Abstract
Background:
Insulin resistance (IR) affects the development of type 2 diabetes mellitus (T2DM), which is also influenced by accumulated fine particle air pollution [particulate matter (PM) with aerodynamic diameter of ()] exposure. Previous experimental and epidemiological studies have proposed several potential mechanisms by which contributes to IR/T2DM, including inflammation imbalance, oxidative stress, and endothelial dysfunction. Recent evidence suggests that the imbalance of the gut microbiota affects the metabolic process and may precede IR. However, the underlying mechanisms of , gut microbiota, and metabolic diseases are unclear.
Objectives:
We investigated the associations between personal exposure to and fasting blood glucose and insulin levels, the IR index, and other related biomarkers. We also explored the potential underlying mechanisms (systemic inflammation and sphingolipid metabolism) between and insulin resistance and the mediating effects between and sphingolipid metabolism.
Methods:
We recruited 76 healthy seniors to participate in a repeated-measures panel study and conducted clinical examinations every month from September 2018 to January 2019. Linear mixed-effects (LME) models were used to analyze the associations between and health data (e.g., functional factors, the IR index, inflammation and other IR-related biomarkers, metabolites, and gut microbiota). We also performed mediation analyses to evaluate the effects of mediators (gut microbiota) on the associations between exposures () and featured metabolism outcomes.
Results:
Our prospective panel study illustrated that exposure to was associated with an increased risk of higher IR index and functional biomarkers, and our study provided mechanistic evidence suggesting that exposure may contribute to systemic inflammation and altered sphingolipid metabolism.
Discussion:
Our findings demonstrated that was associated with the genera of the gut microbiota, which partially mediated the association between and sphingolipid metabolism. These findings may extend our current understanding of the pathways of and IR. https://doi.org/10.1289/EHP9688
Introduction
Insulin resistance (IR) refers to the ability of insulin to stimulate glucose uptake and affects the development of type 2 diabetes mellitus (T2DM) (Dang et al. 2018; Liu 2019; Yang et al. 2018), which contributes to the global burden of diseases and premature death (Health Effects Institute 2019). In China, diabetes mellitus was the 19th leading cause of allage disability adjusted life years (DALYs) in 1990, increasing dramatically to eighth in 2017 (Zhou et al. 2019). Additionally, fine particle air pollution [particulate matter (PM) with aerodynamic diameter of ()] has been ranked fifth among global risk factors for mortality and is the leading environmental health threat worldwide, particularly in developing countries (Health Effects Institute 2019). Most of the disease burden related to comes from chronic diseases, including 20% of deaths from T2DM (Health Effects Institute 2019). Accumulating evidence suggests that exposure to ambient may increase the risk of IR/T2DM (Alderete et al. 2017; Eze et al. 2015; Liu 2019; Rao et al. 2015). Most existing studies have suggested that an inflammatory mechanism may be potentially related to and IR (Kelishadi et al. 2009; Wolf et al. 2016). Some animal studies (Lehrskov and Christensen 2019; Rao et al. 2015; Senn et al. 2002) have reported that particles delivered via the lung’s air–blood barrier into the human circulatory system elevated the response to inflammatory biomarkers, such as interleukin 6, C-reactive protein, and , and these findings were also observed in epidemiological studies (Kelishadi et al. 2009; Lehrskov and Christensen 2019; Pilz et al. 2018; Rao et al. 2015). Additionally, experimental and epidemiological studies have proposed other potential mechanisms of leading to IR/T2DM, such as oxidative stress and endothelial dysfunction (reviewed by Rajagopalan and Brook 2012).
Over the past decade, omics-based approaches, such as metabolomics and microbiomics, have emerged as a powerful approach to identify novel mechanisms and highly sensitive and specific biomarkers (Liang et al. 2018; Zhang et al. 2017a). Mounting evidence suggests a link between metabolites/gut microbiota and IR that may reveal the mechanism underlying IR/T2DM (Harsch and Konturek 2018; Pedersen et al. 2016; Qin et al. 2012; Salim et al. 2014b). An epidemiological study demonstrated that an imbalance in the gut microbiota affects the serum metabolome and insulin sensitivity in a nondiabetic population, and the results were validated in T2DM patients (Pedersen et al. 2016). These findings suggest a potential role of gut microbiota in metabolic disorders, further contributing to the pathogenesis of IR (Pedersen et al. 2016). Recently, metabolomics analyses have been performed in experimental (Du et al. 2020; Zhang et al. 2017b) and epidemiological (GBD 2017 Risk Factor Collaborators 2018) studies to explore the pathophysiological changes under exposure. Those studies evaluated the associations between and lipid metabolites (GBD 2017 Risk Factor Collaborators 2018) and identified biomarkers associated with exposure using metabolomics analysis (Du et al. 2020; Zhang et al. 2017b). Other studies also provide evidence of a link between and the genera of the gut microbiota (Alderete et al. 2018; Liu 2019; Qin et al. 2012). The potential mechanism might be that 5% of particles found in the circulation come from gastrointestinal absorption (Bailey et al. 2020; Dujardin et al. 2020; Feng et al. 2020; Schleh et al. 2012; U.S. EPA 2019) and may be linked to changes in the gut microbiota (Alderete et al. 2018; Liu et al. 2020b; Qin et al. 2012). Evidence from previous studies measured the percentage of inhaled particles in the lungs and characterized the translocation of particles from the respiratory tract (Semmler et al. 2004; Smith et al. 2011, 2014; U.S. EPA 2019). These particles could reach the gastrointestinal tract via many pathways, not only via the air–blood barrier (Smith et al. 2011; U.S. EPA 2019). A study based on a healthy adult population found that 99.95% of the particle deposition in the posterior nose was translocated to the gastrointestinal tract, no matter what the size of the inhaled particles (Smith et al. 2011, 2014). A small percentage of particles may cross cell membranes from their deposition site and distribute throughout the gastrointestinal tract (U.S. EPA 2019). Semmler et al. demonstrated that particles from the peripheral lung were cleared to the gastrointestinal tract via the mucociliary escalator (Semmler et al. 2004). Based on this evidence of gastrointestinal absorption, exposure might be linked to the gut microbiota and metabolic diseases. Therefore, we hypothesized that the genera of the gut microbiota altered by might mediate serum metabolome IR/T2DM.
Here, we conducted a prospective repeated-measures panel study in Shandong Province, China, to further understand the potential mechanism between and IR. This study aimed a) to estimate the association between personal exposure to and fasting blood glucose and insulin, IR index, and other related biomarkers and b) to explore potential underlying mechanisms among exposure, the gastrointestinal tract, and insulin resistance using the microbiome and metabolome.
Methods
Study Participants and Design
The Jinan panel study was launched in the Dianliu community, Shandong Province, from September 2018 to January 2019. Seventy-six healthy seniors of age (37 male and 39 female) were recruited and scheduled to participate in clinical examinations at baseline and subsequently at four repeated visits every month during the study period. The inclusion criteria for the participants were as follows: a) healthy elderly individuals () living in the Dianliu community, b) living at the current residential address for more than 2 y without travel plans during the survey period, and c) voluntary participation with good compliance and good living habits. The exclusion criteria were as follows: a) tobacco use or alcohol abuse; b) auditory disorders or language barriers; c) diagnosed diseases, such as diabetes mellitus, respiratory diseases, cardiovascular and cerebrovascular diseases, cancers or tumors, hyperglycemia, hyperlipidemia, or hypertension; d) fever or infection in the past month; e) use of antibiotics, hormones, anti-inflammatory medications, or other medications in the past month; f) vegetarians or those with abnormal dietary patterns; g) abnormal pulmonary function; and h) a body mass index .
A unique identification number was assigned to each participant at enrollment. The baseline survey was conducted from 13 September 2018 to 21 September 2018, followed by four follow-up visits 30 d apart until January 2019 (visit 1: 13 October 2018 to 22 October 2018; visit 2: 12 November 2018 to 20 November 2018; visit 3: 12 December 2018 to 20 December 2018; visit 4: 11 January 2019 to 19 January 2019). A total of 350 person-visits were collected in all analyses. We also provided consecutive 5-d uniform diets for the participants before health examinations to minimize a potential confounding effect of the daily diet. Each participant was asked to complete a detailed questionnaire including personal information (e.g., sex, age, education status, and occupation) and family information (e.g., annual income of the whole family and cooking and drinking habits) in each round of visits. Additionally, functional indices (e.g., height and weight), blood, and feces were collected and examined at Ankang Clinic at baseline and at four repeated visits. This panel study was approved by the Ethical Review Committee of the National Institute of Environmental Health, Chinese Center for Disease Control and Prevention (NIEH, China CDC; No. 201,816), and written informed consent was obtained from every participant.
Clinical and Biomarker Measurements
Functional factors and the insulin resistance index.
Routine blood measurements were immediately conducted at Ankang Clinical. We measured functional factors, including the fasting blood glucose, total cholesterol (TC), and triglyceride (TG) levels, using a Roche Cobas c702 (Roche Diagnostics) at Dian Diagnostics. We also measured the serum insulin levels using a Millipex Human metabolism-related hormone panel (Merck; HMHEMAG-34K-07). Next, we calculated the IR index, including homeostasis model assessment of insulin resistance (HOMA-IR) and the insulin action index (IAI), using the following equations: a) [fasting insulin (in millimoles per liter)]/22.5; and b) 1/[fasting insulin (in millimoles per liter)].
Inflammation and other IR-related biomarkers.
Blood samples (one without anticoagulation and two with EDTA in a total of per subject) were collected during the clinical examination of all the participants. A blank control was set every 10 samples during the sampling step. All the samples were stored at and then were transported to the National Biobank at the NIEH, China CDC, via a cold-chain shipment. We used a Millipex Human Inflammation Panel (HSTCMAG28SPMX21; Merck) (Scally et al. 2018, 2019), a metabolism-related hormone panel (HMHEMAG-34K-07; Merck) and a cardiovascular disease panel (HCVD3MAG-67K; Merck) to measure inflammation [e.g., interleukin 4 (IL-4), interleukin 6 (IL-6), interleukin 10 (IL-10), interleukin 13 (IL-13), interleukin 23 (IL-23), macrophage inflammatory protein-3 alpha (), and tumor necrosis factor-alpha ()] and other IR-related biomarkers, including monocyte chemotactic protein-1 (MCP-1), peptide YY (PYY), C-reactive protein (CRP), fetuin A, and serum amyloid protein (SAP).
Untargeted metabolomics.
All the tests were conducted according to the manufacturer’s instructions, and samples were stored at . Untargeted metabolomics were detected at Dian Diagnostics. These analytical methods for serum untargeted metabolomics were commonly used in previous studies (de Groot et al. 2020; Koronowski et al. 2019; Labbé et al. 2019). Each sample received was accessioned into the Metabolon Laboratory Information Management System (LIMS) and was assigned a unique identifier by the LIMS that was associated with the original source identifier only. This identifier was used to track all sample handling, tasks, and the results, and the samples (and all derived aliquots) were tracked using the LIMS. Ultrahigh-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) was used to archive and extract raw data files, including bioinformatics, LIMS, data extraction and compound identification, curation, metabolite quantification, and data normalization.
An automated MicroLab STAR system from the Hamilton Company was used to prepare the samples. Quality control samples were added before the first step to assess the data repeatability. Before analysis, the samples were stored overnight under nitrogen using ultrahigh-performance liquid chromatography–mass spectrometry (UPLC-MS). Ion peaks from UPLC-MS were annotated through the human metabolite database and the Discovery HD4 Metabolomics Platform.
Gut microbiota.
Fecal samples were collected using a commercial collection kit (MGIEasy) with a total mass of . This collection kit contained a preservative (MGI Tech) that stabilizes DNA at ambient temperatures, providing a professional tool for precise health research and intestinal microorganism detection. Participants sampled their feces following detailed instructions. After collection, the fecal samples were immediately stored at until analysis.
DNA extraction and 16S rRNA gene testing of distinct regions (16S V4/16S V3-V4/16S V4-V5) were conducted at Novogene (Alderete et al. 2018; Fouladi et al. 2020; Rothschild et al. 2018), and PCR was analyzed using Phusion High-Fidelity PCR Master Mix (New England Biolabs). The TruSeq DNA-PCR-Free Sample Preparation Kit (Illumina) was used to generate sequencing libraries and add index codes. The library quality was assessed using a Qubit 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system and was sequenced using the Illumina HiSeq 2500 platform. The data were rarefied to the average number of reads detected in a single sample.
Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using Uparse (version 7.0.1090; http://drive5.com/uparse/). The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier (version 2.11; http://sourceforge.net/projects/rdp-classifier/) against the Silva (SSU115) 16S rRNA database using a confidence threshold of 70%. Sequence analysis was denoised using Usearch software (version 10; http://drive5.com/usearch). A denoised sequence is called a “ZOTU” (zero-radius OTU). Each ZOTU was screened for further annotation. For each representative sequence, the Greengenes Database (release 13.5; http://greengenes.secondgenome.com/) was used based on the RDP classifier algorithm to annotate taxonomic information.
OTU abundance information was normalized using a standard sequence number corresponding to the sample with the fewest sequences. Subsequent analyses of alpha diversity and beta diversity were all performed based on these output normalized data. The Shannon diversity index and observed OTU levels were used as the alpha diversity metrics. All these indices in our samples were calculated using QIIME (version 1.7.0; http://qiime.org/scripts/assign_taxonomy.html) and displayed using R software (version 2.15.3; R Project). Beta diversity analysis was used to evaluate differences in species complexity of the samples; beta diversity was calculated using unweighted UniFrac and assessed using principal component analysis (PCA) by QIIME (version 1.7.0) with R software (version 3.3.1).
Personal and Meteorological Parameter Measurements
Each participant was required to wear a portable MicroPEM sampler (version 3.2; RTI International) to mimic actual real-time personal exposure. A plastic sampling tube of approximately 40 cm was attached to the sampler and kept near the breathing zone of the participant to mimic actual personal exposures. Each participant was required to wear the sampler for 3 d before the health examinations but was allowed to place the sampler nearby when at home. Researchers suggested that all participants maintain their daily activity during the study period. The real-time concentrations were recorded every 10 s, and 1-min average concentrations of ambient were measured using the MicroPEM. The researchers checked the wearing of MicroPEM regularly to ensure the accuracy of the collected data. The hourly mean concentrations of personal were calculated based on the percentage of effective data in 1 h. Otherwise, it was marked as a missing value. The real-time data from MicroPEM analyzed in this study were all gravimetric corrected data based on the filter gravimetric mass and the sampled air volume. Multiple separate lag period measures according to exposure were computed as the 1-h mean concentrations before the day of the health examinations, including 0–6 h, 0–12 h, 0–24 h, 0–36 h, 0–48 h, and 0–72 h. We also recorded personal temperature and relative humidity data using the MicroPEM. The frequency of measurements and the data cleaning strategy were the same as those for . To adjust for gaseous pollutants, we collected the real-time concentrations of , sulfur dioxide (), nitrogen dioxide (), carbon monoxide (CO), and ozone () from a nearby fixed-site monitoring station during the study period.
Statistical Analyses
Data processing for untargeted metabolomics and gut microbiota.
The peaks of metabolic compounds were quantified using the area under the curve, and data normalization was performed to correct any variation. The OTU abundance information of 16S rRNA genes was normalized using a standard sequence number according to the fewest sequence samples, and species with relative abundance were removed. Alpha diversity and beta diversity were also analyzed in the following steps. The Kruskal-Wallis H test was used to test the significant differences in featured genera among all the study participants.
Descriptive Analysis.
Individual exposure and health data were linked using a unique identification number. Descriptive statistics were conducted for all biomarkers, , and meteorological parameters. Additionally, the means and standard deviations were calculated for continuous variables and proportions for categorical variables.
Statistical models.
Linear mixed-effects (LME) models, considering the correlation within subjects in the panel study design, were used to analyze the associations between and health data (e.g., functional factors, the IR index, inflammation and other IR-related biomarkers, metabolites, and gut microbiota). The associations were also analyzed in the gender-stratified stage. Health indicators with abnormal distributions were log-transformed before statistical analyses. We included several covariates in the models: age (continuous), sex (female or male), BMI (continuous), annual income (continuous), education status (below primary school, primary school, junior school, senior high school, university), cooking and drinking habits (yes or no), blood cotinine level as a marker of passive smoking (continuous), day of the week of the clinical visit, personal temperature and relative humidity data with both 3 degrees of freedom to control for potential lagged and nonlinear effects of weather conditions. The inclusion criteria of the covariates for adjustment in the LME models were the same as those reported previously (Li et al. 2019; Wu et al. 2013; Xu et al. 2019) using the same panel-based study design. We examined exposures to at multiple separate lag periods, including 0–6 h, 0–12 h, 0–24 h, 0–36 h, 0–48 h, and 0–72 h. All the estimates were reported as percent changes with 95% CIs associated with a increase in the concentrations. The Benjamini-Hochberg false discovery rate (FDRB-H) method was used to account for multiple testing to adjust the probability of Type I error (p) value, with considered statistically significant. All analyses were performed using R (version 3.6.1) with the “lme4” package. Sensitivity analyses were performed on model specification by removing income or education from the main model or using the accumulated days or month of the health examinations to replace the day of the week of the clinical visit. We also conducted sensitivity analysis to examine the robustness of our results to the adjustment of concomitant exposure to gaseous pollutants.
We also performed mediation analyses to evaluate the effects of mediators (gut microbiota) on the associations between exposure () and featured metabolism outcomes. This approach divided the total effect of on the measured biomarkers into a direct effect of exposure and a mediation effect accounting for mediators (Albert 2008; Shrout and Bolger 2002). Two LME models were fitted with random intercepts in the single-mediator model, with one modeling the exposure–mediator association and the other modeling the mediator–outcome association. In this study, the direct effect of on sphingolipid metabolism was independent of the effects of the gut microbiota. The indirect or mediation effects quantify whether and how much of the effect of on sphingolipid metabolism is due to or is mediated by the gut microbiota to illustrate the mechanism by which affects sphingolipid metabolism. The proportion mediated is the proportion of the total effect due to a mediator, estimated as the mediation effect of the gut microbiota divided by the total effect of on sphingolipid metabolism. Because the mediation effect can be positive or negative, most studies (Albert 2008; Collins et al. 1998; Liao et al. 2019; Mascha et al. 2013; Shrout and Bolger 2002) agree with these three requirements: a) the total effect must be significant; b) the mediation effect must be significant; and c) the proportion mediated is in the positive direction. This analysis was performed using R (version 3.6.1) with the “mediation” package.
Pathway analysis for metabolites.
To identify the potential pathway related to IR, we conducted pathway analysis for metabolites. A total of 253 significantly differentiated metabolites were identified in serum based on the LME models. These significant metabolites were then uploaded to pathway identification and enrichment analysis modules (MetaboAnalyst 4.0) for statistical, functional, and integrative analysis to further identify the IR-related pathways.
Results
Baseline Description
Table 1 summarizes the demographic statistics of the 76 participants in the Jinan panel study. Of the 76 participants, 39 (51%) were women. The average age and BMI of the subjects were 64.45 () y old and 25.04 , respectively. The descriptive statistics of the repeated measurement biomarkers and potential behavioral risk factors are also provided in Table 1. Table S1 shows more details concerning the study population and examined biomarkers at four repeated visits. Table 2 illustrates the average concentrations measured during the 3 d before health examinations in separate lag periods. On average, the personal concentration over 3 d was . The personal temperature and humidity are also summarized in Table 2.
Table 1.
Descriptive characteristics of 76 study participants at baseline and repeated measurement variables in the Jinan panel from 2018 to 2019.
| Demographic variables | (%) | Range | |
|---|---|---|---|
| Characteristicsa | |||
| Gender | |||
| Male | 37 (48.68) | — | — |
| Female | 39 (51.32) | — | — |
| Age (y) | — | 60–70 | |
| Education | — | 76 | 100 |
| Below primary school | 5 | 5 | 6.58 |
| Primary school | 3 | 3 | 3.95 |
| Junior school | 21 | 21 | 27.63 |
| Senior high school | 33 | 33 | 43.42 |
| University | 14 | 14 | 18.42 |
| Height (cm) | — | 143–178 | |
| Weight (kg) | — | 44.1–85 | |
| BMI () | — | 17.86–28.19 | |
| Income (10,000 RMB) | — | 0–36 | |
| Cotinine () | — | 0.01–84.38 | |
| Drink alcohol | — | 76 | 100 |
| Yes | 2 | 2 | 2.63 |
| No | 74 | 74 | 97.37 |
| Cook | — | 76 | 100 |
| Yes | 65 | 65 | 85.53 |
| No | 11 | 11 | 14.47 |
| Repeated mseasurement variables | Range | ||
|---|---|---|---|
| Functional indicators | |||
| Glucose () | 293 | 4.49–13.05 | |
| TC () | 275 | 2.82–11.39 | |
| TG () | 275 | 0.63–4.28 | |
| Insulin resistance indexes | |||
| Insulin () | 349 | 1.71–26.58 | |
| HOMA-IR | 292 | 0.35–13.31 | |
| IAI | 292 | 0.003–0.13 | |
| Insulin resistance–related biomarkers (inflammation) | |||
| IL-4 () | 350 | 5.84–1131 | |
| IL-6 () | 350 | 0.04–30.73 | |
| IL-10 () | 350 | 0.35–518.01 | |
| IL-13 () | 350 | 0.06–16.14 | |
| IL-23 () | 350 | 1.78–5062 | |
| () | 350 | 2.69–755.73 | |
| () | 350 | 1.05–8.87 | |
| Insulin resistance–related biomarkers (other) | |||
| MCP-1 () | 350 | 25.16–326.03 | |
| PYY () | 350 | 7.26–170.67 | |
| CRP () | 350 | 0.2–153.6 | |
| Fetuin A () | 350 | 93.6–664.4 | |
| SAP () | 350 | 2.8–36.4 | |
Note: —, no data; BMI, body mass index; CRP, C-reactive protein; HOMA-IR, homeostasis model assessment of insulin resistance; IAI, insulin action index; IL, interleukin; IR, insulin resistance; MCP-1, monocyte chemotactic protein-1; MIP, macrophage inflammatory protein; PYY, peptide YY; SAP, serum amyloid protein; SD, standard devitation; TC, total cholesterol; TG, triglyceride; TNF-α, tumor necrosis factor-alpha.
Data are complete for all characteristics.
Table 2.
Descriptive distribution of the personal , temperature, and relative humidity at different lag periods in Jinan during the study period ().
| Variables | Lag (h) | Mean | SD | Min | Median | Max | IQR |
|---|---|---|---|---|---|---|---|
| () | 06 | 49.59 | 67.42 | 2.90 | 38.10 | 993.81 | 30.61 |
| 012 | 58.72 | 53.81 | 8.65 | 45.83 | 609.36 | 38.80 | |
| 024 | 65.67 | 53.34 | 13.02 | 49.15 | 340.64 | 37.21 | |
| 036 | 59.75 | 49.08 | 12.98 | 46.45 | 357.88 | 27.65 | |
| 048 | 60.32 | 49.10 | 12.40 | 48.96 | 337.70 | 27.80 | |
| 072 | 57.11 | 44.87 | 10.98 | 45.27 | 309.55 | 34.58 | |
| Temperature (°C) | 06 | 22.40 | 3.40 | 10.37 | 22.30 | 49.24 | 4.03 |
| 012 | 22.28 | 3.33 | 10.76 | 22.20 | 50.90 | 3.89 | |
| 024 | 21.83 | 3.37 | 11.50 | 21.66 | 51.18 | 3.62 | |
| 036 | 21.91 | 3.34 | 11.16 | 21.66 | 50.41 | 3.76 | |
| 048 | 21.78 | 3.36 | 11.27 | 21.52 | 49.59 | 3.99 | |
| 072 | 21.70 | 3.36 | 11.04 | 21.36 | 47.8 | 3.83 | |
| Relative humidity (%) | 06 | 47.32 | 14.06 | 20.30 | 45.11 | 90.04 | 20.54 |
| 012 | 47.87 | 13.66 | 21.05 | 45.17 | 89.51 | 19.81 | |
| 024 | 47.40 | 13.34 | 21.49 | 44.70 | 91.65 | 16.92 | |
| 036 | 47.18 | 13.21 | 20.39 | 44.62 | 89.22 | 15.09 | |
| 048 | 46.56 | 13.22 | 20.04 | 43.59 | 90.77 | 14.30 | |
| 072 | 45.69 | 12.89 | 20.20 | 42.89 | 87.64 | 14.25 |
Note: IQR, interquartile range; Max, maximum; Min, minimum; SD, standard deviation.
Serum Metabolite Identification
Untargeted metabolomics and pathway analysis.
Untargeted metabolomics profiling (Excel Table S1) revealed that 253 metabolites were significantly () associated with exposure at least one lag after correcting for multiple testing. Specifically, 52 metabolic pathways were analyzed in total. Of those, 11 pathways were significantly identified (). These included metabolic pathways predominantly associated with amino acid metabolism, lipid metabolism, and energy metabolism (Table S2).
Sphingolipid metabolism and IR-index related with .
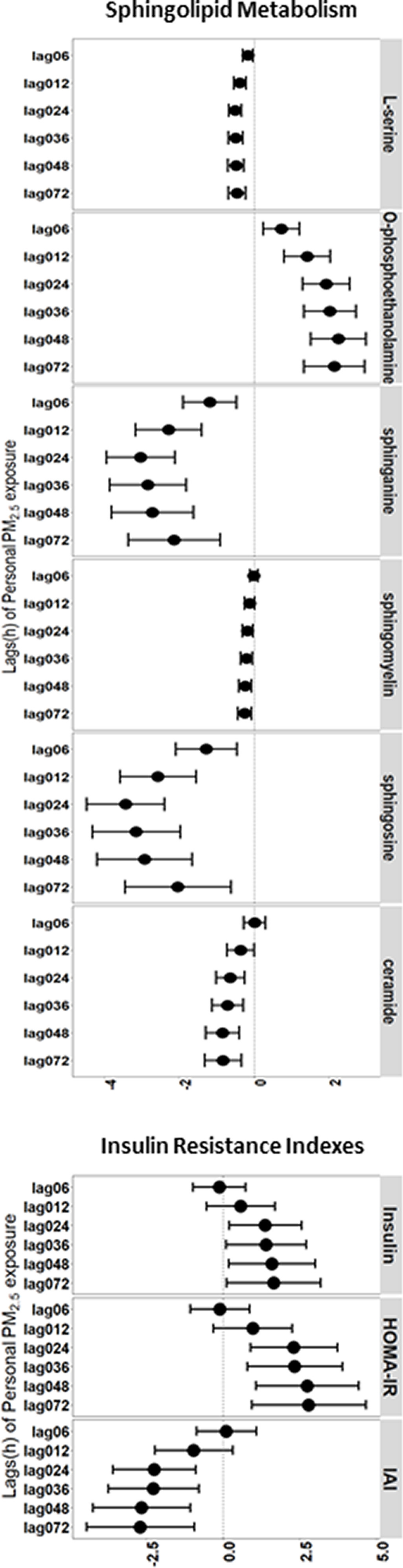
In our study, six featured metabolites of the sphingolipid metabolism pathway (L-serine, O-phosphoethanolamine, sphingasine, sphingomyelin, sphingosine, and ceramide) were significantly associated with exposure (Figure S1). Each increase in the exposure level was associated with a 0.45% [95% confidence interval (CI): , ] decrease in L-serine, a 2.14% (95% CI: , ) decrease in sphingasine, a 0.25% (95% CI: , ) decrease in sphingomyelin, a 2.04% (95% CI: , ) decrease in sphingosine, and a 0.82% (95% CI: , ) decrease in ceramide at a lag of 0–72 h. Additionally, 2.15% (95% CI: 1.34%, 2.96%) increase in O-phosphoethanolamine was observed for each increase in exposure at a lag of 0–72 h (Figure 1; Table S3). The sphingolipid metabolite results of the sex-stratified analysis are shown in Figure S2 and Table S4.
Figure 1.
Percent change in insulin resistance-related biomarkers for a increase in among older Chinese adults in the LME models with 95% conference intervals. Adjusted covariates included age (continuous), sex (female or male), BMI (continuous), annual income (continuous), education status, smoking status, alcohol consumption status, cooking status, day of the week of the clinical visit, temperature, and relative humidity. Numeric data are presented in Table S3.
Similar significant changes were also observed for serum insulin, HOMA-IR and the IAI. For serum insulin and HOMA-IR, each increase in the exposure level was associated with a 1.64% (95% CI: 0.12%, 3.19%) increase in serum insulin and a 2.78% (95% CI: 0.94%, 4.66%) increase in HOMA-IR at a lag of 0–72 h. The IAI decreased by 2.71% (95% CI: , ) according to each increase in exposure at a lag of 0–72 h (Figure 1; Table S3). The IR index results for gender-stratified analysis are shown in Figure S2 and Table S4.
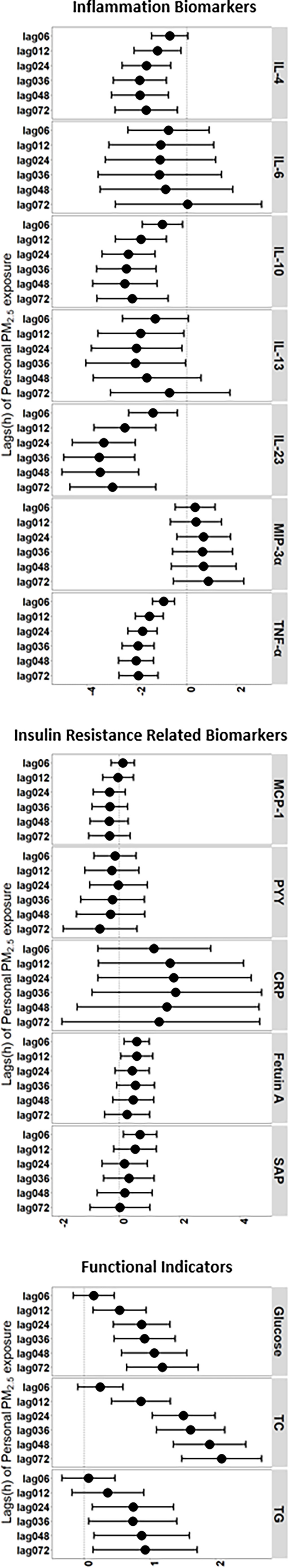
Inflammatory, functional, and other biomarkers related to .
We evaluated four protein biomarkers of the proinflammatory acute phase response and observed fluctuations in IL-6, IL-23, , and associated with elevated exposure periods (Figure 1; Table S3). Similarly, for biomarkers of anti-inflammatory cytokines, exposure was associated with decreases in IL-4, IL-10, and IL-13, particularly at a lag of 0–36 h (Figure 1; Table S3). Higher serum levels of glucose, TC, and TG were also significantly associated with exposure. The associations were present generally at a lag of 0–6 h and remained statistically significant at a lag (Figure 1; Table S3). For other IR-related biomarkers, we observed significantly increasing levels of fetuin A and SAP associated with exposure at a lag of 0–6 h (Figure 1; Table S3). The results of analyses stratified by gender are shown in Figure S2 and Table S4.
Gut Microbiota Sequencing Measurement and Mediation Analysis
The relative abundance distributions of 516 genera were obtained through species annotation. The Shannon and Sobs indexes of the OTU levels were used to evaluate alpha diversity (Figure S3A,B). PCA showed separation in beta diversity among all the participants (Figure S3C). LME models were performed, and 16 featured genera (e.g., Cellulosilyticum, Oribacterium, Shuttleworthia, and Streptococcus; Excel Table S2) were significantly identified () (Figure S4,5). The Kruskal-Wallis H test was used to test the significant differences of 16 featured genera among all the study participants (Table S5). The mediation analysis of the gut microbiota (Oribacterium and Shuttleworthia) regarding the association between personal exposure and sphingolipid metabolites (sphingosine) are shown in Figure 2 and Table S6. A increase in the personal exposure was associated with decreases in sphingolipid metabolism after adjusting for potential confounders. Decreases in the Oribacterium and Shuttleworthia abundance mediated 8.59% (95% CI: 0.01%, 29.34%) and 37.83% (95% CI: 16.42%, 72.90%), respectively, of the total effect of the personal exposure on sphingolipid metabolism at a lag of 0–6 h (Figure 2). Similarly, the Shuttleworthia abundance mediated 8.62% (95% CI: 0.16%, 30.71%) of the total effect of the personal exposure on sphingolipid metabolism with a lag of 0–72 h (Figure 2).
Figure 2.
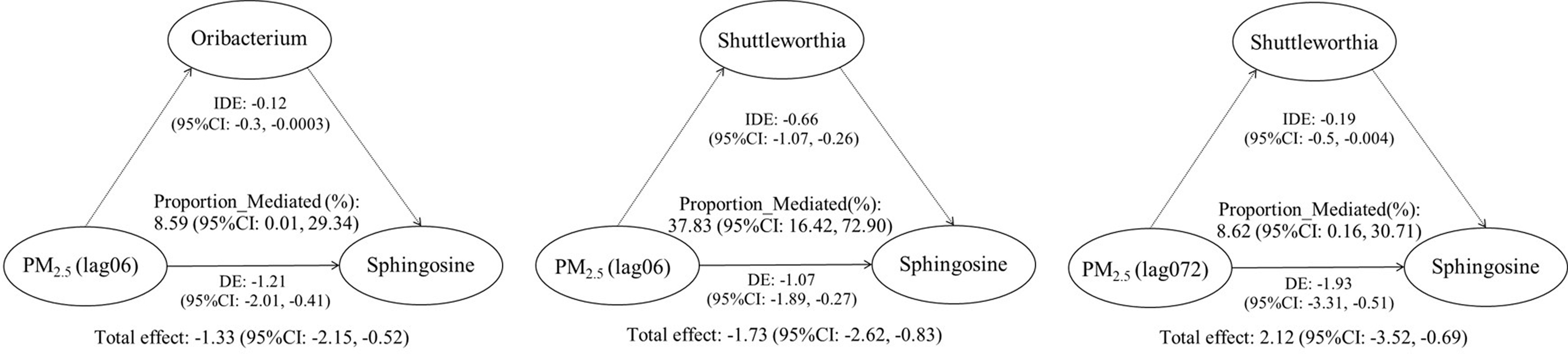
Estimates of the mediation effect of the gut microbiota on the association between exposure to and sphingolipid metabolism among older Chinese adults in the mediation models (per increase in ). Adjusted covariates included age (continuous), sex (female or male), BMI (continuous), annual income (continuous), education status, smoking status, alcohol consumption status, cooking status, day of the week of the clinical visit, temperature, and relative humidity.
Sensitivity Analyses
We conducted sensitivity analyses on model specification by removing income or education from the main model or using the accumulated days or month of the health examinations to replace the day of the week of the clinical visit. Sensitivity analyses showed that our results were consistent and robust (Tables S7 and S8).
Discussion
Our study was a prospective panel study that proposed the hypothesis that the genera of the gut microbiota altered by may mediate serum metabolome IR/T2DM. Exposure to was associated with increased risks of the IR index and functional biomarkers and provided evidence that exposure may contribute to systemic inflammation and altered sphingolipid metabolism (as shown in Figure 3). Given the key role of sphingolipid metabolism in preventing the development of IR, our findings suggest that may change the genera of the gut microbiota, which partially mediates the effects between and sphingolipid metabolism. These findings may extend our current understanding of pathways of and IR.
Figure 3.
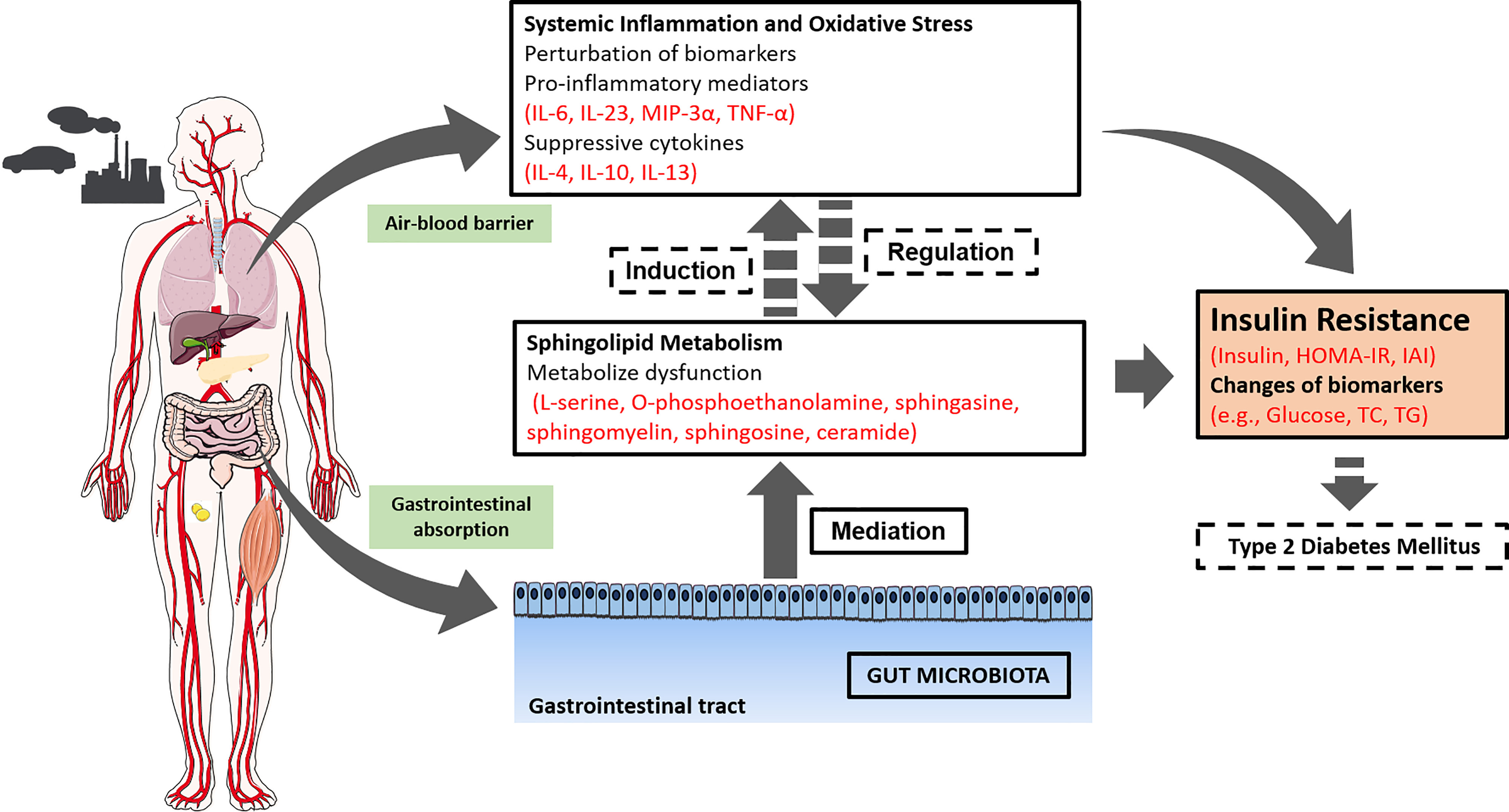
Potential biological pathways by which promotes type 2 diabetes mellitus. Note: The figure represents the effects for which experimental or epidemiologic evidence related to exposure and insulin resistance or type 2 diabetes mellitus is available, and the arrows indicate a proposed relationship between those effects. Solid arrows and lines denote evidence observed in this Jinan panel study, and dotted arrows and lines denote evidence from previous studies.
Insulin resistance is a critical risk factor for T2DM (Kim and Hong 2012). Several epidemiological studies have observed associations between and indicators of glucose and insulin homeostasis (Chen et al. 2016; Liu et al. 2014). Our panel study found increases in circulating glucose, insulin, and HOMA-IR and a decrease in the IAI in collected blood samples after 3 d of exposure. Brook et al. reported increased glucose, insulin, and HOMA-IR among healthy subjects exposed to (Brook et al. 2013). Yang et al. studied the relationship of with diabetes and the glucose homeostasis index in northern China and found that was associated with higher concentrations of glucose and insulin (Yang et al. 2018). The results of those studies were confirmed by our findings. Similar results were also reported in Mexican Americans (Chen et al. 2016) and residents of southern Germany (Wolf et al. 2016). Although the previous studies have linked to an increased risk for developing IR, to our knowledge, very limited epidemiological evidence has been reported for the potential mechanisms of and IR (U.S. EPA 2019).
Human exposure and toxicological studies provide some evidence that systemic inflammation may play a role in metabolic disruption, such as IR and the delivery of 95% PM to the circulatory system, which may cause systemic inflammation from the lung’s air–blood barrier (reviewed by U.S. EPA 2019). These studies indicate that may affect glucose and insulin homeostasis and report positive associations between and insulin or the IR index and systemic inflammation (Chen et al. 2016; Kelishadi et al. 2009; Sun et al. 2009). A cross-sectional study of 374 children showed that air quality is linked to higher CRP and HOMA index values (Kelishadi et al. 2009). A toxicological study also demonstrated that ambient exposure may disturb metabolic progress in healthy mice and exacerbate metabolic disorders in mice with diabetes via glucose tolerance, IR, and systemic inflammation (Pan et al. 2019). Our panel study provides evidence for the perturbation of the serum concentrations of anti-inflammatory cytokines (e.g., IL-4, IL-10, and IL-13) and proinflammatory cytokines (e.g., IL-6, CRP, and ) in association with and IR (Figure 3). To our knowledge, most of the literature has reported significant alterations between and inflammation biomarkers. However, the levels of inflammatory biomarkers did not show consistent changes in response to exposure in different studies. Binisor et al. found increased serum IL-4 levels in diabetic patients likely due to IL-4 resistance in patients with type 2 diabetes (Binisor et al. 2016). Although we observed a decrease in IL-6, which is not consistent with several short-term exposure studies (Liu et al. 2014; Pan et al. 2019), other studies also demonstrate that chronically increased IL-6 might benefit metabolic progress (Harsch and Konturek 2018; Xie et al. 2019).
The evidence for systemic inflammation following short-term exposure is limited, with some studies reporting changes in inflammation biomarkers (Binisor et al. 2016; Rao et al. 2015; Sun et al. 2009), whereas other studies show no changes in these biomarkers (Chen et al. 2016; Li et al. 2017; Pan et al. 2019). The response to acute inflammation is dynamic, and it is technically difficult to measure cytokine levels below the detection limit (Liu et al. 2020a; Xie et al. 2019). Therefore, several factors were related to the inconsistency across those studies, including the transient nature of changes in inflammation biomarkers. These conflicting findings on inflammatory biomarkers revealed the complexity of cytokine molecules in regulating metabolism. Our study suggests an inflammatory mechanism between and IR.
Untargeted metabolomics has emerged as a powerful approach to explore the enrichment pathways between and IR/T2DM by providing new insights into the mechanisms (Jeong et al. 2018; Liang et al. 2018). In our panel study, 52 metabolic pathways were analyzed using 253 metabolites. Specifically, 11 pathways associated with amino acid metabolism, lipid metabolism, and energy metabolism were significantly identified. Consistent with these potential pathways, our untargeted systematic approach confirmed that sphingolipid metabolism was associated with exposure (Zhao et al. 2019), a finding consistent with the significantly increased risk of IR (Chen et al. 2016; Sun et al. 2009; Figure 3). Sphingolipids, such as ceramide and sphingosine, constitute the structure of cell membranes, and both epidemiological studies (Bellini et al. 2015; Lemaitre et al. 2018) and an animal study (Maceyka et al. 2005) suggest an important role for sphingolipids in the development of IR. Ceramides are critical mediators of lipotoxicity and accumulate in insulin-sensitive tissues (Bellini et al. 2015; Zhao et al. 2019). However, limited attention has been allocated to the distinguishing role of sphingolipids in IR. Lemaitre et al. investigated sphingolipids associated with insulin and other biomarkers in a cohort study (Lemaitre et al. 2018). Higher plasma levels of ceramides were associated with increased insulin levels and HOMA-IR, mirroring our findings of decreasing concentrations of ceramides. The reason may be the health effects of personal exposure among the healthy elderly population. Ceramides measured in blood samples may not be consistent with those measured at the tissue level. The results from a mouse model (Xie et al. 2019) showed that exposure to reduced the levels of sphingomyelin and ceramide and sequentially induced the decreased secretion of cytokines, such as and IL-6. This study supported our findings and indicated that metabolic pathways, including sphingomyelin–ceramide signaling, were disrupted. At the metabolic level, exposure to significantly altered sphingolipid metabolism (Zhao et al. 2019), including the observed changes in L-serine, O-phosphoethanolamine, sphingasine, sphingomyelin, sphingosine, and ceramide. The potential mechanism underlying the changes was as follows: Ceramide and key enzymes in the sphingolipid pathway induce the secretion of proinflammatory cytokines, the generation of lipotoxicity, and alterations of RNA splicing (Gulbins and Kolesnick 2003; Maceyka et al. 2005; Meyer zu Heringdorf and Jakobs 2007; Zhao et al. 2019).
The results from animal experiments suggest that environmental pollutants significantly alter the gut microbiota composition and change the processes of metabolism (Salim et al. 2014b). may carry microorganisms, which would induce the proinflammatory response of the immune system, increase intestinal permeability, and lead to the growth of specific microbiota (Wu et al. 2020). These changes in the host environment will alter the genera of the gut microbiota (Ananthakrishnan et al. 2018; Salim et al. 2014b, 2014a). In accordance with previous studies, we observed statistically significant relationships between personal exposure to and the gut microbiota (Alderete et al. 2018; Harsch and Konturek 2018; Qin et al. 2012; Wang et al. 2018).
The gut microbiota could bioactivate inorganic compounds, which may increase the development of chronic diseases (Van de Wiele et al. 2005). Studies have shown that exposure to PM is related to increased intestinal permeability accompanied by intestinal inflammation (Alderete et al. 2017; Arrieta 2006; Salim et al. 2014b). Previous studies have demonstrated that alterations in the gut microbiota may be another crucial mechanism for IR. Pedersen et al. demonstrated that gut microbiota imbalance affects metabolic progress and may further develop into IR (Pedersen et al. 2016), a finding that was consistent with ours. Our analysis also found that 37.83% of the total effect of personal exposure on sphingolipid metabolism could be explained by the mediation effect of the gut microbiota (Shuttleworthia). However, limited information exists on the genus Shuttleworthia, and its function in the gut is unclear. Shuttleworthia is the normal component of the oral microbiome (Downes 2002), and it was demonstrated in a cohort study to be overrepresented in patients who had alcohol use disorder with alcoholic liver disease (Maccioni et al. 2020). These findings suggest that the translocation of Shuttleworthia may be a potential determinant in alcoholic liver disease progression (Maccioni et al. 2020). In this panel study, the mediation results agree with an epidemiological study (Liu 2019) that illustrated the mediation effects of human gut microbiota associated with and IR. These findings revealed that gut microbiota may play an important mediating role in the mechanism of , contributing to IR or the further development of T2DM.
Our study has several strengths. It was a prospective study based on a healthy elderly population, which explored the hypothetical relationship among , potential biomarkers, and the risk of IR. Panel-based study designs are considered an effective method to investigate the associations between and health outcomes (Feng et al. 2020; He et al. 2021; Sarnat et al. 2012; Wu et al. 2011). Second, the availability of data on lifestyle and multiple risk factors allowed us to adjust for many factors that may confound the associations. Third, we provided consecutive 5-d uniform diets for the participants before health examinations to minimize the potential confounding of daily diets, which was seldom taken in previous studies (Liang et al. 2018; Liu 2019; Qin et al. 2012). Fourth, we accurately measured the personal exposure level of instead of fixed-site monitoring stations, providing more reliable exposure data for each subject, avoiding the underestimation of measures of association, and increasing the statistical power of the study. Finally, we introduced omics, such as untargeted metabolomics and microbiomics, and identified potential pathways of IR.
However, our study has some limitations. First, this panel study was conducted in a unique population of healthy elderly individuals, and is highly heterogeneous and varies in chemical constituents; therefore, whether our findings could be generalizable to other populations and different localities or be translated to long-term health impacts remains to be examined. Second, this panel study has limited capacity to shed light on the pathways linking to IR/T2DM, and the findings only allow us to generate the hypothesis that the genera of the gut microbiota altered by may mediate serum metabolome IR/T2DM. Third, we did not use the glucose insulin clamp technique (DeFronzo et al. 1979), which uses the infusion of exogenous insulin and glucose to raise plasma insulin levels and maintain blood glucose at basic steady levels, to measure the levels of insulin and glucose in target tissues because the technique was not available for the healthy elderly population. Instead, we used HOMA-IR, the IAI, and circulating insulin as indices for IR to approximately assess IR status, which was largely driven by insulin and glucose levels and could substitute for clamp experiments (Gall et al. 2010). Fourth, we used confidence-based computational approaches for metabolite annotation, and a large portion of unidentifiable metabolites that may be functionally or mechanistically important was not characterized based on the current knowledge. Finally, we cannot exclude confounding entirely, although we have considered demographic factors, socioeconomic status, and living habits as covariates in the LME models according to current epidemiological studies.
In summary, our panel study of a microbiome and metabolomics analysis of identified potential metabolic pathways in the development of IR. Our analyses suggest that significant changes in the IR index, metabolites of sphingolipids, inflammation, and functional biomarkers are associated with changes in exposure levels. Additionally, may alter the genera of the gut microbiota, which partially mediates the association between and sphingolipid metabolism. Future studies should verify that these potential mechanisms of IR are associated with exposure and improved health in the long term.
Supplementary Material
Acknowledgments
This study was supported by the National Research Program for Key Issues in Air Pollution Control (grant numbers DQGG0401 and DQGG0401-04).
T.L. and X.S. contributed to the concept and design of the study. T.L. developed the analysis methods and guided the data analysis and manuscript writing. L.Z. drafted the manuscript and conducted the statistical analyses. J.F. and S.T. implemented the field study. All the authors contributed to the study design and interpretation of results, critically revised the manuscript, and approved the final version of the paper.
The data sets generated and R codes analyzed during this study will be made available from the corresponding authors on reasonable request.
References
- Albert JM. 2008. Mediation analysis via potential outcomes models. Stat Med 27(8):1282–1304, PMID: , 10.1002/sim.3016. [DOI] [PubMed] [Google Scholar]
- Alderete TL, Habre R, Toledo-Corral CM, Berhane K, Chen Z, Lurmann FW, et al. 2017. Longitudinal associations between ambient air pollution with insulin sensitivity, β-Cell function, and adiposity in Los Angeles Latino children. Diabetes 66(7):1789–1796, PMID: , 10.2337/db16-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderete TL, Jones RB, Chen Z, Kim JS, Habre R, Lurmann F, et al. 2018. Exposure to traffic-related air pollution and the composition of the gut microbiota in overweight and obese adolescents. Environ Res 161:472–478, PMID: , 10.1016/j.envres.2017.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR, et al. 2018. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol 15(1):39–49, PMID: , 10.1038/nrgastro.2017.136. [DOI] [PubMed] [Google Scholar]
- Arrieta MC, Bistritz L, Meddings JB. 2006. Alterations in intestinal permeability. Gut 55(10):1512–1520, PMID: , 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Naik NN, Wild LE, Patterson WB, Alderete TL. 2020. Exposure to air pollutants and the gut microbiota: a potential link between exposure, obesity, and type 2 diabetes. Gut Microbes 11(5):1188–1202, PMID: , 10.1080/19490976.2020.1749754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini L, Campana M, Mahfouz R, Carlier A, Véret J, Magnan C. 2015. Targeting sphingolipid metabolism in the treatment of obesity/type 2 diabetes. Expert Opin Ther Targets 14:. [DOI] [PubMed] [Google Scholar]
- Binisor ID, Moldovan R, Moldovan I, Andrei AM, Banita MI. 2016. Abdominal obesity and type 2 diabetes mellitus are associated with higher seric levels of IL 4 in adults. Curr Health Sci J 42(3):231–237, PMID: , 10.12865/CHSJ.42.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Xu X, Bard RL, Dvonch JT, Morishita M, Kaciroti N, et al. 2013. Reduced metabolic insulin sensitivity following sub-acute exposures to low levels of ambient fine particulate matter air pollution. Sci Total Environ 448:66–71, PMID: , 10.1016/j.scitotenv.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Salam MT, Toledo-Corral C, Watanabe RM, Xiang AH, Buchanan TA, et al. 2016. Ambient air pollutants have adverse effects on insulin and glucose homeostasis in Mexican Americans. Diabetes Care 39(4):547–554, PMID: , 10.2337/dc15-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LM, Graham JJ, Flaherty BP. 1998. An alternative framework for defining mediation. Multivariate Behav Res 33(2):295–312, PMID: , 10.1207/s15327906mbr3302_5. [DOI] [PubMed] [Google Scholar]
- Dang J, Yang M, Zhang X, Ruan H, Qin G, Fu J, et al. 2018. Associations of exposure to air pollution with insulin resistance: a systematic review and meta-analysis. IJERPH 15(11):2593, 10.3390/ijerph15112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, et al. 2020. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69(3):502–512, PMID: , 10.1136/gutjnl-2019-318320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo RA, Tobin JD, Andres R. 1979. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol 237(3):E214–E223, PMID: , 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- Downes J, Munson MA, Radford DR, Spratt DA, Wade WG. 2002. Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 52(Pt 5):1469–1475. PMID: 12361248, 10.1099/00207713-52-5-1469. [DOI] [PubMed] [Google Scholar]
- Du X, Zeng X, Pan K, Zhang J, Song L, Zhou J, et al. 2020. Metabolomics analysis of urine from healthy wild type mice exposed to ambient PM2.5. Sci Total Environ 714:136790, PMID: , 10.1016/j.scitotenv.2020.136790. [DOI] [PubMed] [Google Scholar]
- Dujardin CE, Mars RAT, Manemann SM, Kashyap PC, Clements NS, Hassett LC, et al. 2020. Impact of air quality on the gastrointestinal microbiome: a review. Environ Res 186:109485, PMID: , 10.1016/j.envres.2020.109485. [DOI] [PubMed] [Google Scholar]
- Eze IC, Hemkens LG, Bucher HC, Hoffmann B, Schindler C, Künzli N, et al. 2015. Association between ambient air pollution and diabetes mellitus in Europe and North America: systematic review and meta-analysis. Environ Health Perspect 123(5):381–389, PMID: , 10.1289/ehp.1307823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Cavallero S, Hsiai T, Li R. 2020. Impact of air pollution on intestinal redox lipidome and microbiome. Free Radic Biol Med 151:99–110, PMID: , 10.1016/j.freeradbiomed.2019.12.044. [DOI] [PubMed] [Google Scholar]
- Fouladi F, Bailey MJ, Patterson WB, Sioda M, Blakley IC, Fodor AA, et al. 2020. Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ Int 138:105604, PMID: , 10.1016/j.envint.2020.105604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Beebe K, Lawton KA, Adam K-P, Mitchell MW, Ryals JA, et al. 2010. α-Hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One 5(5):e10883. PMID: , 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2017 Risk Factor Collaborators. 2018. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 10;392(10159):1923–1994, PMID: , 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbins E, Kolesnick R. 2003. Raft ceramide in molecular medicine. Oncogene 22(45):7070–7077, PMID: , 10.1038/sj.onc.1207146. [DOI] [PubMed] [Google Scholar]
- Harsch I, Konturek P. 2018. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into “old” diseases. Med Sci 6(2):32, 10.3390/medsci6020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z-Z, Guo P-Y, Xu S-L, Zhou Y, Jalaludin B, Leskinen A, et al. 2021. Associations of particulate matter sizes and chemical constituents with blood lipids: a panel study in Guangzhou, China. Environ Sci Technol 55(8):5065–5075, PMID: , 10.1021/acs.est.0c06974. [DOI] [PubMed] [Google Scholar]
- Health Effects Institute. 2019. State of Global Air: a special report on global exposure to air pollution and its disease burden. http://www.healthdata.org/news-release/state-global-air-2019-report [accessed 6 June 2020].
- Jeong A, Fiorito G, Keski-Rahkonen P, Imboden M, Kiss A, Robinot N, et al. 2018. Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases. Environ Int 119:334–345, PMID: , 10.1016/j.envint.2018.06.025. [DOI] [PubMed] [Google Scholar]
- Kelishadi R, Mirghaffari N, Poursafa P, Gidding SS. 2009. Lifestyle and environmental factors associated with inflammation, oxidative stress and insulin resistance in children. Atherosclerosis 203(1):311–319, PMID: , 10.1016/j.atherosclerosis.2008.06.022. [DOI] [PubMed] [Google Scholar]
- Kim JH, Hong Y-C. 2012. GSTM1, GSTT1, and GSTP1 polymorphisms and associations between air pollutants and markers of insulin resistance in elderly Koreans. Environ Health Perspect 120(10):1378–1384, 10.1289/ehp.1104406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronowski KB, Kinouchi K, Welz P-S, Smith JG, Zinna VM, Shi J, et al. 2019. Defining the independence of the liver circadian clock. Cell 177(6):1448–1462.e14, PMID: , 10.1016/j.cell.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé DP, Zadra G, Yang M, Reyes JM, Lin CY, Cacciatore S, et al. 2019. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC program. Nat Commun 10(1):4358, PMID: , 10.1038/s41467-019-12298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrskov LL, Christensen RH. 2019. The role of interleukin-6 in glucose homeostasis and lipid metabolism. Semin Immunopathol 41(4):491–499, PMID: , 10.1007/s00281-019-00747-2. [DOI] [PubMed] [Google Scholar]
- Lemaitre RN, Yu C, Hoofnagle A, Hari N, Jensen PN, Fretts AM, et al. 2018. Circulating sphingolipids, insulin, HOMA-IR, and HOMA-B: The Strong Heart Family Study. Diabetes 67(8):1663–1672. PMID: , 10.2337/db17-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Cai J, Chen R, Zhao Z, Ying Z, Wang L, et al. 2017. Particulate matter exposure and stress hormone levels: a randomized, double-blind, crossover trial of air purification. Circulation 136(7):618–627, PMID: , 10.1161/CIRCULATIONAHA.116.026796. [DOI] [PubMed] [Google Scholar]
- Li J, Zhou C, Xu H, Brook RD, Liu S, Yi T, et al. 2019. Ambient air pollution is associated with HDL (high-density lipoprotein) dysfunction in healthy adults. Arterioscler Thromb Vasc Biol 39(3):513–522. PMID: , 10.1161/ATVBAHA.118.311749. [DOI] [PubMed] [Google Scholar]
- Liang D, Moutinho JL, Golan R, Yu T, Ladva CN, Niedzwiecki M, et al. 2018. Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ Int 120:145–154, PMID: , 10.1016/j.envint.2018.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Li Y, Wang X, Zhang B, Xia W, Peng Y, et al. 2019. Prenatal exposure to fine particulate matter, maternal hemoglobin concentration, and fetal growth during early pregnancy: associations and mediation effects analysis. Environ Res 173:366–372, PMID: , 10.1016/j.envres.2019.03.056. [DOI] [PubMed] [Google Scholar]
- Liu T, Chen X, Xu Y, Wu W, Tang W, Chen Z, et al. 2019. Gut microbiota partially mediates the effects of fine particulate matter on type 2 diabetes: evidence from a population-based epidemiological study. Environ Int 130:104882–104811. PMID: , 10.1016/j.envint.2019.05.076. [DOI] [PubMed] [Google Scholar]
- Liu C, Fonken LK, Wang A, Maiseyeu A, Bai Y, Wang T-Y, et al. 2014. Central IKKβ inhibition prevents air pollution mediated peripheral inflammation and exaggeration of type II diabetes. Part Fibre Toxicol 11(1):53, PMID: , 10.1186/s12989-014-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Guo W, Cai Y, Yang H, Li W, Yang L, et al. 2020a. Personal exposure to fine particulate matter and renal function in children: a panel study. Environ Pollut 266(Pt 2):115129, PMID: , 10.1016/j.envpol.2020.115129. [DOI] [PubMed] [Google Scholar]
- Liu Y, Talbot O, Bain JR, Muehlbauer MJ, Hayes MG, Ilkayeva OR, et al. 2020b. Metabolomic and genetic associations with insulin resistance in pregnancy. Diabetologia 63(9):1783–1795, PMID: , 10.1007/s00125-020-05198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccioni L, Gao B, Leclercq S, Pirlot B, Horsmans Y, De Timary P, et al. 2020. Intestinal permeability, microbial translocation, changes in duodenal and fecal microbiota, and their associations with alcoholic liver disease progression in humans. Gut Microbes 12(1):1782157, PMID: , 10.1080/19490976.2020.1782157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, Sankala H, Hait NC, Le Stunff H, Liu H, Toman R, et al. 2005. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. J Biol Chem 280(44):37118–37129, PMID: , 10.1074/jbc.M502207200. [DOI] [PubMed] [Google Scholar]
- Mascha EJ, Dalton JE, Kurz A, Saager L. 2013. Understanding the mechanism: mediation analysis in randomized and nonrandomized studies. Anesth Analg 117(4):980–994, PMID: , 10.1213/ANE.0b013e3182a44cb9. [DOI] [PubMed] [Google Scholar]
- Meyer zu Heringdorf D, Jakobs KH. 2007. Lysophospholipid receptors: signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim Biophys Acta 1768(4):923–940, PMID: , 10.1016/j.bbamem.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pan K, Jiang S, Du X, Zeng X, Zhang J, Song L, et al. 2019. AMPK activation attenuates inflammatory response to reduce ambient PM2.5-induced metabolic disorders in healthy and diabetic mice. Ecotoxicol Environ Saf 179:290–300, PMID: , 10.1016/j.ecoenv.2019.04.038. [DOI] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, et al. 2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535(7612):376–381, PMID: , 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- Pilz V, Wolf K, Breitner S, Rückerl R, Koenig W, Rathmann W. 2018. C-reactive protein (CRP) and long-term air pollution with a focus on ultrafine particles. Int J Hyg Environ Health 221(3):510–518. PMID: , 10.1016/j.ijheh.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418):55–60, PMID: , 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Brook RD. 2012. Air pollution and type 2 diabetes: mechanistic insights. Diabetes 61(12):3037–3045, PMID: , 10.2337/db12-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Patel P, Puett R, Rajagopalan S. 2015. Air pollution as a risk factor for type 2 diabetes. Toxicol Sci 143(2):231–241, PMID: , 10.1093/toxsci/kfu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555(7695):210–215, PMID: , 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Salim SY, Jovel J, Wine E, Kaplan GG, Vincent R, Thiesen A, et al. 2014a. Exposure to ingested airborne pollutant particulate matter increases mucosal exposure to bacteria and induces early onset of inflammation in neonatal IL-10-deficient mice. Inflamm Bowel Dis 20(7):1129–1138, PMID: , 10.1097/MIB.0000000000000066. [DOI] [PubMed] [Google Scholar]
- Salim SY, Kaplan GG, Madsen KL. 2014b. Air pollution effects on the gut microbiota: a link between exposure and inflammatory disease. Gut Microbes 5(2):215–219, PMID: , 10.4161/gmic.27251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnat SE, Raysoni AU, Li W-W, Holguin F, Johnson BA, Luevano SF, et al. 2012. Air pollution and acute respiratory response in a panel of asthmatic children along the U.S.–Mexico border. Environ Health Perspect 120(3):437–444, PMID: , 10.1289/ehp.1003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally C, Abbas H, Ahearn T, Srinivasan J, Mezincescu A, Rudd A, et al. 2019. Myocardial and systemic inflammation in acute stress-induced (takotsubo) cardiomyopathy. Circulation 139(13):1581–1592, PMID: , 10.1161/CIRCULATIONAHA.118.037975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally C, Rudd A, Mezincescu A, Wilson H, Srivanasan J, Horgan G, et al. 2018. Persistent long-term structural, functional, and metabolic changes after stress-induced (takotsubo) cardiomyopathy. Circulation 137(10):1039–1048, PMID: , 10.1161/CIRCULATIONAHA.117.031841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleh C, Semmler-Behnke M, Lipka J, Wenk A, Hirn S, Schäffler M, et al. 2012. Size and surface charge of gold nanoparticles determine absorption across intestinal barriers and accumulation in secondary target organs after oral administration. Nanotoxicology 6(1):36–46, PMID: , 10.3109/17435390.2011.552811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler M, Seitz J, Erbe F, Mayer P, Heyder J, Oberdörster G, et al. 2004. Long-Term clearance kinetics of inhaled ultrafine insoluble iridium particles from the rat lung, including transient translocation into secondary organs. Inhal Toxicol 16(6–7):453–459, PMID: , 10.1080/08958370490439650. [DOI] [PubMed] [Google Scholar]
- Senn JJ, Klover PJ, Nowak IA, Mooney RA. 2002. Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes 51(12):3391–3399, PMID: , 10.2337/diabetes.51.12.3391. [DOI] [PubMed] [Google Scholar]
- Shrout PE, Bolger N. 2002. Mediation in experimental and nonexperimental studies: new procedures and recommendations. Psychol Methods 7(4):422–445, PMID: , 10.1037/1082-989X.7.4.422. [DOI] [PubMed] [Google Scholar]
- Smith JRH, Bailey MR, Etherington G, Shutt AL, Youngman MJ. 2011. An experimental study of clearance of inhaled particles from the human nose. Exp Lung Res 37(2):109–129, PMID: , 10.3109/01902148.2010.518301. [DOI] [PubMed] [Google Scholar]
- Smith JRH, Birchall A, Etherington G, Ishigure N, Bailey MR. 2014. A revised model for the deposition and clearance of inhaled particles in human extra-thoracic airways. Radiat Prot Dosimetry 158(2):135–147, PMID: , 10.1093/rpd/nct218. [DOI] [PubMed] [Google Scholar]
- Sun Q, Yue P, Deiuliis JA, Lumeng CN, Kampfrath T, Mikolaj MB, et al. 2009. Ambient air pollution exaggerates adipose inflammation and insulin resistance in a mouse model of diet-induced obesity. Circulation 119(4):538–546, PMID: , 10.1161/CIRCULATIONAHA.108.799015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2019. Integrated science assessment for particulate matter. https://cfpub.epa.gov/ncea/isa/recordisplay.cfm?deid=347534#tab-3 [accessed 24 May 2020]. [PubMed]
- Van de Wiele T, Vanhaecke L, Boeckaert C, Peru K, Headley J, Verstraete W, et al. 2005. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ Health Perspect 113(1):6–10, PMID: , 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang J, He MZ, Kinney PL, Li T. 2018. A county-level estimate of PM 2.5 related chronic mortality risk in China based on multi-model exposure data. Environ Int 110:105–112, PMID: , 10.1016/j.envint.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Popp A, Schneider A, Breitner S, Hampel R, Rathmann W, et al. 2016. Association between long-term exposure to air pollution and biomarkers related to insulin resistance, subclinical inflammation, and adipokines 65(11):3314–3326. PMID: , 10.2337/db15-1567. [DOI] [PubMed] [Google Scholar]
- Wu S, Deng F, Liu Y, Shima M, Niu J, Huang Q, et al. 2013. Temperature, traffic-related air pollution, and heart rate variability in a panel of healthy adults. Environ Res 120:82–89, PMID: , 10.1016/j.envres.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Wu S, Deng F, Niu J, Huang Q, Liu Y, Guo X. 2011. Exposures to PM2.5 components and heart rate variability in taxi drivers around the Beijing 2008 Olympic Games. Sci Total Environ 409(13):2478–2485, PMID: , 10.1016/j.scitotenv.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Wu Y, Pei C, Wang X, Wang M, Huang D, Wang F, et al. 2020. Effect of probiotics on nasal and intestinal microbiota in people with high exposure to particulate matter ≤ 2.5 μm (PM2.5): a randomized, double-blind, placebo-controlled clinical study. Trials 21(1):850, PMID: , 10.1186/s13063-020-04759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Zhao C, Huang W, Yong T, Chung ACK, He K, et al. 2019. Prenatal exposure to ambient fine particulate matter induces dysregulations of lipid metabolism in adipose tissue in male offspring. Sci Total Environ 657:1389–1397. PMID: 30677905, 10.1016/j.scitotenv.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang T, Liu S, Brook RD, Feng B, Zhao Q, et al. 2019. Extreme levels of air pollution associated with changes in biomarkers of atherosclerotic plaque vulnerability and thrombogenicity in healthy adults: the Beijing AIRCHD study. Circ Res 124(5):e30–e43. PMID: 30661461, 10.1161/CIRCRESAHA.118.313948. [DOI] [PubMed] [Google Scholar]
- Yang BY, Qian ZM, Li S, Chen G, Bloom MS, Elliott M, et al. 2018. Ambient air pollution in relation to diabetes and glucose-homoeostasis markers in China: a cross-sectional study with findings from the 33 Communities Chinese Health Study. Lancet Planet Health 2(2):e64–e73. ,, PMID: , 10.1016/S2542-5196(18)30001-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hu H, Shi Y, Yang X, Cao L, Wu J, et al. 2017b. 1 H NMR-based metabolomics study on repeat dose toxicity of fine particulate matter in rats after intratracheal instillation. Sci Total Environ 589:212–221, PMID: , 10.1016/j.scitotenv.2017.02.149. [DOI] [PubMed] [Google Scholar]
- Zhang S-Y, Shao D, Liu H, Feng J, Feng B, Song X, et al. 2017a. Metabolomics analysis reveals that benzo[a]pyrene, a component of PM 2.5, promotes pulmonary injury by modifying lipid metabolism in a phospholipase A2-dependent manner in vivo and in vitro. Redox Biol 13:459–469, PMID: , 10.1016/j.redox.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhu L, Li R, Wang H, Cai Z. 2019. Omics approach reveals metabolic disorders associated with the cytotoxicity of airborne particulate matter in human lung carcinoma cells. Environ Pollut 246:45–52, PMID: , 10.1016/j.envpol.2018.11.108. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, et al. 2019. Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 394(10204):1145–1158, 10.1016/S0140-6736(19)30427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.