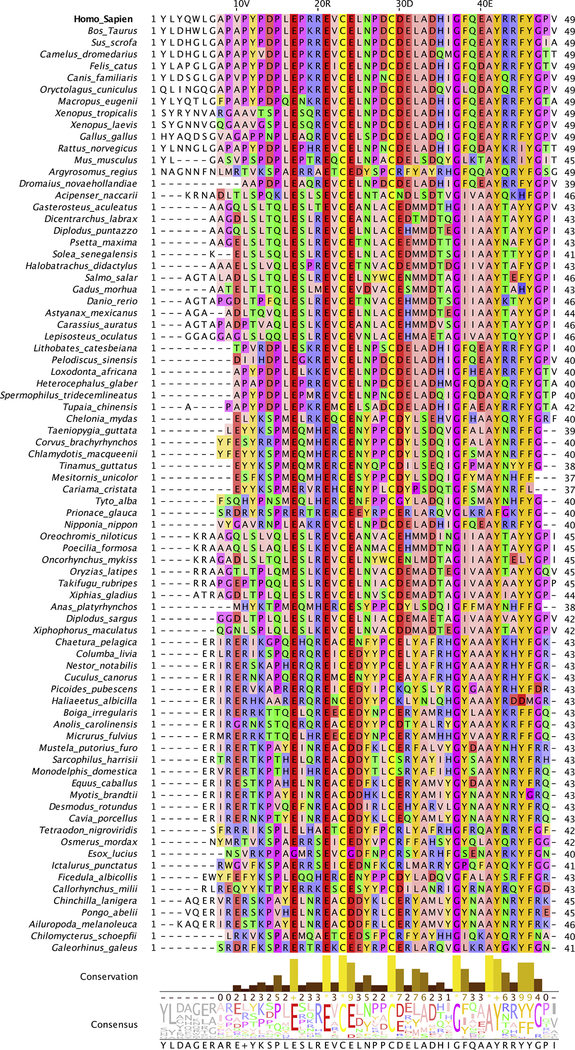
The multiple sequence alignment obtained from HHblits contained 115 homologous sequences from 80 species with a coverage (number of sequences/sequence length) of 2.3. The alignment shows that Ocn is well conserved in different species, especially in the helix H1 containing the three γGLu residues at positions 17, 21 and 24, a single intrachain disulfide bond joining Cys-23 and Cys-29, and Gly-37 present between the H2 and H3 helices (Fig. 1). The residues in the C-terminal (44–49) are also fairly well conserved. The highly flexible/disordered 12-residue N-terminal region shows more sequence variation between different species, with parts of this region missing in some species. The unrooted phylogenetic tree shown in Fig. S1 highlights the evolutionary relationship between the sequences from different species. Ocns belonging to mammalian species, including Homo sapiens, Bos taurus and Sus scrofa, belong to the same clade of the tree, indicating their evolutionary proximity.
Fig. 1.
Multiple sequence alignment of Ocn sequences. Human Ocn alignment with homologous sequences from other species is shown (only single sequences from each species displayed). The residues are colored according to their physio-chemical properties (Zappo coloring scheme), with aliphatic/hydrophobic residues (ILVAM) shown in pink, aromatic residues (FWY) in orange, positively charged residues (KRH) in blue, negatively charged residues (DE) in red, hydrophilic residues (STNQ) in green, conformationally special residues (PG) in purple, and cysteine (C) in yellow. Conservation between the different sequences is visualized as histograms with their heights, and color variation from yellow to brown, reflecting the level of conservation of physico-chemical properties in the alignment. Conserved and identical amino acid columns show the highest score of 11 and are indicated by ‘*’ whereas conserved and similar amino acid columns show a score of 10 and are indicated by ‘+’. Other groupings are indicated by lower scores accordingly. The consensus sequence logo indicates the relative frequency of occurrence of residues per column which can be estimated by its size in the logo. Below the logo, percentage of modal (top) residue for every column is also displayed. If the top value is shared by more than one residue, a ‘+’ symbol is displayed instead.
3.2. Structural features of Ocn
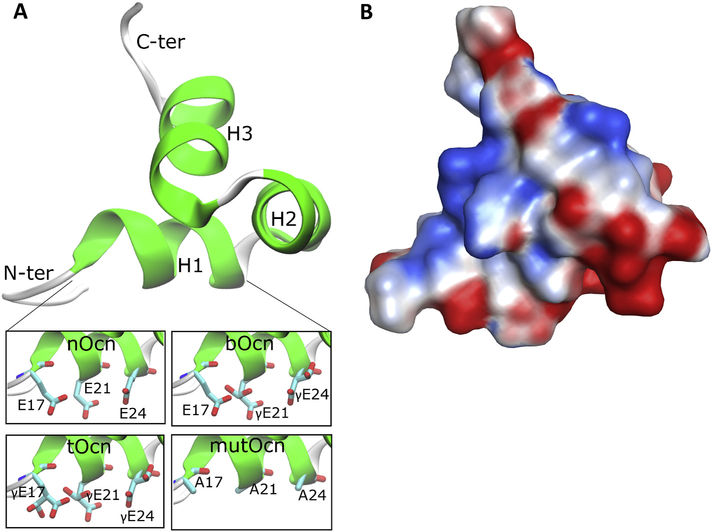
Human (homo sapien) Ocn exhibit sequence identities of 91.8% and 87.8% with bovine (Bos taurus) and porcine (Sus scrofa) Ocn, respectively. The highest scoring homology model, based on bovine Ocn as the template structure, is shown in Fig. 2A and the corresponding secondary structure calculations are provided in Table 2. The disordered 1–12 N-terminus residues were not included in the model. The resulting human Ocn model possesses three helices - H1 (Pro18-Leu25), H2 (Pro27-Ile36), and H3 (Phe38-Tyr46), along with unstructured N- and C-terminal regions, consisting of 5 and 3 residues respectively. H1 and H2 are connected via a β-turn from Leu25 to Asn26 and form a V-shaped arrangement that is stabilized by a disulfide bridge between Cys23 and Cys29. H3 is aligned so as to bisect the V-shape of H1 and H2. The tertiary structure is stabilized by numerous hydrogen bonds involving Arg19 and Val22 in H1, Glu31 and His35 in H2, and Gln39 and Tyr46 in H3, and salt-bridges between Arg20 and Glu24, Arg43 and Val49, and Arg44 and Glu40, respectively. Residues present in H2 and H3, including Leu32, Ile36 and Phe45, form the hydrophobic core of the protein. Ocn thus forms a close globular structure with well conserved interactions keeping the 3-helical domain intact. The different forms of Ocn, used in subsequent MD simulations, were generated using the starting model by adding γ-carboxylation to the glutamate residue at the 17, 21 and 24 positions, along with the mutated form of Ocn where these 3 glutamate residues are mutated to alanine (Fig. 2A, inset).
Fig. 2.
Structural features of Ocn. (A) The starting Ocn homology model is shown in new cartoon representation. The insets at the bottom show the different forms of Ocn (H1 helix) generated by carboxylation of the Glu residues at the 17, 21 and 24 positions in the starting homology model, along with the mutated Ocn generated by mutating these residues to Ala. (B) Electrostatic surface of the Ocn structure generated from the homology model and present in the same orientation as structure in (A). The red color represents electrostatically negative region and blue represents positive region.
Table 2.
Secondary structure calculations for Ocn. Secondary structure predictions for the starting homology model generated are provided. The calculations were carried out with both STRIDE and DSSP.
| Residue | STRIDE prediction | Residue | DSSP prediction |
|---|---|---|---|
| 13–17 | coil | 13–17 | coil |
| 18–25 | α-helix | 18–24 | α-helix |
| 26 | coil | 25–26 | turn |
| 27–36 | α-helix | 27–36 | α-helix |
| 37 | coil | 37 | coil |
| 38–46 | α-helix | 38–46 | α-helix |
| 47–49 | coil | 47–49 | coil |
The electrostatic surface (Fig. 2B) for the Ocn structure shows a wide negatively-charged surface generated by clustering of acidic residues in H1 (Glu/γGlu-17, 21 and 24) and H2 (Asp-28, Asp-30, Glu-31 and Asp-34), while a few positively charged amino acids (Arg-19, 20, 43 and 44) are located on the opposite face of the protein. This creates a differential distribution of positive and negative charges on the surface of Ocn.
3.3. Local conformational differences between different carboxylated forms of Ocn
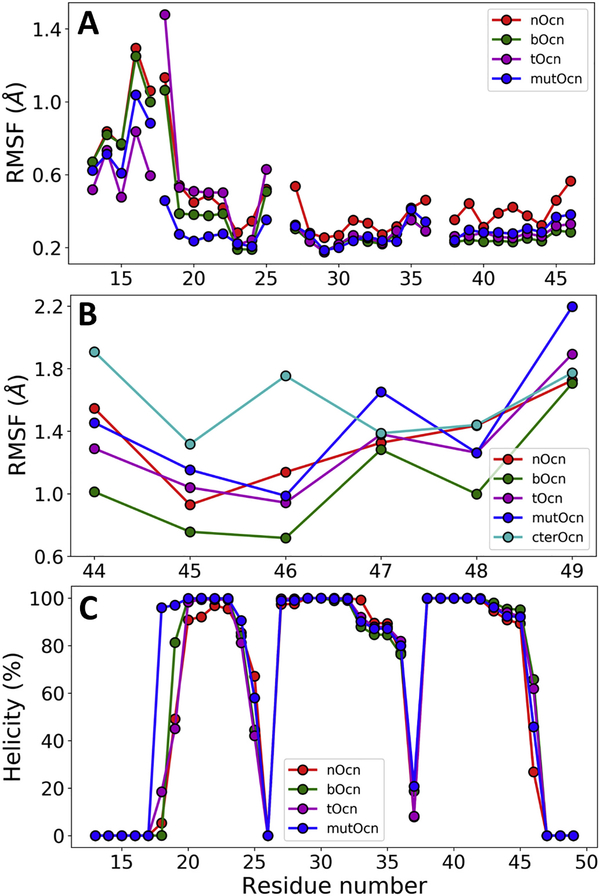
The RMSF of 4 regions in the protein: N-terminal (res 13–17), H1 (res 18–25), H2 (res 27–36) and H3 (res 38–46), calculated separately by independently aligning each region of the structure, are shown in Fig. 3A. The disordered 5 residue N-terminal region remains unstable in all Ocn, with tOcn showing the least variations. In general, nOcn shows the largest fluctuations in the helical regions of the protein. Both bOcn and tOcn display relatively stable H2 and H3 helices, whereas mutOcn shows relatively low variations in all four regions. Fig. 3B shows the RMSF of the active 6 residue C-terminal (res 44–49). bOcn shows the lowest fluctuations in this region. Expectedly, MD simulations of the Ocn-6aa-C shows that the isolated peptide displays the largest RMSF, followed by the mutOcn.
Fig. 3.
Local structural changes in Ocn. (A) RMSF of N-terminal, H1, H2 and H3 helices calculated independently for each region are shown for different forms of Ocn. The coil/turn regions (res 26 and 37) connecting the three helices were not included in the calculations. (B) RMSF of the active 6 residue C-terminal region are shown for different forms of Ocn along with the isolated Ocn-6aa-C simulated independently. (C) Percentage helicity for each residue in different forms of Ocn are shown.
Fig. 3C shows the percentage helicity as calculated by DSSP for each residue in different types of Ocn averaged over the entire simulation. As expected from the RMSF values, bOcn shows higher % helicity for the active C-terminal region, whereas nOcn shows the least. bOcn, and tOcn also display a more structured N-terminal and H1 regions compared to nOcn. Interestingly, mutOcn shows relatively higher helicity compared to the other forms of Ocn. Overall, the results indicate that bOcn displays the most stable local structure.
3.4. Global conformational differences between different Ocn
The RMSD for the protein backbones of nOcn, bOcn and tOcn (excluding the flexible N- and C-terminal regions) over the course of the MD trajectories are shown in Fig. S2. nOcn samples different conformations during the MD, whereas bOcn stabilizes after the initial 60 ns and samples RMSD values close to 0.7 Å for the rest of the trajectory. tOcn starts as a stable structure but the RMSD shows higher variation at around 150 ns and afterwards. The RMSD values of mutOcn are close to the values sampled by bOcn, albeit with higher fluctuations around the value of 0.7 Å. The RMSD results show that bOcn is the most stable form of Ocn in terms of overall conformational changes in the protein.
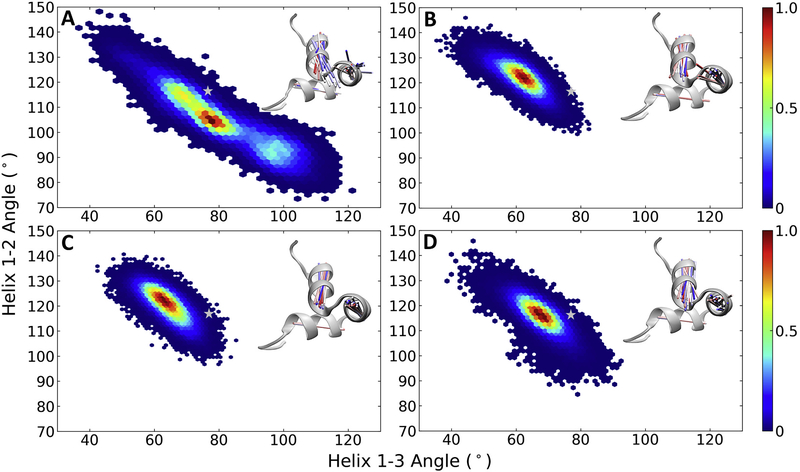
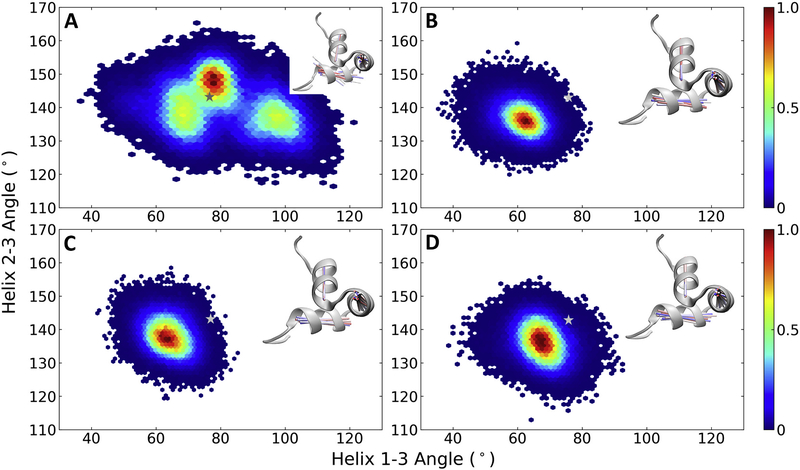
The relationship between inter-helical angles H1-H2 and H1-H3 sampled over the entire trajectory for the different Ocn is shown in Fig. 4. nOcn shows large fluctuations in these angles, with an increase in the H1-H2 angle correlating with a decrease in H1-H3 angle (Fig. 4A). The minimum lies around 100–105° for the H1-H2 angle and 75–80° for H1-H3, which is similar to the starting structure in terms of the H1-H3 angle but less than the original H1-H2 angle. The helical orientation in both bOcn and tOcn remains relatively rigid throughout the simulation, sampling values around minima of 120–125° for the H1-H2 angle and 60–65° for H1-H3 (Fig. 4B and C). The conformational minima sampled by both these forms of Ocn lies away from the values observed in the starting modeled structure of Ocn. mutOcn also stays rigid during the simulation, although sampling slightly different angles compared to bOcn/mutOcn, with a minima around the values of 115–120° for the H1-H2 angle and 65–70° for H1-H3 (Fig. 4D). The corresponding relationship between the inter-helical distances H1-H2 and H1-H3 sampled over the trajectory for different Ocn are shown in Supplementary Fig. S3A, B, C and D. As expected, nOcn shows larger fluctuations compared to other Ocn.
Fig. 4.
Orientational conformational changes in Ocn. The relationship between inter-helical angles H1-H2 and H1-H3 sampled over the entire trajectory are shown for (A) nOcn, (B) bOcn, (C) tOcn and (D) mutOcn. The color bars show the probability distribution of the conformations ranging from blue, representing lowest probability, to red, representing the highest probability. The background in white in each plot represents unsampled space. The inter-helical angles observed in the starting modeled structure is shown as a grey star symbol on top of the plots. The insets in each plot shows the structural variation in this conformational space for structures taken at an interval of 20 ns, in the form of vector lines representing each of the helix, and colored from red (stating structure) to blue (final structure). These structures were superimposed on the common H1 helix used in the calculation of both these angles. The starting structure is shown in grey new cartoon representation for each of the simulated Ocn.
A trend similar to above is also observed in the case of the inter-helical H2-H3 and H1-H3 angles, with nOcn showing conformational diversity and sampling 3 minima, around 145–150°/75–80°, 140°/70°, 140°/100° for the H2-H3/H1-H3 angles, respectively (Fig. 5A). The first minima lies close to the values observed in the starting structure. The H2-H3 angles remain stable throughout the simulation for all other Ocn, sampling minima around the values of 135–140° (Figs. 5B, C and D), different from the starting structure. The relationship between the inter-helical distances H2-H3 and H1-H3 are shown in Supplementary Fig. S4, with nOcn sampling a larger basin compared to other forms of Ocn. Overall, bOcn/tOcn show a relatively rigid 3-helical tertiary structure, whereas nOcn displays a higher flexibity.
Fig. 5.
Orientational conformational changes in Ocn. The relationship between inter-helical angles H2-H3 and H1-H3 sampled over the entire trajectory are shown for (A) nOcn, (B) bOcn, (C) tOcn and (D) mutOcn. The color bars show the probability distribution of the conformations ranging from blue, representing lowest probability, to red, representing the highest probability. The background in white in each plot represents unsampled space. The inter-helical angles observed in the starting modeled structure is shown as a grey star symbol on top of the plots. The insets in each plot shows the structural variation in this conformational space for structures taken at an interval of 20 ns, in the form of vector lines representing each of the helix and colored from red (stating structure) to blue (final structure). These structures were superimposed on the common H3 helix used in the calculation of both these angles. The starting structure is shown in grey new cartoon representation for each of the simulated Ocn.
3.5. Correlation between different regions of Ocn
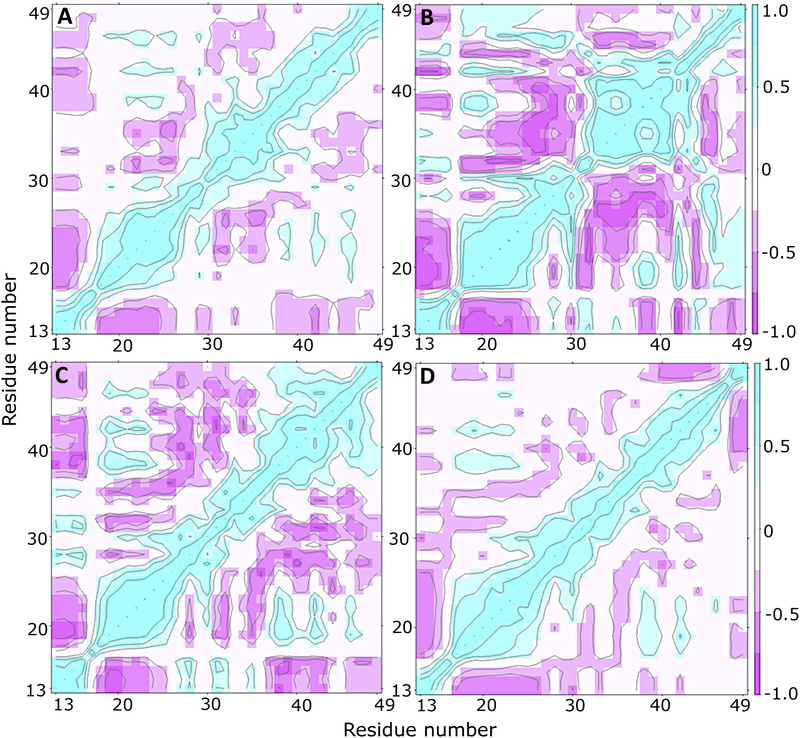
The cross-correlation maps, showing the dynamical relationship between the different regions of Ocn, are shown in Figs. 6A, B, C and D for nOcn, bOcn, tOcn and mutOcn, respectively. nOcn shows moderately negative correlation between the N-terminal and H1 regions as well as between some parts of the H1 and H2 helices. In contrast, bOcn shows strong negative correlation between most regions of the protein, for example, between N-terminal and H1/H3/C-terminal, between H1 and H2 and between H2 and H3, as well as a weak positive correlation between H1 and C-terminal. tOcn also shows strong/moderate negative correlation between most regions and a stronger positive correlation between H3 and C-terminal. mutOcn does not show much correlation between its different regions. Overall, the results imply that different regions in bOcn and tOcn communicate with each other structurally, whereas different regions in both nOcn and mutOcn behave independent of each other. In addition, cross-correlation analysis carried out on 50% (150 ns) and 75% of the trajectory (225 ns) (Fig. S5), shows that the correlation between different regions of the protein has fairly converged over the course of the simulation.
Fig. 6.
Communication between different regions of Ocn. Cross-correlation maps representing residue-wise cross-correlations are shown for (A) nOcn, (B) bOcn, (C) tOcn and (D) mutOcn. Color-code varies between dark to light purple for strong to moderately negative correlation and between light to dark cyan for moderate to strong positive correlation. White regions represent non-correlated regions of the protein.
3.6. Differential interactions of Ocn with Ca2+ ions
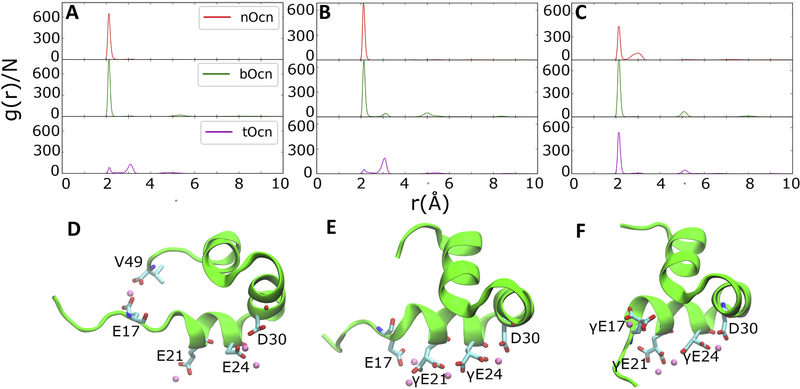
Fig. 7 shows the radial distribution of Ca2+ ions around the carboxylate groups of Glu/ γGlu residues of the different forms of Ocn. It is apparent from the ionic distribution that bOcn shows the highest and sharpest peaks at around 2.1 Å for all the Glu/ γGlu residues, correlating with highest local density of Ca2+ ions. nOcn shows relatively higher peaks for the Glu-17 and Glu-21 residues compared to γGlu-17 and γGlu-21 in tOcn, whereas γGlu-24 in tOcn shows higher local density of Ca2+ ions located at 2.1 Å compared to Glu-24 in nOcn. The two oxygens of the carboxylate groups in Glu/ γGlu are observed to interact with Ca2+ either at the same time, corresponding to a stronger interactions, with Ca2+ peak located at ~2.1 Å, or individually, corresponding to relatively weaker interaction, with Ca2+ peak located at ~3 Å. Additionally, residual broader peaks of smaller density are observed at distances 5 Å and 8 Å in the cases of both bOcn and tOcn, highlighting higher order of Ca2+ distribution around these residues.
Fig. 7.
Ca2+ distribution around Ocn. Radial distribution function calculated from the trajectories of the different carboxylated forms of Ocn for Glu/γGlu residues at positions (A) 17, (B) 21 and (C) 24 positions are shown. D), E), and F) show the snapshots of the nOcn, bOcn and tOcn, respectively, captured along their trajectories. The protein is shown in new cartoon representation (green). The Glu/γGlu residues along with V49 and D30, also involved in interacting with Ca2+, are shown in licorice representation. The Ca2+ are shown as pink van der Waals spheres.
Ca2+ ions coordinating with Glu-17, present on an extended and disordered N-terminal in nOcn, are also found to simultaneously coordinate with the C-terminal of the protein (Val-49; Fig. 7D). This allows nOcn to form an extended structure overall. The other Glu residues coordinate with one, as in the case of Glu-21, or two, as in the case of Glu-24, Ca2+ ions at a time. Glu-24 can also coordinate with additional Ca2+ ions through concurrent coordination of the ion with Asp-30. Glu-17 coordinating with Ca2+ present in a relatively more structured N-terminal in the case of bOcn, is flipped to the opposite side of the other γGlu residues present on the H1 helix (Fig. 7E). This residue remains on the same side in the case of tOcn by sampling a more helical secondary structure (Fig. 7F). The γGlu-21/24 along with Asp-30 in the case of both bOcn and tOcn can concurrently coordinate with the same Ca2+ ion, creating a more ordered structure with the observed orientational stability.
4. Discussion
Previous studies have shown that the three forms of Ocn, which vary in the degree of carboxylation, may also perform different biological functions [15–17]. The structural as well functional consequences of carboxylation in Ocn are still not well understood and are thus important to study further. As the first step of the present study, large-scale sequence comparison of human Ocn with other species was carried out. The results show that the 3-helical domain and the 6-residue C-terminal region of Ocn are both well conserved in different species, indicating conserved structural and functional roles of these regions (Fig. 1). The 3-helical domain assembly plays an important structural role in bone formation [5,6], whereas the C-terminal is one domain that has been shown to function to activate the GPRC6A receptor [57]. Other potential binding domains in the full-length Ocn may also activate GPRC6A. The disordered N-terminal domain, on the other hand, shows larger variations in its sequence as well as length in different species, indicating a more secondary role, for example in stabilizing protein-protein interactions.
Consequently, structural modeling of human Ocn constructed based on the sequentially similar template structure available from bovine Ocn showed that the three α-helices present in the structure form the hydrophobic core of Ocn, which displays a tight globular structure overall (Fig. 2). One of the distinctive structural features of human Ocn is the asymmetric distribution of positive and negative amino acids within the structure, creating positive and negatively charged surfaces on opposite sides of the protein. This may allow Ocn to differentially bind its interacting partners through its opposite surfaces based on the electrostatic properties of the binding interface present on its receptor.
The role of carboxylation on the dynamics and the structure of Ocn was explored using MD simulations. The RMSF calculations carried out independently for different regions of the protein show that the residues in the three helices of nOcn are more mobile, whereas both bOcn/tOcn show less fluctuations in structure (Fig. 3). Since the undecarboxylated form of Ocn is purported to have hormonal functions, the mobility of the helices may permit binding to GPRC6A. Interestingly, bOcn shows the most stable structure for the active 6-residue C-terminal region, both in terms of RMSF and as well as in forming more helical secondary structure. In contrast, the C-terminal of mutOcn shows RMSF values comparable to the isolated Ocn-6aa-C also simulated in the study. This may point to an important secondary role played by the γ-carboxylation of the Glu residues in stabilizing the active C-terminal, which is more pronounced when the carboxylation is present at the 21 and 24 Glu residues, as in the case of bOcn. Additionally, the N-terminal region is observed to be more ordered in tOcn (in terms of RMSF and secondary structure sampled), likely allowing the protein to form a more compact structure in comparison to the other carboxylated forms. The compact nature of tOcn may in turn promote the formation of the functional configuration of Ocn in the extracellular matrix [5]. The N-terminal region also displays high order in the mutOcn, which is expected as the Ala residues (present through Glu/Ala mutations) are known to promote formation of a helical structure [71].
Simulations of the three carboxylated forms of Ocn show bOcn sampling a stable structure throughout the simulation (in terms of calculated RMSD values). nOcn, on the other hand, displays a more dynamic nature, sampling multiple conformations over the course of the simulation. The results are indicative of a possible structural consequence of the different carboxylation states of Ocn. The global conformational dynamics of Ocn was further elucidated in terms of the orientational variations within its 3-helical domain. Large variations observed in the inter-helix angles and distances in nOcn highlight the dynamic nature of the individual helices in this form of Ocn, allowing it to sample unique conformations different from both bOcn and tOcn (Figs. 4 and 5). tOcn and bOcn, on the other hand, display a relatively rigid tertiary structure and sample similar minima in conformational space. All carboxylated forms display propensities for regions in the conformational space away from that observed in the starting modeled structure derived from bovine Ocn, highlighting how small sequential variations between the species can further translate into structural differences. The cross-correlation analysis carried out provides a possible rationale for the stability in the tertiary structure in both tOcn and bOcn. Most helical as well as N- and C-terminal regions in these two forms are more strongly correlated (Fig. 6). The behavior of nOcn, on the other hand, is similar to mutOcn, with both showing the least correlations between different regions in the structure, and consequently larger structural variations observed.
The presence of different negative charges in H1 helix in different forms of Ocn and their differential interactions with Ca2+ ions may create an interplay of attractive/repulsive forces surrounding this helix, that likely result in the orientational differences observed in the 3-helical tertiary structure of the of different forms of Ocn. Compared to nOcn, the relatively higher local densities of Ca2+ ions observed in the vicinity of Glu/γGlu residues in bOcn indicate that these ions are strongly confined in their position and pack around these residues in a more ordered manner, forming strong short-range electrostatic interactions (Fig. 7). This may further promote the orientational rigidity observed in the structure. γGlu residues present in both bOcn and tOcn also show residual peaks, diagnostic of long-range ordering in the Ca2+ ion distribution around these conserved acidic residues in the H1 helix, further promoting stability of these forms of Ocn. Additionally, the same Ca2+ ions are observed to coordinate with both γGlu-21 and γGlu-24, and also with γGlu-24 and Asp-28 (present in the H2 helix), in these forms of Ocn. These doubly coordinating Ca2+ ions may better shield the negative charges present on neighboring γGlu residues in bOcn and tOcn or between negative charges present on H1 and H2 helices, in turn stabilizing these systems more than the singly coordinating Ca2+ ion observed in the case of Glu residue in nOcn. A previous study using steered MD simulations also showed an increased affinity (30 fold) of Ca2+ ions for tOcn compared to nOcn [37], likely due to the concurrent coordination of the these Ca2+ ions with multiple γGlu in tOcn compared to single coordination in nOcn.
The overall low Ca2+ densities observed around the Glu/γGlu residues in tOcn, also displaying a rigid orientational structure as bOcn, may be due to the shorter timescale of the simulations and limited sampling of the Ca2+ ions around the negatively charged residues located on helix H1. tOcn shows a greater affinity/interaction with the Ca2+ ions in hydroxyapatite crystals [72], as these ions are confined in the crystals compared to their diffusive nature in the extracellular solution environment. Thus, in bones, tOcn may display stronger interactions with Ca2+, in turn playing a constitutive role in affecting bone and mineral crystal properties. Decarboxylation of γGlu-17 in tOCn may prevent the protein from correctly packing and matching with the Ca2+ in the hydroxyapatite crystal [6], encouraging disengagement of bOc from bone mineral, binding of soluble Ca2+, and release from bone into circulation where it displays important endocrine functions.
The carboxylation state of the active form of Ocn that interacts with the cell surface receptor GPRC6A has remained controversial, leading to uncertainty over their exact biological functions. Here, bOcn has been shown to be the most stable form of Ocn, both in terms of the helical orientations in the tertiary structure, as well as the stability of the active C-terminal binding to the allosteric site of the receptor GPRC6A [57]. It is possible that the rigid bOcn is involved in cell receptor interactions, with the stable C-terminal better able to bind in the conserved allosteric binding pocket of the receptor (following the conformation selection binding model). In contrast, the higher modulation of the helical orientation in nOcn may allow the protein to better fit the flexible C-terminal in the allosteric pocket (following an induced fit model of binding) or allow binding of other regions of nOcn to other domains in GPRC6A required for activation.
Indeed, recently, a murine circulating pentadecapeptide, named metabolitin, was shown to bind and activate the G-protein coupled receptor GPRC6A, successfully affecting downstream signaling leading to an improvement in non-alcoholic fatty liver disease and insulin resistance in mice [31]. Metabolitin is flanked by potential convertase cleavage sites of Ocn and is sequentially identical to the first 15 N-terminal Ocn residues in mice. Out of these, residues 13–18 are conserved between mouse and human Ocn sequences, making it likely that small N-terminal peptides from Ocn may in fact bind and activate GPRC6A in humans as well. This further leads to the possibility that the full length Ocn may display multiple modes of interaction with GPRC6A, through the active C-terminal region and also through the flexible N-terminal region in which the carboxylated Glu17 may be directly involved in forming interactions and binding to GPRC6A. It can be further hypothesized that the carboxylation at Glu21/24 residues may play a similar allosteric role in modulating the structure and affinity of the N-terminal for GPRC6A similar to what is observed for the C-terminal region.
5. Conclusions
The present study highlights differences observed in the conformational dynamics of Ocn that can be connected to its carboxylation status. The γ-carboxylation status of Glu-residues in Ocn has dramatic consequences on MD-simulated Ocn structure and dynamics, with the differences observed in the structures and dynamics of the different forms of Ocn possibly resulting in differences in their biological activity. Ca2+ ions are observed to differentially interact with the different Ocn forms, likely playing an important role in modulating the conformational landscape of the protein and its subsequent function. tOcn plays an important role in bone-mineral binding, whereas soluble bOcn/nOcn may function as active hormones for male fertility and energy metabolism. The regulation of equilibrium between these different forms of Ocn may in turn govern the structural and endocrine functions of this protein. Future functional studies quantifying the binding affinity and activation of the proposed cellular receptor with different forms of Ocn will help to shed light on its specific mechanism of action. Moreover, modification of the Ocn sequence or its carboxylation state may provide the blueprint for developing high-affinity peptides targeting its cellular receptor GPRC6A, with therapeutic potential for treatment of metabolic disorders.
Supplementary Material
Acknowledgement
This work was supported by 1R01DK120567–01 from the National Institutes of Health.
Footnotes
Declaration of Competing Interest
The authors declare that there is no conflict of interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbagen.2020.129809.
References
- [1].Hauschka PV, Lian JB, Gallop PM, Direct identification of the calcium binding amino acid, γ carboxyglutamate, in mineralized tissue, Proc. Natl. Acad. Sci. U. S. A 72 (1975) 3925–3929, 10.1073/pnas.72.10.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Price PA, Otsuka AS, Poser JW, Kristaponis J, Raman N, Characterization of a γ carboxyglutamic acid containing protein from bone, Proc. Natl. Acad. Sci. U. S. A 73 (1976) 1447–1451, 10.1073/pnas.73.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Poser JW, Esch FS, Ling NC, Price PA, Isolation and sequence of the vitamin K-dependent protein from human bone. Undercarboxylation of the first glutamic acid residue, J. Biol. Chem 255 (1980) 8685–8691. [PubMed] [Google Scholar]
- [4].Laizé V, Martel P, Viegas CSB, Price PA, Cancela ML, Evolution of matrix and bone γ-carboxyglutamic acid proteins in vertebrates, J. Biol. Chem 280 (2005) 26659–26668, 10.1074/jbc.M500257200. [DOI] [PubMed] [Google Scholar]
- [5].Hauschka PV, Lian JB, Cole DE, Gundberg CM, Osteocalcin and matrix Gla protein: Vitamin K-dependent proteins in bone, Physiol. Rev 69 (1989) 990–1047, 10.1152/physrev.1989.69.3.990. [DOI] [PubMed] [Google Scholar]
- [6].Poser JW, Price PA, A method for decarboxylation of gamma-carboxyglutamic acid in proteins. Properties of the decarboxylated gamma-carboxyglutamic acid protein from calf bone, J. Biol. Chem 254 (1979) 431–436. [PubMed] [Google Scholar]
- [7].Price PA, Nishimoto SK, Radioimmunoassay for the vitamin K-dependent protein of bone and its discovery in plasma, Proc. Natl. Acad. Sci. U. S. A 77 (1980) 2234–2238, 10.1073/pnas.77.4.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Boivin G, Morel G, Lian JB, Anthoine-Terrier C, Dubois PM, Meunier PJ, Localization of endogenous osteocalcin in neonatal rat bone and its absence in articular cartilage: effect of warfarin treatment, Virchows Arch. A Pathol. Anat. Histopathol 417 (1990) 505–512, 10.1007/BF01625731. [DOI] [PubMed] [Google Scholar]
- [9].Hauschka PV, Reid ML, Timed appearance of a calcium-binding protein containing γ-carboxyglutamic acid in developing chick bone, Dev. Biol 65 (1978) 426–434, 10.1016/0012-1606(78)90038-6. [DOI] [PubMed] [Google Scholar]
- [10].Price PA, Lothringer JW, Baukol SA, Hari Reddi A, Developmental appearance of the vitamin K-dependent protein of bone during calcification. Analysis of mineralizing tissues in human, calf, and rat, J. Biol. Chem 256 (1981) 3781–3784. [PubMed] [Google Scholar]
- [11].McKee MD, Farach-Carson MC, Butler WT, Hauschka PV, Nanci A, Ultrastructural immunolocalization of noncollagenous (osteopontin and osteocalcin) and plasma (albumin and α2HS-glycoprotein) proteins in rat bone, J. Bone Miner. Res 8 (1993) 485–496, 10.1002/jbmr.5650080413. [DOI] [PubMed] [Google Scholar]
- [12].Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS, Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix, J. Cell. Physiol 143 (1990) 420–430, 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- [13].Garnero P, Biomarkers for osteoporosis management: utility in diagnosis, fracture risk prediction and therapy monitoring, Mol. Diagnosis Ther 12 (2008) 157–170, 10.1007/BF03256280. [DOI] [PubMed] [Google Scholar]
- [14].Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G, Increased bone formation in osteocalcin-deficient mice, Nature. 382 (1996) 448–452, 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- [15].Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G, Endocrine regulation of energy metabolism by the skeleton, Cell 130 (2007) 456–469, 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, Ducy P, Karsenty G, Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism, Cell 142 (2010) 296–308, 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ferron M, Hinoi E, Karsenty G, Ducy P, Osteocalcin differentially regulates β cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 5266–5270, 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kanazawa I, Yamaguchi T, Yamamoto M, Yamauchi M, Kurioka S, Yano S, Sugimoto T, Serum osteocalcin level is associated with glucose metabolism and atherosclerosis parameters in type 2 diabetes mellitus, J. Clin. Endocrinol. Metab 94 (2009) 45–49, 10.1210/jc.2008-1455. [DOI] [PubMed] [Google Scholar]
- [19].Foresta C, Strapazzon G, De Toni L, Gianesello L, Calcagno A, Pilon C, Plebani M, Vettor R, Evidence for osteocalcin production by adipose tissue and its role in human metabolism, J. Clin. Endocrinol. Metab 95 (2010) 3502–3506, 10.1210/jc.2009-2557. [DOI] [PubMed] [Google Scholar]
- [20].Oury F, Sumara G, Sumara O, Ferron M, Chang H, Smith CE, Hermo L, Suarez S, Roth BL, Ducy P, Karsenty G, Endocrine regulation of male fertility by the skeleton, Cell. 144 (2011) 796–809, 10.1016/j.cell.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pi M, Kapoor K, Wu Y, Ye R, Senogles SE, Nishimoto SK, Hwang DJ, Miller DD, Narayanan R, Smith JC, Baudry J, Quarles LDA, Structural and functional evidence for testosterone activation of GPRC6A in peripheral tissues, Mol. Endocrinol 29 (2015) 1759–1773, 10.1210/me.2015-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Karsenty G, Oury F, Regulation of male fertility by the bone-derived hormone osteocalcin, Mol. Cell. Endocrinol 382 (2014) 521–526, 10.1016/j.mce.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Oury F, Khrimian L, Denny CA, Gardin A, Chamouni A, Goeden N, Huang YY, Lee H, Srinivas P, Gao XB, Suyama S, Langer T, Mann JJ, Horvath TL, Bonnin A, Karsenty G, XMaternal and offspring pools of osteocalcin influence brain development and functions, Cell. 155 (2013) 228, 10.1016/j.cell.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oury F, Ferron M, Huizhen W, Confavreux C, Xu L, Lacombe J, Srinivas P, Chamouni A, Lugani F, Lejeune H, Kumar TR, Plotton I, Karsenty G, Osteocalcin regulates murine and human fertility through a pancreas-bone-testis axis, J. Clin. Invest 123 (2013) 2421–2433, 10.1172/JCI65952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Min P, Xu F, Ye R, Nishimoto SK, Williams RW, Lu L, Quarles DL, Role of GPRC6A in regulating hepatic energy metabolism in mice, Sci. Rep 10 (2020) 7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mera P, Laue K, Ferron M, Confavreux C, Wei J, Galán-Díez M, Lacampagne A, Mitchell SJ, Mattison JA, Chen Y, Bacchetta J, Szulc P, Kitsis RN, De Cabo R, Friedman RA, Torsitano C, McGraw TE, Puchowicz M, Kurland I, Karsenty G, Osteocalcin Signaling in Myofibers is necessary and sufficient for optimum adaptation to exercise, Cell Metab. (2016), 10.1016/j.cmet.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mera P, Laue K, Wei J, Berger JM, Karsenty G, Osteocalcin is necessary and sufficient to maintain muscle mass in older mice, Mol. Metab (2016), 10.1016/j.molmet.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pi M, Kapoor K, Ye R, Nishimoto SK, Smith JC, Baudry J, Quarles LD, Evidence for osteocalcin binding and activation of GPRC6A in β-cells, Endocrinology. 157 (2016) 1866–1880, 10.1210/en.2015-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pi M, Wu Y, Quarles LD, GPRC6A mediates responses to osteocalcin in β-cells in vitro and pancreas in vivo, J. Bone Miner. Res (2011), 10.1002/jbmr.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sabek OM, Nishimoto SK, Fraga D, Tejpal N, Ricordi C, Gaber AO, Osteocalcin effect on human β-cells mass and function, Endocrinology (2015), 10.1210/EN.2015-1143. [DOI] [PubMed] [Google Scholar]
- [31].Teng B, Huang C, Cheng C-L, Udduttula A, Yu X-F, Liu C, Li J, Yao Z-Y, Long J, Miao L-F, Zou C, Chu J, Zhang JV, Ren P-G, Newly identified peptide hormone inhibits intestinal fat absorption and improves NAFLD through its receptor GPRC6A, J. Hepatol (2020), 10.1016/j.jhep.2020.02.026. [DOI] [PubMed] [Google Scholar]
- [32].Nishimoto SK, Waite JH, Nishimoto M, Kriwacki RW, Structure, activity, and distribution of fish osteocalcin, J. Biol. Chem 278 (2003) 11843–11848, 10.1074/jbc.M211449200. [DOI] [PubMed] [Google Scholar]
- [33].Hauschka PV, Carr SA, Calcium-Dependent α-Helical Structure in Osteocalcin, Biochemistry. 21 (1982) 2538–2547, 10.1021/bi00539a038. [DOI] [PubMed] [Google Scholar]
- [34].Sakamoto R, Okamoto Y, Conformational studies of poly-L-alanine in solution, Nippon Kagaku Zassi. 90 (1969) 753–756, 10.1246/nikkashi1948.90.8_753. [DOI] [Google Scholar]
- [35].Delmas PD, Stenner DD, Romberg RW, Riggs BL, Mann KG, Immunochemical studies of conformational alterations in bone γ-carboxyglutamic acid containing protein, Biochemistry 23 (1984) 4720–4725, 10.1021/bi00315a030. [DOI] [PubMed] [Google Scholar]
- [36].Dowd TL, Rosen JF, Li L, Gundberg CM, The three-dimensional structure of bovine calcium ion-bound osteocalcin using 1H NMR spectroscopy, Biochemistry. 42 (2003) 7769–7779, 10.1021/bi034470s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cristiani A, Maset F, De Toni L, Guidolin D, Sabbadin D, Strapazzon G, Moro S, De Filippis V, Foresta C, Carboxylation-dependent conformational changes of human osteocalcin, Front. Biosci. - Landmark 19 (2014) 1105–1116, 10.2741/4270. [DOI] [PubMed] [Google Scholar]
- [38].Hoang QQ, Sicheri F, Howard AJ, Yang DSC, Bone recognition mechanism of porcine osteocalcin from crystal structure, Nature. 425 (2003) 977–980, 10.1038/nature02079. [DOI] [PubMed] [Google Scholar]
- [39].Frazão C, Simes DC, Coelho R, Alves D, Williamson MK, Price PA, Cancela ML, Carrondo MA, Structural evidence of a fourth Gla residue in fish osteocalcin: biological implications, Biochemistry. 44 (2005) 1234–1242, 10.1021/bi048336z. [DOI] [PubMed] [Google Scholar]
- [40].Dowd TL, Rosen JF, Mints L, Gundberg CM, The effect of Pb2+ on the structure and hydroxyapatite binding properties of osteocalcin, Biochim. Biophys. Acta - Mol. Basis Dis 1535 (2001) 153–163, 10.1016/S0925-4439(00)00094-6. [DOI] [PubMed] [Google Scholar]
- [41].Malashkevich VN, Almo SC, Dowd TL, X-ray crystal structure of bovine 3 Glu-osteocalcin, Biochemistry. 52 (2013) 8387–8392, 10.1021/bi4010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gössl M, Mödder UI, Atkinson EJ, Lerman A, Khosla S, Osteocalcin expression by circulating endothelial progenitor cells in patients with coronary atherosclerosis, J. Am. Coll. Cardiol 52 (2008) 1314–1325, 10.1016/j.jacc.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Benayahu D, Shamay A, Wientroub S, Osteocalcin (BGP), gene expression, and protein production by marrow stromal adipocytes, Biochem. Biophys. Res. Commun 231 (1997) 442–446, 10.1006/bbrc.1997.6116. [DOI] [PubMed] [Google Scholar]
- [44].Thiede MA, Smock SL, Petersen DN, Grasser WA, Thompson DD, Nishimoto SK, Presence of messenger ribonucleic acid encoding osteocalcin, a marker of bone turnover, in bone marrow megakaryocytes and peripheral blood platelets, Endocrinology. 135 (1994) 929–937, 10.1210/endo.135.3.8070388. [DOI] [PubMed] [Google Scholar]
- [45].Fleet JC, Hock JM, Identification of osteocalcin mrna in nonosteoid tissue of rats and humans by reverse transcription—polymerase chain reaction, J. Bone Miner. Res 9 (1994) 1565–1573, 10.1002/jbmr.5650091009. [DOI] [PubMed] [Google Scholar]
- [46].Remmert M, Biegert A, Hauser A, Söding J, HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment, Nat. Methods 9 (2012) 173–175, 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- [47].Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ, Jalview version 2-a multiple sequence alignment editor and analysis workbench, Bioinformatics. 25 (2009) 189–1191, 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tobergte DR, Curtis S, MOE Molecular Operating Environment, 2013, 10.1017/CBO9781107415324.004. [DOI]
- [49].MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M, All-atom empirical potential for molecular modeling and dynamics studies of proteins, J. Phys. Chem. B 102 (1998) 3586–3616, 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [50].Labute P, The generalized born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area, J. Comput. Chem 29 (2008) 1693–1698, 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- [51].Kabsch W, Sander C, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features, Biopolymers. 22 (1983) 2577–2637, 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- [52].Frishman D, Argos P, Knowledge-based protein secondary structure assignment, Proteins Struct. Funct. Bioinforma 23 (1995) 566–579, 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- [53].Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD, CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comput. Chem 31 (2010) 671–690, 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J. Comput. Chem 29 (2008) 1859–1865, 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- [55].NRC NRC, Mineral Tolerance of Animals, 2005, 10.17226/11309. [DOI] [Google Scholar]
- [56].Pi M, Faber P, Ekema G, Jackson PD, Ting A, Wang N, Fontilla-Poole M, Mays RW, Brunden KR, Harrington JJ, Quarles LD, Identification of a novel extracellular cation-sensing G-protein-coupled receptor, J. Biol. Chem 280 (2005) 40201–40209, 10.1074/jbc.M505186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pi M, Kapoor K, Ye R, Nishimoto SK, Smith JC, Baudry J, Quarles LD, Evidence for Osteocalcin Binding and Activation of GPRC6A in β-Cells, 2016, 10.1210/en.2015-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Humphrey W, Dalke A, Schulten K, VMD: Visual molecular dynamics, J. Mol. Graph 14 (1996) 33–38, 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [59].Feller SE, Zhang Y, Pastor RW, Brooks BR, Constant pressure molecular dynamics simulation: the Langevin piston method, J. Chem. Phys 103 (1995) 4613–4621, 10.1063/1.470648. [DOI] [Google Scholar]
- [60].Herce HD, Garcia AE, Darden T, The electrostatic surface term: (I) periodic systems, J. Chem. Phys 126 (2007), 10.1063/1.2714527. [DOI] [PubMed] [Google Scholar]
- [61].Darden T, York D, Pedersen L, Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems, J. Chem. Phys 98 (1993) 10089–10092, 10.1063/1.464397. [DOI] [Google Scholar]
- [62].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K, Scalable molecular dynamics with NAMD, J. Comput. Chem 26 (2005) 1781–1802, 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cochran JJ, Cox LA, Keskinocak P, Kharoufeh JP, Smith JC, Dai Y-H, Nonlinear conjugate gradient methods, in: Wiley Encycl. Oper. Res. Manag. Sci, 2011, 10.1002/9780470400531.eorms0183. [DOI] [Google Scholar]
- [64].Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD, Bio3d: an R package for the comparative analysis of protein structures, Bioinformatics. 22 (2006) 2695–2696, 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- [65].R. R Development Core Team, R: A Language and Environment for Statistical Computing, 2011, 10.1007/978-3-540-74686-7. [DOI]
- [66].Bakan A, Meireles LM, Bahar I, ProDy: protein dynamics inferred from theory and experiments, Bioinformatics. 27 (2011) 1575–1577, 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hunter JD, Matplotlib: a 2D graphics environment, Comput. Sci. Eng 9 (2007) 99–104, 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- [68].DeLano WL, Pymol: an open-source molecular graphics tool, CCP4 Newsl. Protein Crystallogr 40 (2002) 82–92. [Google Scholar]
- [69].Christopher JA, Swanson R, Baldwin TO, Algorithms for finding the axis of a helix: fast rotational and parametric least-squares methods, Comput. Chem 20 (1996) 339–345, 10.1016/0097-8485(95)00075-5. [DOI] [PubMed] [Google Scholar]
- [70].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC, GROMACS: Fast, flexible, and free, J. Comput. Chem 26 (2005) 1701–1718, 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- [71].Rohl CA, Fiori W, Baldwin RL, Alanine is helix-stabilizing in both template-nucleated and standard peptide helices, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 3682–3687, 10.1073/pnas.96.7.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee AJ, Hodges S, Eastell R, Measurement of osteocalcin, Ann. Clin. Biochem 37 (2000) 432–446, 10.1177/000456320003700402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.