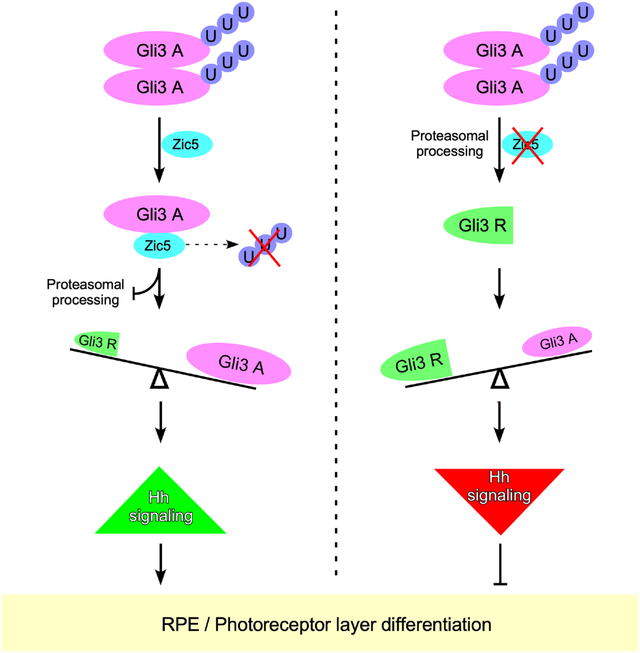
Summary
The Zic family of zinc finger transcription factors play critical roles in multiple developmental processes. Using loss of function studies, we find that Zic5 is important for the differentiation of Retinal Pigmented Epithelium (RPE) and the rod photoreceptor layer through suppressing Hedgehog (Hh) signaling. Further, Zic5 interacts with the critical Hh signaling molecule, Gli3, through the zinc finger domains of both proteins. This Zic5/Gli3 interaction disrupts Gli3/Gli3 homodimerization, resulting in Gli3 protein stabilization via a reduction in Gli3 ubiquitination. During embryonic Hh signaling, the activator form of Gli is normally converted to a repressor form through proteosome-mediated processing of Gli3, and the ratio of Gli3 repressor to full length (activator) form of Gli3 determines the Gli3 repressor output required for normal eye development. Our results suggest Zic5 is a critical player in regulating Gli3 stability for the proper differentiation of RPE and rod photoreceptor layer during Xenopus eye development.
Keywords: Zic5, eye differentiation, RPE, rod photoreceptor, Hedgehog signaling, Gli3
Graphical Abstract
eTOC blurb:
Sun et al. reveal a critical role for Zic5 in regulating the differentiation of retinal pigmented epithelium (RPE) and rod photoreceptor layer by interacting and stabilizing Gli3, which functions predominantly as a repressor for the Hedgehog (Hh) signaling pathway during Xenopus development.
Introduction
Development of the vertebrate eye is a complex process that is regulated by an evolutionarily conserved molecular network. Eye formation begins with the determination of the eye field in the anterior neuroectoderm by neural induction and patterning at the onset of gastrulation (Chow and Lang, 2001; Sinn and Wittbrodt, 2013). The eye field is specified by expression of several eye field transcription factors (EFTFs) including Pax6, Rx1/Rax, Otx2, Six3, Six6 and Lhx2 (Andreazzoli et al., 1999; Casarosa et al., 1997; Hirsch and Harris, 1997; Lagutin et al., 2003; Matsuo et al., 1995; Pannese et al., 1995; Porter et al., 1997; Zuber et al., 2003). A consequence of this specification is bilateral expansion and bisection of the eye field, where a pair of optic vesicles form bilaterally through evagination of the ventral diencephalon and grow towards the overlying surface ectoderm during neurulation. As the optic vesicles grow and reach the surface ectoderm, the lens placode is induced (Chow and Lang, 2001; Sinn and Wittbrodt, 2013; Zagozewski et al., 2014). Following invagination of the dorsal region of the optic vesicle, the optic vesicle collapses and is transformed into the optic cup (OC) with two distinct layers. The outer layer of the OC that gives rise to the RPE is specified by bHLH transcription factor Mitf. The inner layer develops into the neural retina. The RPE continues to grow as a single layer of pigmented epithelial cells surrounding the entire neural retina (NR). The neural retina will undergo neurogenesis to generate neurons and glia, and eventually develops into a mature retina. During retina maturation, retinal neurons are added to the periphery of the retina by differentiation of retinal stem and progenitor cells that reside in the ciliary marginal zone (CMZ) (Chow and Lang, 2001; Fischer et al., 2014; Sinn and Wittbrodt, 2013).
In vertebrates, retinal neurogenesis gives rise to six classes of neurons and one class of glia which laminate into three cellular layers (Stenkamp, 2015; Zagozewski et al., 2014). The innermost layer of the retina is ganglion cell layer (GCL) that is composed of retinal ganglion cells. The outermost layer, also known as outer nuclear layer (ONL), which includes the cell bodies of rod and cone photoreceptors that detect photons and convey this information by chemical signaling. Between the GCL and ONL, the cell bodies of bipolar cells, horizontal cells and amacrine cells compose the inner nuclear layer (INL) (Stenkamp, 2015). All the different types of retinal cells are generated by differentiation of retinal progenitors, which are regulated by a complex interplay of various signaling pathways including Hedgehog (Hh), Notch and Wnt signaling pathways (Koenig et al., 2016; Kubo et al., 2005; Lad et al., 2009; Levine et al., 1997; Pfirrmann et al., 2016; Stenkamp and Frey, 2003; Yamaguchi et al., 2005). Among all these signaling pathways, the Hh pathway has been shown to play an important role in photoreceptor differentiation in Drosophila and zebrafish eye development (Dominguez and Hafen, 1997; Shkumatava et al., 2004; Stenkamp and Frey, 2003; Stenkamp et al., 2000). In addition, Hh signaling has been shown to be required for the proper RPE differentiation in vertebrate systems (Dakubo et al., 2008; Perron et al., 2003).
Hh binding to its receptor Patched (PTCH) leads to the de-repression of Smoothened (SMO) and activates the Gli protein, a transcriptional effector of the Hh pathway (Carballo et al., 2018; Kong et al., 2019). In vertebrates, both Gli2 and Gli3 proteins mediate Hh signals and can function as activators or repressors of Hh target gene expression (Bai et al., 2002; Mo et al., 1997; Persson et al., 2002). In the absence of Hh, Gli proteins are phosphorylated and proteolytically processed into transcriptional repressors by the proteasome. In response to activation of Hh signaling, Gli proteins translocate into the nucleus and function as transcriptional activators that induce expression of Hh target genes such as Gli1 and Ptch1 (Jia et al., 2002; Ou et al., 2002; Zhou et al., 2015). Interestingly, recent evidence reveals that the activator form of Gli3 is dispensable for proper eye morphogenesis in mice, while the repressor form of Gli3 is essential for eye patterning and morphogenesis (Wiegering et al., 2019). Indeed, a critical balance in the ratio of the Gli3 repressor to activator determines a proper transcriptional response to Hh (Chang et al., 2012; Wang et al., 2007; Zhou et al., 2015). However, the molecular mechanisms that regulate the balance between Gli3 activator and the repressor is still largely unknown.
Zic family proteins are transcriptional regulators that contain a highly conserved C2H2 zinc finger motif. In vertebrates, five Zic genes (Zic1–5) have been identified and investigated during embryonic development (Grinberg and Millen, 2005; Houtmeyers et al., 2013). In mice, Zic1 determines the cerebellar folia pattern principally through the regulation of cell proliferation in the external germinal layer (Aruga et al., 1998). Zic2 is required for neural crest formation and hindbrain patterning during mouse development (Elms et al., 2003). Moreover, Zic1 and Zic2 activate Myf5 expression in mouse somites during myogenesis (Pan et al., 2011). Zic3 functions in the earliest stages of left–right (LR) body axis formation, which is associated with human situs abnormalities (Gebbia et al., 1997). In zebrafish, Zic1 and Zic4 regulate roof plate specification and hindbrain ventricle morphogenesis (Elsen et al., 2008). Zic5-deficient mutant mice display neural tube defects and hypoplasia of cephalic neural crest derivatives (Inoue et al., 2004). Among all the Zic genes, Zic2 and Zic5 show robust expression in the eye vesicle (Fujimi et al., 2006; Nakata et al., 2000). In mice, Zic2 has been shown to contribute to RGC subtype identity by directing the retinal axon projection at the optic chiasm midline (Herrera et al., 2003). However, the role of Zic5 in eye development is still unclear.
In this study, we investigate the function of Zic5 in eye development using Xenopus embryos. Using a combination of loss-of-function and replacement experiments in vivo, we show that Zic5 is a major regulator for the differentiation of RPE and rod photoreceptor layer in the developing eye through regulating the Hh pathway. Further evidence shows that Zic5 interacts with Gli3 through their zinc finger domains, and this Zic5/Gli3 interaction is important for Gli3 stability through reducing its ubiquitination level, which acts to balance the ratio of Gli3 repressor to Gli3 activator in Hh signaling. Taken together, our data reveal a role and action of Zic5 in regulating the Hh pathway through stabilizing Gli3 during Xenopus eye development.
Results
Zic5 is essential for normal eye development in the Xenopus embryo
The first step in determining a possible role for Zic5 during eye development was to examine the expression pattern of Zic5 during eye development using whole-mount in situ hybridization (WISH). The expression of Zic5 was detected in the eye field at the mid-neurula stage. At the early tailbud stage, Zic5 was clearly expressed in the optic vesicles. As the embryos progress to the optic cup stage, expression of Zic5 was enriched at the ciliary marginal zone (CMZ) (Figure 1A). To investigate whether Zic5 makes a significant contribution to eye development, a translation blocking morpholino (MO) for Zic5 was designed. The blocking efficiency of this MO was tested by Western blotting lysates from embryos injected with mRNA encoding wild type Zic5 or a morpholino-resistant (MOR) form of Zic5 lacking most of 5’UTR sequences (Figure 1B). To knock down Zic5 in developing eyes, we performed targeted injection of Zic5 MO into the D1.1.1 blastomere, a major contributor (>50%) to the retina population in 32-cell stage embryos (Figure 1C) (Moore et al., 2004). Since exogenous expression of Zic5 throughout an embryo early in development causes major disruptions to several morphogenetic events, we decided to circumvent this problem using an inducible Zic5-GR (glucocorticoid receptor) fusion construct (Figure S1A). This construct allowed us to temporally regulate expression of a functional Zic5 in response to dexamethasone treatment (Figure S1B), and therefore determine a specific time frame in which to perform rescue experiments in Zic5 morphants (Figure S1C). Embryos injected with Zic5 MO into a D1.1.1 blastomere at the 32-cell stage displayed striking eye development defects with severe pigment loss at later stages, although the eye size was not affected (Figure 1D). This phenotype was rescued by co-injection of Zic5-GR MO-resistant (Zic5-GR MOR) mRNA along with subsequent dexamethasone treatment (Figure 1D). To further validate the Zic5 loss-of-function phenotype in the eye, we employed a knockout strategy using CRISPR/Cas9 (Figure S1D). Similarly, Zic5 CRISPR-targeted embryos also showed pigment defects in the eye without affecting eye size (Figure 1E). These data suggest that Zic5 is required for proper eye development.
Figure 1. Zic5 is required for normal eye development in Xenopus.
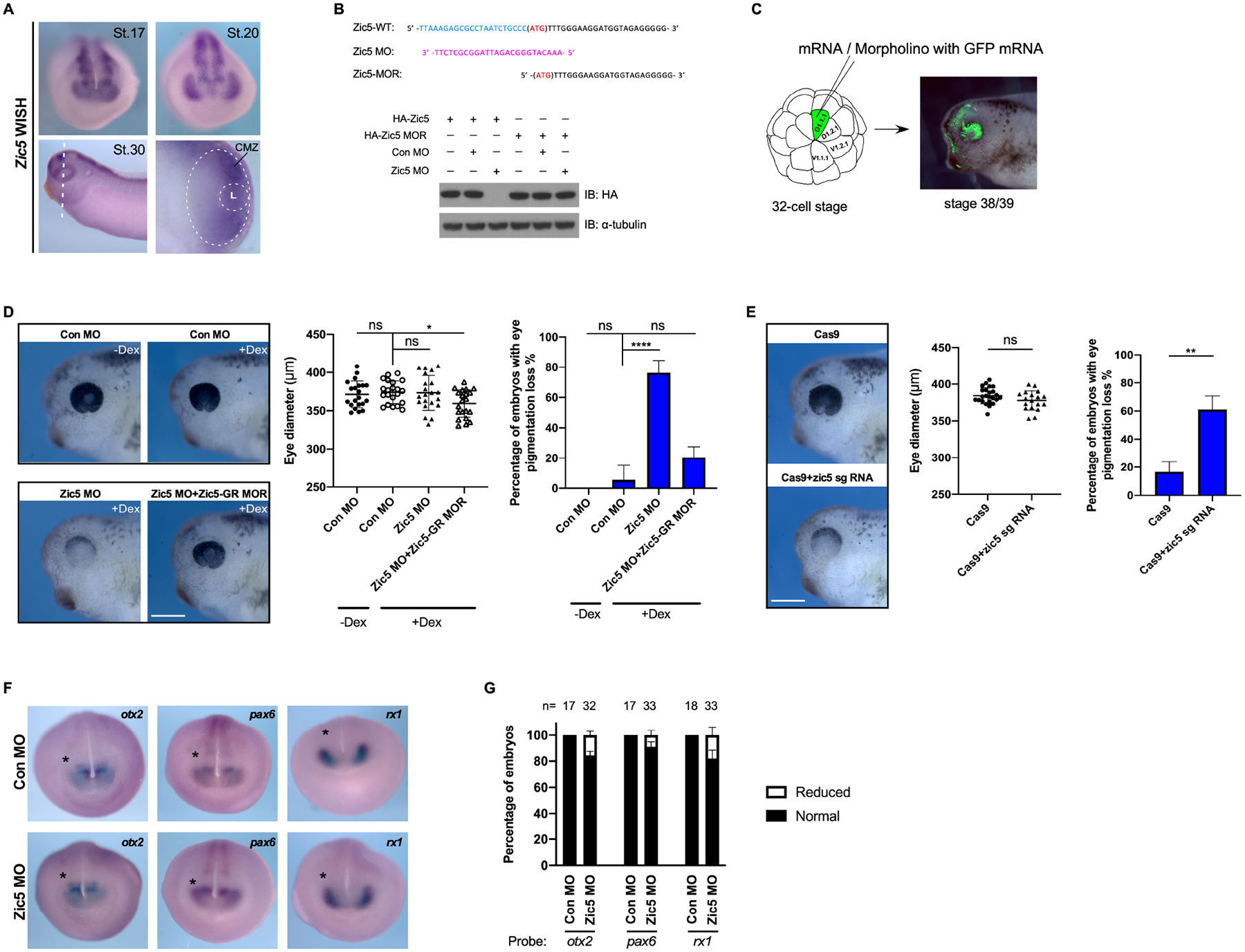
(A) Whole-mount in situ hybridization with Xenopus Laevis Zic5 probe at indicated stages. The white dotted oval outlines the retina. The smaller dotted circle outlines the lens. L, Lens.
(B) Zic5 MO efficiently blocks exogenous Zic5-WT expression, while the injection of the MO-resistant mutant (Zic5-MOR) mRNA is not affected by the MO. Above the blot is a depiction of the wild-type Zic5 nucleotide sequence near the ATG start codon, the Zic5 MO sequence, and the MO-resistant mRNA sequence.
(C) Scheme for microinjection into the D.1.1.1 blastomere at the 32-cell stage. The D.1.1.1 blastomere is a major contributor to retina and the lineage (green) can be traced at tadpole stages.
(D) Knockdown of Zic5 causes pigmentation loss in the eye. Eye diameter was quantified with one-way ANOVA (Dunnett’s multiple comparisons test). Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups. Scale bar, 400 μm. Percentage of embryos with eye pigmentation loss was quantified with one-way ANOVA (Dunnett’s multiple comparisons test). Histograms represent means ± s.d from three biological repeats, *, P<0.05, ****, P<0.0001, ns: no statistical differences between the groups.
CRISPR/Cas9 knockout of Zic5 in F0 embryos results in pigmentation loss in the eye. Eye diameter was quantified with unpaired t test. Scatterplots represent means ± s.d from three biological repeats. ns: no statistical differences between the groups. Scale bar, 400 μm. Percentage of embryos with eye pigmentation loss was quantified with unpaired t test. Histograms represent means ± s.d from three biological repeats, **, P<0.01.
(E) Control morphants and Zic5 morphants were analyzed by whole-mount in situ hybridization with eye filed probes Rx1, Pax6 and Otx2 at stage 18.
(F) Histograms represent the percentage of embryos with normal or reduced expression of Rx1, Pax6 and Otx2 from two biological repeats.
Zic5 is a major regulator for the differentiation of RPE and rod photoreceptor layer in Xenopus developing eyes
To investigate whether knockdown of Zic5 impairs eye induction and specification, eye field marker genes Otx2, Pax6 and Rx1 were examined by WISH. The expression of all three marker genes were largely not affected in the absence of Zic5 (Figures 1F and 1G), indicating that Zic5 is not involved in early eye induction and specification. We then tested whether Zic5 plays a role in eye differentiation by examining the RPE marker RPE65, the rod photoreceptor layer marker rhodopsin, and the inner nuclear layer marker islet1 (Kha et al., 2019). Interestingly, knockdown of Zic5 significantly reduced RPE65 and rhodopsin expression in the eye, which was restored by introducing Zic5-GR MOR along with subsequent dexamethasone treatment (Figures 2A and 2B; Movies S1–S3). In contrast, we detected no obvious difference in Islet1 expression between control morphants and Zic5 morphants (Figure S2A). In addition, the lamination of retina was largely not affected by Zic5 knockdown (Figure S2B). These data suggest a specific involvement of Zic5 in the differentiation of RPE and rod photoreceptor layer, but not the inner nuclear layer. Similarly, CRISPR/Cas9 genetic disruption of Zic5 also showed a dramatic reduction in RPE65 and rhodopsin expression (Figures 2C and 2D). To exclude the possibility that the reduction of RPE65 and rhodopsin expression resulted from cell apoptosis, we examined cleaved-caspase3. No significant apoptosis was detected in the RPE and photoreceptor layer in the absence of Zic5 (Figure S3A). It has also been reported that when retinal cells fail to exit from the cell cycle and continue to proliferate, defects in retinal differentiation are observed (Yamaguchi et al., 2005). To address the possibility that the Zic5 MO-mediated defects in the differentiation of RPE and rod photoreceptor layer might be the consequence of a blockade of cell cycle exit, we examined EdU incorporation. The EdU labeling of dividing cells revealed that a large number of retinal progenitor cells remain mitotic in the CMZ and only a few retinal cells with EdU incorporation were found in the INL layer of control embryos (Figure S3B). Knockdown of Zic5 did not affect the EdU incorporation when compared to control morphants (Figure S3B), indicating that the impairment of RPE and rod photoreceptor layer differentiation in the absence of Zic5 did not result from the failure of cell cycle exit. Taken together, these data suggest that Zic5 is major regulator for RPE and photoreceptor layer differentiation rather than the regulation of cell cycle or cell apoptosis.
Figure 2. Zic5 is a major regulator for the differentiation of RPE and the rod photoreceptor layer.
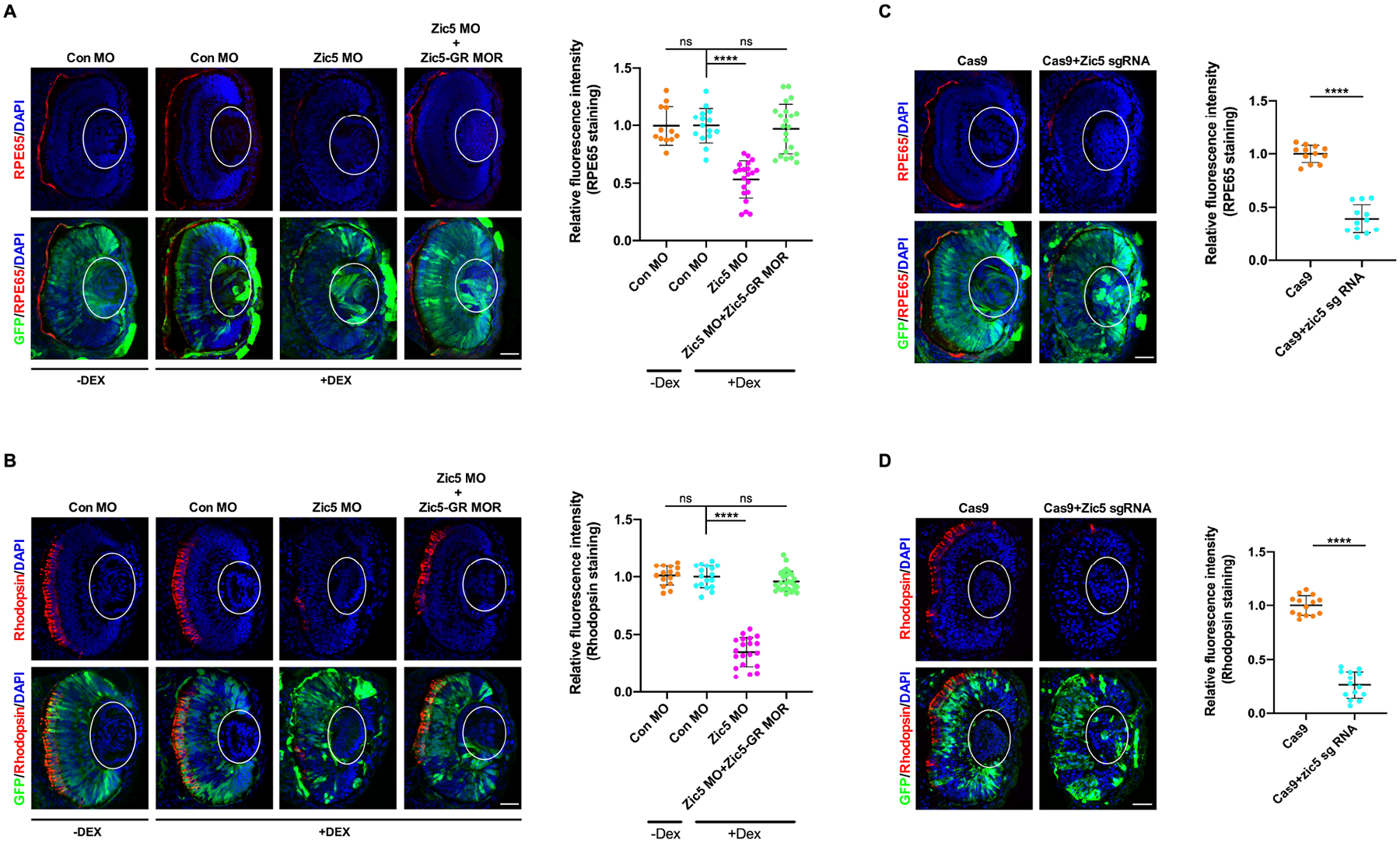
(A) Knockdown of Zic5 impairs RPE layer differentiation as marked by RPE65 immunostaining. The lens is outlined with white oval. Quantification of relative RPE65 fluorescence intensity with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001. Scatterplots represent means ± s.d from three biological repeats. ns: no statistical differences between the groups. Scale bar, 40μm
(B) Knockdown of Zic5 impairs rod photoreceptor layer differentiation as marked by Rhodopsin immunostaining. Quantification of relative rhodopsin fluorescence intensity with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001. Scatterplots represent means ± s.d from three biological repeats. ns: no statistical differences between the groups. Scale bar, 40μm
(C) Zic5 knockout impairs RPE layer differentiation as marked by RPE65 immunostaining. Quantification of relative RPE65 fluorescence intensity with unpaired t test, ****, P < 0.0001, scale bar, 40μm. Scatterplots represent means ± s.d from three biological repeats.
(D) Zic5 knockout impairs rod photoreceptor layer differentiation as marked by Rhodopsin immunostaining. Quantification of relative rhodopsin fluorescence intensity with unpaired t test, ****, P < 0.0001, scale bar, 40μm. Scatterplots represent means ± s.d from three biological repeats.
Zic5 regulates differentiation of RPE and rod photoreceptor layer through the Hh pathway
The differentiation of RPE and rod photoreceptor layers has been shown to be regulated by different signaling pathways such as Wnt, Notch and Hedgehog signaling (Dominguez and Hafen, 1997; Kubo et al., 2005; Lad et al., 2009; Levine et al., 1997; Perron et al., 2003; Yamaguchi et al., 2005). Hh signaling is required for RPE differentiation as well as to promote rod photoreceptor differentiation both in vitro and in vivo (Dominguez and Hafen, 1997; Levine et al., 1997; Perron et al., 2003; Shkumatava et al., 2004; Stenkamp and Frey, 2003; Stenkamp et al., 2000). The constitutive activity of Notch transgenic mice enhances RPE cell proliferation, whereas Notch signaling negatively regulates photoreceptor layer differentiation (Nelson et al., 2007; Schouwey et al., 2011). Treatment with a Wnt inhibitor reduces the number of the RPE cells, while Wnt3a promotes the RPE differentiation and suppresses neural retina generation (Eiraku et al., 2011). To test whether these signaling pathways are involved in Zic5-mediated regulation of the differentiation of RPE and rod photoreceptor layer, real-time quantitative PCR was performed using dissected eyes. MO-mediated knockdown of Zic5 significantly decreased the expression of Hh target genes Gli1and Patched1, which was restored by Zic5-GR MOR plus dexamethasone treatment (Figure 3A). However, the expression of CyclinD1, a Wnt target gene in the eye (El Yakoubi et al., 2012), was not dramatically altered in the absence of Zic5 (Figure 3A). Interestingly, the expression of known Notch target genes Hes1 and Hes5 was also decreased (Figure 3A). It has been reported that in chick retinal explants, Hes1 and Hes5 are Hh signaling targets in retinal progenitor cells independent of Notch (Wall et al., 2009). Indeed, inhibition of Hh signal in the eye by cyclopamine treatment decreases both Hes1 and Hes5 expression (Figure S4A). These results indicate Zic5 may regulate Hh signaling in the developing eyes.
Figure 3. Zic5 regulates Hh signaling in the developing eyes.
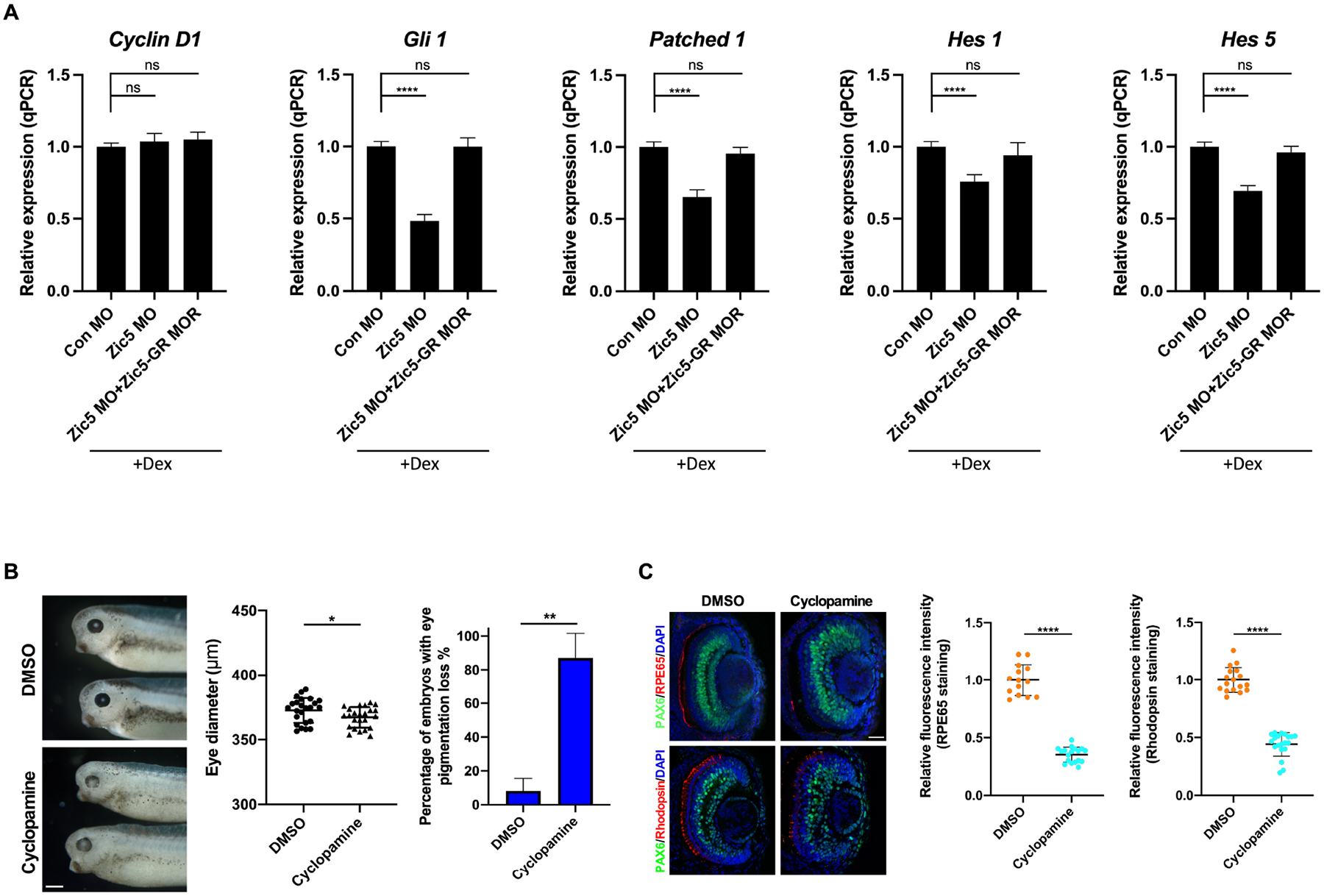
(A) Quantitative PCR analysis of indicated genes expression using dissected eyes at stage 38. Quantification of normalized fold expression of indicated genes with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001. Histograms represent means ± s.d. ns: no statistical differences between the groups.
(B) Inhibition of Hh pathway causes eye pigmentation loss. Eye diameter was quantified with unpaired t test, scale bar, 400 μm. Scatterplots represent means ± s.d from three biological repeats, *P < 0.05. Percentage of embryos with eye pigmentation loss was quantified with unpaired t test. Histograms represent means ± s.d from three biological repeats, **, P<0.01.
(C) Embryos were treated with DMSO or cyclopamine from stage 18–19 and then sectioned and immunostained with indicated antibodies at stage 39. Quantification of relative RPE65 fluorescence intensity with unpaired t test, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ****P < 0.0001. Quantification of relative rhodopsin fluorescence intensity with unpaired t test, scale bars, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ****P < 0.0001.
To further confirm a role for Hh signaling in the differentiation of RPE and the rod photoreceptor layer in Xenopus eyes, embryos were treated with cyclopamine from late neurula stage, when optic vesicles have formed bilaterally. Phenotypically, cyclopamine treated embryos displayed significant eye development defects, including a striking loss of pigment with only a modest decrease in eye size when compared to the DMSO treated control group (Figure 3B), resembling the Zic5 depletion phenotype. In eye sections, the expression of RPE65 and rhodopsin were also significantly reduced with cyclopamine treatment (Figure 3C), indicating a requirement of Hh signaling during the differentiation of RPE and the rod photoreceptor layer during Xenopus eye development. Although examination of endogenous Zic5 protein levels is precluded due to lack of Zic5 specific antibodies for the amphibian, we found that Zic5 mRNA levels were not altered by either cyclopamine or purmorphamine treatment, indicating that expression of Zic5 transcripts is not regulated by Hh signaling (Figure S4B). Together, these results suggest that Zic5 regulates the differentiation of RPE and the rod photoreceptor layer through the Hh pathway.
Zic5 is co-expressed with Glis in developing Xenopus eyes
Hh signaling is mediated intracellularly by the Ci/Gli family of transcription factors. In vertebrates, there are three homologs of the Drosophila Cubitus interruptus (Ci) known as Gli1-3, which are all expressed in the developing eyes (Perron et al., 2003). Gli1 functions exclusively as an activator. Gli2 and Gli3 are bifunctional, acting as transcriptional activators in their full-length forms and repressors in their truncated forms. As transcription factors, members (Zic1 and Zic2) of the Zic family have been shown to physically interact with all three Glis in vitro (Koyabu et al., 2001). This begged the question of whether Zic5 regulates the Hh signaling pathway through an interaction with the Glis. We first examined the temporal and spatial expression pattern of Zic5 and all three Glis in developing eyes using hybridization chain reaction (HCR), which provides a much higher resolution when compared to the traditional in situ hybridization method (Anderson et al., 2020; Choi et al., 2018). At the optic vesicle stage, the expression of Zic5 overlaps with that of Gli2 and Gli3 in both presumptive neural retina (pNR) and presumptive retinal pigmented epithelium (pRPE) which is marked by Mitf (Figure 4A), whereas Gli1 is only expressed in the pRPE and not the pNR at this stage (Figure 4A). As eye development continues to the optic cup stage, Zic5 is expressed in the entire NR and robust expression is detected in the CMZ, where retinal stem cells reside. All Glis are expressed in both the NR and peripheral RPE with enhanced expression at the CMZ (Figure 4B). At a later stage (stage 39), Zic5 continues to be co-expressed with all Glis in the CMZ (Figure S5).
Figure 4. Gli3 functions as a repressor for Hh signaling in Xenopus eye development.
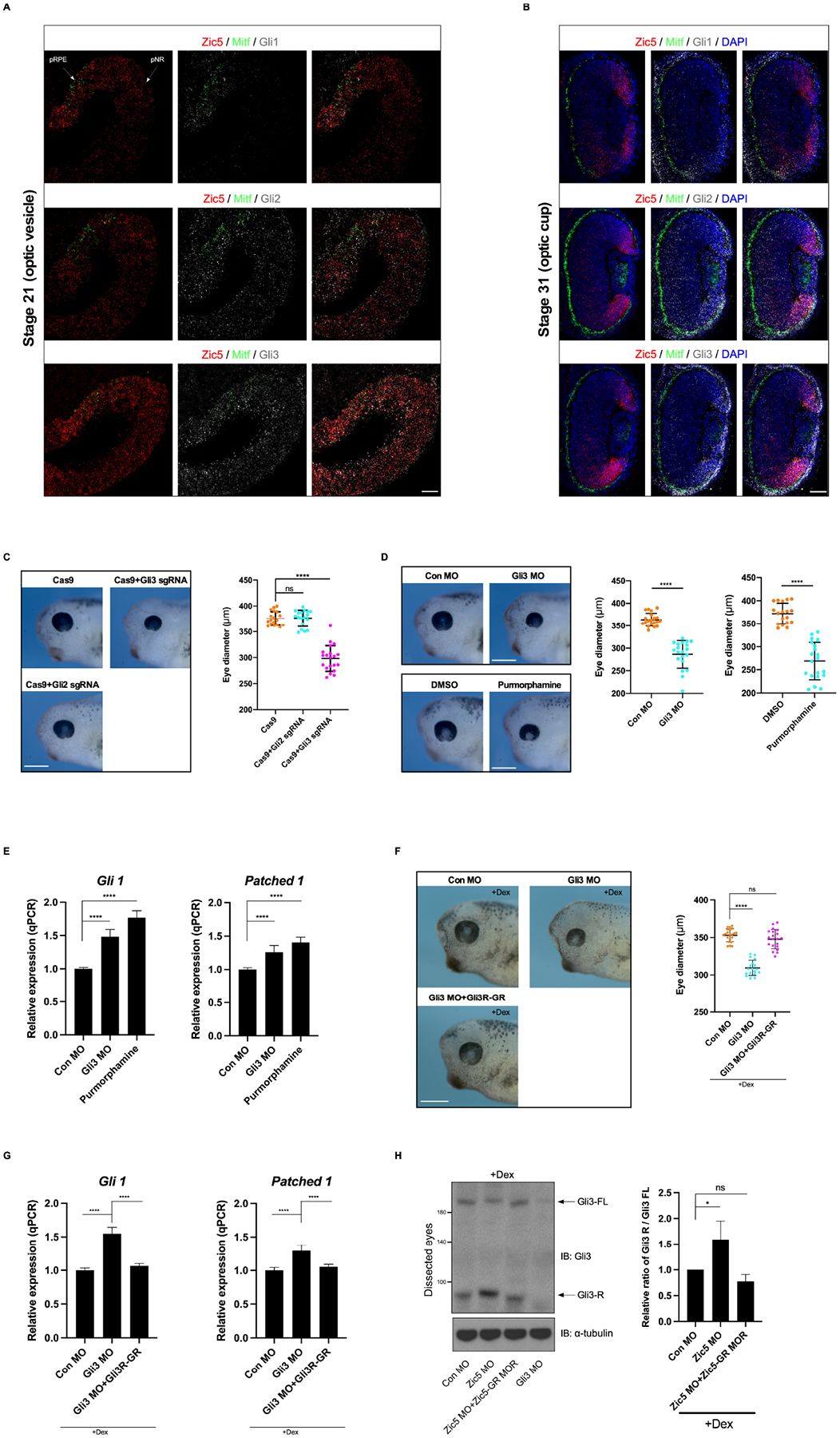
(A) Section HCR analysis was performed using embryos from stage 21 (optic vesicle stage) with indicated probe sets. White arrows indicate pre-retinal pigmented epithelium (pRPE) and pre-neural retina (pNR). Mitf probes were used as a RPE marker. Scale bar, 40 μm.
(B) Section HCR analysis was performed using embryos from stage 31 (optic vesicle stage) with indicated probe sets. Scale bar, 40 μm.
(C) Gli3 knockout reduces eye size. Eye diameter was quantified with one-way ANOVA (Dunnett’s multiple comparisons test). ****, P < 0.0001. Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups. Scale bar, 400 μm.
(D) Gli3 knockdown or activation of Hh pathway by purmorphamine decreases eye size. Eye diameter was quantified with unpaired t test, ****, P < 0.0001, scale bars, 400 μm. Scatterplots represent means ± s.d from three biological repeats.
(E) Quantitative PCR analysis of the expression of Gli1 and Patched1 from dissected eyes injected with control MO or Gli3 MO or treated with Purmorphamine. Quantification of normalized fold expression of indicated genes with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001. Histograms represent means ± s.d.
(F) Indicated MOs or RNA was injected with GFP RNA into D1.1.1 blastomere at the 32-cell stage. The eye phenotypes were analyzed at stage 37. Eye diameter was quantified with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001, scale bar, 400 μm. ns: no statistical differences between the groups. Scatterplots represent means ± s.d from three biological repeats.
(G) Quantitative PCR analysis of the expression of Gli1 and Patched1 from dissected eyes injected with control MO or Gli3 MO or Gli3 MO plus Gli3R-GR mRNA. Quantification of normalized fold expression of indicated genes with one-way ANOVA (Dunnett’s multiple comparisons test), ****, P < 0.0001. Histograms represent means ± s.d.
(H) Indicated MOs and RNA were injected into D1.1.1 blastomere at 32-cell stage. Embryos were treated with dexamethasone (4μg/ml) from stage 16 for rescue purpose. Approximately, 46 Eyes were dissected at stage 35 for each sample, lysed and immunoblotted with anti-Gli3 N-terminal antibody. Quantification of relative ratio of Gli3R/Gli3FL of western bolt from (F) with one-way ANOVA (Dunnett’s multiple comparisons test), *P < 0.05. Histograms represent means ± s.d from three biological repeats. ns: no statistical differences between the groups.
Gli3 functions as a repressor for Hh signaling in Xenopus eye development
To investigate whether Gli2 and Gli3 are involved in eye development in Xenopus embryos, Gli2 or Gli3 genes were disrupted using CRISPR/Cas9 technology (Figure S6A and S6B). Gli3 crispants exhibited a small eye phenotype without impairment of retinal pigmentation (Figure 4C). Interestingly, Gli2 crispants did not display striking morphological defects in the eyes (Figure 4C). The lack of an eye phenotype in Xenopus Gli2 crispants is supported by a previous report examining Gli2 KO mice (Furimsky and Wallace, 2006), leading us to conclude that Gli2 is dispensable for eye development in Xenopus embryos. To further validate the phenotype displayed by the Gli3 crispants, we employed Gli3 morpholino oligonucleotides (Gli3 MO). Similar to the crispants, the Gli3 morphants also displayed a small eye phenotype with normal eye pigmentation (Figure 4D). A recent study showed that the Gli3 repressor, but not the Gli3 activator, is essential for mouse eye patterning and morphogenesis (Wiegering et al., 2019). To test whether the Gli3 also functions as a repressor for Hh signal during Xenopus eye development, embryos were treated with purmorphamine, an established Hh signal activator (El Yakoubi et al., 2012). Similar to the Gli3 knockout or knockdown phenotype, embryos display smaller eyes without a loss in eye pigmentation after purmorphamine treatment (Figure 4D). Moreover, both Gli3 morphant and purmorphamine treated embryos enhance the expression of Gli1 and Patched1 (Hh target genes) in dissected eyes (Figure 4E), suggesting a repressor role of Gli3 in Xenopus developing eyes. Since our data strongly suggested that Gli3 functions predominantly as a repressor for Hh signaling, we performed the rescue experiment by using the repressor form of Gli3 (Gli3R) fused to a GR. This construct allows temporal regulation of expression of a functional Gli3R in response to dexamethasone treatment (Figure S7A). Co-injection of Gli3R-GR with Gli3 MO along with subsequent dexamethasone treatment successfully rescues the eye phenotype (Figure 4F). Using qPCR on dissected eyes, we found the expression of Hh target genes Gli1 and Patched1 were enhanced by Gli3 knockdown, whereas Gli3R-GR restored their expression (Figure 4G). Thus, Gli3 but not Gli2 is involved in proper eye development in Xenopus and functions primarily as a repressor for Hh signaling.
Zic5 regulates differentiation of RPE and the rod photoreceptor layer through stabilizing Gli3
Gli3 exists as both a full-length form, which is as a transcriptional activator, and a proteolytically processed form that acts as a transcriptional repressor. Gli3 processing is complete in most tissues and therefore functions exclusively as a strong repressor for the Hh pathway (Wu et al., 2017). Altering the ratio of Gli3 repressor to activator leads to different transcriptional responses of Hh target genes (Chang et al., 2012; Wang et al., 2007). Since Zic5 knockdown inhibits the Hh pathway target genes Gli1 and Patched1 in vivo (Figure 3A), we tested whether the Gli3 repressor and activator levels were altered in the absence of Zic5. For each sample, eyes from several dozen embryos were dissected and lysed, and Gli3 N-terminal antibody was used to detect both the endogenous Gli3 full length (Gli3FL; activator) and processed form (Gli3R; repressor) in Western blots. The relative ratio of Gli3R/Gli3FL was increased upon knockdown of Zic5 in the eyes (Figure 4H; Figure S7B). Dexamethasone treatment of Zic5 morphant embryos that were also injected with Zic5-GR MOR mRNA restored the ratio of Gli3R/Gli3FL (Figures 4H). To exclude the possibility that the endogenous Gli3 mRNA levels were altered by knockdown of Zic5, qPCR was performed using dissected eyes from control morphants and Zic5 morphants. The result showed that knockdown of Zic5 does not affect the expression levels of Gli3 (Figure S7C). Taken together, these data may suggest an important role of Zic5 in balancing the ratio of Gli3R/Gli3FL in Hh signaling, which is critical for the differentiation of RPE and rod photoreceptor layer.
To determine whether Gli3 is involved in the differentiation of RPE and rod photoreceptor layer, Gli3 MO-mediated knockdown or CRISPR/Cas9-mediated Gli3 knockout was performed. The expression of both the RPE differentiation marker RPE65 and the rod photoreceptor marker rhodopsin were not affected when Gli3 was depleted (Figures 5A–5D). Interestingly, rhodopsin was also found ectopically expressed in the inner nuclear layer in the absence of Gli3 (Figures 5C–5E). Given that Zic5 knockdown enhances the ratio of Gli3R/Gli3FL, and Gli3 functions predominantly as a repressor in the developing eyes, we tested whether knockdown of Gli3 would rescue the expression of those markers impaired by Zic5 depletion. Indeed, knockdown of both Gli3 and Zic5 in the eye restored expression of RPE65 and rhodopsin when compared to Zic5 morphants (Figures 5F and 5G). Taken together, these data suggest that Zic5 regulates the differentiation of RPE and rod photoreceptor layer through Gli3.
Figure 5. Zic5 regulates differentiation of RPE and rod photoreceptor layer through Gli3.
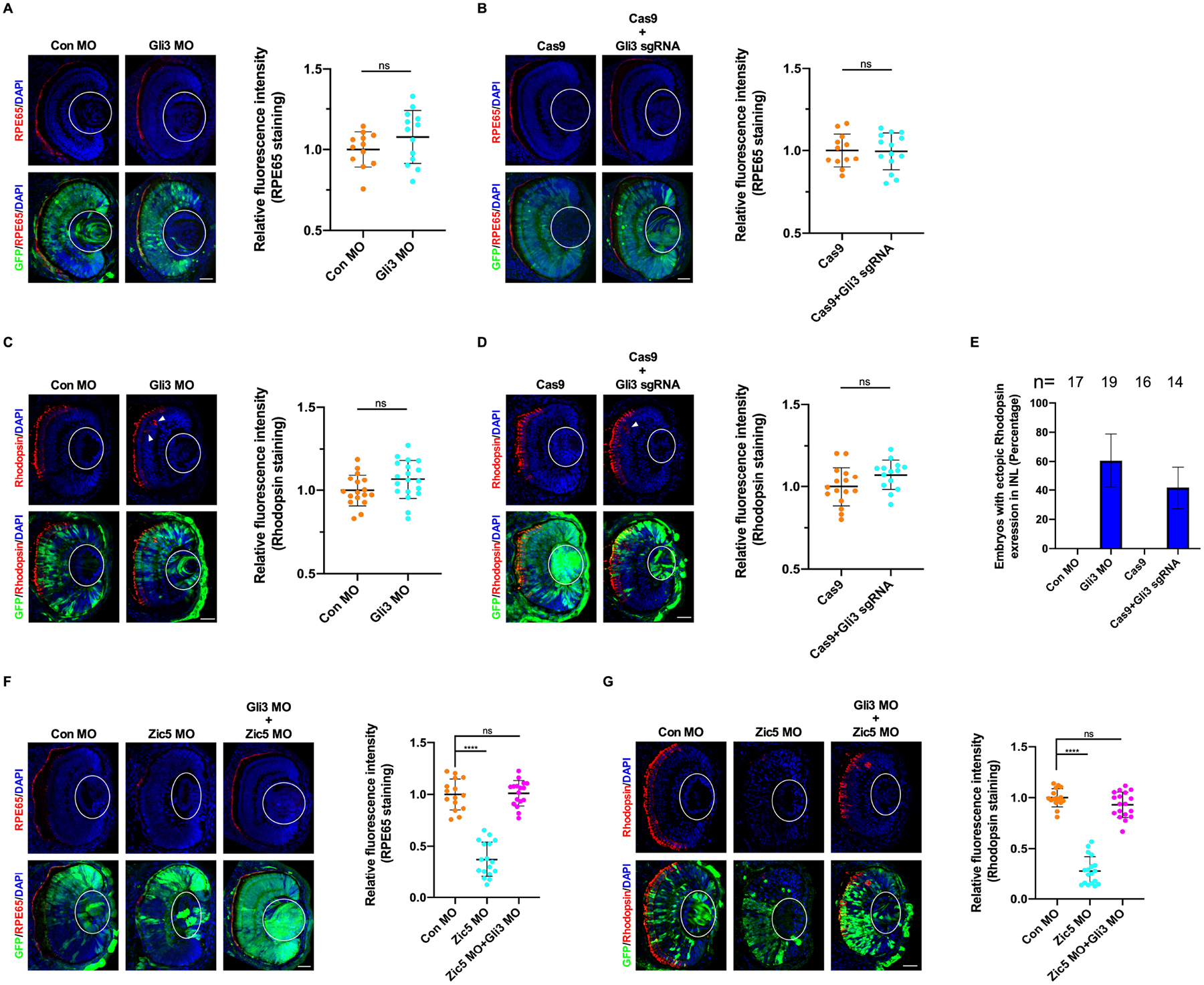
(A) Gli3 knockdown impairs RPE layer differentiation. Quantification of relative RPE65 fluorescence intensity with unpaired t test, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups.
(B) Gli3 knockout impairs RPE layer differentiation. Quantification of relative RPE65 fluorescence intensity with unpaired t test, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups.
(C) Gli3 knockdown impairs rod photoreceptor layer differentiation. Arrowhead indicates ectopic rhodopsin expression. Quantification of relative rhodopsin fluorescence intensity with unpaired t test, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups.
(D) Gli3 knockout impairs rod photoreceptor layer differentiation. Arrowhead indicates ectopic rhodopsin expression. Quantification of relative rhodopsin fluorescence intensity with unpaired t test, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats, ns: no statistical differences between the groups.
(E) Histograms represent the percentage of embryos from (C and D) with ectopic rhodopsin expression in the inner nuclear layer (INL) from three biological repeats.
(F) Gli3 knockdown rescues RPE layer differentiation impaired by Zic5 knockdown. Quantification of relative RPE65 fluorescence intensity with one-way ANOVA (Dunnett’s multiple comparisons test), ****P < 0.0001, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats.
(G) Gli3 knockdown rescues rod photoreceptor layer differentiation impaired by Zic5 knockdown. Quantification of relative rhodopsin fluorescence intensity with one-way ANOVA (Dunnett’s multiple comparisons test), ****P < 0.0001, scale bar, 40 μm. Scatterplots represent means ± s.d from three biological repeats.
Zic5 binds and stabilizes Gli3
Having established a connection between Zic5 and Hh signaling via Gli3, we were interested in determining how Zic5 is mechanistically linked to Gli3. It has been reported using a GST pulldown assay that Zic1 and Zic2 physically interact with all three Glis through their 3rd to 5th zinc finger (ZF) domains (Koyabu et al., 2001). Since Zic5 shares highly conserved ZF domains with Zic1 and Zic2, we examined whether Zic5 interacts with Gli3. Co-IP analysis from HEK293T cells showed that endogenous Gli3 but not Gli2 was detected in the GFP-Zic5 immune-complexes (Figure 6A). In addition, endogenous Zic5 was also found in Gli3 immune-complexes in HEK293T cells (Figure 6B), indicating that an endogenous interaction between Zic5 and Gli3 may exist. We tested whether the 3rd to 5th ZF domains in Zic5 and Gli3 are critical for their interaction. A Zic5 deletion mutant (Zic5ΔZF3−5) and a Gli3 deletion mutant (Gli3ΔZF3−5) were generated (Figure 6C). Co-IP analysis from HEK293T lysates showed that the 3rd-5th ZF domains are required for a Zic5/Gli3 interaction (Figures 6D and 6E). Moreover, in Xenopus embryos, overexpression of wild type Zic5 but not Zic5ΔZF3−5 associated with endogenous Gli3 in Co-IP experiments (Figure 6F). Since we determined that the Zic5/Gli3 interaction can occur, we examined the subcellular localization of Gli3 proteins in different contexts using HEK293T cells that were generated to stably express Flag-tagged Gli3 (Gli3-Flag). Cell immunostaining showed that Gli3 was located predominantly in the cytoplasm (Figure 6G). Transient overexpression of Zic5 resulted in Gli3 protein being translocated to the nucleus, whereas Zic5ΔZF3–5 overexpression failed to cause this translocation (Figure 6G), indicating that Zic5/Gli3 interaction is critical for Gli3 nuclear translocation. Since the ZF domain was originally identified as a DNA-binding motif (Krishna et al., 2003), we tested whether the Zic5/Gli3 interaction relies upon DNA-binding using Zic5-GR. Co-IP analysis showed that Zic5-GR also interacts with Gli3 (Figure 6H). Accordingly, overexpression of Zic5-GR enhances Gli3 protein localization to the cytoplasm, since this is the expected localization of the Zic5-glucocorticoid receptor fusion protein in the absence of dexamethasone (Figure 6G). Taken together, these data suggest that Zic5 interacts with Gli3 and promotes nuclear translocation of Gli3 through the 3rd-5th ZF domains independent of the DNA-binding function.
Figure 6. Zic5 interacts with Gli3.
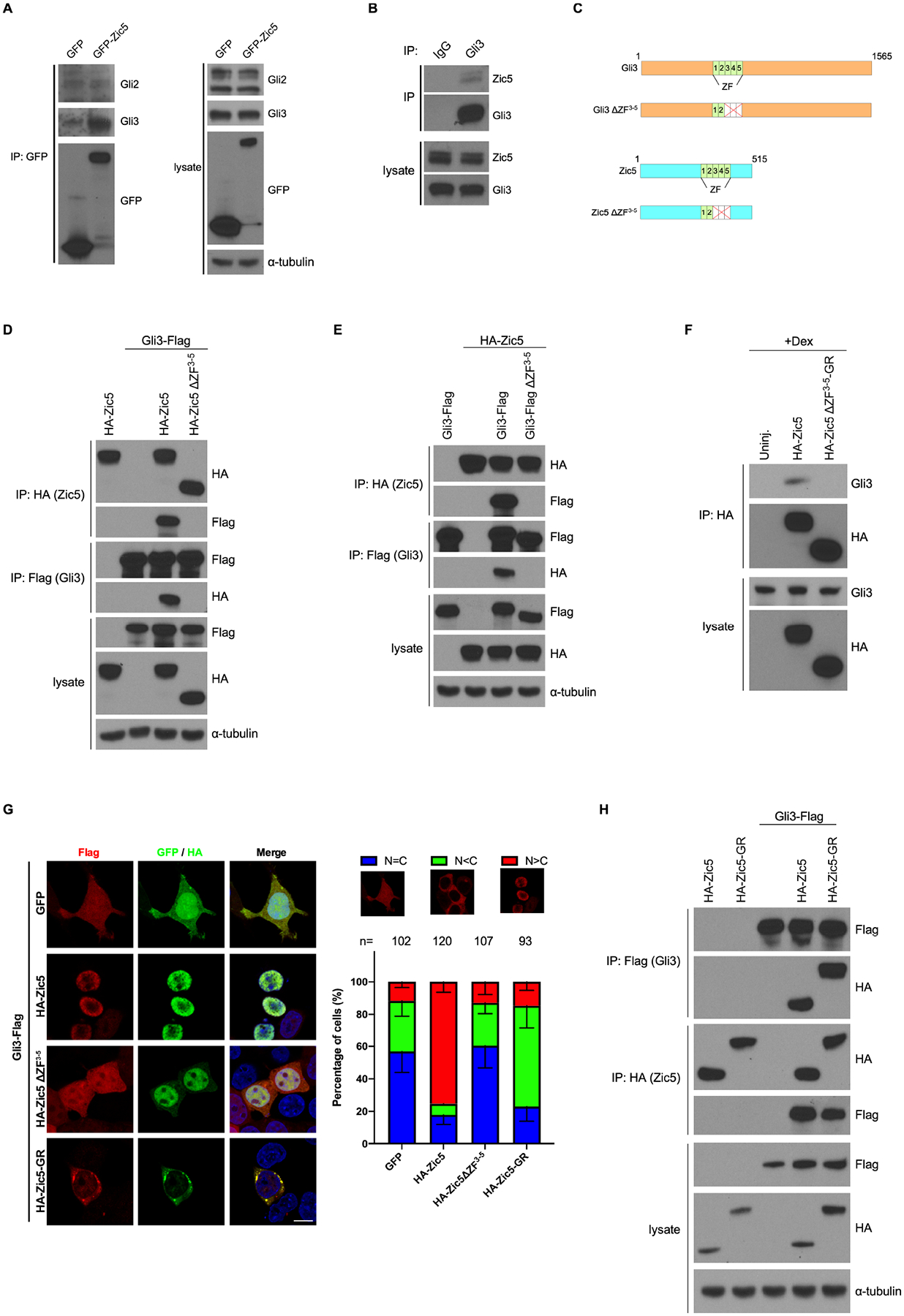
(A) GFP or GFP-Zic5 DNA was transfected in HEK293T cells. Co-IP using GFP antibody shows exogenous Zic5 interacts with endogenous Gli3 but not Gli2.
(B) Zic5 interacts with Gli3 endogenously in HEK293T cell
(C) Illustration of the 3rd-5th ZF domains deletion mutants for Zic5 and Gli3.
(D and E) Zic5 interacts with Gli3 through the 3rd-5th ZF domains.
(F) Co-IPs using HA antibody on lysates from chopped embryo heads injected with indicated RNA shows that Zic5 interacts with endogenous Gli3 through the 3rd-5th ZF domains.
(G) Cell immunofluorescence assays in HEK293T cells stably expressing Gli3-flag with indicated plasmids transfected. Histograms represent the percentage of cells with indicated Gli3-flag subcellular localization. scale bar, 10 μm.
(H) Co-IPs using the indicated antibody on lysates of HEK293T cells transfected with indicated plasmids shows that Zic5-GR interacts with Gli3.
One possible mechanism for the observed increased ratio of Gli3R/Gli3FL in response to Zic5 knockdown in embryonic eyes (Figures 4H; Figure S7B) is through enhancement of proteosome processing of the full length Gli3 protein. Thus, to test whether Zic5 may stabilize Gli3, we used the amenable HEK293T cell system. Knockdown of Zic5 using a specific siRNA in HEK293T cells resulted in an increased ratio of Gli3R/Gli3FL (Figure 7A), suggesting a requirement for Zic5 in stabilizing Gli3 in cells. To test this concept, we examined Gli3 protein stability in a cycloheximide chase assay. In the presence of wild-type Zic5, Gli3 was stabilized, while expression of Zic5ΔZF3−5 made a negligible contribution to Gli3 protein stabilization (Figure 7B), suggesting that the Zic5/Gli3 interaction is important for Gli3 stability. We then examined whether Zic5 stabilizes Gli3 by regulating its ubiquitination level using a cell-based ubiquitination assay. We generated a cell line that stably expresses Myc-tagged Gli3 (Gli3-myc), and overexpressed wild-type Zic5 or the ΔZF3−5 mutant. Over-expression of wild type Zic5 but not Zic5ΔZF3−5 dramatically decreases Gli3 ubiquitination in the presence of MG132 (Figure 7C). Conversely, Gli3 ubiquitination was significantly increased upon Zic5 knockdown (Figure 7D). Moreover, an in vivo assay was also performed using several dozen eyes per sample. Depletion of Zic5 in the Xenopus eyes also enhanced endogenous Gli3 ubiquitination, which could be rescued by co-injection of Zic5-GR MOR mRNA (Figure 7E). Similarly, Gli3 ubiquitination was dramatically increased when embryos were treated with cyclopamine, whereas purmorphamine treatment reduced Gli3 ubiquitination level (Figure 7F). Collectively, these data indicate that Zic5 binds and stabilizes Gli3 through reducing its ubiquitination level.
Figure 7. Zic5 stabilizes Gli3 through reducing its ubiquitination level.
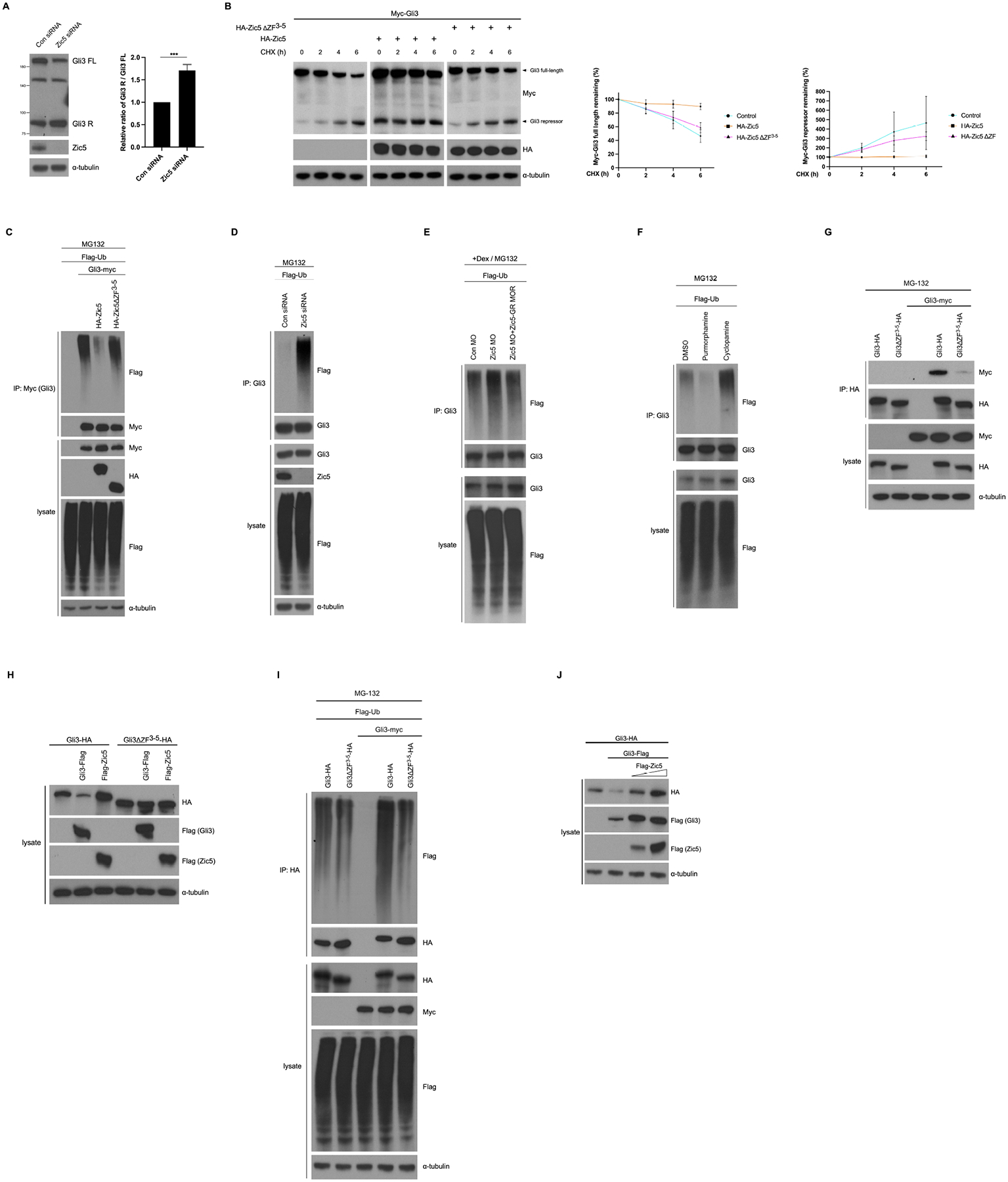
(A) Western blot analysis using the indicated antibodies on lysates from HEK293T cells transfected with control siRNA or Zic5 siRNA. Quantification of relative ratio of Gli3R/Gli3FL of western bolt with unpaired t test, ***P < 0.001. Histograms represent means ± s.d from three biological repeats.
(B) Zic5 stabilizes Gli3 in HEK293T cell by CHX assay. The relative levels of Gli3 full-length and repressor were quantified and normalized against α-tubulin from three independent experiments.
(C) Zic5 overexpression decreases Gli3 ubiquitination in HEK293T cell.
(D) Zic5 knockdown in HEK293T cells increases Gli3 ubiquitination.
(E) Zic5 knockdown in Xenopus eyes increases Gli3 ubiquitination.
(F) Cyclopamine treatment increases Gli3 ubiquitination and purmorphamine treatment decreases Gli3 ubiquitination in Xenopus eyes.
(G) Gli3/Gli3 interaction depends on 3rd-5th ZF domains.
(H) HEK293T cells were transfected with indicated plasmids and harvested 48h after transfection. Cells were lysed and immunoblotted with indicated antibodies.
(I) Gli3/Gli3 interaction increases its ubiquitination.
(G) HEK293T cells were transfected with indicated plasmids and harvested 48h after transfection. Cells were lysed and immunoblotted with indicated antibodies.
The ZF domain dependent interaction between Zic5 and Gli3 raised the question of whether Gli3 could homo-dimerize through 3rd-5th ZF domains. To assess this possibility, we expressed Gli3 constructs using two different tags, myc or hemaglutinin (HA) in HEK293T cells. Co-IP analysis showed myc-tagged Gli3 strongly interacted with Gli3-HA but weakly interacted with Gli3ΔZF3−5-HA (Figure 7G), indicating the ZF domains are required for Gli3 self-interaction to form a homodimer. To test whether Gli3 self-interaction affects its protein stability, we overexpressed Gli3-Flag or Zic5-Flag in Gli3-HA and Gli3ΔZF3−5-HA stable cell lines. Surprisingly, unlike Zic5-Flag, which enhanced Gli3-HA protein levels, Gli3-Flag overexpression dramatically reduced Gli3-HA levels (Figure 7H). As expected, the Gli3ΔZF3–5-HA protein was not affected by either Zic5-Flag or Gli3-Flag overexpression (Figure 7H). Accordingly, Gli3-HA displayed a higher ubiquitination level when Gli3-myc was overexpressed. In contrast, the ubiquitination level of Gli3ΔZF3−5-HA was largely unaltered (Figure 7I). These data indicate that Gli3/Gli3 self-interaction forms an unstable homodimer which was highly ubiquitinated. Next, we examined whether the mechanism by which Zic5 may affect Gli3 stability was by interfering with the Gli3/Gli3 interaction, perhaps competing with the unstable homodimer, resulting in a more stable heterodimer with Zic5. Gli3-HA and Gli3-Flag were expressed in HEK293T cells and increasing amounts of Zic5 were introduced (Figure 7J). Gli3-HA levels were reduced by Gli3-Flag overexpression, while introducing Flag-Zic5 restored Gli3-HA levels in a dose dependent manner (Figure 7J). Taken together, these cell biochemistry data support a possible model for Zic5-mediated Gli3 stabilization in which Zic5 interacts with Gli3 through their ZF domains, which in turn reduces the amount of less stable Gli3/Gli3 homodimer in favor of a more stable Gli3/Zic5 heterodimer. Collectively, our results suggest a positive role for Zic5 in regulating Hh signaling by stabilizing Gli3, which is important for the differentiation of RPE and rod photoreceptor layer during Xenopus eye development.
Discussion
The function of Zic genes in vertebrate embryonic development has been widely studied during the past three decades. However, such studies have been mostly focused on their roles in neural crest and neural tube development (Aruga et al., 1998; Elms et al., 2003; Nakata et al., 2000; Nakata et al., 1997; 1998). Functional investigation of Zic genes during eye development has not been clearly addressed. Among the five Zic genes, Zic2 and Zic5 are strongly expressed in the developing eye (Fujimi et al., 2006; Garcia-Frigola and Herrera, 2010). It has been shown that Zic2 is necessary and sufficient for the specification of retinal ganglion cells that project ipsilaterally at the optic chiasm midline (Herrera et al., 2003). In the case of Zic5, one of the least studied Zic transcription factors, a role in the developing eye is still unclear. In this study, we revealed a requirement of Zic5 in the differentiation of RPE and rod photoreceptor layer during eye development in Xenopus embryos. Mechanistically, we found that Zic5 regulates Hh signaling through stabilizing Gli3.
Using third generation in situ hybridization chain reaction (HCR) which offers high sensitivity and resolution (Choi et al., 2018), we found Zic5 is expressed both in the progenitors of RPE and NR at optic vesicle stage (Figure 4A). During the transition from the optic vesicle to optic cup stage, when retinal progenitors undergo retinal neurogenesis and generate six classes of neurons and one class of glia (Stenkamp, 2015; Zagozewski et al., 2014), the expression of Zic5 becomes enriched at the CMZ (Figure 4B). At a late eye stage (stage 39) when retinal differentiation is completed, Zic5 is only expressed at the CMZ (Figure S5). Given that Zic5 is not expressed in the mature rod photoreceptor, it is very likely that Zic5 is involved in the determination of a retinal progenitor to adopt a rod photoreceptor cell fate rather than a mature photoreceptor. Moreover, no significant differences were observed in Islet1 expression in Zic5 morphants, strongly indicating that the inner nuclear layer is not affected by Zic5 function.
In Gli3 morphant, we found the ectopic expression of rhodopsin in the INL (Figure 5E). One possibility is that ectopic expression of rhodopsin in Gli3 morphant eyes may result from premature differentiation of retinal progenitors into rod photoreceptor cells, due to improper activation of Hh signaling in pNR (pre-neural retina) at the optic vesicle stage. From the HCR data (Figure 4A and 4B), we observe that the Hh signal was only activated in pRPE but not the pNR (as evidenced by Gli1 expression) at the optic vesicle stage. However, Gli3 was expressed in both pRPE and pNR. Therefore, it is likely that Gli3 was predominantly in its repressor form which suppresses the Hh signal in pNR. When Gli3 was depleted, Hh was prematurely activated in pNR, which promotes retinal progenitors to differentiate into rod photoreceptor cells.
Hh signaling is required for proper eye differentiation (Dominguez and Hafen, 1997; Perron et al., 2003; Stenkamp and Frey, 2003), and Gli3 functions predominantly as a repressor for the Hh pathway, while the full length activator form seems to be functionally dispensable for eye development (Wiegering et al., 2019). That raises a question of whether the Gli3 repressor can regulate the activation of the Hh pathway. Indeed, studies have shown that the Gli3 repressor determines Hh pathway activation and is required for the response to a Smoothened antagonist, glasdegib, in acute myeloid leukemia (Chaudhry et al., 2017). Similarly, in articular cartilage, the Gli3 repressor is a key regulator of Hh signals. Altered Gli3 repressor to activator ratio in mutant cartilage results in high Hh signaling, subsequently leading to osteoarthritis symptoms (Chang et al., 2012). In addition, a proper balance between the Gli3 activator and repressor specifies limb digit number and identity (Wang et al., 2007). It is commonly believed that Hh signaling shapes the transcriptional response by altering the ratio of repressor to activator functions of the Gli3 proteins. A low level of Hh signaling requires a high ratio of Gli3 repressor to activator. Conversely, a low Gli3 repressor/activator ratio enhances Hh signaling. In this study, we found knockdown of Zic5 increases the Gli3 repressor/activator ratio both in cells and developing eyes, suggesting a positive role for Zic5 in the Hh pathway by regulating Gli3 stability.
Like other Zic family members, Zic5 shares similar ZF domains which are involved in DNA-binding and transcriptional activity (Mizugishi et al., 2001). In this study, we found the Zic5 binds Gli3 through their 3rd-5th ZF domains, and this interaction is important for Gli3 stabilization. Moreover, the Zic5/Gli3 interaction through the 3rd-5th ZF domains is independent of the DNA-binding function (Figure 6H). However, we could not exclude the ZF domains in Zic5 also contribute to its transcriptional activity during Xenopus eye development. Indeed, it has been shown that the ZF domains from Zic1, Zic2 and Zic3 bind Gli1 binding sequence (Gli-BS) by electrophoretic mobility shift assay-based target selection and mutational analysis. However, the binding affinity was much lower than that of Gli3-ZF domains, and Zics-ZF domains do not compete with Gli3-ZF for the target sequence (Mizugishi et al., 2001). In a reporter assay, Zic1 enabled the Gli3 proteins to participate in transcriptional regulation through ZF domains association (Koyabu et al., 2001).
Both Gli and Zic family proteins are transcriptional regulators that contain highly conserved C2H2 zinc finger domains (Koyabu et al., 2001). Although the C2H2 zinc finger domains were originally identified as DNA-binding domains and are generally assumed to have DNA-binding capabilities, a number of structural and functional studies suggested a critical and more widespread role for these domains in protein-protein interactions (Brayer and Segal, 2008). C2H2 domains utilize many different regions for protein associations including the α -helix, β-sheets and the linker regions. However, DNA-binding of C2H2 domains only use a binding surface comprised of a small number of amino acids invariably located in the N-terminal region of the α-helix (Brayer and Segal, 2008). A ZF domain-mediated protein interaction has been found with some wellknown DNA-binding proteins. For example, Sp1 interacts with p300 through ZF domains, and this interaction results in acetylation of Sp1 (Suzuki et al., 2000). Additionally, GST-pull down assays have shown Sp1 ZF domains were able to interact with BRG1, BAF170, and BAF155 (Kadam et al., 2000). YY1 can interact with ATFa2 or YAF2 using ZF domains 1 and 2 (Kalenik et al., 1997; Zhou et al., 1995). FOG1 has been shown to interact with GATA1 using ZF domains 1, 5, 6, and 9 (Fox et al., 1999). In the case of Gli proteins, although they have been shown to interact with Zic1 and Zic2 through their 3rd to 5th ZF domains via a GST-pulldown assay (Koyabu et al., 2001), the functional mechanism underlying this ZF domain-mediated Gli/Zic interaction is still unknown. In this study, we found that the similar ZF domains also mediated a Zic5 and Gli3 interaction independent of DNA-binding (Figures 6D – 6H). Interestingly, Gli3 is able to homodimerize using the same ZF domains (Figure 7G). This homocomplex is highly ubiquitinated and leads to Gli3 instability (Figures 7H and 7I). However, Zic5 is capable of displacing this Gli3/Gli3 homocomplex to a Gli3/Zic5 heterocomplex that stabilizes Gli3 (Figure 7J). In summary, our findings demonstrate that Zic5 is a critical regulator of Hh signaling by stabilizing Gli3, which functionally influences the differentiation of RPE and the rod photoreceptor layer during Xenopus eye development.
Limitations of the Study
While our data showed that Zic5 regulates Gli3 protein stability in a cycloheximide chase assay in HEK293T cell, we fail to perform this assay using dissected eyes after several attempts were made. The eyes usually disassociate in 1.5–2 hour upon CHX treatment, precluding the success of the experiments. In addition, in this study, we showed Gli3 could bind itself and form Gli3/Gli3 homocomplex by Co-IP experiments using different tagged Gli3 DNA constructs in HEK293T cell. However, approaches to examine the endogenous Gli3/Gli3 homocomplex are currently undeveloped. Moreover, whether Gli3/Gli3 homocomplex and Gli3/Zic5 heterocomplex display different three-dimensional (3D) structures and whether the 3D structure of these proteins complex affects protein stability and functions are still unclear. Thus, future structure analysis of these proteins complex needs to be studied.
STAR★Methods
LEAD CONTACT
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Ira O. Daar (daari@mail.nih.gov).
MATERIALS AVAILABILITY
This study did not generate new unique reagents.
DATA AND CODE AVAILABILITY
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Xenopus laevis
Wild-type Xenopus laevis were obtained from Nasco, USA. All experimental procedures were specifically approved by the Animal Care and Use Committee of the National Cancer Institute-Frederick (ASP #18–433) in compliance with AAALAC guidelines.
Human HEK293T cell
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal bovine serum (R&D Systems) and 1% penicillin/streptomycin (ThermoFisher) at 37°C with 5% CO2.
METHOD DETAILS
Plasmids
Xenopus Zic5 was cloned by PCR from stage 15 embryo cDNA, verified by DNA sequencing, and subcloned into pCS2-HA and pDEST-663 (Protein Expression Laboratory, Frederick National Lab for Cancer Research) to express Flag tagged protein. Xenopus Gli3 was cloned by PCR from stage 15 embryo cDNA, verified by DNA sequencing, and subcloned into pDEST-663 (Protein Expression Laboratory, Frederick National Lab for Cancer Research) to express Flag, HA or Myc tagged protein. Mutants of Zic5 (ΔZF3−5) and Gli3 (ΔZF3−5) were generated using the Quick-Change II Site-Directed Mutagenesis Kit.
Xenopus Embryo, morpholinos and microinjection
Xenopus embryos were obtained by standard methods (Moody, 2000). Capped sense RNAs were transcribed using the mMessage mMachine SP6 kit (ThermoFisher). For the targeted injection into the developing eyes, embryos were injected with mRNAs or morpholinos into D1.1.1 blastomere at 32-cell stage. The following mRNAs were used: GFP (100 pg); Zic5-GR-MOR (100 pg); Gli3R-GR (50 pg). Protein activity of GR chimeric constructs was induced by incubating the embryos in 4 μg/ml dexamethasone (Sigma-Aldrich) from stage 16–17. The morpholinos were obtained from Gene Tools with the following sequences. 4ng Zic5 MO or 1ng Gli3 MO was injected into D1.1.1 blastomere at 32-cell stage. sgRNA sequences were designed and evaluated for the specificity by the ZiFiT Targeter program website (http://zifit.partners.org/ZiFiT/ChoiceMenu.aspx) and CRISPRscan website (https://www.crisprscan.org). The 5’ oligonucleotide sequence (5’-GCAGCTAATACGACTCACTATA ~20 nt~ GTTTTAGAGCTAGAAATA-3’) containing each specific sgRNA sequence (20 nucleotides) and the 3’ common oligonucleotide sequence (5’-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’) were synthesized, annealed and PCR-amplified. sgRNAs were in vitro transcribed using the MEGAscript T7 Transcription Kit (ThermoFisher). For CRISPR/Cas9-mediated knockout, 300pg Zic5 sgRNA, 300pg Gli2 sgRNA or 300 pg Gli3 sgRNA plus 1ng Cas9 protein (PNA Bio Inc.) was injected at one cell stage. The target site for Zic5 sgRNA: GGGGAGTTGGGGAGTGACCC; The target site for Gli2 sgRNA: GGTGAGCAGTACAGTCAATC; The target site for Gli3 sgRNA: GGTAGGGAACTGAGGGTTC.
Experimental procedures were specifically approved by the Animal Care and Use Committee of the National Cancer Institute-Frederick (ASP #18–433) in compliance with AAALAC guidelines.
In situ Hybridization and HCR
Whole-mount in situ hybridization were performed by standard methods using Digoxigenin-labeled antisense RNA probes for Zic5 (Nakata et al., 2000), Pax6, Rx1 and Otx2 (Lee et al., 2006). Embryos were injected with GFP mRNA to indicate the injected side. HCR probe sets were designed by Molecular Instruments. Xenopus embryos were collected at desired stages and then processed for HCR following the HCR v3.0 protocol for whole-mount zebrafish embryos and larvae provided by Molecular Instruments. HCR images were taken by confocal microscopy (Zeiss LSM880).
Immunofluorescence and confocal microscopy
Embryos were fixed with MEMFA (4% formaldehyde in 1× MEM salt) 4°C overnight and then dehydrated with 100% methanol. The following primary antibodies were used: Rabbit anti-GFP (Novus biologicals), rabbit anti-cleaved Caspase-3 (Cell Signaling Technology), mouse anti-Rhodopsin (Millipore), mouse anti-RPE65 (Invitrogen), rabbit anti-Pax6 (Biolegend), mouse anti-islet1 (DSHB). The secondary antibodies used were Alexa Fluor–488 or Alexa Fluor–594 conjugated Goat anti-rabbit IgG or anti-mouse IgG (Invitrogen). Embryos were embedded in 4% low melting agarose gel and were sectioned with a thickness of 60 μm with the vibratome (LEICA VT 1200S). The samples were mounted and imaged using Zeiss LSM880 laser scanning confocal microscope.
Cell culture and transfection
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal bovine serum (R&D Systems) and 1% penicillin/streptomycin (ThermoFisher) at 37°C with 5% CO2. For DNA transfection, cells were transfected using X-tremeGENE 9 DNA Transfection Reagent (Roche) at 40–50% confluency. For siRNA transfection, cells were transfected using RNAiMAX Reagent (ThermoFisher) at 30–40% confluency. Non-Targeting (control) siRNA and Zic5 siRNA were purchased from Dharmacon. Target sequence for control siRNA: AUGUAUUGGCCUGUAUUAG. Target sequence for Zic5 siRNA: CCAAUAGCAGUGAUCGGAA.
Immunoprecipitation and immunoblotting
HEK293T cells were lysed in lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM phenylmethyl sulphonyl fluoride (PMSF, ThermoFisher) and protease inhibitor cocktail (Roche). The cell lysates were sonicated and cleared by centrifugation at 13,000 g for 10 min at 4 °C. IPs were performed at 4 °C for 6–8 h with the following agarose beads: Anti-HA-agarose (Sigma-Aldrich), Anti-flag-agarose (Sigma-Aldrich), Anti-myc-agarose (Sigma-Aldrich), GFP-Trap affinity resin (Chromotek). Western blot analysis was performed using anti-Flag-HRP-conjugated (1:5,000, Sigma-Aldrich), anti-HA-HRP-conjugated (1:5,000, Sigma-Aldrich), anti-myc-HRP-conjugated (1:5,000, Sigma-Aldrich), anti-GFP-HRP-conjugated (1:5,000, Rockland Immunochemicals), goat anti-Gli2 (R&D Systems), goat anti-Gli3 (R&D Systems), rabbit anti-Gli3 (Novus biologicals), goat anti-Zic5 (Novus biologicals) and mouse anti-alpha-tubulin-HRP-conjugated (Proteintech). Secondary antibodies used were goat anti-rabbit-HRP-conjugated (Cell Signaling Technology) and mouse anti-goat-HRP-conjugated (Santa Cruz Biotechnology).
Ubiquitination assay
HEK293T cells were transfected with a Flag-ubiquitin construct together with the indicated plasmids. Proteasomal inhibitor MG132 (Sigma) at 20 μM was added 6 h before cells harvesting. At 48 h after transfection, cells were harvested and lysed with denaturing buffer (1.5% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 2mM sodium orthovanadate, 50 mM sodium fluoride, and protease inhibitors) at 95°C for 10 min. The lysates were then diluted 10-fold with regular lysis buffer and subject to immunoprecipitation and western blot analysis. For detecting ubiquitination of endogenous Gli3 in Xenopus embryos, Flag-ubiquitin mRNA was injected at one cell stage, followed by targeted injection of the indicated morpholino or RNA into D1.1.1 blastomere at 32-cell stage. At stage 19, MG132 was added in culture medium at a final concentration of 40 μM. After 12h MG132 treatment, eyes were dissected and lysed with denaturing buffer (1.5% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 2mM sodium orthovanadate, 50 mM sodium fluoride, and protease inhibitors) at 95°C for 15 min. The lysates were then diluted 10-fold with regular lysis buffer and subject to immunoprecipitation by Gli3 C-terminal antibody and western blot analysis.
Pharmacological treatments
Cyclopamine (50μM; Sigma-Aldrich) and purmorphamine (50μM; Sigma-Aldrich) were applied to the embryo culture medium from stage 18–19.
Reverse transcription and real-time PCR
Reverse transcription was carried out by using a SuperScript™ IV First-Strand Synthesis System (ThermoFisher). The PCR reactions were performed with SsoAdvanced Universal SYBR Green Supermix (BIO-RAD) using CFX96 Touch Real-Time PCR Detection System (BIO-RAD). ODC is used for normalization. Primers and sequences can be found in Table S1.
Quantification and statistical analysis
All experiments were performed blinded with order of testing randomized. ImageJ was used for all analysis. Datasets were compared by unpaired two tailed t-test or one-way ANOVA using the Prism 8 software (GraphPad Software). Differences were considered significant when p was <0.05. Error bars indicate s.d.
Supplementary Material
Movie S1. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with control MO.
Movie S2. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with Zic5 MO.
Movie S3. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with Zic5 MO plus Zic5GR-MOR mRNA.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Novus biologicals | Cat#NB600-308 RRID:AB_10003058 |
| Rabbit polyclonal anti-cleaved Caspase-3 | Cell Signaling Technology | Cat#9661 RRID:AB_2341188 |
| Mouse monoclonal anti-Rhodopsin (4D2) | Millipore | Cat#MABN15 RRID:AB_10807045 |
| Mouse monoclonal anti-RPE65 | Thermo Fisher Scientific | Cat#MA1-16578 RRID:AB_2181003 |
| Rabbit polyclonal anti-Pax6 | Biolegend | Cat#901301 |
| Mouse monoclonal anti-islet1 | Developmental Studies Hybridoma Bank | Cat#39.4D5 RRID:AB_2314683 |
| Alexa Fluor 488-conjugated Goat anti-rabbit | Thermo Fisher Scientific | Cat#A11034 RRID:AB_2576217 |
| Alexa Fluor 594-conjugated Goat anti-mouse | Thermo Fisher Scientific | Cat#A11005 RRID:AB_2534073 |
| Rat monoclonal anti-HA-Peroxidase (clone 3F10) | Roche | Cat#12013819001 RRID:AB_390917 |
| Mouse monoclonal anti-FLAG M2-Peroxidase | Sigma-Aldrich | Cat#A8592 RRID:AB_439702 |
| Mouse monoclonal anti-Myc-Peroxidase | Millipore | Cat#16-212 RRID:AB_11213335 |
| Goat Polyclonal anti-Zic5 | Novus biologicals | Cat# NBP1-50469 RRID:AB_10012659 |
| Goat Polyclonal anti-Gli2 | R&D System | Cat# AF3635 RRID: AB_2111902 |
| Goat Polyclonal anti-Gli3 | R&D System | Cat# AF3690 RRID:AB_2232499 |
| Chemicals, peptides, and recombinant proteins | ||
| Dexamethasone | Sigma-Aldrich | Cat# D4902 |
| Purmorphamine | Sigma-Aldrich | Cat#540220 |
| Cyclopamine | Sigma-Aldrich | Cat#239803 |
| Cas9 protein with NLS | PNA Bio | Cat#CP01 |
| Critical commercial assays | ||
| Ambion mMessage Machine kit | Thermo Fisher Scientific | Cat#AM1340 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| Sso Advanced Universal SYBR Green Supermix | Bio-Rad | Cat#1725275 |
| MEGAscript T7 transcription kit | Thermo Fisher Scientific | Cat#AM1333 |
| SuperScript IV First-Strand Synthesis System | Thermo Fisher Scientific | Cat#18091050 |
| Experimental models: cell lines | ||
| Human: HEK293 cells | ATCC | CRL-11268 |
| Experimental models: organisms/strains | ||
| Wild type Xenopus laevis | Nasco | N/A |
| Oligonucleotides | ||
| Control MO: 5′-CTAAACTTGTGGTTCTGGCGGATA-3′ | Gene Tools | N/A |
| Zic5 MO: 5′- AAACATGGGCAGATTAGGCGCTCTT-3′ | Gene Tools | N/A |
| Gli3 MO: 5′-TAGTGCTACGGGACTGGGCTTCCAT-3′ | Gene Tools | N/A |
| Primers for realtime PCR, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| PCS2-HA-xZic5 | This paper | N/A |
| PCS2-HA-xZic5-GR | This paper | N/A |
| PCS2-HA-xZic5 ΔZF3−5 | This paper | N/A |
| pDEST-xGli3-Myc | This paper | N/A |
| pDEST-xGli3-HA | This paper | N/A |
| pDEST-xGli3 ΔZF3−5-HA | This paper | N/A |
| pDEST-xGli3-Flag | This paper | N/A |
| pDEST-xGli3 ΔZF3−5-Flag | This paper | N/A |
| PCS2-xGli3-GR | ||
| Software and algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/download.html | N/A |
| Prism 8 | GraphPad | N/A |
| siRNAs | ||
| Non-Targeting (control) siRNA | Horizon Discovery | Cat#D-001810-03-05 |
| Zic5 siRNA | Horizon Discovery | Cat#J-013668-20-0002 |
Highlights:
Zic5 regulates differentiation of RPE and rod photoreceptor layer through Hh pathway
Zic5 is co-expressed with Gli3 in developing Xenopus eyes
Gli3 functions as a repressor for Hh signaling in Xenopus eye development
Zic5 binds and stabilizes Gli3
Acknowledgements
This work was supported by funding from the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Reference
- Anderson MJ, Magidson V, Kageyama R, and Lewandoski M (2020). Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. Elife 9. ARTN e55608 10.7554/eLife.55608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Angeloni D, Menna E, and Barsacchi G (1999). Role of XRx1 in Xenopus eye and anterior brain development. Development 126, 2451–2460. [DOI] [PubMed] [Google Scholar]
- Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, and Mikoshiba K (1998). Mouse Zic1 is involved in cerebellar development. J Neurosci 18, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, and Joyner AL (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761. [DOI] [PubMed] [Google Scholar]
- Brayer KJ, and Segal DJ (2008). Keep your fingers off my DNA: Protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys 50, 111–131. 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- Carballo GB, Honorato JR, de Lopes GPF, and Spohr T (2018). A highlight on Sonic hedgehog pathway. Cell Commun Signal 16, 11. 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, and Barsacchi G (1997). XRx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev 61, 187–198. 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- Chang CF, Ramaswamy G, and Serra R (2012). Depletion of primary cilia in articular chondrocytes results in reduced Gli3 repressor to activator ratio, increased Hedgehog signaling, and symptoms of early osteoarthritis. Osteoarthritis Cartilage 20, 152–161. 10.1016/j.joca.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry P, Singh M, Triche TJ, Guzman M, and Merchant AA (2017). GLI3 repressor determines Hedgehog pathway activation and is required for response to SMO antagonist glasdegib in AML. Blood 129, 3465–3475. 10.1182/blood-2016-05-718585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145. ARTN dev165753 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RL, and Lang RA (2001). Early eye development in vertebrates. Annu Rev Cell Dev Biol 17, 255–296. 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Mazerolle C, Furimsky M, Yu C, St-Jacques B, McMahon AP, and Wallace VA (2008). Indian hedgehog signaling from endothelial cells is required for sclera and retinal pigment epithelium development in the mouse eye. Dev Biol 320, 242–255. 10.1016/j.ydbio.2008.05.528. [DOI] [PubMed] [Google Scholar]
- Dominguez M, and Hafen E (1997). Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev 11, 3254–3264. 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, and Sasai Y (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56. 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- El Yakoubi W, Borday C, Hamdache J, Parain K, Tran HT, Vleminckx K, Perron M, and Locker M (2012). Hes4 controls proliferative properties of neural stem cells during retinal ontogenesis. Stem Cells 30, 2784–2795. 10.1002/stem.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms P, Siggers P, Napper D, Greenfield A, and Arkell R (2003). Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol 264, 391–406. 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Choi LY, Millen KJ, Grinblat Y, and Prince VE (2008). Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Dev Biol 314, 376–392. 10.1016/j.ydbio.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Bosse JL, and El-Hodiri HM (2014). Reprint of: the ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp Eye Res 123, 115–120. 10.1016/j.exer.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, and Crossley M (1999). Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. Embo J 18, 2812–2822. DOI 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimi TJ, Mikoshiba K, and Aruga J (2006). Xenopus Zic4: conservation and diversification of expression profiles and protein function among the Xenopus Zic family. Dev Dyn 235, 3379–3386. 10.1002/dvdy.20906. [DOI] [PubMed] [Google Scholar]
- Furimsky M, and Wallace VA (2006). Complementary Gli activity mediates early patterning of the mouse visual system. Dev Dynam 235, 594–605. 10.1002/dvdy.20658. [DOI] [PubMed] [Google Scholar]
- Garcia-Frigola C, and Herrera E (2010). Zic2 regulates the expression of Sert to modulate eye-specific refinement at the visual targets. Embo J 29, 3170–3183. 10.1038/emboj.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, et al. (1997). X-linked situs abnormalities result from mutations in ZIC3. Nat Genet 17, 305–308. 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- Grinberg I, and Millen KJ (2005). The ZIC gene family in development and disease. Clin Genet 67, 290–296. 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, Brown S, and Mason CA (2003). Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell 114, 545–557. 10.1016/s0092-8674(03)00684-6. [DOI] [PubMed] [Google Scholar]
- Hirsch N, and Harris WA (1997). Xenopus Pax-6 and retinal development. J Neurobiol 32, 45–61. [PubMed] [Google Scholar]
- Houtmeyers R, Souopgui J, Tejpar S, and Arkell R (2013). The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell Mol Life Sci 70, 3791–3811. 10.1007/s00018-013-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hatayama M, Tohmonda T, Itohara S, Aruga J, and Mikoshiba K (2004). Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev Biol 270, 146–162. 10.1016/j.ydbio.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, and Jiang J (2002). Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416, 548–552. 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, and Emerson BM (2000). Functional selectivity of recombinant mammalian SWI/SNF subunits. Gene Dev 14, 2441–2451. DOI 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalenik JL, Chen DG, Bradley ME, Chen SJ, and Lee TC (1997). Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Research 25, 843–849. DOI 10.1093/nar/25.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kha CX, Guerin DJ, and Tseng KA (2019). Using the Xenopus Developmental Eye Regrowth System to Distinguish the Role of Developmental Versus Regenerative Mechanisms. Front Physiol 10, 502. 10.3389/fphys.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig KM, Sun P, Meyer E, and Gross JM (2016). Eye development and photoreceptor differentiation in the cephalopod Doryteuthis pealeii. Development 143, 3168–3181. 10.1242/dev.134254. [DOI] [PubMed] [Google Scholar]
- Kong JH, Siebold C, and Rohatgi R (2019). Biochemical mechanisms of vertebrate hedgehog signaling. Development 146. ARTN dev166892 10.1242/dev.166892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyabu Y, Nakata K, Mizugishi K, Aruga J, and Mikoshiba K (2001). Physical and functional interactions between Zic and Gli proteins. J Biol Chem 276, 6889–6892. DOI 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, and Grishin NV (2003). Structural classification of zinc fingers: survey and summary. Nucleic Acids Res 31, 532–550. 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo F, Takeichi M, and Nakagawa S (2005). Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes. Development 132, 2759–2770. 10.1242/dev.01856. [DOI] [PubMed] [Google Scholar]
- Lad EM, Cheshier SH, and Kalani MY (2009). Wnt-signaling in retinal development and disease. Stem Cells Dev 18, 7–16. 10.1089/scd.2008.0169. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, and Oliver G (2003). Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev 17, 368–379. 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Bong YS, Moore KB, Soria K, Moody SA, and Daar IO (2006). Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat Cell Biol 8, 55–63. 10.1038/ncb1344. [DOI] [PubMed] [Google Scholar]
- Levine EM, Roelink H, Turner J, and Reh TA (1997). Sonic hedgehog promotes rod photoreceptor differentiation in mammalian retinal cells in vitro. J Neurosci 17, 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, and Aizawa S (1995). Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev 9, 2646–2658. 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, and Mikoshiba K (2001). Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem 276, 2180–2188. 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, et al. (1997). Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124, 113–123. [DOI] [PubMed] [Google Scholar]
- Moody SA (2000). Cell lineage analysis in Xenopus embryos. Methods Mol Biol 135, 331–347. 10.1385/1-59259-685-1:331. [DOI] [PubMed] [Google Scholar]
- Moore KB, Mood K, Daar IO, and Moody SA (2004). Morphogenetic movements underlying eye field formation require interactions between the FGF and ephrinB1 signaling pathways. Dev Cell 6, 55–67. 10.1016/s1534-5807(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Nakata K, Koyabu Y, Aruga J, and Mikoshiba K (2000). A novel member of the Xenopus Zic family, Zic5, mediates neural crest development. Mech Dev 99, 83–91. 10.1016/s0925-4773(00)00480-9. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, and Mikoshiba K (1997). Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc Natl Acad Sci U S A 94, 11980–11985. 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, and Mikoshiba K (1998). Xenopus Zic family and its role in neural and neural crest development. Mech Dev 75, 43–51. 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, and Reh TA (2007). Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol 304, 479–498. 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen YJ, and Chien CT (2002). Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 16, 2403–2414. 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Gustafsson MK, Aruga J, Tiedken JJ, Chen JC, and Emerson CP Jr. (2011). A role for Zic1 and Zic2 in Myf5 regulation and somite myogenesis. Dev Biol 351, 120–127. 10.1016/j.ydbio.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, and Boncinelli E (1995). The Xenopus homologue of Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development 121, 707–720. [DOI] [PubMed] [Google Scholar]
- Perron M, Boy S, Amato MA, Viczian A, Koebernick K, Pieler T, and Harris WA (2003). A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development 130, 1565–1577. 10.1242/dev.00391. [DOI] [PubMed] [Google Scholar]
- Persson M, Stamataki D, te Welscher P, Andersson E, Bose J, Ruther U, Ericson J, and Briscoe J (2002). Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev 16, 2865–2878. 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirrmann T, Jandt E, Ranft S, Lokapally A, Neuhaus H, Perron M, and Hollemann T (2016). Hedgehog-dependent E3-ligase Midline1 regulates ubiquitin-mediated proteasomal degradation of Pax6 during visual system development. Proc Natl Acad Sci U S A 113, 10103–10108. 10.1073/pnas.1600770113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, et al. (1997). Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124, 2935–2944. [DOI] [PubMed] [Google Scholar]
- Schouwey K, Aydin IT, Radtke F, and Beermann F (2011). RBP-Jkappa-dependent Notch signaling enhances retinal pigment epithelial cell proliferation in transgenic mice. Oncogene 30, 313–322. 10.1038/onc.2010.428. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Fischer S, Muller F, Strahle U, and Neumann CJ (2004). Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131, 3849–3858. 10.1242/dev.01247. [DOI] [PubMed] [Google Scholar]
- Sinn R, and Wittbrodt J (2013). An eye on eye development. Mech Dev 130, 347–358. 10.1016/j.mod.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL (2015). Development of the Vertebrate Eye and Retina. Prog Mol Biol Transl Sci 134, 397–414. 10.1016/bs.pmbts.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL, and Frey RA (2003). Extraretinal and retinal hedgehog signaling sequentially regulate retinal differentiation in zebrafish. Dev Biol 258, 349–363. 10.1016/s0012-1606(03)00121-0. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Frey RA, Prabhudesai SN, and Raymond PA (2000). Function for hedgehog genes in zebrafish retinal development. Developmental Biology 220, 238–252. DOI 10.1006/dbio.2000.9629. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kimura A, Nagai R, and Horikoshi M (2000). Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA-binding. Genes Cells 5, 29–41. DOI 10.1046/j.1365-2443.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, and Wallace VA (2009). Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol 184, 101–112. 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ruther U, and Wang B (2007). The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev Biol 305, 460–469. 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering A, Petzsch P, Kohrer K, Ruther U, and Gerhardt C (2019). GLI3 repressor but not GLI3 activator is essential for mouse eye patterning and morphogenesis. Dev Biol 450, 141–154. 10.1016/j.ydbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Wu FJ, Zhang Y, Sun B, McMahon AP, and Wang Y (2017). Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol 24, 252–280. 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, and Masai I (2005). Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development 132, 3027–3043. 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Zagozewski JL, Zhang Q, and Eisenstat DD (2014). Genetic regulation of vertebrate eye development. Clin Genet 86, 453–460. 10.1111/cge.12493. [DOI] [PubMed] [Google Scholar]
- Zhou QJ, Gedrich RW, and Engel DA (1995). Transcriptional Repression of the C-Fos Gene by Yy1 Is Mediated by a Direct Interaction with Atf/Creb. J Virol 69, 4323–4330. DOI 10.1128/Jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yao X, Li S, Xiong Y, Dong X, Zhao Y, Jiang J, and Zhang Q (2015). Deubiquitination of Ci/Gli by Usp7/HAUSP Regulates Hedgehog Signaling. Dev Cell 34, 58–72. 10.1016/j.devcel.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, and Harris WA (2003). Specification of the vertebrate eye by a network of eye field transcription factors. Development 130, 5155–5167. 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with control MO.
Movie S2. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with Zic5 MO.
Movie S3. Related to Figure 2. View of whole eye immunostained with rhodopsin injected with Zic5 MO plus Zic5GR-MOR mRNA.
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


