Abstract
Background
In May, 2021, the delta (B.1.617.2) SARS-CoV-2 variant became dominant in the UK, superseded by the omicron (B.1.1.529) variant in December, 2021. The delta variant is associated with increased transmissibility compared with the alpha variant, which was the dominant variant in the UK between December, 2020, and May, 2021. To understand transmission and the effectiveness of interventions, we aimed to investigate whether the delta variant generation time (the interval between infections in infector–infectee pairs) is shorter—ie, transmissions are happening more quickly—than that of the alpha variant.
Methods
In this epidemiological analysis, we analysed transmission data from an ongoing UK Health Security Agency (UKHSA) prospective household study. Households were recruited to the study after an index case had a positive PCR test and genomic sequencing was used to determine the variant responsible. By fitting a mathematical transmission model to the data, we estimated the intrinsic generation time (which assumes a constant supply of susceptible individuals throughout infection) and the household generation time (which reflects realised transmission in the study households, accounting for susceptible depletion) for the alpha and delta variants.
Findings
Between February and August, 2021, 227 households consisting of 559 participants were recruited to the UKHSA study. The alpha variant was detected or assumed to be responsible for infections in 131 households (243 infections in 334 participants) recruited in February–May, and the delta variant in 96 households (174 infections in 225 participants) in May–August. The mean intrinsic generation time was shorter for the delta variant (4·7 days, 95% credible interval [CI] 4·1–5·6) than the alpha variant (5·5 days, 4·7–6·5), with 92% posterior probability. The mean household generation time was 28% (95% CI 0–48%) shorter for the delta variant (3·2 days, 95% CI 2·5–4·2) than the alpha variant (4·5 days, 3·7–5·4), with 97·5% posterior probability.
Interpretation
The delta variant transmits more quickly in households than the alpha variant, which can be attributed to faster depletion of susceptible individuals in households and a possible decrease in the intrinsic generation time. Interventions such as contact tracing, testing, and isolation might be less effective if transmission of the virus occurs quickly.
Funding
National Institute for Health Research, UK Health Security Agency, Engineering and Physical Sciences Research Council, and UK Research and Innovation.
Introduction
In May, 2021, the delta (B.1.617.2) SARS-CoV-2 variant became dominant in the UK1 and globally.2 This variant presents a higher risk of severe disease compared with previous variants,2, 3 although vaccination is still protective.4 The delta variant led to an increase in the growth rate of COVID-19 cases in the UK, outcompeting other variants.1, 5 This growth was attributed to increased transmissibility, with a study5 suggesting that the delta variant was 43–68% more transmissible5 than the alpha (B.1.1.7) variant, which was first identified in a sample from September, 2020.6
One way to characterise transmission of a SARS-CoV-2 variant is to measure its speed and strength.7, 8 Speed refers to how quickly the variant grows at the population-level, which is measured by the exponential growth rate and is inferable from disease incidence data.8 Strength reflects the variant's transmissibility8 and is typically measured by the time-dependent reproduction number (ie, the number of people that each infected person is expected to infect). The generation time (ie, the time between infection events in infector–infectee pairs) determines the relationship between a variant's speed and its strength8, 9, 10, 11 and is an input used in models to estimate the reproduction number from case notification data.12, 13 In principle, an increased growth rate of COVID-19 cases, as observed for the delta variant, is attributable to increased transmissibility, a shorter generation time, or both of these factors.8, 14
Previous studies have estimated the SARS-CoV-2 generation time,15, 16, 17, 18, 19, 20 with most estimates using data collected early in the COVID-19 pandemic. A household study18 from the UK Health Security Agency (UKHSA) indicated that the generation time of SARS-CoV-2 became shorter between September and November, 2020, compared with the earlier months. However, although this period coincided with emergence of the alpha variant, that variant was responsible for infections in only two households in the UKHSA study, and the delta variant had not yet emerged.18 Therefore, in this epidemiological analysis we compared the effect of these variants on the SARS-CoV-2 generation time using data from an ongoing household study in the UK.
Research in context.
Evidence before this study
We searched PubMed, Google Scholar, and medRxiv for articles published in English from database inception to Dec 7, 2021, with the search terms “COVID-19” or “SARS-CoV-2” and “generation time” or “generation interval”. We identified 15 studies directly estimating the SARS-CoV-2 generation time (ie, the time between infection events in infector–infectee pairs). Almost all analyses used data from the early stages of the pandemic; published analyses based on data collected from December, 2019, to April, 2020, found central mean generation time estimates of 3·44–7·50 days. However, the emergence of novel variants of concern (eg, the alpha [B.1.1.7], delta [B.1.617.2], and omicron [B.1.1.529] variants) can be expected to have changed the characteristics of transmission, such as the generation time, since the beginning of the pandemic. To our knowledge, the effect of different variants on the SARS-CoV-2 generation time has not been compared previously.
Added value of this study
We combined state-of-the-art mathematical modelling techniques for generation time estimation with transmission data from a household study in England. Because the household study included sequencing of PCR-positive cases, we could assess and compare the generation time for individuals infected by the alpha and delta variants. Our study shows that the generation time within households is shorter for the delta variant than the alpha variant (ie, the delta variant is transmitted more quickly).
Implications of all the available evidence
The inferred transmissibility of SARS-CoV-2 and the effectiveness of interventions depends on the speed of transmission. Our results suggest that the generation time in households is shorter for the delta variant than the alpha variant, which has implications for epidemiological analyses (eg, estimation of the reproduction number). The finding that the generation time can differ between SARS-CoV-2 variants highlights the need for further investigations to estimate the generation time of the omicron variant.
Methods
Study design and data
In this epidemiological analysis, we analysed transmission data from an ongoing prospective household study conducted by the UKHSA (appendix 1). Because the study began in January, 2021, and data were available from February to September, 2021, we could analyse the transition from alpha to delta becoming the dominant variant. Households were recruited to the study after an index case had a positive PCR test. The data contain PCR test results (including three tests taken as part of the study), symptom onset dates, ages, and vaccination statuses of participating household members (appendix 1). Genomic sequencing was used to determine the variant responsible for PCR-confirmed infections. Where sequencing data were unavailable, we assumed that the alpha variant was responsible for infections in households in which the index case first tested positive before May, 2021, and the delta variant was responsible for infections in households in which the index case first tested positive after May, 2021 (data from three unsequenced households recruited during May, 2021, were excluded from our analysis).1 A sensitivity analysis was done involving only households in which sequencing was performed.
A full description of the household study protocol is provided in appendix 2 (p 2). The household surveillance protocol was approved by the UKHSA Research Ethics and Governance Group as part of the portfolio of the UKHSA enhanced surveillance activities in response to the COVID-19 pandemic. Verbal informed consent for participation in the study (including data analyses to understand characteristics of household transmission) was obtained by the nurses from household members, who could decline participation in the household study at any time. Consent for children was obtained from a parent or legal guardian. Only anonymised data were provided to non-UKHSA authors.
Mathematical modelling and statistical analysis
We estimated the generation time by fitting a mechanistic model, motivated by compartmental modelling (appendix 2 pp 2–9), to the household transmission data. In this model, each infection is divided into three successive stages (the duration of time in each stage followed independent distributions): latent, presymptomatic and infectious, and symptomatic and infectious.17, 18 Unlike standard methods for estimating the generation time,15, 16 this approach links an infected host's infectivity profile to the time at which they develop symptoms.
Data augmentation Markov chain Monte Carlo (MCMC) was used to fit the transmission model to the data18, 21 with the following assumptions: (1) an incubation period distribution with a mean of 5·8 days (SD 3·1; obtained from a meta-analysis carried out before the alpha and delta variants emerged);22 (2) entirely asymptomatic, infected hosts being 35% as infectious as those who develop symptoms;23 (3) reduced susceptibility of vaccinated individuals compared with unvaccinated individuals (ie, the probability of transmission between an infectious individual and a susceptible individual is lower if the susceptible individual is vaccinated), using previous estimates;24 (4) no difference in infectiousness between infected individuals with different vaccination statuses; and (5) each household transmission cluster originating from a single primary case, with no further infections introduced into the household from the outside. We did sensitivity analyses to investigate the robustness of our results to these assumptions.
The following four model parameters were estimated for each variant in the parameter fitting procedure: the mean latent period (ie, the time from being infected to becoming infectious), as a proportion of the mean incubation period; the mean symptomatic infectious period; the relative infectiousness of presymptomatic compared with symptomatic infectious hosts; and the overall transmissibility parameter β0, which represents the expected number of household transmissions generated by a single, non-asymptomatic infector in an unvaccinated and otherwise entirely susceptible household, assuming that following each transmission, the infectee is removed and replaced by another susceptible individual.
Trace plots and comparisons between prior and posterior distributions of fitted model parameter values are shown in appendix 2 (p 10). The prior distributions used and central estimates of posterior distributions with 95% credible intervals (CIs) are given in appendix 2 (p 24) for each variant. To test the fitting procedure, we also conducted a simulation study, in which we used the central parameter estimates for each variant to generate synthetic data and refitted our household transmission model to those data (appendix 2 p 11).
The intrinsic generation time distribution (the distribution assuming the number of susceptible individuals remains constant throughout infection—ie, supposing that whenever a transmission occurs, the infectee is immediately removed from the population and replaced with another susceptible individual), which provides a generalised estimate independent of household size, was calculated for each variant from assumed and estimated model parameter values (appendix 2 pp 4–5). The household generation time distribution (the distribution of realised generation times within households, accounting for depletion of susceptible individuals)25 was also estimated for each variant (appendix 2 p 8). The household generation time is shorter on average than the intrinsic generation time because infected individuals might run out of susceptible individuals to infect within households.9, 25 This effect is often caused by multiple infected hosts competing to infect the available susceptible individuals (with only the first potential transmission being realised).26, 27 Consequently, realised transmissions occur faster within households than the intrinsic generation time distribution suggests.
We calculated the posterior distributions of the mean and SD of the intrinsic and household generation time distributions for each variant. We also combined the estimates obtained in each step of the MCMC procedure (after a burn-in period to allow the parameter inference procedure to identify the relevant region of parameter space and thinning to reduce autocorrelation between estimates used from different iterations) to estimate the entire household generation time distribution (appendix 2 p 13).
By comparing estimates of mean intrinsic and household generation times for the alpha and delta variants obtained in each iteration of the MCMC procedure, we calculated posterior estimates of percentage reduction in these quantities due to the delta variant (compared with the alpha variant). We also calculated the posterior probability that each quantity was smaller for the delta variant than the alpha variant (ie, the proportion of MCMC iterations in which the estimated mean intrinsic or household generation time was shorter for the delta variant). Household generation times were also compared on the basis of vaccination status, date, and age. All analyses were performed in MATLAB (version R2021b).
Role of the funding source
EM, PW, and NJA are employees of the UKHSA. The UKHSA was involved in the design of the household study, and collecting, accessing, and verifying the data. Other funders had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit the paper for publication.
Results
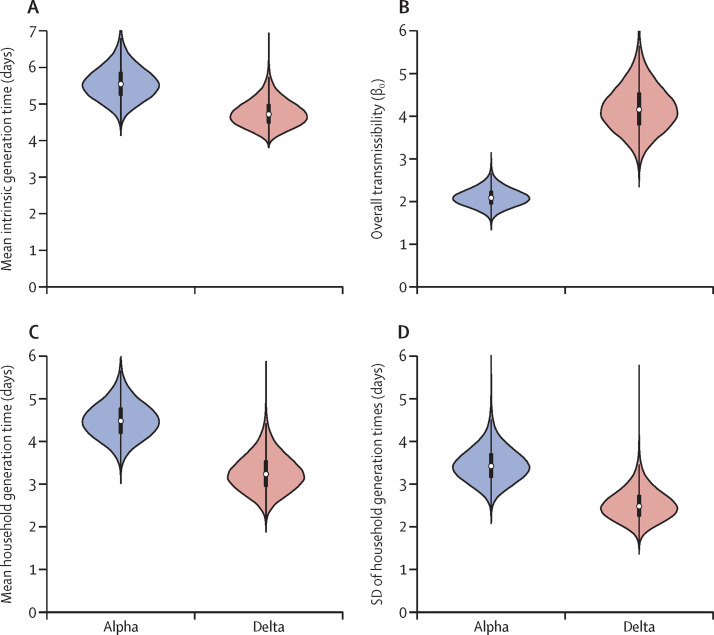
Between February and August, 2021, 227 households and 559 participants were recruited to the UKHSA study (table ; shown by vaccination status and age in appendix 2 pp 21–22). The alpha variant was detected or assumed to be responsible for infections in 131 households (243 infections in 334 participants) recruited in February–May, and the delta variant in 96 households (174 infections in 225 participants) in May–August. The mean intrinsic generation time was estimated to be 4·7 days (95% CI 4·1–5·6) for the delta variant and 5·5 days (4·7–6·5) for the alpha variant (figure 1A ). Comparing the posterior estimates suggests that the mean intrinsic generation time is 15% (95% CI −7% to 31%) shorter for the delta variant than the alpha variant, with 92% posterior probability that the delta variant has the shorter mean intrinsic generation time of the two variants (figure 1A). Our results also indicate higher transmissibility within households for the delta variant than for the alpha variant (figure 1B). Estimates of the mean household generation time for the two variants (figure 1C) are shorter than the corresponding intrinsic generation time estimates (figure 1A). The mean household generation time was estimated to be 28% (95% CI 0–48%) shorter for the delta variant (3·2 days, 95% CI 2·5–4·2) than the alpha variant (4·5 days, 3·7–5·4), with 97·5% posterior probability that the delta variant has the shorter mean household generation time of the two variants.
Table.
Number of households, participants, and infections by variant
| Recruitment months, 2021 | Number of households (n=227) | Number of participants (n=559) | Number of infections (n=417) | ||
|---|---|---|---|---|---|
| Alpha | February–May | 131 | 334 | 243 | |
| Confirmed | February–May | 116 | 300 | 223 | |
| Assumed | February–April | 15 | 34 | 20 | |
| Delta | May–August | 96 | 225 | 174 | |
| Confirmed | May–June | 27 | 61 | 55 | |
| Assumed | June–August | 69 | 164 | 119 | |
In each household, the variant responsible for infections was either determined by genomic sequencing or assumed on the basis of the month in which the household index case first tested positive. Only a single variant was detected in each sequenced household (ie, no household had detected infections with both the alpha [B.1.1.7] and delta [B.1.617.2] variants). Sequencing data from July, 2021, onwards were not available at the time of our analysis, although the delta variant was dominant in this period.1
Figure 1.
The effect of variant on the intrinsic and household generation times
Violin plots indicate posterior estimates for the alpha and delta variants. (A) The mean intrinsic generation time (the mean generation time if the supply of susceptible individuals remains constant throughout infection). (B) Overall transmissibility, β0 (the expected number of household transmissions generated by a single, non-asymptomatic infector in an unvaccinated and otherwise entirely susceptible household, assuming that following each transmission, the infectee is removed and replaced by another susceptible individual). (C) The mean household generation time (the mean realised generation time accounting for depletion of susceptible individuals in households). (D) The SD of household generation times. Central estimates and 95% credible intervals are shown in appendix 2 (pp 24–25). Posterior estimates of the SD for the intrinsic generation time distribution, and posterior estimates of fitted model parameters, are compared between variants in appendix 2 (p 12).
We combined the estimates obtained in each step of the MCMC procedure to estimate the entire household generation time distribution for the alpha and delta variants, which showed that household delta variant transmissions typically occur earlier during SARS-CoV-2 infection than transmissions with the alpha variant (appendix 2 p 13). We also show posterior estimates of the mean and SD of the household serial interval distribution (ie, the period between symptom onset times in infector–infectee pairs; appendix 2 p 14). Similarly to the household generation time, we found a shorter mean household serial interval for the delta variant (1·8 days, 95% CI 1·0–2·4) than the alpha variant (3·5 days, 2·7–4·1). For both variants, the mean household serial interval was shorter than the mean household generation time—by 1·0 days (0·2–1·8) for the alpha variant and 1·5 days (0·7–2·4) for the delta variant—but the household serial interval had a higher SD than the household generation time.
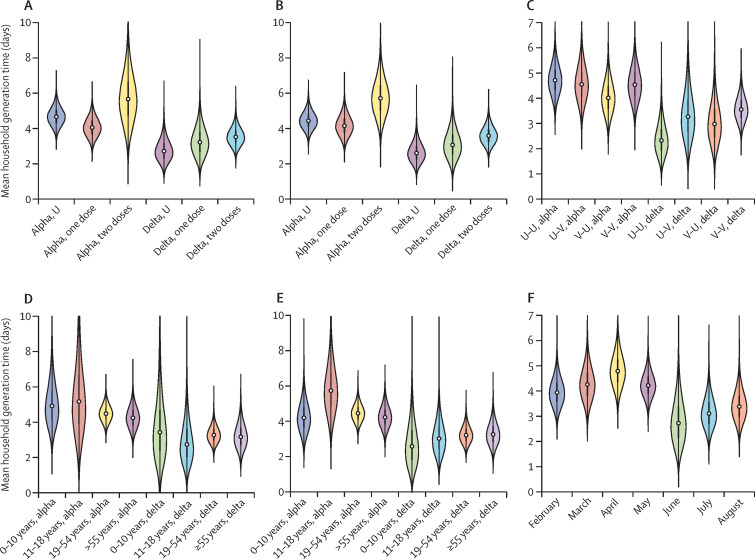
We also explored the effects of vaccination (figure 2A–C ), age (figure 2D–E), and month in which the index case first tested positive (figure 2F) on household generation times. We estimated the generation times associated with infectors and infectees with different vaccination statuses and considered different combinations of infector–infectee pairs. The effect of variant on the household generation time was larger than that of vaccination status (figure 2A–C). For both variants, there was no clear effect of the infector or infectee age on the generation time (figures 2D–E). A reduction in household generation time occurred during April–June, 2021, probably because the delta variant became dominant in the UK during that timeperiod (figure 2F).1
Figure 2.
The effect of different factors on household generation times
Violin plots indicate posterior estimates of the mean household generation time depending on the vaccination status of the infector and the variant (A), the vaccination status of the infectee and the variant (B), the combination of vaccination statuses in infector–infectee pairs (eg, U–V corresponds to transmissions from unvaccinated infectors to vaccinated infectees; vaccinated defined here as individuals who have received either one or two doses) and the variant (C), the age of the infector and the variant (D), the age of the infectee and the variant (E), and the month in 2021 in which the household index case first tested positive (F). U=unvaccinated. V=vaccinated.
Finally, we conducted sensitivity analyses considering the robustness of our results to the modelling assumptions (appendix 2 pp 15–20). Assuming a shorter incubation period28 than used in our main analysis22 for both variants or only the delta variant (appendix 2 p 15), led to a larger difference in mean intrinsic generation time between variants than shown in figure 1A (eg, in the sensitivity analysis in which both variants had a shorter incubation period, the estimated mean intrinsic generation time was 27% [95% CI 5–44%] shorter for the delta variant than the alpha variant). Conversely, when we only considered data from households in which genomic sequencing was conducted (appendix 2 p 20), we did not find a difference in the mean intrinsic generation time between variants; however, posterior distributions of estimated quantities for the delta variant in this sensitivity analysis were wide because sequencing data were unavailable from households recruited after June, 2021, by which time the delta variant was dominant.1 Nonetheless, in both of these sensitivity analyses, our finding of a shorter household generation time for the delta variant than for the alpha variant remained unchanged. This finding was robust to the different infectiousness levels of asymptomatic, infected hosts (appendix 2 p 16) and vaccinated, infected hosts (appendix 2 p 17) and to multiple virus introductions into households (appendix 2 pp 18–19).
Discussion
Novel SARS-CoV-2 variants possess different transmission characteristics to the coronavirus that originally emerged in Wuhan, China.5, 14 A key characteristic is the generation time, which measures the speed of transmission.17, 25, 29 To estimate the generation time, analysing datasets in which infector–infectee pairs can be determined or estimated is necessary.17, 25, 29 Household studies provide an opportunity to conduct such analyses.18, 21 In our analysis of data from a UKHSA household study, we found that the mean intrinsic generation time was slightly shorter for the delta variant than the alpha variant, but with substantial overlap in the CIs between the variants. For each variant, realised household generation times were shorter on average than intrinsic generation times, because the household generation time accounts for depletion of susceptible individuals within households (potential transmissions with longer generation times are then less likely to occur). We found a larger difference in the mean household generation time between the variants than expected from the estimated difference in intrinsic generation time alone. This finding is attributable to the increased transmissibility of the delta variant in households, leading to susceptible individuals being infected quickly, thereby being unavailable for infection later (thus shortening the average period between realised transmissions). The effect of variant on the household generation time was greater than the effects of other factors, including age and vaccination status.
To our knowledge, this is the first study to compare the generation time for different SARS-CoV-2 variants directly, and we are not aware of other generation time estimates for the alpha variant. An estimate for the delta variant based on 55 transmission pairs was stated in China CDC Weekly in 2021,28 although the methods underlying the estimate were not presented. Nonetheless, their mean generation time estimate of 2·9 days28 lies within the CI of our estimate of the mean household generation time for the delta variant (3·2 days, 95% CI 2·5–4·2) and is shorter than the lower limit of the 95% CI of our estimate for the alpha variant (4·5 days, 3·7–5·4). This observation supports our finding that the delta variant is transmitted more quickly than the alpha variant in households. Because we conducted our analysis before November, 2021, we did not analyse the generation time for the newly emerged omicron (B.1.1.529) variant. However, our finding of a different mean household generation time for the alpha versus the delta variant highlights the need for generation time estimates to be updated for the omicron variant when suitable data become available.
Other studies have estimated the serial interval (the period between symptom onset times in infector–infectee pairs) for the delta variant.30, 31, 32 Recently, preliminary studies have also estimated the serial interval33 and related test-to-test interval34 for the omicron variant. Although the serial interval is sometimes used as a proxy for the generation time, these two quantities might not follow the same distribution29—eg, the generation time always has positive values, whereas the serial interval can be negative if presymptomatic transmission is possible. The serial interval distribution often has a higher SD than the generation time distribution,29 and those distributions might not have the same mean value.18 A shorter mean serial interval than generation time (as we found for household transmission in our analysis) can be attributed to presymptomatic transmission, because individuals with longer incubation periods might generate more transmissions on average. This effect, which is captured in our mathematical modelling approach, leads to an expected incubation period that is longer for infectors than infectees in transmission pairs, and therefore, a shorter mean serial interval than generation time18 (since the difference between the generation time and the serial interval is equal to the difference between the incubation periods of the infector and the infectee29).
The viral load trajectories of individuals with infections due to the alpha and delta variants have also previously been estimated.35, 36, 37 Although estimating how infectiousness varies during infection using viral load data might be possible,38 the timing of realised transmissions depends on factors additional to inherent infectiousness, such as behaviour (eg, individuals might be less likely to transmit after developing symptoms and self-isolating) and the availability of susceptible individuals. Our approach explicitly accounts for changes in transmission risk following symptom onset, and the household generation time accounts for depletion of susceptible individuals within households.
Generation time estimates underlie a range of epidemiological analyses, including inference of the time-varying reproduction number12, 13 in different regions or countries and the relative transmissibility of different variants.5, 14 Such analyses often neglect temporal changes in the generation time. Furthermore, our results highlight that the reproduction number can affect the realised generation time (specifically, higher transmissibility of the delta variant leads to faster transmission within households). This finding underlines the importance of using up-to-date generation time estimates when estimating reproduction numbers. Overestimation of the mean generation time generally leads to reproduction number estimates that are too high in a growing epidemic, and too low in a declining epidemic.10 When analysing the transmissibility advantage of the delta variant over the alpha variant, neglecting a potentially shorter generation time for the delta variant would lead to its transmissibility advantage being overestimated.8
Because depletion of susceptible contacts might be less important in determining the timing of transmissions occurring outside households than within households, we expect the overall generation time distribution (accounting for transmissions across all settings) to lie between our household and intrinsic estimates. Therefore, we expect that inferring the delta variant transmissibility advantage over the alpha variant from incidence data using our household generation time estimates would give a lower bound for the transmissibility advantage (because the observed increased growth rate of the delta variant is explained partly by a substantial generation time reduction). By contrast, inferring the transmissibility advantage of the delta variant using intrinsic generation time estimates would give an upper bound.
We note that our estimates of overall transmissibility (β0) of the alpha and delta variants cannot be directly compared with previous estimates of the delta variant's transmission advantage (eg, an estimate of 43–68% inferred from incidence data5). The value of β0 represents the expected number of household transmissions generated by a single, non-asymptomatic infector in an unvaccinated and entirely susceptible household, assuming that following each transmission, the infectee is removed and replaced by another susceptible individual. As a result, β0 reflects intrinsic transmissibility rather than realised transmission. The intrinsic transmission advantage of the delta variant will only be realised if a sufficient number of susceptible individuals is available for each infector.
Limitations of this epidemiological modelling study include the model assumptions and the extent to which data are representative of the wider population. Although we analysed household data, transmission characteristics might differ between settings, which motivated us to estimate both the household and intrinsic generation times. The intrinsic generation time estimate, which does not account for depletion of susceptible individuals in households, can be conditioned to specific contact networks and transmission settings (eg, households, schools, workplaces, and social contact networks). We also conducted sensitivity analyses, confirming that our finding of a shorter household generation time for the delta variant than for the alpha variant was robust to important modelling assumptions.
In summary, our analysis indicates that the delta variant is transmitted more quickly than the alpha variant in households. This finding has implications for interventions, because measures such as contact tracing are less effective if transmission of the virus occurs quickly. Epidemiological models need to be updated to reflect the generation time of variants driving transmission and assessed further as the characteristics of SARS-CoV-2 transmission continue to change.
Data sharing
All data generated or analysed during this study are available in this Article and the appendices. Data and code for reproducing our results are available at https://github.com/will-s-hart/variant_generation_times.
Declaration of interests
We declare no competing interests. WSH, SF, and RNT report participation in discussions of the UK Government's Scientific Pandemic Influenza Group on Modelling, Operational subgroup.
Acknowledgments
Acknowledgments
Data collection for this study was done by the UK Health Security Agency (UKHSA; an executive agency of the UK Department of Health), as part of the COVID-19 response. WSH was funded by an Engineering and Physical Sciences Research Council Excellence Award for his doctoral studies (EP/R513295/1). EM receives support from the National Institute for Health Research (NIHR) Health Protection Research Unit in Immunisation at the London School of Hygiene & Tropical Medicine in partnership with the UKHSA (NIHR200929). SF receives funding from the Wellcome Trust (210758/Z/18/Z). RNT receives support from UK Research and Innovation (EP/V053507/1). We thank Matt Keeling for helpful discussions about this research. We also thank household members who took part in this study, the UKHSA nursing staff who recruited and followed up the households, the laboratory staff who tested the swabs, and administrative staff who arranged for the delivery and collection of testing kits from households.
Contributors
WSH, SF, and RNT conceptualised this study. WSH and RNT contributed to the literature search. All authors had full access to all the data in the study and accept responsibility for the decision to submit for publication. EM and PW have accessed and verified all the data in the study. WSH contributed to the formal analysis, investigation, validation, and visualisation. WSH wrote and tested the MATLAB computing code used in our analysis. WSH, SF, NJA, and RNT contributed to the methodology. SF and RNT were responsible for project administration. PKM, SF, and RNT supervised the study. WSH and RNT wrote the original draft. All authors reviewed and edited the manuscript.
Supplementary Materials
References
- 1.Public Health England SARS-CoV-2 variants of concern and variants under investigation in England. Sept 17, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1018547/Technical_Briefing_23_21_09_16.pdf
- 2.WHO COVID-19 weekly epidemiological update, edition 58. Sept 21, 2021. https://apps.who.int/iris/handle/10665/345456
- 3.Twohig KA, Nyberg T, Zaidi A, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22:35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Public Health England SARS-CoV-2 variants of concern and variants under investigation in England. March 11, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/972247/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf
- 7.Dushoff J, Park SW. Speed and strength of an epidemic intervention. Proc Biol Sci. 2021;288 doi: 10.1098/rspb.2020.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SW, Bolker BM, Funk S, et al. Roles of generation-interval distributions in shaping relative epidemic strength, speed, and control of new SARS-CoV-2 variants. medRxiv. 2021 doi: 10.1101/2021.05.03.21256545. published online May 5. (preprint). [DOI] [Google Scholar]
- 9.Fraser C. Estimating individual and household reproduction numbers in an emerging epidemic. PLoS One. 2007;2:e758. doi: 10.1371/journal.pone.0000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parag KV, Thompson RN, Donnelly CA. Are epidemic growth rates more informative than reproduction numbers? medRxiv. 2021 doi: 10.1101/2021.04.15.21255565. published online June 4. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gostic KM, McGough L, Baskerville EB, et al. Practical considerations for measuring the effective reproductive number, Rt. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbott S, Hellewell J, Thompson RN, et al. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112. [Google Scholar]
- 14.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferretti L, Wymant C, Kendall M, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368 doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganyani T, Kremer C, Chen D, et al. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.17.2000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart WS, Maini PK, Thompson RN. High infectiousness immediately before COVID-19 symptom onset highlights the importance of continued contact tracing. eLife. 2021;10 doi: 10.7554/eLife.65534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart WS, Abbott S, Endo A, et al. Inference of SARS-CoV-2 generation times using UK household data. medRxiv. 2021 doi: 10.1101/2021.05.27.21257936. published online May 30. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun K, Wang W, Gao L, et al. Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. Science. 2021;371 doi: 10.1126/science.abe2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu S, Wang W, Wang Y, et al. Infectivity, susceptibility, and risk factors associated with SARS-CoV-2 transmission under intensive contact tracing in Hunan, China. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson NM, Cummings DAT, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 22.McAloon C, Collins Á, Hunt K, et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buitrago-Garcia D, Egli-Gany D, Counotte MJ, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pouwels KB, Pritchard E, Matthews PC, et al. Effect of delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SW, Champredon D, Dushoff J. Inferring generation-interval distributions from contact-tracing data. J R Soc Interface. 2020;17 doi: 10.1098/rsif.2019.0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenah E, Lipsitch M, Robins JM. Generation interval contraction and epidemic data analysis. Math Biosci. 2008;213:71–79. doi: 10.1016/j.mbs.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q-H, Ajelli M, Aleta A, Merler S, Moreno Y, Vespignani A. Measurability of the epidemic reproduction number in data-driven contact networks. Proc Natl Acad Sci USA. 2018;115:12 680–12 685. doi: 10.1073/pnas.1811115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Xiao J, Deng A, et al. Transmission dynamics of an outbreak of the COVID-19 delta variant B.1.617.2—Guangdong province, China, May–June 2021. China CDC Wkly. 2021;3:584–586. doi: 10.46234/ccdcw2021.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehtinen S, Ashcroft P, Bonhoeffer S. On the relationship between serial interval, infectiousness profile and generation time. J R Soc Interface. 2021;18 doi: 10.1098/rsif.2020.0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pung R, Mak TM, Kucharski AJ, Lee VJ. Serial intervals in SARS-CoV-2 B.1.617.2 variant cases. Lancet. 2021;398:837–838. doi: 10.1016/S0140-6736(21)01697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang M, Xin H, Yuan J, et al. Transmission dynamics and epidemiological characteristics of delta variant infections in China. medRxiv. 2021 doi: 10.1101/2021.08.12.21261991. published online Aug 13. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu S, Kim D, Lim J-S, Ali ST, Cowling BJ. Serial interval and transmission dynamics during SARS-CoV-2 delta variant predominance, South Korea. Emerg Infect Dis. 2022;28:407–410. doi: 10.3201/eid2802.211774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Jo J, Lim J-S, Ryu S. Serial interval and basic reproduction number of SARS-CoV-2 omicron variant in South Korea. medRxiv. 2021 doi: 10.3390/v14030533. https://www.medrxiv.org/content/10.1101/2021.12.25.21268301v1 published online Dec 25. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott S, Sherratt K, Gerstung M, Funk S. Estimation of the test to test distribution as a proxy for generation interval distribution for the omicron variant in England. medRxiv. 2022 https://www.medrxiv.org/content/10.1101/2022.01.08.22268920v1 published online Jan 10. (preprint). [Google Scholar]
- 35.Kissler SM, Fauver JR, Mack C, et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med. 2021;385:2489–2491. doi: 10.1056/NEJMc2102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 delta variant. Nat Commun. 2022;13:460. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (alpha), B.1.315 (beta), and B.1.617.2 (delta) Clin Infect Dis. 2021 doi: 10.1093/cid/ciab721. published online Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart WS, Maini PK, Yates CA, Thompson RN. A theoretical framework for transitioning from patient-level to population-scale epidemiological dynamics: influenza A as a case study. J R Soc Interface. 2020;17 doi: 10.1098/rsif.2020.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are available in this Article and the appendices. Data and code for reproducing our results are available at https://github.com/will-s-hart/variant_generation_times.