Abstract
Ticks are of great menace to animal and human health. They serve as vectors to both animals and human pathogens including Rickettsia species. Tick-borne rickettsiosis in West Africa remains incompletely understood. We determined the prevalence of tick infestation among small ruminants and molecularly described a clinically significant spotted fever Rickettsia massiliae from Rhipicephalus ticks collected from North-Central, Nigeria. A total of 352 small ruminants comprising of 152 sheep and 200 goats that were brought for slaughter at the major small ruminant slaughterhouse in Ilorin were examined for the presence of ticks. The collected Rhipicephalus species were subjected to molecular studies to detect and characterize Rickettsia massiliae. Of the small ruminants examined, 21 sheep and 46 goats were infested with ticks representing 13.82% and 23.00% respectively. Eight and nine different species of ticks were detected in sheep and goats respectively, with Rhipicephalus (Boophilus) decoloratus being the most prevalent tick species in both sheep and goats. There was a significant difference (p <0.01) in the prevalence of the different tick species collected in sheep and in goats. Based on the PCR amplification of the 23S-5S intergenic spacer (IGS), only 2 of the 142 Rhipicephalus tick samples screened for R. massiliae were positive (1.41%; 95% CI = 0.39–4.99). Rickettsia massiliae was detected from Rhipicephalus turanicus collected from sheep. Sequences obtained from the PCR carried out by amplifying Rickettsia 23S-5S IGS showed 99–100% close identity with members of the R. massiliae group. This study has for the first time confirmed the presence of spotted fever group Rickettsia massiliae from feeding ticks in Nigerian small ruminants. Further investigations to determine the possible pathogenic role of human R. massiliae infection in Nigeria would be beneficial.
Introduction
Small ruminants (sheep and goat) rearing is one of the most important aspects of agriculture in most parts of the world [1]. This has contributed greatly to the growth and development of many economies worldwide [1, 2]. Small ruminants are a major component of the ruminant industry in Nigeria, with an estimated population of sheep and goats at 22.1 million and 34.5 million, respectively [3, 4]. Small ruminants maintain an available economic and ecological niche in Nigerian agriculture, as they represent about one-third of the country’s agricultural gross domestic product [3, 5]. Sheep and goats are very vital sources of protein to man in terms of meat and milk in both developed and developing economies [4, 6], they are also useful for the provision of manure, skin, and for household income and socio-cultural purposes [4, 7].
Ticks are the second most important arthropod parasite after mosquitoes, that affect mammals and birds [8, 9]. They are ranked as the most economically important ectoparasites of livestock in the tropics, including sub-Saharan Africa [10, 11]. A number of tick species can act as vectors of pathogens causing a number of tick-borne diseases, which causes a serious impairment to the health, welfare, production, and reproduction of ruminants including sheep and goats [9, 12].
Tick-borne rickettsiosis is among the oldest known vector-borne zoonotic diseases in the world [13]. Although still largely neglected, human rickettsiosis is the second most frequent cause of febrile illnesses after malaria in travelers returning from sub-Saharan Africa [14]. Several human tick-borne Rickettsia are classified as members of the spotted fever groups (SFG). So far, the recognized species of the SFG include: R. conorii, the agent of Mediterranean spotted fever; R. rickettsii, agent of Rocky Mountain spotted fever; R. africae, agent of African tick bite fever; R. sibirica, agent of Siberian tick typhus. Others include: R. slovaca, R. honei, R. japonica, R. australis, R. akari, R. felis, R. helvetica, R. parkeri, R. peacockii as well as the newly emerging human pathogen: R. massiliae and its closely related species that make up the R. massiliae group which consists of R. massiliae, Rickettsia sp. Bar29, R. rhipicephali, R. motanensis and R. aeschlimannii [13, 15–17].
Rickettsia massiliae, although first isolated from ticks in Marseille, is a confirmed human pathogen [16]. Subsequently, human cases of spotted fever group R. massiliae infection have been reported in Europe [18]; and South America [19]. Members of the R. massiliae group have been established to be some of the Rickettsia species that are naturally resistant to rifampicin by a Phe-to-Leu mutation within the rpoB gene [20]. In Europe and Africa, members of the Rhipicephalus tick complex are the documented vectors of R. massiliae [21]. This tick transmitting zoonotic pathogens has been reported to cause fever, palpable purpuric rash on the upper and lower extremities, skin lesion (eschar) on the right leg in humans [19]. Chorioretinitis with macular involvement was also reported in human cases with R. massiliae [22].
Limited molecular studies exist describing ticks and associated Rickettsia spp. affecting cattle and dogs in Nigeria [23, 24]. In fact, only a single study reported R. massiliae in questing Rhipicephalus evertsi ticks collected from vegetations in Southern Nigeria [25]. In contrast, no such studies have been conducted on feeding ticks among small ruminants from northern Nigeria. We report our findings in small ruminants from North-Central Nigeria using sensitive molecular methods to identify the tick-borne Rickettsia species present in small ruminants.
Materials and methods
Study location
Ethical approval for the study was granted by the University of Ilorin ethical review committee (Approval number: UERC/ASN/2018/1387).
This study was conducted at the Ipata municipal abattoir in Ilorin, Kwara State, North-Central Nigeria. This abattoir is one of the largest small ruminant abattoirs in the sub-region.
Collection and identification of ticks
Between May and August 2019, 352 apparently healthy small ruminants (152 sheep and 200 goats) were screened for the presence of ticks. Two hundred and forty adult ticks were collected from 67 small ruminants (21 sheep and 46 goats). Ticks were collected at the point of slaughter with the use of forceps thorough body search. The collected ticks were placed in 70% ethanol and transported to the laboratory. All of the ticks were morphologically identified by previous reported standard keys (based on: size, mouthparts, scutum, presence or absence of pale rings on legs, presence or absence eyes, etc.) as documented by Mathison and Pritt [26] and Walker et al. [27].
Deoxyribonucleic acid (DNA) extraction and PCR amplification
Genomic DNA was extracted from each tick that was morphologically identified as Rhipicephalus species (been the predominant tick species; n = 142) using the QIAamp tissue DNA kit (Qiagen, Hilden, Germany), according to the protocols prescribed by the manufacturers. Before the extraction of DNA, the individual tick was washed with phosphate-buffered saline (PBS), then air-dried for about 10 min on tissue paper and the tick was separately sliced into small pieces by using a sterile scalpel blade, afterward, it was manually homogenized with a sterile micro pestle, and resuspended in 200 μl of lysis buffer and 20 μl of proteinase K and incubated overnight at 56°C with continuous gentle shaking. The eluted ticks DNA was screened for the presence of Rickettsia by polymerase chain reaction (PCR) targeting the 350 bp DNA non-coding region of the Rickettsia 23S-5S intergenic spacer (IGS), and the PCR preparations were done as described by Kakumanu et al. [28] with modifications whereby only the secondary primer sets were used in a final volume of 25 μl. DNA from R. hoogstraali served as positive control while nuclease-free water was used as negative control in all PCR reactions. The set of oligonucleotide primers, the targeted fragment, thermocyclic protocols, and amplicon sizes (bp) for this study are summarized in Table 1.
Table 1. Rickettsia massiliae oligonucleotide primers used for PCR amplification and its thermocyclic protocols.
| Primer Name | Primer sequence (5′–3′) | Fragment | Thermocyclic protocols | Amplicon size (bp) |
|---|---|---|---|---|
| RCK/23-5N1F | TGTGGAAGCACAGTAATGTGTG | 23S-5S IGS | ID: 95°C / 10 min | 350 |
| (No of repeat: 1) | ||||
| RCK/23-5N1R | TCGTGTGTTTCACTCATGCT | D: 94°C / 30 sec | ||
| A: 56°C / 30 sec | ||||
| E: 72°C / 90 sec | ||||
| (No of cycles 35) | ||||
| FE: 72°C / 10 min | ||||
| HT°: 4°C |
Themocyclic protocol is as used for this study.
ID = initial denaturing of DNA, D = denaturing of DNA, A = annealing of primers, E = extension of DNA, FE = finial extension of DNA, HT° = holding temperature, bp = base pair.
The resultant amplicons were electrophoresed and visualized on 1.5% agarose gel that was stained with ethidium bromide ethidium to check the quality of amplification.
Gel purification and confirmation of the 23S-5S intergenic spacer (IGS) region of Rickettsia groups
Representative positive amplicons from the PCR were purified for sequencing using the Qiaquick® PCR purification kit (Qiagen®) according to the manufacturer’s instructions. Sequencing of the PCR products and confirmation of the 23S-5S intergenic spacer (IGS) region of Rickettsia groups was carried out at DBS Genomics, Durham.
Data and phylogenetic analyses
The Statistical Package for Social Sciences (SPSS) version 23.0 (SPSS Inc., Chicago, Illinois) was used for the statistical analysis. The prevalence and corresponding 95% confidence interval (CI) were used to measure the level of infestation among sheep and goats. The Chi-square (χ2) test was used to evaluate the level of each tick species infestation on sheep and goats. Statistical significance was set at p < 0.05. Sequences obtained from this study were searched for homologous sequences in the GenBank® using BLASTn (www.ncbi.nlm.nih.gov/BLAST). Sequences were aligned by using the Clustal W program. Based on these alignments, nucleotide alignments were performed and phylogenetic analyses were conducted in MEGA version 7.0 (https://www.megasoftware.net/). The phylogenetic tree was constructed by the Maximum Likelihood method, 1000 replicates bootstrap. The sequence obtained from this study has been deposited in GenBank® under the accession number OK350078.
Results
Of the 152 sheep and 200 goats sampled, 21 sheep and 46 goats were infested with ticks, representing 13.82% and 23.00% respectively. Sixty-seven of the total 352 small ruminants sampled were infested with ticks. The prevalence of tick infestation was significantly higher in goats compared to sheep (χ2 = 4.73; p = 0.03) (Table 2).
Table 2. Prevalence of tick infestation among small ruminants in Ilorin, Nigeria.
| Small ruminants | Number sampled | Number infested | Prevalence (%) | 95% CI |
|---|---|---|---|---|
| Sheep | 152 | 21 | 13.82 | 9.00–20.00 |
| Goats | 200 | 46 | 23.00 | 17.56–29.21 |
| Total | 352 | 67 | 19.03 | 15.19–23.39 |
CI = Confidence interval.
χ2 (Chi Square value) = 4.73.
DF (Degrees of Freedom) = 1.
p-value = 0.03.
Eight different tick species (belonging to 3 genera) were detected in sheep, while 9 species (belonging to 4 genera) were detected in goats. Eighty-one ticks were collected from sheep, while 159 ticks were collected from goats.
Rhipicephalus species were the most numerous species infesting sheep (55/81; 67.90%; 95% CI = 57.17–77.37) and goats (87/159; 54.72%; 95% CI = 46.93–62.33). In sheep, Rhipicephalus (Boophilus) decoloratus (n = 22; 27.16%) was the most prevalent tick species, followed by Amblyomma variegatum (n = 15; 18.52%), while Rhipicephalus lunulatus was the least prevalent with a 2.47%. Rhipicephalus (Boophilus) decoloratus and Amblyomma variegatum were the most prevalent tick species in goats, while Rhipicephalus evertsi was the least prevalent. There was a significant difference (p <0.01) in the prevalence of the different tick species collected in sheep and in goats (Table 3).
Table 3. Diversity and prevalence of tick species infesting small ruminants in Ilorin, Nigeria.
| Tick species | Number (%) | χ 2 | DF | p-value |
|---|---|---|---|---|
| Sheep (n = 81) | ||||
| Rhipicephalus turanicus | 5 (6.17) | 32.61 | 7 | <0.01¥ |
| Rhipicephalus sanguineus | 12 (14.81) | |||
| Rhipicephalus lunulatus | 2 (2.47) | |||
| Rhipicephalus evertsi | 5 (6.17) | |||
| Rhipicephalus (Boophilus) decoloratus | 22 (27.16) | |||
| Rhipicephalus (Boophilus) geigyi | 9 (11.11) | |||
| Amblyomma variegatum | 15 (18.52) | |||
| Hyalomma rufipes | 11 (13.58) | |||
| Goats (n = 159) | ||||
| Rhipicephalus turanicus | 2 (1.26) | 167.10 | 8 | <0.01¥ |
| Rhipicephalus sanguineus | 27 (16.98) | |||
| Rhipicephalus lunulatus | 3 (1.89) | |||
| Rhipicephalus evertsi | 1 (0.63) | |||
| Rhipicephalus (Boophilus) decoloratus | 51 (32.08) | |||
| Rhipicephalus (Boophilus) geigyi | 3 (1.89) | |||
| Amblyomma variegatum | 39 (24.53) | |||
| Hyalomma truncatum | 15 (9.43) | |||
| Hyalomma rufipes | 18 (11.32) |
n = Number of ticks collected in each small ruminant.
Χ2 = Chi square.
DF = Degrees of Freedom.
¥ = Significant at p < 0.05.
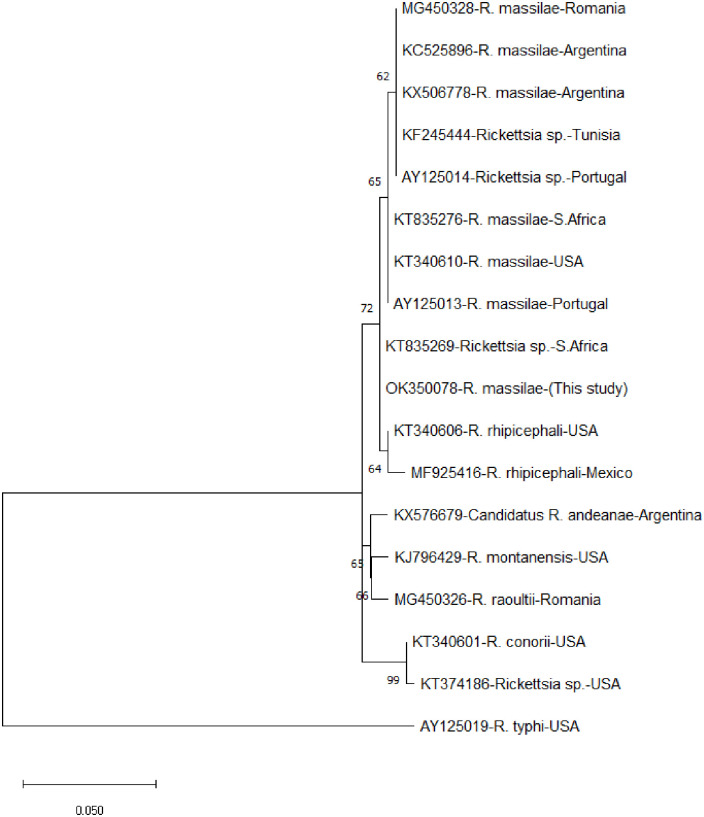
Based on the PCR amplification of the 23S-5S IGS, 2 of the 5 Rhipicephalus turanicus tick species collected from sheep were positive for R. massiliae (40.00%; 95% CI = 11.76–76.93). The molecular prevalence of R. massiliae in relation to the total number of Rhipicephalus tick (142) screened was 1.41%; 95% CI = 0.39–4.99. Sequences obtained from the PCR carried out by amplifying Rickettsia 23S-5S intergenic spacer (IGS) showed 99% close identity with members R. massiliae group. Phylogenetic analysis carried out based on the IGS showed that samples from this study clustered together with other reported R. massiliae available in the gene bank (Fig 1).
Fig 1. Molecular phylogenetic analysis inferred by Maximum Likelihood of Rickettsia massiliae based on the 350bp partial 23S-5S IGS sequences of Rickettsia species taken from the NCBI database and sequence amplified from Rhipicephalus species tick collected from Nigerian small ruminants.
Discussion
The overall ticks prevalence of 19.03% we recorded among small ruminants in this study is higher than the 8.10% total prevalence reported among small ruminants in Makurdi, North-Central Nigeria [29] and the 14.85% reported in Uli, southeast Nigeria [5]. Although a higher total prevalence of tick infestation among small ruminants has been documented in Pakistan (51.02%) [30] and Ethiopia (79.70%) [31]. The differences could be attributed to seasonal, environmental, and ecological factors.
A higher prevalence of tick infestation was observed in goats compared to sheep. This may be linked to the more roaming nature of goats compared to sheep, making goats more exposed to questing ticks in vegetations. It could also be due to the high wool level of sheep, making it difficult for ticks to attach on them.
Ticks are usually present in vegetations, awaiting suitable host(s) to attach onto [32], thus goats become more vulnerable due to their more roaming nature. Higher prevalence of tick infestation has been reported in goats compared to sheep in Nigeria [33] and outside Nigeria (Pakistan) [30].
Eight different species of ticks were detected among sheep in our study. This number is higher than the two, five, and six different species of ticks detected among sheep in Nigeria by Alayande et al. [34], Kaze et al. [33] and Maidala [35] respectively. Lower number of tick species (4) have been reported to infest sheep in Ethiopia [36]. In like manner, a lower number of tick species was reported among goats in Nigeria [33, 34, 35] and outside Nigeria (Ethiopia) [31] compared to the number (9) we observed in our study. These observations may suggest that tick infestation is of concern among small ruminants in the study area.
Rhipicephalus was the most predominant genus of ticks infesting sheep and goats, with Rhipicephalus (Boophilus) decoloratus being the most prevalent species among both animal species in our study. Rhipicephalus species has been reported to be the most prevalent genus infesting small ruminants in Nigeria [1, 29, 33, 35] and Ethiopia [37]. The high prevalence of Rhipicephalus species among other tick genera was the reason we used the genus in our molecular study of Rickettsia massilae.
The 23S-5S IGS is a conserved, non-coding region of DNA that is useful in Rickettsia molecular taxonomy [28]. Here, we are able to confirm the presence of Rickettsia massilae in ticks collected from domestic small ruminants. These tick-borne pathogens appear common in South Africa and have been reported from Cameroon [38], Central African Republic [39], and Ivory Coast [40].
The 40.00% molecular prevalence of R. massiliae among Rhipicephalus turanicus ticks collected from sheep in our study is far higher than the 3.51% of Rickettsia massiliae reported among Rhipicephalus turanicus ticks collected from sheep in China [41]. This significant level of high molecular prevalence of R. massiliae among Rhipicephalus turanicus collected from sheep in our study, calls for great public health concerns. Rickettsia massiliae is a known pathogenic Rickettsia causing spotted fever in humans [42].
Rickettsia massiliae was detected only from Rhipicephalus turanicus collected from sheep in the study area. In a similar vein, Wei et al. [41] and Ereqat et al. [43] detected R. massiliae in Rhipicephalus turanicus infesting sheep in China and the Palestinian territories respectively. Rhipicephalus turanicus is a three-host tick [27], with both transovarial and transstadial transmission of R. massiliae reported in the tick vector [44]. This shows there may be possibilities of R. massiliae infection in other animals infested by this tick species during the developmental stages of its lifecycle. There was a close relationship seen with the R. massiliae of this study and that seen in South Africa, suggesting a universal spread of the organism.
Rickettsia massiliae, strain Bar29, has been previously reported to be detected in engorged female Rhipicephalus turanicus tick collected in Corsica [44] and from many other members of the Rhipicephalus ticks from different parts of the world [21]. Rickettsia massiliae was previously isolated from questing R. evertsi ticks in cattle grazing areas from Southern Nigeria [25]. However, Ricketssia massiliae has not been reported in ticks infesting highly domesticated sheep and goats kept in Nigeria.
Detection of R. massiliae in feeding tick on sheep is of zoonotic concern. This is of significance because, sheep are one of the most widespread animals kept by humans in sub-Saharan Africa owing to their high fertility, short generation interval, adaptation to harsh environments, and are considered as a source of investments for rural households [45]. They are mostly kept as free-range and often tethered at night close to human dwellings. This provides a suitable environment for both contacts with disease vectors such as ticks thus facilitating their transfer of infections including Rickettsia massiliae to humans.
Conclusions
Rhipicephalus species was the most prevalent tick genus infesting small ruminants in the study area, with Rhipicephalus (Boophilus) decoloratus being the most prevalent species in both sheep and goats. This study has for the first time confirmed the presence of the spotted fever group Rickettsia massiliae from feeding ticks in Nigerian small ruminants. Rickettsia massiliae was detected in two Rhipicephalus turanicus ticks collected from sheep. So far it is not recognized as a potential human pathogen in Nigeria and is not likely to be considered during the evaluation of clinical cases. Confirmed life-threatening human cases elsewhere in the world emphasize the need to consider this diagnosis and investigate among clinical patients most at risk. Further investigations with more extensive tick samples as well as human studies determining the possible pathogenic role of human R. massiliae infection in Nigeria would be beneficial.
Supporting information
(XLSX)
Acknowledgments
We would also like to acknowledge Mr. O. Akintola for providing technical support for the project.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This study was supported by the Africa Research Excellence Fund Fellowship. Grant Number: MRF-157-0022-F-ELELU. The grant recipient is Dr. Nusirat Elelu.
References
- 1.Adang KL, Ayuba K, Yoriyo KP. Ectoparasites of Sheep (Ovis aries L.) and Goats (Capra hirus L.) in Gombe, Gombe State, Nigeria. Pak J Biol Sci. 2015; 18(5): 224–231. [Google Scholar]
- 2.Sertse T, Wossene A. Effect of ectoparasites on quality of pickled skins and their impact on the tanning industries in Amhara regional state, Ethiopia. Small Rumin Res. 2007; 69: 55–61. [Google Scholar]
- 3.Lawal-Adebowale OA. Dynamics of ruminant livestock management in the context of the Nigerian Agricultural System. In: Javed K. Livestock Production. IntechOpen Book Series: London, SW7 2QJ, UK; 2012. Pp. 61–82. [Google Scholar]
- 4.Ola-Fadunsin SD, Ibitoye EB. A retrospective evaluation of parasitic conditions and their associated risk factors in sheep and goats in Osun state, Nigeria. Sokoto J Vet Sci. 2017; 15(3): 15–24. [Google Scholar]
- 5.Obi ZC, Anyaegbunam L, Orji MN. Ectoparasitosis, a challenge in sheep and goat production in Uli, Anambra state, Nigeria. Int J Fauna Biol Stud. 2014; 1(5): 27–29. [Google Scholar]
- 6.Wesongah JO, Chemilitti FD, Wesongah L, Munga P, Ngare P, Munilla GA. Trypanosomiasis and other parasitic diseases affecting sheep and goats production in two group ranches, Narok district, Kenya. Pak Vet J. 2003; 14(3): 133–141. [Google Scholar]
- 7.Okorafor UP, Obebe OO, Unigwe CR, Atoyebi TJ, Ogunleye OK. Studies on the gut parasites of small ruminants reared in some selected farms in Ido local government area, Oyo state, Nigeria. Appl Res J. 2015; 1(3): 153–159. [Google Scholar]
- 8.Ebrahimzade E, Fattahi R, Ahoo MB. Ectoparasites of stray dogs in Mazandaran, Gilan and Qazvin provinces, north and center of Iran. J Arthropod Dis. 2016; 10: 364–369. [PMC free article] [PubMed] [Google Scholar]
- 9.Ola-Fadunsin SD, Sharma RSK, Abdullah DA, Gimba FI, Abdullah FFJ, Sani RA. The molecular prevalence, distribution and risk factors associated with Babesia bigemina infection in Peninsular Malaysia. Ticks Tick-borne Dis. 2021; 12: 101653. doi: 10.1016/j.ttbdis.2021.101653 [DOI] [PubMed] [Google Scholar]
- 10.Uilenberg G. International collaborative research: significance of tickborne hemoparasitic diseases to world animal health. Vet Parasitol. 1995; 57: 19–41. doi: 10.1016/0304-4017(94)03107-8 [DOI] [PubMed] [Google Scholar]
- 11.Lorusso V, Picozzi K, De Bronsvoort BMC, Majekodunmi A, Dongkum C, Balak G, et al. Ixodid ticks of traditionally managed cattle in central Nigeria: where Rhipicephalus (Boophilus) microplus does not dare (yet?). Parasites Vectors. 2013; 6: 171–180. doi: 10.1186/1756-3305-6-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorusso V, Wijnveld M, Majekodunmi AO, Dongkum C, Fajinmi A, Dogo AG, et al. Tick-borne pathogens of zoonotic and veterinary importance in Nigerian cattle. Parasites Vectors. 2016; 9: 217–229. doi: 10.1186/s13071-016-1504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005; 18: 719–756. doi: 10.1128/CMR.18.4.719-756.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensenius M, Davis X, Sonnenburg F, Schwartz E, Keystone JS, Leder K, et al. Multicenter GeoSentinel analysis of rickettsial diseases in international travelers, 1996–2008. Emerg Infect Dis. 2009; 15: 1791–1798. doi: 10.3201/eid1511.090677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003; 12: 5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitale G, Mansuelo S, Rolain JM, Raoult D. Rickettsia massiliae human isolation. Emerg Infect Dis. 2006; 12(1): 174–175. doi: 10.3201/eid1201.050850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazihan W, Dong Z, Guo L, Rizabek K, Askar D, Gulzhan K, et al. Molecular detection of spotted fever group rickettsiae in ticks parasitizing pet dogs in Shihezi City, northwestern China. Exp Appl Acarol. 2019; 77: 73–81 doi: 10.1007/s10493-018-00337-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oteo JA, Portillo A. Tick-borne rickettsioses in Europe. Ticks Tick-borne Dis. 2012; 3: 271–278. doi: 10.1016/j.ttbdis.2012.10.035 [DOI] [PubMed] [Google Scholar]
- 19.García-García JC, Portillo A, Núnez MJ, Santibáñez S, Castro B, Oteo JA. A patient from Argentina infected with Rickettsia massiliae. Am J Trop Med Hyg. 2010; 82(4): 691–692. doi: 10.4269/ajtmh.2010.09-0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolain JM, Maurin M, Vestris G, Raoult D. In vitro susceptibilities of 27 rickettsiae to 13 antimicrobials. Antimicrob Agents Chemother. 1998; 42: 1537–1541. doi: 10.1128/AAC.42.7.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eremeeva ME, Bosserman EA, Demma LJ, Zambrano ML, Blau DM, Dasch GA. Isolation and identification of Rickettsia massiliae from Rhipicephalus sanguineus ticks collected in Arizona. Appl Environ Microbiol. 2006; 72(8): 5569–5577. doi: 10.1128/AEM.00122-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parola P, Socolovschi C, Jeanjean L, Bitam I, Fournier P, Sotto A, et al. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl Trop Dis. 2008; 2(11): e338. doi: 10.1371/journal.pntd.0000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorusso V, Gruszka KA, Majekodunmi A, Igweh A, Welburn SC, Picozzi K. Rickettsia africae in Amblyomma variegatum ticks, Uganda and Nigeria. Emerg Infect Dis. 2013; 19(10): 1705–1707. doi: 10.3201/eid1910.130389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamani J, Baneth G, Mumcuoglu KY, Waziri NE, Eyal O, Guthmann Y, et al. Molecular detection and characterization of tick-borne pathogens in dogs and ticks from Nigeria. PLoS Negl Trop Dis. 2013; 7(3): p.e2108. doi: 10.1371/journal.pntd.0002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reye AL, Arinola OG, Hubschen JM, Muller CP. Pathogen prevalence in ticks collected from the vegetation and livestock in Nigeria. Appl Environ Microbiol. 2012; 78: 2562–2568. doi: 10.1128/AEM.06686-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathison BA, Pritt BS. Laboratory Identification of Arthropod Ectoparasites. Clin Microbiol Rev. 2014; 27(1): 48–67. doi: 10.1128/CMR.00008-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker AR, Bouattour A, Camicas J-L, Estrada-Peña A, Horak IG, Latif AA, et al. Ticks of Domestic Animals in Africa: a Guide to Identification of Species. Bioscience Reports: Edinburgh Scotland, U.K; 2014. pp. 1–221. [Google Scholar]
- 28.Kakumanu ML, Ponnusamy L, Sutton HT, Meshnick SR, Nicholson WL., Apperson, C.S. Development and validation of an improved PCR method using the 23S-5S intergenic spacer for detection of rickettsiae in Dermacentor variabilis ticks and tissue samples from humans and laboratory animals. J Clin Microbiol. 2016; 54: 972–979. doi: 10.1128/JCM.02605-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ofukwu RA, Ogbaje CI, Akwuobu CA. Preliminary study of the epidemiology of ectoparasite infestation of goats and sheep in Makurdi, north central Nigeria. Sokoto J Vet Sci. 2008; 7(2): 22–26. [Google Scholar]
- 30.Rehman A, Nijhof AM, Sauter-Louis C, Schauer B, Staubach C, Conraths FJ. Distribution of ticks infesting ruminants and risk factors associated with high tick prevalence in livestock farms in the semiarid and arid agro-ecological zones of Pakistan. Parasites Vectors. 2017; 10: 190–204. doi: 10.1186/s13071-017-2138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammed K, Admasu P. Prevalence of Ixodid Ticks in Small Ruminants in Selected Districts of Fafen Zone, Eastern Ethiopia. Eur J Appl Sci. 2015; 7(2): 50–55. [Google Scholar]
- 32.Mathisson DC, Kross SM, Palmer MI, Diuk-Wasser MA. Effect of vegetation on the abundance of tick vectors in the Northeastern United States: a review of the literature. J Med Entomol. 2021; 20(10): 1–8. doi: 10.1093/jme/tjab098 [DOI] [PubMed] [Google Scholar]
- 33.Kaze PD, Dogo GA, Tanko J, Bialla M, Kogi CA. Epidemiology of Ectoparasites Infestation in Jos North, Plateau State, Nigeria. Saudi J Med Pharm Sci. 2017; 3(3): 206–210. [Google Scholar]
- 34.Alayande MO, Mayaki AM, Lawal MD, Abubakat A, Kassu M, Talabi AO. Pattern of Ticks and Lice Infestation on Small Ruminants in Sokoto, Sokoto State. Niger J Anim Sci. 2016; 18(1): 183–189. [Google Scholar]
- 35.Maidala A. A Survey of Cattle, Sheep and Goat Ticks Infestation in Katagum Local Government Area of Bauchi State, Nigeria. Int J Agric Earth Sci. 2015; 1(8): 1–5. [Google Scholar]
- 36.Zewdu S, Tsegaye T, Agerie A. Ectoparasites Prevalence in Small Ruminants in and around Sekela, Amhara Regional State, Northwest Ethiopia. J Vet Med. 2015; Volume 2015, Article ID 216085, 6 pages doi: 10.1155/2015/216085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yishak I, Tsegalem A, Befekadu UW. Epidemiological study of ectoparasite infestation of small ruminants in Sodo Zuria District, Southern Ethiopia. J Vet Med Anim Health. 2015; 7(4): 140–144. [Google Scholar]
- 38.Essbauer S, Hofmann M, Kleinemeier C, Wölfel S, Matthee S. Rickettsia diversity in southern Africa: A small mammal perspective. Ticks Tick-borne Dis. 2018; 9: 288–301. doi: 10.1016/j.ttbdis.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 39.Dupont HT, Cornet JP, Raoult D. Identification of rickettsiae from ticks collected in the Central African Republic using the polymerase chain reaction. Am J Trop Med Hyg. 1994; 50: 373–380. doi: 10.4269/ajtmh.1994.50.373 [DOI] [PubMed] [Google Scholar]
- 40.Berrelha J, Briolant S, Muller F, Rolain JM, Marie JL, Pages F, et al. Rickettsia felis and Rickettsia massiliae in Ivory Coast, Africa. Clin Microbiol Infect. 2009; 15(Supplement 2): 215–252. doi: 10.1111/j.1469-0691.2008.02273.x [DOI] [PubMed] [Google Scholar]
- 41.Wei QQ, Guo LP, Wang AD, Mu LM, Zhang K, Chen CF, et al. The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasites Vectors. 2015; 8: 631–634. doi: 10.1186/s13071-015-1242-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013; 26(4): 657–702. doi: 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ereqat S, Nasereddin A, Al-Jawabreh A, Azmi K, Harrus S, Mumcuoglu K, et al. Molecular Detection and Identification of Spotted Fever Group Rickettsiae in Ticks Collected from the West Bank, Palestinian Territories. PLoS Negl Trop Dis. 2016; 10(1): e0004348. doi: 10.1371/journal.pntd.0004348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto K, Ogawa M, Brouqui P, Raoult D, Parola P. Transmission of Rickettsia massiliae in the tick, Rhipicephalus turanicus. Med Vet Entomol. 2005; 19: 263–270. doi: 10.1111/j.1365-2915.2005.00569.x [DOI] [PubMed] [Google Scholar]
- 45.Abunna F, Tsedeke E, Kumsa B, Megersa B, Regassa A, Debela E. Abomasal Nematodes: Prevalence in Small Ruminants Slaughtered at Bishooftu Town, Ethiopia. The Internet J Vet Med. 2008; 7(1): 6 pages. [Google Scholar]