ABSTRACT
Severe Community Acquired Pneumonia (SCAP) challenges public health globally. Considerable improvements in molecular pathogen testing emerged in the last few years. Our prospective study combinedly used traditional culture, antigen tests, PCR and mNGS in SCAP pathogen identification with clinical outcomes. From June 2018 to December 2019, we conducted a multi-centre prospective study in 17 hospitals of SCAP patients within 48 hours of emergency room stay or hospitalization in China. All clinical data were uploaded into an online database. Blood, urine and respiratory specimens were collected for routine culture, antigen detection, PCR and mNGS as designed appropriately. Aetiology confirmation was made by the local attending physician group and scientific committee according to microbiological results, clinical features, and response to the treatment. Two hundred seventy-five patients were included for final analysis. Combined detection methods made identification rate up to 74.2% (222/299), while 14.4% (43/299) when only using routine cultures and 40.8% (122/299) when not using mNGS. Influenza virus (23.2%, 46/198), S. pneumoniae (19.6%, 39/198), Enterobacteriaceae (14.6%, 29/198), Legionella pneumophila (12.6%, 25/198), Mycoplasma pneumoniae (11.1%, 22/198) were the top five common pathogens. The in-hospital mortality of patients with pathogen identified and unidentified was 21.7% (43/198) and 25.9% (20/77), respectively. In conclusion, early combined detection increased the pathogen identification rate and possibly benefitted survival. Influenza virus, S. pneumoniae, Enterobacteriaceae was the leading cause of SCAP in China, and there was a clear seasonal distribution pattern of influenza viruses. Physicians should be aware of the emergence of uncommon pathogens, including Chlamydia Psittaci and Leptospira.
Keywords: Severe community acquired pneumonia, mNGS, combined detection, multi-centre study, Chlamydia Psittaci
Introduction
Community-acquired pneumonia (CAP) is a leading global burden of infectious disease [1]. Severe CAP (SCAP) represents the most critical one, in 17–21% of hospitalized CAP patients [2–4]. The in-hospital mortality of SCAP is 30.6%–60% [3,5,6] in Europe according to the severity of hypoxemia and up to 51% in Pakistan [7]. In the EPIC study [2], S. pneumoniae, S. aureus and Enterobacteriaceae were detected more frequently in ICU patients. In a single-centre retrospective study in Singapore [8], Influenza virus and S. pneumoniae were the most common pathogens isolated. However, CAP is an extraordinarily heterogeneous illness in responsible pathogens [9], and most prior studies were conducted in developed countries. On the other hand, considerable improvements in the sensitivity, availability, and affordability of molecular pathogen testing in the past decade now influence our understanding of the causes of SCAP [10,11]. The aim of our study was to describe the clinical features of adult patients with SCAP and assess the benefit of combined detection, which may help make more precise clinical judgement.
Methods
Study design and patients enrolment
From 1 June 2018 to 31 December 2019, a prospective multi-centre observational study was conducted in 17 hospitals from 10 different regions of mainland China (Figure S1). A scientific committee was set up to study design, CRF design, study coordination and data audit.
All adult hospitalized patients (age above 18), with SCAP diagnosed according to the guidelines by the Chinese Thoracic Society (Version 2016), were screened [12]. Briefly, SCAP was defined as CAP patients who met any of the major criteria: (1) requiring tracheal intubation and mechanical ventilation; (2) septic shock, and still in need of vasoactive drugs after active fluid resuscitation or ≥ 3 minor criteria: (1) respiratory rate (RR) ≥ 30 bpm; (2) oxygenation index ≤ 250 mmHg (1 mmHg = 0.133 kPa); (3) infiltrates in multiple lung lobes; (4) disturbance of consciousness and (or) disorientation; (5) blood urea nitrogen (BUN) ≥ 7.14 mmol/L; (6) systolic blood pressure (SBP) < 90 mmHg, requiring active fluid resuscitation. Patients were excluded if any one of the following criteria was met: (1) patients with bronchiectasis or confirmed active tuberculosis; (2) a history of hospitalization within 2 weeks of enrolment or those who cannot be excluded for hospital-acquired pneumonia (HAP); (3) over 48 hours of emergency observation room stay or hospitalized or mechanical ventilation before enrolment; (4) patients in immunosuppressive condition, including HIV-positive, granulocyte deficiency for more than 10 days, post-bone marrow stem cell transplantation, post-solid organ transplantation, current glucocorticoid shock therapy, current immunosuppressive therapy, or solid tumours receiving chemoradiotherapy; (5) patients or their relatives unable to understand and implement the study.
Written informed consent was obtained from all patients or their relatives. This study was approved by the Ruijin Hospital Ethics Committee, Shanghai Jiaotong University School of Medicine (permit number: Ruijin Hospital Ethics Committee 2017-186) and was approved by the local hospital ethics committees in the other 16 centres.
Data collection
Baseline characteristics, including age, sex, underlying diseases, medication using history and epidemiology history, were collected, and Pneumonia Severity Index (PSI) and CURB-65 were assessed on enrolment. Laboratory tests results, including blood routine and biochemical test, imaging findings and management, including antibiotic treatment and organ support, were extracted during hospitalization. Clinical outcomes, including hospital length of stay, ICU length of stay, in-hospital mortality and 30-day mortality and total direct medical cost, were analysed. Electronic case report forms were uploaded into an online database (https://cfu.rjh.com.cn) by trained clinical research coordinators. Data audit was performed at an interval of 3 months.
Specimen collection
Blood samples, mid-stream urine (MSU) samples, nasopharyngeal/oropharyngeal (NP/OP) swabs, and qualified lower respiratory tract specimens (LRS), including sputum, tracheal aspirate, protected specimen brush (PSB) specimen or bronchoalveolar lavage fluid (BALF), were obtained from patients as soon as possible after the presentation and preferred to be collected before the antimicrobial therapy began. A gram stain test on sputum and tracheal aspirate was performed within 2 hours after specimen collection for quality control in local labs. Qualified samples met the following criteria: < 10 squamous epithelial cells and ≥ 25 white blood cells per low-power field. Convalescent serums were also collected 14–21 days after enrolment. Samples were stored at −80°C until transferred to the central lab.
Pathogen detection
In local labs, routine bacterial/fungal cultures and polymerase chain reaction (PCR) for virus detection, including influenza virus, were performed according to microbiological guidance. S. pneumoniae was isolated by a specific selective medium obtained from the study committee. Urine-antigen tests were performed to detect S. pneumoniae and Legionella pneumophila (Figure S2).
All samples stored in local hospitals were transferred to the central lab in China Medical University affiliated to Shengjing Hospital by cold-chain transportation for temperature-controlled demands. In the central lab, Legionella was isolated from LRS on buffered charcoal yeast extract agar. Pathogen-specific antibody tests were performed on paired serum samples (acute-phase on enrolment and convalescent-phase serum specimens) to detect Mycoplasma pneumoniae, Chlamydia pneumoniae and Legionella pneumoniae. Microbiological next-generation sequencing (mNGS) for 6975 pathogens was performed on blood samples and LRS in all enrolled patients by the Beijing Genomics Institute (BGI).
Aetiology confirmation
Microbiological results evaluation followed the following rules: (1) Positive bacterial culture from blood, qualified LRS or pleural fluid [13]; (2) Detection of positive respiratory virus in LRS or OP/NP swabs by real-time PCR [14]; (3) Positive urinary antigen for L. pneumophila or S. pneumoniae; (4) Mycoplasma pneumoniae, Chlamydia pneumoniae and Legionella pneumoniae were considered positive if a pathogen-specific antibody titre was increased by a factor of four or more between the acute-phase serum specimen and the convalescent-phase serum specimen. (5) NGS results were considered positive if bacteria (Mycobacteria excluded) or virus (species level) whose coverage rate scored 10-fold greater than that of any other microbes, and for fungi whose coverage rate scored 5-fold higher than that of any other fungus [15,16]. According to Miao’s study [17], Mycobacterium tuberculosis (MTB) was considered positive when at least 1 read was mapped to the species or genus level. For Nontuberculous mycobacteria (NTM), when the mapping read number (genus or species level) was in the top 10 in the bacteria list, it was considered positive. If the criteria were not met, physicians decided contaminants based on clinical features, radiographic findings and regimen.
The responsible pathogen for SCAP was identified as follows: (1) The final aetiology result was assessed by the local attending physician group led by the principal investigator in each hospital-based on microbiological results, clinical features, and response to the treatment. (2) When there was a controversy, the scientific committee discussed and made the final decision.
Statistical analysis
Continuous variables were expressed as median with interquartile range (IQR), and categorical variables were reported as frequency and percentages (%). The categorical variables were analysed by the Pearson Chi-square test. Data analyses and figures were done using IBM SPSS Statistics (Version 25.0) and R software (Version 3.6.0).
Results
Study population
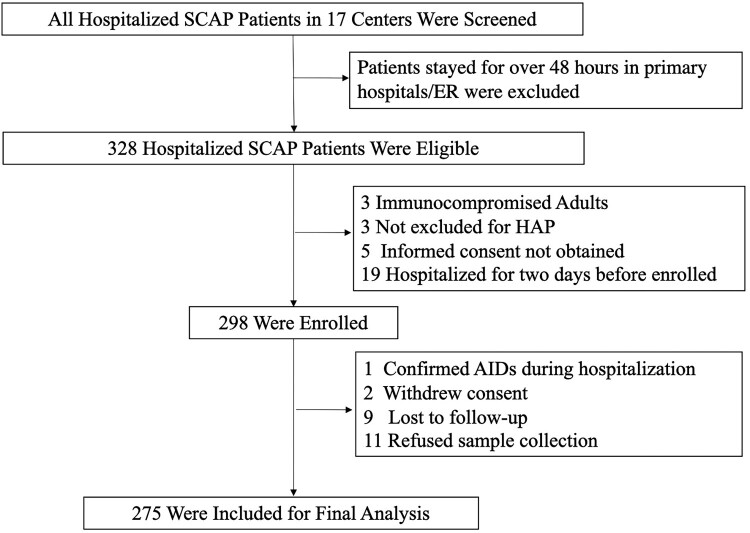
Three hundred twenty-eight adult patients with severe community-acquired pneumonia from 17 hospitals (Figue S1) were eligible for screening (Figure 1). Of which, 298 (90.8%) met the criteria for enrolment. Twenty (6.7%) patients were excluded because of data missing and three (1.0%) confirmed as Acquired Immune Deficiency Syndrome or withdrawn their consent. Eventually, 275 (92.3%) SCAP cases were included for the final analysis. The median age of SCAP patients was 61 years old (interquartile range, 49–69) (Table 1). Twenty-seven (9.8%) patients were over 80 years old, 199 (72.4%) were male, and 166 (60.4%) suffered from different underlying diseases, hypertension (59/275, 21.5%) and diabetes (54/275, 19.6%) were the most common underlying conditions. 124 (45.1%) of patients were current smokers. Fever, cough and dyspnoea were found in 235 (85.5%), 234 (85.1%) and 183 (66.5%) of patients on admission, which were the three most common symptoms. The highest temperature on admission was 39°C (interquartile range, 38.3–39.5). The duration from illness onset to admission was 5 days (interquartile range, 3–7). 183 (66.5%) scored 4 in Pneumonia Severity Index on admission. Consolidation, ground-glass opacity and pleural effusion were found in 194 (70.6%), 179 (65.1%) and 121 (44.0%) patients on enrolment, respectively.
Figure 1.
Screening, Eligibility, and Enrolment of Adult Patients with Severe Community-Acquired Pneumonia. SCAP stands for severe community-acquired pneumonia. ER stands for emergency room. HAP stands for hospital acquired pneumonia. AIDS stands for acquired immune deficiency syndrome.
Table 1.
Baseline Characteristics, Clinical Manifestation and Radiographic Findings of Adult Patients with Severe Community-Acquired Pneumonia in the Study on Admission.
| Characteristics On Admission | All Patients |
|---|---|
| Age – yr – median (IQR) | 61(49-69) |
| 18–64 yr | 170/275(61.8%) |
| 65–79 yr | 78/275(28.4%) |
| ≥ 80 yr | 27/275(9.8%) |
| Male – no. (%) | 199/275(72.4%) |
| Any underlying Disease – no. (%) | 166/275(60.4%) |
| COPD | 20/275(7.3%) |
| Diabetes Mellitus | 54/275(19.6%) |
| Hypertension | 59/275(21.5%) |
| Chronic renal disease | 11/275(4.0%) |
| Malignant | 18/275(6.5%) |
| Chronic heart disease | 25/275(9%) |
| Cerebrovascular disease | 13/275(4.7%) |
| Current Smoker – no. (%) | 124/275(45.1%) |
| Current Alcoholic – no. (%) | 58/275(21.1%) |
| Duration from illness onset to admission – days – median (IQR) | 5(3-7) |
| Clinical manifestation – no. (%) | |
| Fever | 235/275(85.5%) |
| Cough | 234/275(85.1%) |
| Dyspnoea | 183/275(66.5%) |
| Tiredness, dizziness or headache | 68/275(24.7%) |
| Shiver | 64/275(23.3%) |
| Myalgia | 35/275(12.7%) |
| Sore throat | 30/275(10.9%) |
| Nausea | 18/275(6.5%) |
| Haemoptysis | 17/275(6.2%) |
| Emesis | 14/275(5.1%) |
| Diarrhoea | 13/275(4.7%) |
| Abdominal pain | 8/275(2.9%) |
| Abnormal auscultation | 223/275(81.1%) |
| White phlegm | 106/275(38.5%) |
| Purulent respiratory secretion | 87/275(31.6%) |
| Cyanosis | 11/275(4%) |
| Rash | 7/275(2.5%) |
| Highest temperature – °C | 39.0 (38.3–39.5) |
| Pulse rate – per min | 98(86-115) |
| Respiration rate – per min | 26(20-31) |
| CURB-65 Score – no. (%) | |
| 0–1 | 82/275(29.8%) |
| 2 | 86/275(31.3%) |
| 3–5 | 107/275(38.9%) |
| Pneumonia Severity Index | |
| Median | 110 |
| Interquartile range | 80–136 |
| Risk class – no. (%) | |
| 1–3 | 92/275(33.5%) |
| 4 | 104/275(37.8%) |
| 5 | 79/275(28.7%) |
| Radiographic finding – no. (%) | |
| Consolidation | 194/275(70.6%) |
| Ground-glass opacity | 179/275(65.1%) |
| Pleural effusion | 121/275(44%) |
Abnormal auscultation including rale or consolidation sign. COPD stands for chronic obstructive pulmonary disease. The CURB-65 severity score measures confusion, urea, respiratory rate, and blood pressure at age 65 years or older. The pneumonia severity index is based on sex, age, nursing home status, mental status, heart rate, respiratory rate, blood pressure, temperature, selected underlying medical conditions, laboratory values, and the presence or absence of pleural effusion.
Responsible pathogens identified for SCAP
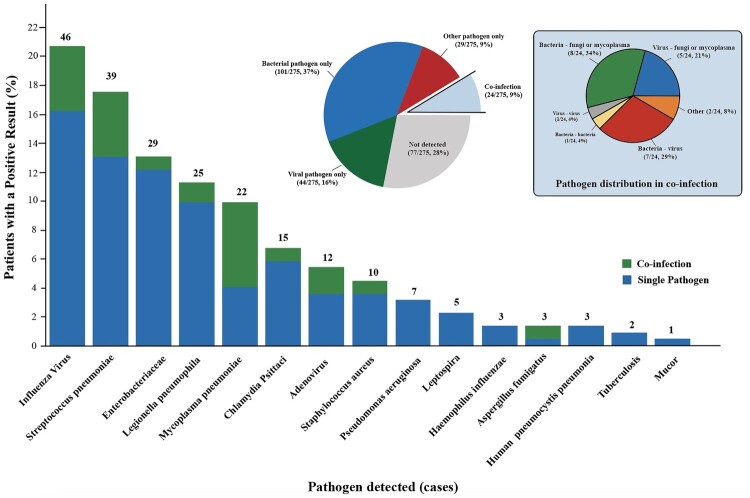
Of 275 eligible patients, only 77 (28.0%) failed to find responsible pathogens (Figure 2). In 198 patients whose responsible pathogen was confirmed, a single pathogen caused 87.9% (174 of 198) cases of SCAP, including bacteria only 58.1% (101 of 174) and virus only 25.3% (44 of 174). 12.1% (24 of 198) cases were co-infection, of which 33.3% (8 of 24) were bacteria and fungi or mycoplasma co-infection, 29.2% (7 of 24) were bacteria and virus co-infection, and 20.8% (5 of 24) were virus and fungi or mycoplasma co-infection. The most common pathogens identified were influenza virus (46/222, 20.7%), S. pneumoniae (39/222, 17.6%) and Enterobacteriaceae (29/222, 13.1%), including 25 (86.2%) K. pneumoniae and 2 (6.9%) Escherichia coli. Other common pathogens identified including Legionella pneumophila (25/222, 11.3%), Mycoplasma pneumoniae (22/222, 9.9%), Chlamydia Psittaci (15/222, 6.8%), adenovirus (12/222, 5.4%), S. Aureus (10/222, 4.5%), and Pseudomonas aeruginosa (7/222, 3.2%). Of 22 Mycoplasma pneumoniae, 13 (59.0%) cases were co-infection.
Figure 2.
Responsible Pathogen identified in Adult Patients with Severe Community-Acquired Pneumonia. Figure 2 shows the numbers (above the bars) and percentages (y-axis) of specific pathogens detected. A total of 222 pathogens were detected in 275 patients, of which 198 patients got positive aetiology results. The big pie chart in the middle shows the proportions of bacterial only, viral only, other pathogens only (including fungi, mycoplasma, Chlamydia Psittaci and Leptospira), not detected and co-infection. The small pie chart on the left shows the detailed proportions of co-infection.
Adenovirus was more commonly found in young patients with a median age of 33 (interquartile range, 30–34) (Table S1). 22 (81.5%) and 6 (85.7%) of patients identified with Enterobacteriaceae and Pseudomonas aeruginosa had underlying diseases. For Chlamydia Psittaci, consolidation was seen radiographically on admission in 12 (92.3%) patients.
Comparison of pathogens for SCAP in different regions, seasons and sex
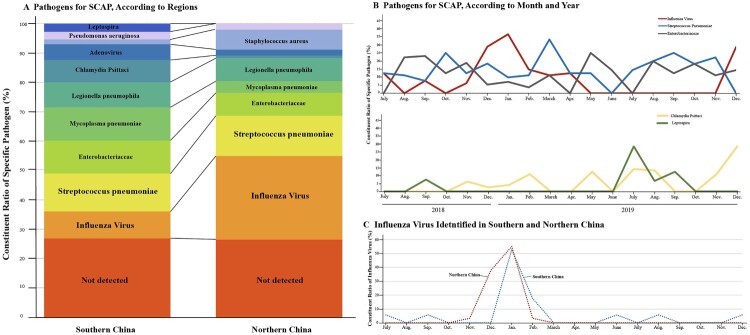
Geographically, study centres were divided into Southern and Northern China according to their location to the Qinling-Huaihe Line [18,19], which differs two regions from each other in climate. 177 (64.4%) cases were enrolled from Southern China, while 98 (35.6%) were from northern China. The constituent ratio of positive diagnostic results, S. pneumoniae and Enterobacteriaceae showed not much difference from the two regions (Figure 3(A)). However, Influenza viruses were more commonly found in the north (South vs. North:9.6% [17/177] cases vs. 29.6% [29/98]) and got a peak in the winter season (Figure 3(B)). Compared to S. pneumoniae, Enterobacteriaceae showed a small peak in summer. On the other hand, Chlamydia Psittaci and Leptospira had an increase in 2019. Leptospira peaked in the hottest day in the south of China possibly due to underwater activities (contact history recalled from patients). Influenza virus emerged a month earlier in northern China than the South due to the earlier temperature drop in the North and ended the same time in early spring (Figure 3(C)). Mycoplasma pneumoniae was more frequently identified in female SCAP patients (Figure S3).
Figure 3.
Comparison of Pathogen Detected in Different Regions and Seasons. Panel A shows the proportions of a specific pathogen in southern (177 patients) and northern China (98 patients), which are geographically divided from the Qinling-Huaihe Line. Panel B shows the number of patients in whom influenza virus, S. pneumoniae, Enterobacteriaceae, Chlamydia Psittaci and Leptospira were detected according to month and year. Panel C shows the number of patients in whom influenza virus was detected according to month and geography. SCAP denotes severe community-acquired pneumonia.
Performance of different diagnostic tools
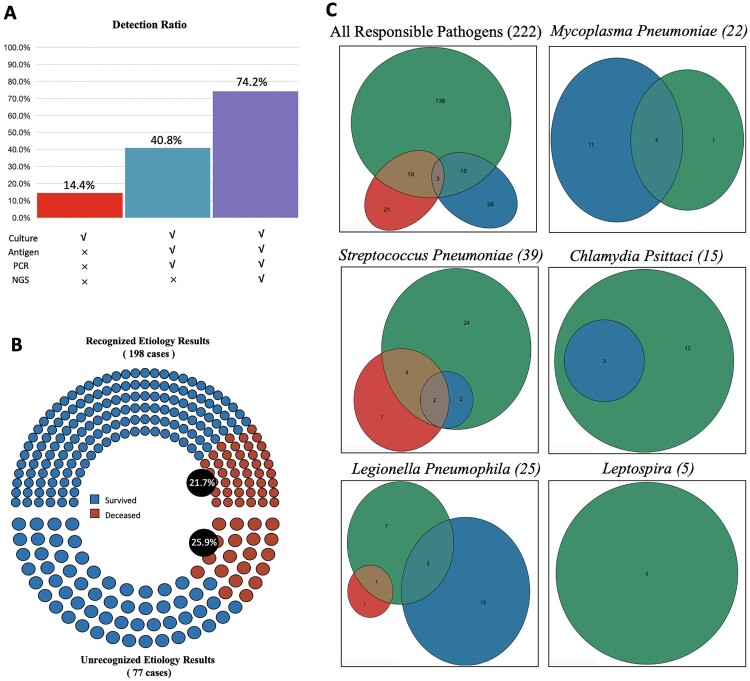
A combination of routine bacterial/fungal cultures, blood/urine-serological tests and molecular pathogen testing, including polymerase chain reaction (PCR) and mNGS, made the positive pathogen identification rate up to 74.2% (222/299) (Figure 4(A)). However, the positive rate was 14.4% (43/299) when only using routine cultures and 40.8% (122/299) when not using mNGS. In-hospital mortality of SCAP patients with recognized positive aetiology results was 21.7% (43/198), and it was 25.9% (20/77) in the patients whose aetiology was unclear, but the difference was not statistically significant (P = .451) (Figure 4(B)). The use of antigen test identified Legionella pneumophila in 16 patients, Mycoplasma pneumoniae in 15 patients and chlamydia psittaci in 3 patients; the number increased to 24, 22 and 15, respectively when using molecular detection methods in addition (Figure 4(C)). All five Leptospira caused SCAP were initially recognized by molecular tools and then confirmed by a serological test.
Figure 4.
Performance of Different Diagnostic Tools. Panel A shows the presume positive rate with different detection methods combination. Only used traditional culture detected 14.4% (43 of 299) pathogens. Culture, antigen test and PCR detected 40.8% (122 of 299). Culture, antigen test, PCR and NGS detected 74.2% (222 of 299). Note all 278 patients enrolled were used combined methods and 24 were detected for two pathogens responsible. PCR stands for a polymerase chain reaction. NGS stands for next-generation sequencing. Panel B shows in-hospital mortality of patients with or without positive aetiology results. Venn diagrams in Panel C show specific pathogens detected according to different detected methods. Green for molecular detection methods, including PCR and mNGS. Red for traditional culture. Blue for antigen tests. The numbers in the circles stand for cases.
Laboratory findings
Blood routine and biochemistry findings of all SCAP patients on admission were collected. The median of C-reaction protein was 90 mg/L (reference range, < 10) (Table 2). For bacterial infection, the median of procalcitonin was 2.8 μg/L (reference range, < 0.5) and lactate dehydrogenase increased to 701 IU/L (reference range, 98–192) for virus infection. (Table S2). The white blood cell count was increased as expected. The average neutrophil percentage increased to 86.1% (reference range, 50–70), and the average lymphocyte percentage dropped to 8.1% (reference range, 20–40). In 177 (64.4%) patients with T cell percentage tests, the CD3 and CD8 positive cells dropped to 20.2% (reference range, 21–29). Urine routines showed 48.3% (133/275) of patients had proteinuria.
Table 2.
Laboratory Findings on Admission.
| Reference Range | On Admission | |
|---|---|---|
| Blood Routine (n = 275) | ||
| Red blood cell count - ×1012/L | 4.09–5.74 × 1012/L | 4.07 (3.5–4.54) |
| White blood cell count - ×109/L | 3.97–9.15 × 109/L | 9.58 (5.57–14.01) |
| Neutrophil count - ×109/L | 2.00–7.00 × 109/L | 7.9 (4.31–12.52) |
| Lymphocyte count - ×109/L | 0.80–4.00 × 109/L | 0.7 (0.48–1.09) |
| Platelet count - ×109/L | 85–303 × 109/L | 152 (105–223) |
| Haemoglobin - g/L | 131–172 g/L | 125 (104–139) |
| Neutrophil percentage - % | 50.0%–70.0% | 86.1 (77.2–91.2) |
| Lymphocyte percentage - % | 20.0–40.0 | 8.1 (5–14) |
| Eosinophil percentage - % | 0.5%–5.0% | 0 (0–0.4) |
| Basophilic granulocyte percentage - % | < 1.0 | 0.1 (0–0.2) |
| Monocyte percent - % | 3.0%–10.0% | 4.5 (2.4–6.6) |
| Blood biochemistry (n = 275) | ||
| Alanine transaminase - IU/L | 10–64IU/L | 36 (23–63) |
| Aspartate aminotransferase - IU/L | 8–40IU/L | 50 (26–89) |
| Total bilirubin - μmol/L | 4.7–24 μmol/L | 12.3 (8.5–19.8) |
| Direct bilirubin - μmol/L | 0–6.8 μmol/L | 5.1 (3.3–10.3) |
| Lactate dehydrogenase - IU/L | 98–192IU/L | 430 (251–698) |
| Blood Urea Nitrogen - mmol/L | 2.5–7.1 mmol/L | 7.7 (4.9–11.7) |
| Serum creatinine - μmol/L | 62–115 μmol/L | 61 (8–89) |
| Serum albumin - g/L | 35–55 g/L | 30 (27–34) |
| Blood sodium - mmol/L | 130–147 mmol/L | 137 (134–140) |
| Blood potassium - mmol/L | 3.50–5.10 mmol/L | 3.75 (3.43–4.2) |
| Blood chlorinum - mmol/L | 96–108 mmol/L | 103 (99–106) |
| Blood phosphorus - mmol/L | 0.80–1.60 mmol/L | 0.92 (0.69–1.13) |
| Blood glucose - mmol/L | 3.90–6.10 mmol/L | 7.49 (6–10.6) |
| Creatine kinase - IU/L | 22–269IU/L | 136 (50–367) |
| D-dimer - mg/L | <0.55 mg/L | 3.64 (1.68–21) |
| Fibrinogen - g/L | 1.8–3.5g/L | 5.4 (3.9–7.9) |
| Prothrombin time - s | 10.0–16.0 s | 13.8 (12.3–15.4) |
| Activated partial thromboplastin time - s | 22.3–38.7 s | 35.9 (30.9–43.1) |
| C-reactive protein - mg/L | < 10 mg/L | 90 (19–152) |
| Procalcitonin - ug/L | < 0.5 ug/L | 1.2 (0.3–9) |
| T cell Percentage (n = 177) | ||
| CD3+ T cell percentage - % | 64.0–76.0% | 62.5 (51.4–73.3) |
| CD3 + CD4+ T cell percentage - % | 30.0–40.0% | 36.8 (28.2–46) |
| CD3 + CD8+ T cell percentage - % | 21.0–29.0% | 20.2 (15–28.2) |
| Arterial blood gas (n = 270) | ||
| pH | 7.35–7.45 | 7.43 (7.38–7.47) |
| PaO2 - mmHg | 80–100 mmHg | 71 (60–86) |
| PaCO2 - mmHg | 35–45 mmHg | 35 (31–41) |
| HCO3- - mmol/L | 22.0–27.0 mmol/L | 23.2 (20.4–25.9) |
| BE - mmol/L | −3–3 mmol/L | 0 (−4-3) |
| Urine routines - no. (%) | ||
| Haematuria | 116/275 (42.2%) | |
| Proteinuria | 133/275 (48.3%) | |
Outcome and mortality
The median length of ICU and hospital stay was 7 (interquartile range, 2–14) and 14 (interquartile range, 9–22) days, respectively (Table 3). Among 275 SCAP patients included in the final analysis, 81 (29.4%) cases were supported by invasive mechanical ventilation and 7(2.5%) patients used extracorporeal membrane oxygenation during hospitalization. The 30-day mortality was 18.9% (52/275) and 63 patients (22.9%) died in hospital. The total median cost per patient was 7064 (interquartile range, 3804–13904) US dollar.
Table 3.
Clinical Outcomes and Medical Cost.
| Variables | (N = 275) |
|---|---|
| Length of hospital stay - days - median (IQR) | 14 (9–22) |
| Length of ICU stay - days - median (IQR) | 7 (2–14) |
| Non-invasive mechanical ventilation - no. (%) | 43/275 (15.6%) |
| Length of non-invasive mechanical ventilation - days - median (IQR) | 7 (2–12) |
| Invasive mechanical ventilation - no. (%) | 81/275 (29.4%) |
| Length of invasive mechanical ventilation - days - median (IQR) | 10 (5–14) |
| Extracorporeal membrane oxygenation - no. (%) | 7/275 (2.5%) |
| Length of ECMO - days - median (IQR) | 10 (2–15) |
| 30-day mortality - no. (%) | 52/275 (18.9%) |
| In-hospital mortality - no. (%) | 63/275 (22.9%) |
| Medical cost - US dollar per patient - median (IQR) | 7064 (3804–13904) |
ICU denotes for intensive care unit.
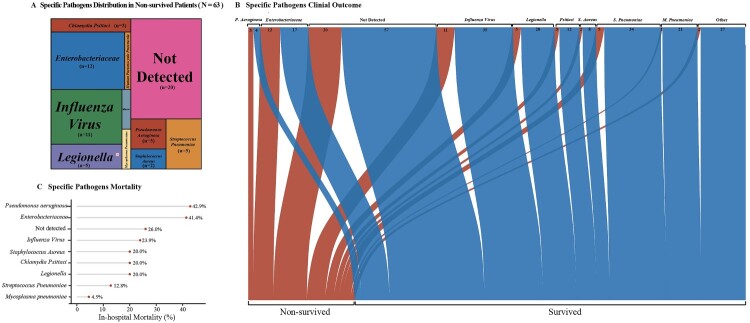
Among 63 non-survived SCAP patients, 20 (31.7%) was without recognized aetiology results (Figure 5(A)). 34 (87.2%), 21 (95.5%), 20, 12 and 8 (80.0%) of S. pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, Chlamydia Psittaci and S. Aureus patients recovered during hospitalization, respectively (Figure 5(B)). However, the in-hospital mortality of Pseudomonas aeruginosa, Enterobacteriaceae and Influenza viruses was 42.9%, 41.4% and 23.9%, respectively (Figure 5(C)). In-hospital mortality was higher in patients without the detection of responsible pathogens (Unidentified vs. Identified: 25.9% [20/77] cases vs. 21.7% [43/198] cases).
Figure 5.
Pathogen-Specific Outcome in Adult Patients with Severe Community-Acquired Pneumonia.The tree map in Panel A shows the proportions of specific pathogen in non-survived patients. The area stands for the quantity of hospital death patients the specific pathogen caused. The Sankey diagram in Panel B shows the different outcomes of specific pathogens. The width of the arrows is proportional to the flow rate (survived in blue or non-survived in red). The numbers above are cases. Panel C shows the in-hospital mortality (%) of pathogens.
Discussion
Our prospective study of adult hospitalized patients with SCAP conducted in 17 teaching hospitals in ten cities in the South and North of mainland China shed light on the aetiology in the new era of emerging combined detection technology.
To the best of our knowledge, the identification rate of responsible pathogens in our study is among the highest in similar studies, and the identification of responsible pathogen by the scientific committee made the result clinically reliable. Bacteria were identified in 37% of the patients, in which S. pneumoniae (19.7% of 198 patients) remained the most frequent pathogen as bacteria aetiology. S. Aureus and Pseudomonas aeruginosa were identified in 5% and 3.5% in SCAP patients, respectively. However, compared with a single-centre study among SCAP in Europe [3] and CAP patients in ICU in the US [2], the detection rate of Enterobacteriaceae (14.6% of 198 patients) was higher in China. Consistent with data from China Antimicrobial Surveillance Network [20] (CHINET, www.chinets.com), K. pneumoniae was the most frequently isolated bacteria in respiratory samples in 2019 (18.53% of 101714 samples).
Respiratory viruses play an increasing role in CAP, especially in SCAP. In our study, the Influenza Virus (23.3% of 198 patients) was the top pathogen identified in SCAP and related to the winter season, the mortality of which was 23.9%. Adenovirus was the second contributing virus for SCAP, more frequent in younger patients (the median age was 33 yrs). In immunocompetent individuals, adenovirus infections were mostly mild and self-limiting [21], but severe and even fatal courses were seen in several types in young patients [22,23]. However, we did not find other viruses responsible for CAP [2] like human rhinovirus or human metapneumovirus in SCAP in China. Those respiratory viruses may tend to mild to moderate pneumonia. The management strategy of SCAP should involve the rapid detection of respiratory viruses such as influenza and adenovirus.
Bacteria with fungi or mycoplasma were the top in co-infection. In a Spanish cohort in 148 ICUs from 2005 to 2015 [24], influenza virus with co-infection increased from 11.4% to 23.4% and was associated with increased ICU mortality. Nevertheless, the situation in China was the same as Spain in the rate of influenza with co-infection (21.7%, 10 of 46), but they all survived, which may be due to the precise diagnosis of co-infection and antibiotic administration.
In our study Chlamydia Psittaci is an important emerging aetiology of SCAP, contributing to 7.5% of the cases. Bacteria of gram-negative intracellular bacteria Chlamydiaceae family got zoonotic potential, and psittacosis was usually reported sporadically around the world. In 1980, the proportion of CAP cases caused by Chlamydia Psittaci was 1.9% [25]. A systematic study revealed Chlamydia Psittaci was responsible for 1.03% (95% CI 0.79%–1.30%) of CAP according to 57 pieces of research before 2012 [26]. However, in 2014, Lagae et. al reported a re-emerging of Chlamydia psittaci in chickens in Belgium [27], and the number of labs reported positive Chlamydia Psittaci in Belgium doubled in 2017 [28]. Along with our data, a retrospective single centre study [29] from 2019 to 2020 reported 16 cases of severe psittacosis pneumonia and Li et al. reported the first family outbreak of psittacosis in China under COVID-19 in December 2020 [30].
Awareness of the emergence of psittacosis is still low among clinicians and the general public due to under-diagnosis. Chlamydia Psittaci infection that often lacks typical clinical and imaging manifestation made the diagnosis of psittacosis difficult [31], all 15 unrelated cases in our cohort were identified by mNGS in high sensitivity and specificity, which helped physicians find the responsible pathogen in time and changed to a strong regimen for targeted pathogen, avoiding deaths because of not being correctly diagnosed [32].
mNGS showed advantages in the rapid detection of atypical pathogens such as Chlamydia Psittaci and Leptospira. However, it missed 38.2% (85 of 222) of responsible pathogens, especially Influenza virus, for which real-time PCR showed higher sensitivity. mNGS also has the disadvantages of confusing explanations of the reports. Although culture and antigen tests require time and show low sensitivity, they are more accessible. These findings prompted us to inspect the impact of combined detection, including traditional culture, antigen tests and molecular methods on clinical outcomes and cost-effectiveness.
SCAP mortality ranged from over 30% to 60% [3,5–7] according to different groups. However, the in-hospital mortality of SCAP patients (22.9%) in our study was much lower, which was possibly contributed by our higher identification rate of responsible pathogens in time. Likewise, we found aetiology diagnosis benefited patients in mortality (Unidentified vs. Identified in-hospital mortality rate: 25.9% vs. 21.7% P = .451). As far as we are concerned, the initial empiric treatment of patients in our cohort covered the most common responsible pathogens, and overall in-hospital mortality was low among studies of SCAP, which is a possible explanation of the non-significant difference between the two groups. However, early recognition of responsible pathogens would facilitate early de-escalation, therefore avoid unnecessary broad-spectrum antibiotics, adverse effects, and some other detrimental consequences. On the other hand, early recognition of less-common pathogens, such as Chlamydia Psittaci and Leptospira, helped physicians change to the appropriate strong targeted regimen in time. To this extent, we recommend all SCAP patients conduct combined detection for pathogen if it’s accessible as soon as they are hospitalized.
Most SCAP patients were elderly men over 65 years old, which is consistent with other studies for CAP conducted in Spain [33] and America [2]. We found elderly people more vulnerable to SCAP, and older age has also been reported as an independent factor for mortality in SARS-CoV [34], MERS [35] and COVID-19 [36]. In the elderly the immune response is blunted due to the decline in several components of the immune senescence and shifting to a chronic pro-inflammatory status [37]. The age-associated imbalance between misplaced and extracellular inflammatory DNA sequences and anti-inflammatory DNA leads to inflamed-ageing derangement [38]. On the other hand, our study, consistent with others [5,39], showed higher morbidity in men. In contrast, an in-vivo study showed sex differences in the susceptibility to SARS-CoV were accompanied by increased accumulation of inflammatory monocyte macrophages and neutrophils in the lungs of male mice [40].
Hypertension and diabetes mellitus increased susceptibility in SCAP, which they were consistent with our previous finding of SARS-CoV-2 in China [41] and Petrakisin’s in Greece [42]. Moreover, 45% of SCAP patients in our cohort were current smokers. A large European CAP cohort showed 30.1% of patients were smokers with undiagnosed diabetes [39]. Family physicians should be aware of the risk of hyperglycaemia and the benefit of lifestyle interventions, including smoking cessation.
Mild CD8 positive T cells decrease instead of CD4 positive cells around expected peak expansion (8 days following the initiation of infection) [43]. CD8 positive T cells, including cytotoxic T lymphocytes, are mediators important for defending against viral, bacterial and protozoal infections. Smith and colleagues found IL-10 directly restricted CD8 positive cells activation through the modification of cell surface glycosylation and allowed the establishment of chronic infection [44]. Our clinical findings could pave the way for further understanding of the natural immune response to SCAP.
Our study is not without limitations. First, we conducted a one-and-a-half years of prospective study instead of 2-year-round as planned because of the pandemic of COVID-19. Since we had enrolled enough patients ahead of schedule, the leading investigator and the scientific committee agreed to stop enrolment on 31 December 2019 because of the unclear situation due to the pandemic. Metlay and colleagues proposed that it was likely the relevant bacterial pathogens in patients with COVID-19 and pneumonia were the same as in previous patients with CAP [45]. How the pandemic has influenced the aetiology of SCAP globally remains unclear and needs further study. Although we analysed a relatively large group of SCAP patients, we were caught in a dilemma of higher recruitment or avoiding HAP/VAP bias. As we designed and mentioned in the Methods, patients who stayed for over two days in primary hospitals before admission to tertiary hospitals (17 centres) were excluded after screening. Second, we did not observe the long-term prognosis of SCAP patients considering the unpredictable pandemic perturbation.
In conclusion, our multi-centre prospective study using all accessible detection methods combined on SCAP with a high identification rate depicted that influenza virus, S. pneumoniae, Enterobacteriaceae were the leading cause of SCAP in China with seasonal prevalence. More public health efforts should focus on elderly adults for the prevention of SCAP and smoking cessation. In the new era of emerging molecular technologies, the positive rate of atypical and typical pathogens increased. Combined detection methods can accurately identify and distinguish responsible pathogens and might benefit in survival for SCAP.
Supplementary Material
Acknowledgements
We thank all the patients who participated in this study, the clinical site investigators (Zhao JY, Shi MM, Jiao Y, Liang S, Zhou F, Shang LH, Wang QY, Cheng Q, Zhang T, Liu JX, Li RR, Li X, Wang GD, Zhang YQ, Rui YW, Zhang L, Xu LN, Zhang SN, Yao YK, Jiang YQ et al.), and Zuo Ming from Ruijin Hospital who maintained the online database. We also thank Zhejiang Medicine, who helped in logistics.
Funding Statement
This work was supported by Severe Pneumonia Cohort and Biological Sample Data Repository (SHDC2020CR5010), Cultivation Project of Shanghai Major Infectious Disease Research Base (20dz2210500), Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases, Shanghai (20dz2261100), and National Innovative Research Team of High-level Local Universities in Shanghai
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Lozano R, Naghavi M, Foreman K, et al. . Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012 Dec 15;380(9859):2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S, Self WH, Wunderink RG, et al. . Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015 Jul 30;373(5):415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cilloniz C, Ferrer M, Liapikou A, et al. . Acute respiratory distress syndrome in mechanically ventilated patients with community-acquired pneumonia. Eur Respir J. 2018 Mar;51(3):1702215. [DOI] [PubMed] [Google Scholar]
- 4.Ferrer M, Travierso C, Cilloniz C, et al. . Severe community-acquired pneumonia: Characteristics and prognostic factors in ventilated and non-ventilated patients. PLoS one. 2018;13(1):e0191721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mongardon N, Max A, Bougle A, et al. . Epidemiology and outcome of severe pneumococcal pneumonia admitted to intensive care unit: a multicenter study. Critical Care (London, England). 2012 Aug 15;16(4):R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz H, Norby GO, Dahl V, et al. . Five-year mortality in patients treated for severe community-acquired pneumonia - a retrospective study. Acta Anaesthesiol Scand. 2017 Apr;61(4):418–426. [DOI] [PubMed] [Google Scholar]
- 7.Khawaja A, Zubairi AB, Durrani FK, et al. . Etiology and outcome of severe community acquired pneumonia in immunocompetent adults. BMC Infect Dis. 2013 Feb 20;13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quah J, Jiang B, Tan PC, et al. . Impact of microbial aetiology on mortality in severe community-acquired pneumonia. BMC Infect Dis. 2018 Sep 4;18(1):451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metlay JP, Waterer GW, Long AC, et al. . Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious diseases society of america. Am J Respir Crit Care Med. 2019 Oct 1;200(7):e45–e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murdoch DR. How recent advances in molecular tests could impact the diagnosis of pneumonia. Expert Rev Mol Diagn. 2016;16(5):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cilloniz C, Liapikou A, Torres A.. Advances in molecular diagnostic tests for pneumonia. Curr Opin Pulm Med. 2020 May;26(3):241–248. [DOI] [PubMed] [Google Scholar]
- 12.Cao B, Huang Y, She DY, et al. . Diagnosis and treatment of community-acquired pneumonia in adults: 2016 clinical practice guidelines by the Chinese thoracic society, Chinese medical association. Clin Respir J. 2018 Apr;12(4):1320–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith DE, Aksamit T, Brown-Elliott BA, et al. . An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007 Feb 15;175(4):367–416. [DOI] [PubMed] [Google Scholar]
- 14.Qu JX, Gu L, Pu ZH, et al. . Viral etiology of community-acquired pneumonia among adolescents and adults with mild or moderate severity and its relation to age and severity. BMC Infect Dis. 2015 Feb 22;15:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langelier C, Zinter MS, Kalantar K, et al. . Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med. 2018 Feb 15;197(4):524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlaberg R, Chiu CY, Miller S, et al. . Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017 Jun;141(6):776–786. [DOI] [PubMed] [Google Scholar]
- 17.Miao Q, Ma Y, Wang Q, et al. . Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018 Nov 13;67(suppl_2):S231–S240. [DOI] [PubMed] [Google Scholar]
- 18.Shuangshuang L, Junping Y, Saini Y, et al. . Spatiotemporal variability of heat waves and influencing factors in the Qinling-Huaihe region, 1960–2016. Progr Geogr; 37(4):504–514. [Google Scholar]
- 19.Yang MA, Fan X, Sun B, et al. . Ancient DNA indicates human population shifts and admixture in northern and southern China. Science. 2020 Jul 17;369(6501):282–288. [DOI] [PubMed] [Google Scholar]
- 20.Hu F, Wang M, Zhu D, et al. . CHINET efforts to control antimicrobial resistance in China. J Glob Antimicrob Resist. 2020 Jun;21:76–77. [DOI] [PubMed] [Google Scholar]
- 21.Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev. 2014 Jul;27(3):441–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carr MJ, Kajon AE, Lu X, et al. . Deaths associated with human adenovirus-14p1 infections, Europe, 2009-2010. Emerg Infect Dis. 2011 Aug;17(8):1402–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou F, Wang Y, Liu Y, et al. . Disease severity and clinical outcomes of community-acquired pneumonia caused by non-influenza respiratory viruses in adults: a multicentre prospective registry study from the CAP-China network. Eur Respir J. 2019 Aug;54(2):1802406. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Loeches I, Schultz MJ, Vincent JL, et al. . Increased incidence of co-infection in critically ill patients with influenza. Intensive Care Med. 2017 Jan;43(1):48–58. [DOI] [PubMed] [Google Scholar]
- 25.Krech T, Wegmann T, Martin H, et al. . [Etiology of atypical pneumonias. A serological study on 1494 patients]. Schweiz Med Wochenschr. 1986 Jan 4;116(1):2–7. [PubMed] [Google Scholar]
- 26.Hogerwerf L, De Gier B, Baan B, et al. . Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017 Nov;145(15):3096–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagae S, Kalmar I, Laroucau K, et al. . Emerging Chlamydia psittaci infections in chickens and examination of transmission to humans. J Med Microbiol. 2014 Mar;63(Pt 3):399–407. [DOI] [PubMed] [Google Scholar]
- 28.Rybarczyk J, Versteele C, Lernout T, et al. . Human psittacosis: a review with emphasis on surveillance in Belgium. Acta Clin Belg. 2020 Feb;75(1):42–48. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Q, Shen W, Zou Y, et al. . Sixteen cases of severe pneumonia caused by Chlamydia psittaci in South China investigated via metagenomic next-generation sequencing. J Med Microbiol. 2021 Nov;70(11):001456. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Li S, Tan W, et al. . Metagenomic next-generation sequencing in the family outbreak of psittacosis: the first reported family outbreak of psittacosis in China under COVID-19. Emerg Microbes Infect. 2021 Dec;10(1):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cilloniz C, Torres A, Niederman M, et al. . Community-acquired pneumonia related to intracellular pathogens. Intensive Care Med. 2016 Sep;42(9):1374–1386. [DOI] [PubMed] [Google Scholar]
- 32.Knittler MR, Sachse K.. Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog Dis. 2015 Feb;73(1):1–15. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Primack BA, Mor MK, et al. . Processes of care and outcomes for community-acquired pneumonia. Am J Med. 2011 Dec;124(12):1175.e9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi KW, Chau TN, Tsang O, et al. . Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann Intern Med. 2003 Nov 4;139(9):715–723. [DOI] [PubMed] [Google Scholar]
- 35.Hong KH, Choi JP, Hong SH, et al. . Predictors of mortality in Middle East respiratory syndrome (MERS). Thorax. 2018 Mar;73(3):286–289. [DOI] [PubMed] [Google Scholar]
- 36.Zhou F, Yu T, Du R, et al. . Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar 28;395(10229):1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butcher SK, Lord JM.. Stress responses and innate immunity: aging as a contributory factor. Aging Cell. 2004 Aug;3(4):151–160. [DOI] [PubMed] [Google Scholar]
- 38.Storci G, Bonifazi F, Garagnani P, et al. . The role of extracellular DNA in COVID-19: clues from inflamm-aging. Ageing Res Rev. 2020 Dec 13;66:101234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen AV, Faurholt-Jepsen D, Egelund GB, et al. . Undiagnosed diabetes mellitus in community-acquired pneumonia: A prospective cohort study. Clin Infect Dis. 2017 Nov 29;65(12):2091–2098. [DOI] [PubMed] [Google Scholar]
- 40.Channappanavar R, Fett C, Mack M, et al. . Sex-Based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. 2017 May 15;198(10):4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Ling Y, Bai T, et al. . COVID-19 with different severities: A multicenter study of clinical features. Am J Respir Crit Care Med. 2020 Jun 1;201(11):1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrakis V, Panagopoulos P, Papazoglou D, et al. . Diabetes mellitus and hypertension as major risk factors of mortality from COVID-19 pneumonia. Exp Clin Endocrinol Diabetes. 2020 Dec 9. [DOI] [PubMed] [Google Scholar]
- 43.Wong P, Pamer EG.. CD8 T cell responses to infectious pathogens. Annu Rev Immunol. 2003;21:29–70. [DOI] [PubMed] [Google Scholar]
- 44.Smith LK, Boukhaled GM, Condotta SA, et al. . Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity. 2018 Feb 20;48(2):299–312.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metlay JP, Waterer GW.. Treatment of community-acquired pneumonia during the coronavirus disease 2019 (COVID-19) pandemic. Ann Intern Med. 2020 Aug 18;173(4):304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.