Abstract
A PCR protocol was developed to detect Helicobacter pylori in human stool specimens. This protocol was based on the association of a magnetic immuno-PCR assay with a technique to remove inhibitors (agarose-embedded DNA preparation). Of the 47 H. pylori-positive and 57 H. pylori-negative patients included in this study, 38 were positive and 66 were negative by this new protocol. The sensitivity, specificity, and predictive values for a positive or a negative result were 80.9% (95% confidence interval [CI], 66.3 to 90.4), 100% (95% CI, 92.1 to 100), 100% (95% CI, 88.6 to 100), and 86.4% (95% CI, 75.2 to 93.2), respectively.
The high clinical relevance of Helicobacter pylori infection has stimulated the development of numerous diagnostic methods. These methods can be classified as invasive, (i.e., those that require an endoscopy to detect H. pylori directly in gastric biopsy specimens) or noninvasive (i.e., based on the study of various samples (serum, breath air, urine, etc.)), which indirectly indicate the presence of H. pylori.
Stool specimens constitute a sample of easy, noninvasive access and consequently of high potential interest for the development of a direct method of H. pylori detection. PCR is a powerful technique for the detection of target DNA in various clinical specimens, but its application to stool specimens has been limited due to the presence of substances inhibiting the reaction. Numerous attempts to detect H. pylori by PCR in stool samples have been made, with controversial results (5, 12, 17; N. P. Mapstone, F. A. Lewis, D. S. Tompkins, D. A. F. Lynch, A. T. R. Axon, M. F. Dixon, and P. Quirke, Letter, Lancet 341:447, 1993).
Difficulty in eliminating PCR inhibitors from stool specimens has been extensively reported (4, 5, 8, 12). Some authors have claimed to be successful, but usually only after applying complex procedures that are difficult to implement routinely.
The aim of this study was to develop a PCR protocol to detect H. pylori in human stool specimens that is simple enough to be used routinely. For this purpose, we associated a magnetic immuno-PCR assay (MIPA) with a technique to remove inhibitors that was recently described (11).
Clinical samples.
One hundred four consecutive, untreated, dyspeptic patients (69 male, 35 female; mean age, 50 years; range, 17 to 87 years) who were consulting gastroenterologists for dyspepsia in Bordeaux, France, were included in the study. Exclusion criteria were as follows: H. pylori eradication treatment in the previous 6 months; consumption of antibiotics in the previous month; or consumption of antisecretory drugs, bismuth salts, or sucralfate in the previous 2 weeks. A history of coagulopathy or other disorders that are contraindications for endoscopy and/or biopsy sampling was also a reason for exclusion. For each patient, biopsies were taken for culture, histological examination, urease test (UT), and PCR. A urea breath test (UBT) and a serological test were also performed as previously described (9).
A patient was classified as being H. pylori positive on the basis of (i) a positive culture; or, in the case of a negative culture, (ii) both a positive histological examination and a positive UT (CLOtest; TriMed Specialities, Osborne Park, Western Australia); or (iii) positive noninvasive tests (serology and UBT) and biopsy PCR. Stool specimens were collected within 1 week of the time of endoscopy in sterile containers and kept at −80°C until analysis.
Immunomagnetic separation of H. pylori from human feces and DNA extraction.
Stool specimens were suspended at 1.5:5 (wt/vol) for solid and semisolid samples and 1.5:5 (vol/vol) for liquid in phosphate-buffered saline and incubated overnight, under agitation at room temperature. The suspension was then filtered through three layers of cotton gauze and used for immunomagnetic separation of H. pylori. Briefly, magnetic uncoated beads (Dynabeads M-450; Dynal, Oslo, Norway) were coated with rabbit anti-H. pylori immunoglobulin (Dako, Glostrup, Denmark) at a concentration of 5 μg of antibody for 107 Dynabeads according to the procedure recommended by Dynal. A 60-μl volume of coated Dynabeads was mixed with 1 ml of fecal suspension and incubated at 4°C with continuous shaking for 2 h. The coated Dynabeads were recovered by magnetic force with a Dynalmagnet and then suspended in the lysis buffer of the QIAamp tissue kit (Qiagen, Hilden, Germany) for DNA extraction (9). Twenty microliters of a proteinase solution (20 mg/ml) was then added, followed by incubation at 56°C for 2 h. A second buffer provided in the kit was added, and the sample was incubated at 70°C for 10 min. Next, 200 μl of ethanol was added, and the suspension was loaded on the QIAamp spin column followed by a centrifugation at 6,000 × g for 1 min. The QIAamp spin column was placed in a 2-ml collection microtube, and the tube containing the filtrate was discarded. The column material was washed twice (250 μl each) with the first washing buffer and twice (250 μl each) with the second washing buffer provided in the kit. Finally, the DNA was eluted with 100 μl of distilled water preheated to 70°C (2 × 50 μl).
PCR.
PCR was performed as described previously (7). Briefly, reactions were carried out in a volume of 50 μl with a combination of 10× PCR buffer [670 mM Tris-HCl (pH 8.8), 160 mM (NH4)SO2, 0.1% Tween]; 1.5 mM MgCl2; 200 μM (each) nucleotides dATP, dTTP, dGTP, and dCTP; and 3 U of Taq polymerase (Eurobio, Les Ulis, France), with 5 μl of purified sample and with specific primers (HPU1 and HPU2) for the ureA gene of H. pylori (1) at 0.4 μM each. PCR consisted of 35 cycles of 1 min at 94°C, 1 min at 45°C, and 1 min at 72°C with a first cycle of 5 min at 95°C and a final cycle of 5 min at 72°C. Bands were visualized on a 1% agarose gel stained with ethidium bromide. The size of the PCR product obtained when using the HPU1 and HPU2 primers was 411 bp. In each experiment, a positive control consisting of 50 ng of reference strain DNA and a negative control (distilled water) were performed.
Removal of PCR inhibitors.
The PCR inhibitors present in stool samples were removed by using an agarose-embedded DNA preparation described previously (9, 11). Briefly, 1 volume of 1.6% melted agarose was added to each DNA aliquot; these were then poured into molds (Disposable Plug Molds; 1.5 by 10 by 5 mm; Bio-Rad, Richmond, Calif.) until solidification. Agarose blocks were removed from molds and washed in Tris-EDTA (10 ml per block) overnight with gentle shaking. A second wash with 5 ml of distilled water was performed (2 h with gentle shaking). PCR was subsequently carried out with DNA-containing agarose slices as templates.
Statistics.
Standard methods were used to calculate sensitivity, specificity, predictive values of positive and negative results, and the 95% confidence intervals (CI) of these values.
Ethics.
This study received the approval of the Ethics Committee of Bordeaux. The informed consent of the patients was also obtained.
According to the case definition, 47 of the 104 patients tested were H. pylori positive, and 57 were negative. The five patients for which doubtful results were found based on invasive tests were evaluated according to the results of noninvasive tests and biopsy PCR (Table 1). Two were considered H. pylori positive, and three were considered H. pylori negative.
TABLE 1.
Diagnostic test results for the five patients for whom the results of invasive tests for Helicobacter pylori diagnosis were doubtful
| Patient | Result bya:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology
|
Urease test
|
Culture
|
PCR with biopsy
|
UBT | Serology | PCR with feces | |||||
| Antrum | Fundus | Antrum | Fundus | Antrum | Fundus | Antrum | Fundus | ||||
| 1 | − | + | − | ND | − | ND | + | ND | + | + | + |
| 2 | − | + | − | ND | − | ND | + | ND | + | + | + |
| 3 | − | − | + | ND | − | ND | − | ND | − | − | − |
| 4 | − | − | + | ND | − | ND | − | ND | − | − | − |
| 5 | + | + | − | ND | − | ND | − | ND | − | − | − |
+, positive result; −, negative result; ND, not determined.
According to this new PCR protocol to detect H. pylori in human feces (Fig. 1), 38 of the 47 H. pylori-positive patients were H. pylori positive (giving 9 false negatives), and all 57 H. pylori-negative patients were also H. pylori negative with the new protocol (no false positive). The sensitivity, specificity, and predictive values for a positive or a negative result were 80.9% (95% CI, 66.3 to 90.4), 100% (92.1 to 100), 100% (88.6 to 100), and 86.4% (75.2 to 93.2), respectively. Most of the cases (n = 32) were detected before agarose blocks were performed. However, six additional cases were detected with this method. The overall correlation between biopsy PCR and stool PCR was good (90 of 104). There were 11 cases positive only by biopsy PCR, and there were 3 cases positive only by stool PCR.
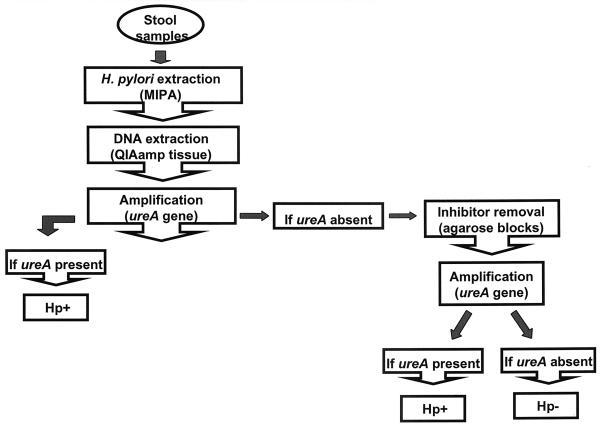
FIG. 1.
Stool sample processing method used in this study.
Detection of H. pylori from stool specimens as a diagnostic tool for detecting H. pylori infection is attractive because it is noninvasive. However, H. pylori has never been reliably detected from stools (3). The first reason is probably the lack of viability of these bacteria in an environment of competing flora under normal conditions in the bowel. In addition, selective culture media for the isolation of H. pylori are frequently contaminated with other, faster-growing gram-negative bacteria. The only positive attempts reported used feces obtained from patients experiencing spontaneous (15) or induced diarrhea (13).
Similarly, detection of H. pylori in stool specimens by using PCR after standard extraction methods has proved to be difficult (5, 12, 17; Mapstone et al., Letter), producing erratic results due to a variety of fecal inhibitors, such as acid polysaccharides, metabolic products, and large amounts of irrelevant DNA (8).
In this study, we developed a new PCR protocol to detect H. pylori in human feces. For this purpose, a MIPA was associated with an agarose-embedded DNA preparation to remove inhibitors (9). Successful amplification and specific detection of H. pylori DNA directly from stool samples in most of the infected patients indicate that MIPA, in association with the agarose-embedded DNA block method, is a promising procedure to detect H. pylori in stool specimens, but requires further development before it will be suitable for routine clinical use, because the sensitivity obtained is not sufficient. However, it is easier to apply than previously published methods (4, 5; Mapstone et al., Letter), avoiding the use of nested PCR. While nested PCR is supposed to increase sensitivity, it also has some limits, especially in routine use. The high risk of contamination makes it difficult to implement, and for this reason, such a procedure has not been recommended in the past (2).
Another stool test, based on the detection of H. pylori antigens to diagnose infection, is also currently available (16). However, despite the fact that this H. pylori antigen stool assay is reliable and easy to perform, it only indicates the presence or absence of H. pylori based on the detection of antigens the exact character of which is unknown. In contrast, PCR can provide additional information, principally related to the presence of pathogenicity factors (cagA or vacA alleles) or susceptibility to macrolides, antibiotics commonly used to treat this infection (6, 14). This subject will be of great interest for future investigations.
REFERENCES
- 1.Clayton C L, Kleanthous H, Coates P J, Morgan D D, Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Helicobacter pylori Study Group. Guidelines for clinical trials in Helicobacter pylori infection. Gut. 1997;41(Suppl. 2):8–13. [PubMed] [Google Scholar]
- 3.Kelly S M, Pitcher M C L, Farmery S M, Gibson G R. Isolation of Helicobacter pylori from feces of patients with dyspepsia in the United Kingdom. Gastroenterology. 1994;107:1671–1674. doi: 10.1016/0016-5085(94)90806-0. [DOI] [PubMed] [Google Scholar]
- 4.Lantz P-G, Matsson M, Wadström T, Rådstrom P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods. 1997;28:159–167. [Google Scholar]
- 5.Makristathis A, Pasching E, Schütze K, Wimmer M, Rotter M L, Hirschl A M. Detection of Helicobacter pylori in stool specimens by PCR and antigen enzyme immunoassay. J Clin Microbiol. 1998;36:2772–2774. doi: 10.1128/jcm.36.9.2772-2774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Mégraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteiro L, Birac C, Mégraud F. Detection of Helicobacter pylori in gastric biopsy by polymerase chain reaction. In: Lee A, Mégraud F, editors. Helicobacter pylori: techniques for clinical diagnosis and basic research. W. B. London, England: Saunders Co., Ltd.; 1995. pp. 112–120. [Google Scholar]
- 8.Monteiro L, Bonnemaison D, Vekris A, Petry K G, Bonnet J, Vidal R, Cabrita J, Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteiro L, Gras N, Vidal R, Cabrita J, Mégraud F. Detection of Helicobacter pylori DNA in human feces by PCR: DNA stability and removal of inhibitors. J Microbiol Methods. 2001;45:87–94. doi: 10.1016/s0167-7012(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro L, de Mascarel A, Sarasqueta A M, Bergey B, Barberis C, Talby P, Roux D, Shouler L, Goldfain D, Lamouliatte H, Mégraud F. Diagnosis of Helicobacter pylori infection: non-invasive methods compared to invasive methods and evaluation of two new tests. Am J Gastroenterol. 2001;96:353–358. doi: 10.1111/j.1572-0241.2001.03518.x. [DOI] [PubMed] [Google Scholar]
- 11.Moreira D. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res. 1998;26:3309–3310. doi: 10.1093/nar/26.13.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notarnicola M, Russo F, Cavallini A, Bianco M, Jirillo E, Pece S, Leoci C, di Matteo G, di Leo A. PCR identification of Helicobacter pylori DNA in faeces from patients with gastroduodenal pathology. Med Sci Res. 1996;24:785–787. [Google Scholar]
- 13.Parsonnet J, Shmuely H, Haggerty T. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA. 1999;23:2240–2245. doi: 10.1001/jama.282.23.2240. [DOI] [PubMed] [Google Scholar]
- 14.Pina M, Occhialini A, Monteiro L, Doermann H-P, Mégraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas J E, Gibson G R, Darboe M K, Dale A, Weaver L T. Isolation of Helicobacter pylori from human feces. Lancet. 1992;340:1194–1195. doi: 10.1016/0140-6736(92)92894-l. [DOI] [PubMed] [Google Scholar]
- 16.Vaira D, Malfertheiner P, Mégraud F, Axon A T. Diagnosis of Helicobacter pylori infection by HpSA test. Lancet. 1999;354:1732. doi: 10.1016/s0140-6736(05)76722-2. [DOI] [PubMed] [Google Scholar]
- 17.van Zwet A A, Thijs J C, Kooistra-Smid A M D, Schirm J, Snijder J A M. Use of PCR with feces for detection of Helicobacter pylori infections in patients. J Clin Microbiol. 1994;32:1346–1348. doi: 10.1128/jcm.32.5.1346-1348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]