Abstract
Cathepsin K (CatK) is a target for the treatment of osteoporosis, arthritis, and bone metastasis. Peptidomimetics with a cyanohydrazide warhead represent a new class of highly potent CatK inhibitors; however, their binding mechanism is unknown. We investigated two model cyanohydrazide inhibitors with differently positioned warheads: an azadipeptide nitrile Gü1303 and a 3-cyano-3-aza-β-amino acid Gü2602. Crystal structures of their covalent complexes were determined with mature CatK as well as a zymogen-like activation intermediate of CatK. Binding mode analysis, together with quantum chemical calculations, revealed that the extraordinary picomolar potency of Gü2602 is entropically favoured by its conformational flexibility at the nonprimed-primed subsites boundary. Furthermore, we demonstrated by live cell imaging that cyanohydrazides effectively target mature CatK in osteosarcoma cells. Cyanohydrazides also suppressed the maturation of CatK by inhibiting the autoactivation of the CatK zymogen. Our results provide structural insights for the rational design of cyanohydrazide inhibitors of CatK as potential drugs.
Keywords: Cathepsin K, protease inhibitor, cyanohydrazide warhead, azadipeptide nitrile, structure
1. Introduction
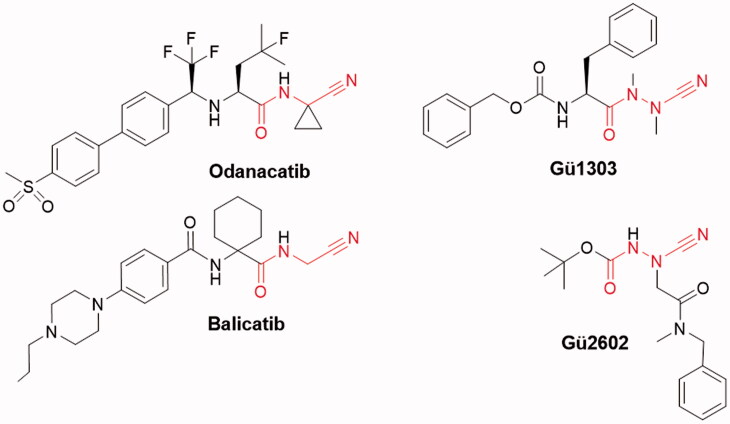
Cathepsin K (CatK) is one of the most investigated cysteine cathepsins, both by academia and pharmaceutical companies. It is expressed in high levels in osteoclasts, where it serves as the principal protease involved in bone remodelling. It has been validated as a therapeutic target for osteoporosis, an increasing health problem in the modern world1–4. This disorder is caused by progressive loss of bone mass due to excessive activity of osteoclastic CatK. Several anti-remodelling inhibitors of CatK, such as odanacatib or balicatib5–7 (Figure 1), have been developed for the treatment of osteoporosis but have not yet been approved. CatK has also been implicated in the pathophysiology of two common forms of arthritis, osteoarthritis and rheumatoid arthritis8,9. In bone and cartilage disorders, CatK functions as a potent collagen degrading enzyme with the unique ability to cleave the triple helix of collagen molecules at multiple locations, an activity that is unparalleled among human collagenases10,11. This activity is induced by glycosaminoglycans, foremost chondroitin-4-sulphate, which mediates the formation of a complex between CatK and the collagen substrate10,12,13.
Figure 1.
Chemical structures of peptidomimetic inhibitors of cathepsin K with a reactive nitrile functionality. The warhead is indicated in red. The dipeptide nitriles odanacatib and balicatib were developed as osteoporosis drugs. The azadipeptide nitrile Gü1303 and 3-cyano-3-aza-β-amino acid Gü2602 contain the cyanohydrazide warhead.
Further, there is increasing evidence that CatK is a pro-tumorigenic protease that plays an important role in processes associated with tumour growth, invasion, and metastasis of cancer cells and their interactions with the tumour microenvironment (for review see14,15). Its complex action includes direct degradation of collagen and other extracellular matrix proteins, e.g. in bone metastases, and indirect affecting of the signalling pathways16–19. In glioma, CatK can regulate cancer stem-like cell mobilisation and proteolytically modulate levels of chemokines and growth factors20. Thus, clinical research is investigating CatK as a marker for diagnosis and survival prognosis in metastatic cancer and as a target for anticancer inhibitors21,22.
At the protein level, CatK activity is regulated by endogenous protein inhibitors23,24 and by zymogen activation24–26. CatK is synthesised as an inactive zymogen (procathepsin) in which the N-terminal propeptide blocks the active site27,28. Activation to the mature, active form occurs upon proteolytic removal of the N-terminal propeptide (also termed the “activation peptide”). This process was shown to be autocatalytic and bimolecular; it is triggered by acidic pH and also enhanced by interaction with chondroitin-4-sulfate25,29. The activation pathway of CatK includes an activation intermediate with a partially processed propeptide25, which has not been studied in detail so far, and its spatial structure remains unknown.
Peptidomimetics with a reactive nitrile functionality have attracted particular attention as potent inhibitors of CatK and other cysteine cathepsins. The electrophilic nitrile warhead allows for covalent interaction with the catalytic cysteine nucleophile, leading to the reversible formation of a covalent thioimidate adduct. Representatives of such CatK inhibitors are dipeptide nitriles odanacatib and balicatib5–7 (Figure 1). An exchange of the α-CH moiety of the P1 amino nitrile by a nitrogen atom led to azadipeptide nitriles with the cyanohydrazide warhead forming a stabilised isothiosemicarbazide adduct30–32. They have been introduced as a class of efficient covalent-reversible inhibitors of human cysteine cathepsins, including CatK, and their homologs from parasites and pathogens30–34. Compared to their parent carbapeptide analogs, bioactive azapeptides can possess improved potency, selectivity, and pharmacokinetics35–38. The 3-cyano-3-aza-β-amino acid derivatives represent another scaffold bearing the cyanohydrazide warhead39. They were designed to position the warhead centrally in the peptidomimetic inhibitor molecule for extended interactions of inhibitor substructures with the non-primed and primed binding regions of the target enzymes. These compounds were found to be exceptionally potent, in particular towards CatK39.
The exact binding mode of peptidomimetics with the cyanohydrazide warhead to CatK had not been characterised so far. To do this, we investigated two model cyanohydrazide inhibitors selective for CatK with high potency in the subnanomolar to picomolar range, namely the azadipeptide nitrile Gü130330,31 and the 3-cyano-3-aza-β-amino acid derivative Gü260239 (Figure 1). The crystal structures of their complexes with mature CatK and the activation intermediate of CatK were determined and functional properties in vitro and in cells were described. The present structures and inhibitor interaction data provide a footing for the rational design of next generation cyanohydrazide inhibitors of CatK as potential therapeutics.
2. Materials and methods
2.1. Materials
Inhibitors Gü1303 and Gü2602 and the CatK activity-based probe 25 were synthesised as described previously30,39,40.
2.2. Expression and purification of the recombinant zymogen of cathepsin K
Human CatK (Uniprot accession number P43235) was expressed in the X-33 strain of the methylotrophic yeast Pichia pastoris (Thermo Fisher). A gene coding for the zymogen form of CatK was purchased from GenScript and recloned into expression plasmid pPICZαA (Thermo Fisher) using XhoI and NotI restriction sites. Transformation of P. pastoris cells and protein expression were carried out as described previously41,42. The yeast medium containing the recombinant CatK zymogen was centrifuged (2,500 g for 10 min), and the supernatant was lyophilised and dissolved in 20 mM MES pH 6.0 (to 10% of the original volume). The protein solution was then desalted over a Sephadex G-25 column equilibrated with the same buffer. The CatK zymogen was purified using chromatography on Mono S (HR 5/5 column) equilibrated with 50 mM sodium acetate pH 5.5, and eluted by a linear gradient of 2 M NaCl. The purified protein was concentrated to 2 mg/ml using an Amicon Ultracel-10k centrifugal filter device (Millipore).
2.3. Activation of the cathepsin K zymogen and preparation of inhibitor complexes
The purified CatK zymogen (75 µM) was activated by incubation in 0.1 M sodium acetate pH 4.0 containing 2.5 mM DTT, 1 mM EDTA, and 0.3 M NaCl under an argon atmosphere at room temperature. The zymogen-like activation intermediate iCatK was obtained after 30 min of incubation, and fully activated mature enzyme mCatK after 75 min. Activation was terminated by the addition of 6-fold molar excess of the inhibitor Gü1303 or Gü2602, followed by incubation under argon atmosphere for 3 h at room temperature. The activation and inhibition were monitored with a kinetic activity assay using the fluorogenic substrate Cbz-Gly-Pro-Arg-AMC (Cbz, benzyloxycarbonyl; AMC, 7-amino-4-methylcoumarin) and Laemmli-SDS-PAGE. The processing sites were identified by N-terminal protein sequencing after electroblotting of Laemmli-SDS-PAGE gels to a PVDF membrane using a Procise 494 cLC protein sequencer (Applied Biosystems) and by peptide mapping using mass spectrometry (LC-MS/MS) on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) coupled to a UHPLC system. The LC–MS/MS data were processed with Bioworks software (Thermo). The complexes were buffer-exchanged into 20 mM sodium acetate pH 5.5 containing 2.5 mM DTT and 0.25 M NaCl, and concentrated to 3.5 mg/ml for mCatK and 5 mg/ml for iCatK using an Amicon Ultracel-10k centrifugal filter device; the inhibitors were maintained during buffer exchange and concentration in a 6-fold molar excess to mCatK/iCatK in the mixture.
2.4. Autoactivation assay with the cathepsin K zymogen and inhibitors
The CatK zymogen (2.8 µM) was incubated at room temperature in the presence or absence of the inhibitor (10 µM Gü1303 or Gü2602) in 150 µL of 0.1 M sodium acetate pH 4.0 containing 2.5 mM DTT and 0.3 M NaCl for 0, 30 and 240 min. The reaction was terminated by the addition of E-64 (10 µM final concentration), followed by acetone precipitation; the reaction mixture was separated by Laemmli-SDS-PAGE.
2.5. Protein crystallisation and data collection
Crystals were obtained by the vapour diffusion technique in hanging drops at 18 °C. Drops consisted of 1 µl of the protein-inhibitor complex, 0.15 µl 1:100 diluted seed stock and 1 µl of the reservoir solution. The drops were equilibrated over the following reservoir solutions: (1) 10% PEG 8000, 20% ethylene glycol, 0.02 M sodium L-glutamate, 0.02 M DL-alanine, 0.02 M glycine, 0.02 M DL-lysine HCl, 0.02 M DL-serine, 0.1 M MES/imidazole pH 6.5 for iCatK-Gü1303 complex; (2) 10% PEG 8000, 20% ethylene glycol, 0.02 M sodium formate, 0.02 M ammonium acetate, 0.02 M trisodium citrate, 0.02 M sodium potassium L-tartrate, 0.02 M sodium oxamate, 0.1 M MES/imidazole pH 6.5 for iCatK-Gü2602 complex; (3) 12.5% PEG 1000, 12.5% PEG 3350, 12.5% MPD, 0.03 M sodium nitrate, 0.03 M disodium hydrogen phosphate, 0.03 M ammonium sulphate, 0.1 M MES/imidazole pH 6.5 for mCatK-Gü1303 complex; and (4) 10% PEG 8000, 20% ethylene glycol, 0.02 M sodium formate, 0.02 M ammonium acetate, 0.02 M trisodium citrate, 0.02 M sodium potassium L-tartrate, 0.02 M sodium oxamate, 0.1 M MES/imidazole pH 6.5 for mCatK-Gü2602 complex. Crystals were flash-cooled by plunging them into liquid nitrogen, and diffraction data from the crystals of mCatK complexes were collected at 100 K on a MicroMax-007 HF Microfocus rotating anode X-ray generator equipped with a PILATUS 300 K detector (Rigaku). Data from the crystals of iCatK complexes were collected at 100 K on an MX 14.1 beamline operated by Helmholtz-Zentrum Berlin at the BESSY II electron storage ring in Berlin-Adlershof, Germany43. All diffraction data was processed using the XDS suite of programs44. Crystal parameters, data collection statistics, and final refinement statistics are in Table S1.
2.6. Structure determination, refinement and analysis
The structures of the mCatK-Gü1303/Gü2602 complexes and the main domain of iCatK in complex with Gü2602 were solved by molecular replacement with the program MolRep45 from the CCP4 program suite46 using the structure of human cathepsin K (PDB code: 7NXM)40 as a search model. The propeptide domain structure in iCatK-Gü2602 was built using the de novo model building program Buccaneer47. The structure of the iCatK-Gü1303 complex was solved by molecular replacement using the structure of iCatK-Gü2602 as a search model. Model refinement was carried out using the program REFMAC 5.5, interspersed with manual adjustments using Coot. The geometric restraints for ligands were constructed by the program AceDRG48. The quality of the final models was validated with MolProbity49. The final refinement statistics are given in Table S1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes: 7QBL, 7QBN, 7QBM, and 7QBO for mCatK-Gü2602, mCatK-Gü1303, iCatK-Gü2602, and iCatK-Gü1303, respectively. Inhibitor interactions were analysed using the programs CONTACT46 and PLIP50. The distance cut-offs were set to 3.3 Å for hydrogen bonds and 4.2 Å for contacts. The nonpolar interactions represent contacts between two hydrophobic atoms defined as carbon atoms having carbon or hydrogen atoms as neighbours. All figures showing structural representations were prepared with the PyMOL Molecular Graphics System, version 1.40 (Schrödinger, LLC).
2.7. Molecular modelling
The X-ray structure of the mCatK-Gü2602 complex was used for molecular modelling. Hydrogen atoms were added to the protein by the Reduce and Leap programs in AMBER 1451. The Asp, Glu, Lys, Arg, and His residues were charged, with the exception of Asp82 and His162. Hydrogen atoms of the inhibitor were added manually using the PyMOL. The ff14SB force field51 was used for the protein, while the GAFF force field51 and RESP charges at HF/6-31G* level were used for the ligand. The molecular dynamics/quenching (MD/Q) technique was used to search for possible conformations of Gü2602 in the complex. The simulations were performed using AMBER 14. During the simulations, the N-benzyl-N-methylformamide segment of the inhibitor was relaxed; specifically, the C, O, and N heavy atoms in this segment and all H atoms of the inhibitor were relaxed while the rest of the system was frozen. The structures were collected every 1 ps in two independent runs with two orientations of the relaxed inhibitor segment (i.e. the heavy atoms as above). The simulations were 10 ps long at 600 K using 1 fs time step and a Berendsen thermostat. All the obtained structures were optimised (i.e. residues within 6 Å of the inhibitor were relaxed) by using the corrected semiempirical quantum mechanical (SQM) PM6-D3H4 method52,53. The environment was described by the COSMO implicit solvent model54,55. The SQM calculations were done by Cuby456 and MOPAC201657. Residues further than 6 Å from the inhibitor were frozen during the optimisation.
2.8. Cathepsin K activity and inhibition assays
Enzymatic activity of mCatK was measured using a kinetic continuous assay with the fluorogenic substrate Cbz-Gly-Pro-Arg-AMC (Bachem). The assay was performed in a 96-well microplate format in a total assay volume of 100 µl at 37 °C. The assay mixture contained an aliquot of mCatK (e.g. from the autoactivation assay) and 20 µM Cbz-Gly-Pro-Arg-AMC in 0.1 M sodium acetate pH 5.5 containing 2.5 mM DTT, 0.15 M NaCl, 0.1% PEG 6000, and 1 mM EDTA. The kinetics of the product release were continuously monitored in an Infinite M1000 microplate reader (Tecan) at excitation and emission wavelengths of 360 and 465 nm, respectively. The Michaelis-Menten kinetic parameters were determined by measuring the rate of hydrolysis of the substrate (0–100 μM) using the same assay with mCatK (0.4 nM); the Km value obtained by nonlinear regression using GraFit software was 17.4 µM.
Inhibition measurements were performed analogously. mCatK (0.42 nM) was added to a mixture of the fluorogenic substrate Cbz-Gly-Pro-Arg-AMC (20 µM) and an inhibitor (0–100 nM) in 0.1 M sodium acetate pH 5.5 containing 2.5 mM DTT, 0.15 M NaCl, 0.1% PEG 6000, and 1 mM EDTA. The substrate hydrolysis was monitored for 40 min. For the slow-binding inhibitor Gü1303, an observed first-order rate constant kobs was calculated at each inhibitor concentration by fitting the progress curve to the equation P = vst + (vi - vs)(1 - exp(kobst))/kobs + d, where P is the product formation, vs is the steady-state velocity, t is the reaction time, vi is the initial velocity, and d is offset. The apparent inhibition constant Ki′ was determined by non-linear regression using equation vs/v0 = 1/(1 + [I]/Ki′). The true inhibition constants Ki were calculated using the Cheng Prusoff equation Ki = Ki′/(1 + [S]/Km), where [S] is the substrate concentration and Km is the Michaelis constant. The apparent second-order rate constant kon′ was determined by fitting to the linear equation kobs = k′on [I] + koff, and the true constant kon was calculated by correction kon = kon′(1 + [S]/Km). The fast-binding inhibitor Gü2602 showed linear progress curves, and the apparent inhibition constant Ki′ was determined by non-linear regression using the Morrison equation for tight binding inhibition58 with GraphPad Prism software. The concentration of mCatK was determined by active site titration as described previously59 with E-64 used as the titrant60. The final concentration of DMSO in the assay systems did not exceed 1.5%.
2.9. Osteosarcoma cell imaging
Imaging of cathepsin K in human bone osteosarcoma cells (U-2 OS) was done as described previously using the CatK activity-based probe 2540. U-2 OS cells were cultivated in McCoy’s 5 A medium supplemented with 10% FBS, 2 mM glutamax (L-alanyl-L-glutamine), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. Fifty thousand cells were seeded into a well of a 12-well plate and allowed to attach. After 24 h, 1 μM probe was added, and cells were incubated for 16 h. Cells were detached using 0.25% trypsin-EDTA solution ( Merck), collected by centrifugation (500 g for 5 min), washed with PBS, resuspended in 100 μl of loading buffer, and heated at 100 °C for 10 min. The competitive labelling was performed after the preincubation of cells with 1 μM Gü1303 or Gü2602 for 3 h. The samples (30 μl) were separated on 4–12% Bis-Tris polyacrylamide gels (Thermo Fisher Scientific). The gels were visualised using a Typhoon RGB imager (GE Healthcare Life Sciences) with excitation at 635 nm and emission at 660 nm (long pass filter).
3. Results
3.1. Cyanohydrazides are potent inhibitors of cathepsin K activity and zymogen activation both in vitro and in cells
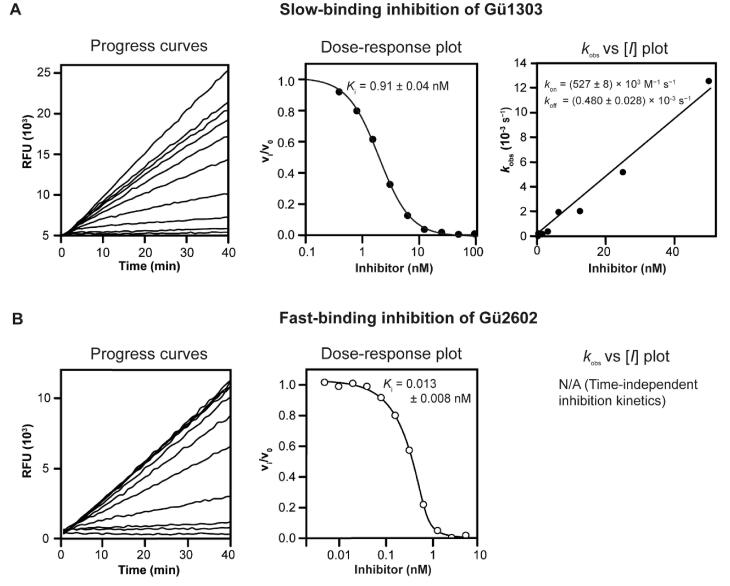
Functional properties of the azadipeptide nitrile inhibitor Gü130330 and the 3-cyano-3-aza-β-amino acid inhibitor Gü260239 were characterised using an in vitro kinetic assay with the mature form of recombinant human cathepsin K (mCatK) and the fluorogenic substrate Cbz-Gly-Pro-Arg-AMC. Subnanomolar values of the inhibition constant Ki were determined: Notably, we found a Ki of 0.91 nM for Gü1303 and an almost two orders of magnitude lower Ki of 0.013 nM for Gü2602 (Table 1). A detailed analysis of the kinetic behaviour showed non-linear progress curves that indicated time-dependent inhibition for Gü1303 typical for slow-binding inhibitors (Figure 2(A)). This allowed for the calculation of the second-order rate constant of inactivation kon of 527 × 103 M−1s−1, thus demonstrating the extraordinary potency of Gü1303 (Table 1). In contrast, linear progress curves were obtained for Gü2602 that are characteristic of fast-binding inhibitors (Figure 2(B)).
Table 1.
Inhibition of human mature cathepsin K.
| Compound | mCatK inhibitiona |
||
|---|---|---|---|
|
Ki (nM) |
kon (103 M−1 s−1) |
koff (10−3 s−1) |
|
| Gü2602 | 0.013 ± 0.008 | n.d.b | n.d.b |
| Gü1303 | 0.91 ± 0.04 | 527 ± 8 | 0.48 ± 0.03 |
aThe inhibition parameters were measured using a kinetic activity assay with the fluorogenic peptide substrate Cbz-Gly-Pro-Arg-AMC at pH 5.5. bn.d.: not determined for linear progress curves.
Figure 2.
Different inhibition kinetics of mature cathepsin K with Gü1303 and Gü2602. Progress curves show the hydrolysis of the fluorogenic substrate Cbz-Gly-Pro-Arg-AMC by mCatK at pH 5.5 in the presence of increasing inhibitor concentrations. (A) Gü1303 exhibited a time-dependent inhibition characterised by non-linear progress curves typical of slow-binding kinetics. (B) Linear progress curves obtained for Gü2602 are characteristic of fast-binding inhibitors. In dose–response plots, the derived steady-state reaction velocities were plotted against inhibitor concentration, and the inhibition constants Ki were obtained after correction by the Cheng-Prusoff and Morrison equations (see Materials and Methods). In the kobs versus [I] plot, the first-order rate constants kobs from the time-dependent progress curves were plotted against inhibitor concentrations to show a linear dependence.
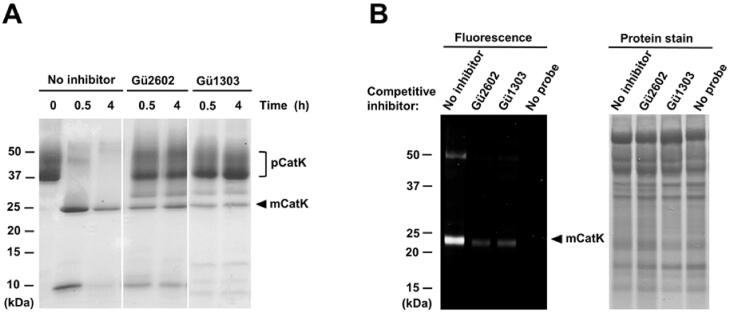
The mature enzyme mCatK is generated during the acidic autocatalytic processing of an inactive precursor, the cathepsin K zymogen (pCatK), and this activation process is associated with the proteolytic removal of the propeptide domain from pCatK24–26. We examined the effect of Gü1303 and Gü2602 on the autoactivation of recombinant pCatK induced by acidic pH using an in vitro assay with SDS-PAGE visualisation of the processing forms pCatK and mCatK (bands of ca 37 kDa and ca 24 kDa, respectively) (Figure 3(A)). Inhibitors were tested under conditions providing full conversion to mCatK and the cleaved propeptide (a band of ca 10 kDa). Both inhibitors substantially suppressed the autoactivation processing of pCatK that resulted in approximately a 10–15% conversion compared to uninhibited conditions. Also, both inhibitors were able to suppress the autodegradation of the generated mCatK observed under prolonged incubation (compare 0.5 and 4 h experiments, Figure 3(A)).
Figure 3.
The inhibitors Gü1303 and Gü2602 suppress the autocatalytic activation of the cathepsin K zymogen and target cathepsin K in cells. (A) The zymogen of cathepsin K (pCatK) was incubated in the presence and absence of inhibitor (10 µM) at pH 4.0, and the generation of mature cathepsin K (mCatK) was analysed at the indicated times. The reaction mixture was resolved by SDS-PAGE and visualised by protein staining. The positions of pCatK and mCatK are indicated; note mass heterogeneity of pCatK due to glycosylation61. (B) The U-2 OS cells were pre-treated with inhibitor (1 µM) for 3 h, followed by 24 h incubation with a fluorescent activity-based probe specific for cathepsin K40 (1 μM); quenching of the labelling reaction by competitive inhibition was analysed. Cell lysates were resolved by SDS-PAGE and visualised by fluorescence imaging (left) and protein staining (right). The position of mCatK is indicated. In control experiments, the probe or inhibitor was omitted.
Finally, we investigated the interaction of the inhibitors with CatK using a cell-based assay with the human osteosarcoma cell line U-2 OS, which has an enhanced expression level of CatK62. For CatK imaging, we used a fluorescent activity-based probe that binds specifically and irreversibly to the active site of CatK40. Competition of the probe and inhibitor was monitored by SDS-PAGE and in-gel fluorescence. As shown in Figure 3(B), both reversible inhibitors Gü1303 and Gü2602 strongly diminished the mCatK labelling, demonstrating that these cyanohydrazides are cell-permeable compounds that effectively interact with the active form of CatK in the lysosomal/endosomal system.
In conclusion, the cyanohydrazide inhibitors were demonstrated to inhibit mCatK with subnanomolar potency and different binding kinetics, to suppress the generation of mCatK from its zymogen, and to effectively target active mCatK in the cell context.
3.2. Crystallography of complexes of two cyanohydrazide inhibitors with mature cathepsin K and its zymogen-like activation intermediate
Human mature cathepsin K (mCatK) and a zymogen-like activation intermediate of cathepsin K (iCatK) were crystallised in complexes with two cyanohydrazide inhibitors, the azadipeptide nitrile Gü1303 and the 3-cyano-3-aza-β-amino acid Gü2602 (Figures 4 and 5). The crystal structures of mCatK complexes with Gü1303 and Gü2602 were determined by molecular replacement using the structure of uncomplexed mCatK as a template. Both complexes crystallised in the orthorhombic space group P21212 with one molecule in the asymmetric unit (Table S1). The structure of the mCatK-Gü1303 complex was refined using data to resolution 1.55 Å, and to 2.00 Å for mCatK-Gü2602. The final crystallographic models contained mCatK residues Ala1 to Met215; additional N-terminal residues derived from the propeptide, Gly-Arg (98p–99p, propeptide numbering), were visible in the mCatK-Gü1303 structure (Figure 5). A comparison of both mCatK complexes did not reveal any significant differences in protein structure (a backbone r.m.s.d. of 0.51 Å, a value within the range observed for different crystal structures of identical proteins).
Figure 4.
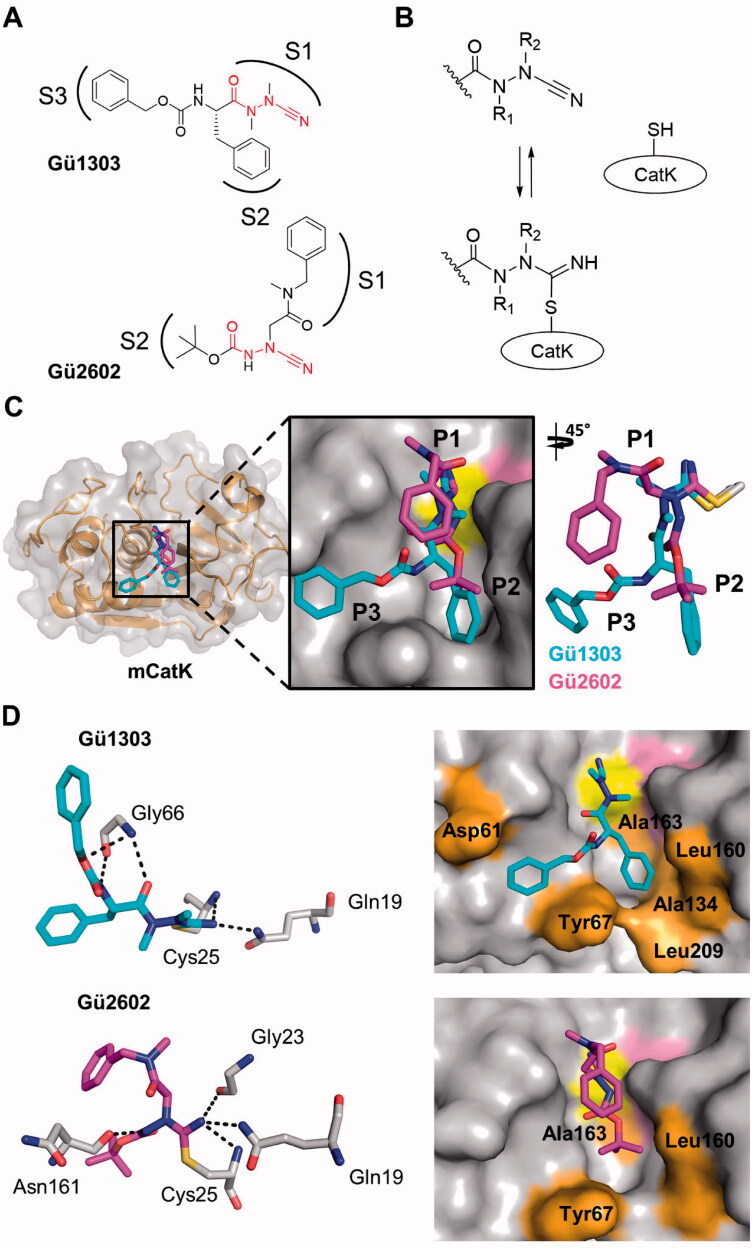
Binding mode of the cyanohydrazide inhibitors Gü1303 and Gü2602 in the active site of mature cathepsin K. (A) Chemical structure of the azadipeptide nitrile inhibitor Gü1303 and 3-cyano-3-aza-β-amino acid inhibitor Gü2602; the binding subsites (S) are marked, and the cyanohydrazide warheads are in red. (B) Reactive warheads form a covalent reversible bond with the thiol of the catalytic cysteine residue of the enzyme; R1 and R2 are substituents on the N atoms of the warheads. (C) The zoomed-in view of the mCatK active site shows a superposition of the inhibitors bound to the S1 to S3 subsites (corresponding inhibitor positions P1 to P3 are indicated). mCatK is displayed in surface representation (grey); highlighted are the catalytic residues Cys25 (yellow) and His162 (pink). Inhibitors are shown in stick representation with carbon atoms in cyan for Gü1303 and magenta for Gü2602; heteroatoms have standard colour coding (O, red; N, blue; S, yellow). (D) Interaction of the inhibitors with active site residues of mCatK. Left panels: the hydrogen bond network formed between inhibitors and mCatK residues (dashed black lines). Inhibitors are coloured as in (C), and interacting enzyme residues are in grey; the side chain of the covalently linked catalytic cysteine residue Cys25 is depicted. Right panels: the surface representation of the mCatK active site shows enzyme residues (highlighted in orange) forming nonpolar interactions with the inhibitors (in stick representation); both inhibitors are in the same orientation.
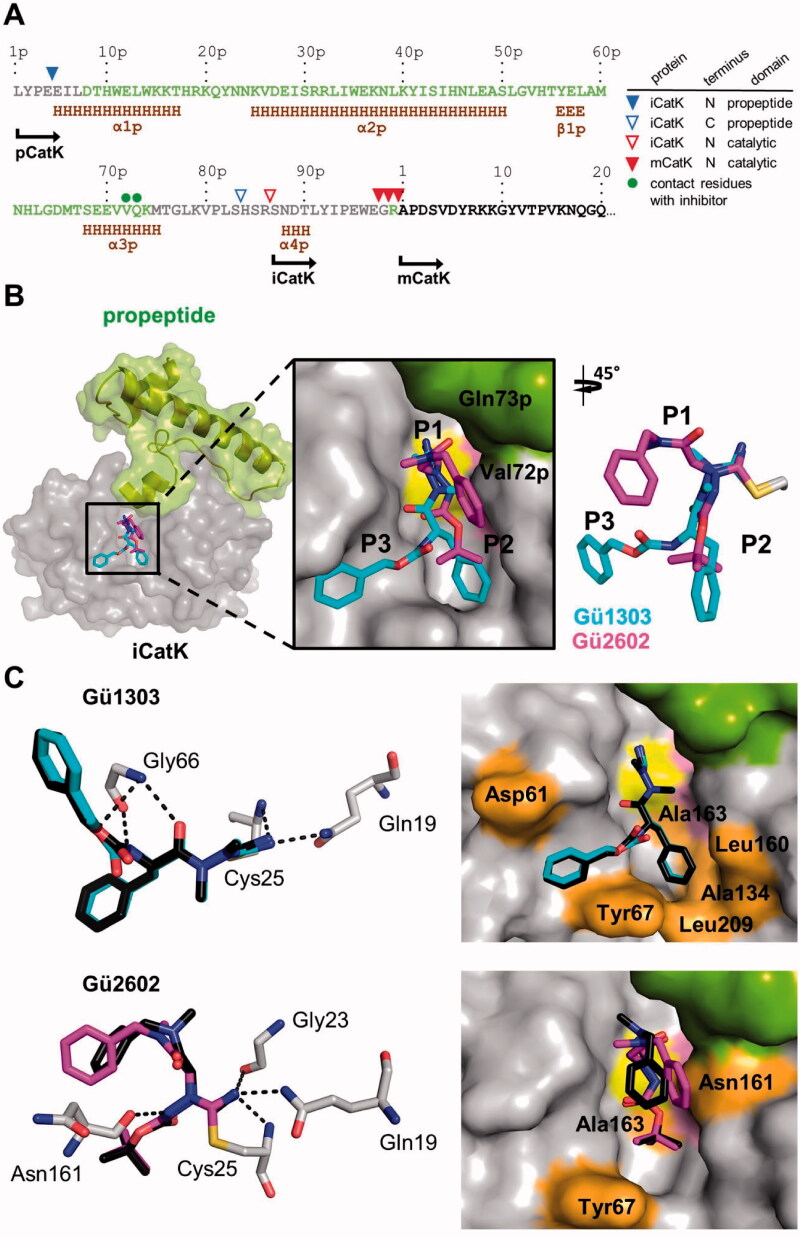
Figure 5.
Binding mode of the cyanohydrazide inhibitors Gü1303 and Gü2602 in the active site of the activation intermediate of cathepsin K (iCatK). (A) The amino acid sequence of the full-length propeptide domain and the N-terminus of mature CatK (mCatK) are shown with secondary structure elements (H, α-helix; E, β-strand) (data absent in the iCatK structure are derived from the intact zymogen pCatK; PDB entry: 1BY8). Propeptide residues present or absent in the final crystallographic models of iCatK are in green or grey, respectively, residues of the catalytic domain are in black. The triangles above the sequence line indicate the N- and C-termini of the residual propeptid domain of iCatK, the N-termini of iCatK and mCatK catalytic domains as determined by the Edman sequencing and mass spectrometry (see the inset legend). The green dots show two residues of the propeptid of iCatK that form contacts with Gü2602. The position of the N-termini is indicated for pCatK and catalytic domains of iCatK/mCatK. (B) The zoomed-in view of the iCatK active site shows a superposition of Gü1303 and Gü2602 bound to the S1 to S3 subsites (corresponding inhibitor positions P1 to P3 are indicated). iCatK is displayed in surface representation, the catalytic domain is highlighted in grey, the residual propeptide domain in green, and the catalytic residues Cys25 and His162 in yellow and pink, respectively. Inhibitors are shown in stick representation with carbon atoms in cyan for Gü1303 and magenta for Gü2602; heteroatoms have a standard colour coding (O, red; N, blue; S, yellow). (C) Interaction of the inhibitors with the iCatK active site residues. Left panels: the hydrogen bond network formed between inhibitors and iCatK residues with the (dashed black lines). Inhibitors are coloured as in (B), and interacting enzyme residues are in grey; the side chain of the covalently linked catalytic cysteine residue Cys25 is depicted. Superimposed (black) are the same inhibitors from the structures of their complexes with mCatK (Figure 4). Right panels: the surface representation of the iCatK active site shows enzyme residues forming nonpolar interactions (highlighted in orange) with the inhibitors (in stick representation); both inhibitors are in the same orientation. The propeptide domain residues are highlighted in green.
iCatK was produced by limited autocatalytic processing of the zymogen pCatK at acidic pH. This resulted in fragmentation of the propeptide with cleavage sites identified after Glu4p, Ser83p, and Arg86p residues by Edman sequencing and mass spectrometry peptide mapping analysis (Figure 5(A)). The obtained iCatK comprised two non-covalently bound domains (chains), namely the residual propeptide domain (Glu5p to Ser83p) and the main domain (Ser87p to Met215), including the C-terminal part of the propeptide and mCatK. The structure of the iCatK-Gü2602 complex was solved by a combination of molecular replacement for the main domain (see previous paragraph) and automated model building by fragment-fitting technique for the residual propeptide. The structure of the iCatK-Gü1303 complex was solved by molecular replacement based on the iCatK-Gü2602 structure. Both iCatK complexes crystallised in the tetragonal space group P43212 containing one molecule in the asymmetric unit (Table S1). The final crystallographic model of the iCatK-Gü1303 complex contained propeptide residues Asp8p to Gln73p and main domain residues Pro2 to Met215. The model of the iCatK-Gü2602 complex contained residues Asp8p to Lys74p and Arg99p to Met215. The structures were refined using data to resolution 1.90 Å for iCatK-Gü1303 and 1.88 Å for iCatK-Gü2602. A comparison of iCatK complexes did not reveal any significant differences in protein structure (a backbone r.m.s.d. of 0.28 Å).
The residual propeptide domain of iCatK is folded in a similar manner to the intact propeptide in the structure of the zymogen pCatK (PDB: 1BY8) and bound at the same position (a backbone r.m.s.d. of 0.81 and 0.48 Å for the propeptide domains and catalytic domains, respectively). However, we observed a slightly different orientation of the α2p helix in iCatK complexes, which is rotated by approximately 10° compared to pCatK. The propeptide segment that blocks the active site in pCatK (downstream of the α3p helix, Figure 5) is proteolytically removed or flexible in iCatK, and therefore the active site cleft of iCatK becomes accessible for inhibitors. However, the residual propeptide domain partially occludes the primed region of the active site, in particular the S1’ subsite of iCatK is occupied.
3.3. Interaction of cyanohydrazide inhibitors with the active site of cathepsin K
3.3.1. Binding mode of the azadipeptide nitrile Gü1303 to mature and zymogen-like cathepsin K
The active site cleft of mCatK contains the catalytic triad residues Cys25, His162, and Asn182. Gü1303 is bound in a substrate-like orientation, and its P1 to P3 residues occupy the S1 to S3 subsites of mCatK (Figure 4(A,C)). The cyanohydrazide warhead reacts with the thiol group of the catalytic Cys25, forming a covalent isothiosemicarbazide adduct through the connection to the C-atom of the nitrile moiety (Figure 4(B)). Azadipeptides such as Gü1303 are atropochiral molecules due to the restricted rotation around the methylated N–N axis, and, in the unbound state, they preferentially adopt the E-configuration of the respective CO–NMe bond63,64 (Figure 4(B)). However, a Z-configuration at the CO–NMe bond was observed in Gü1303 bound to mCatK, suggesting an E- to Z-conformational change in Gü1303 upon binding to the enzyme, most likely due to a "configurational selection" that we recently reported for an azadipeptide nitrile inhibitor of the protease SmCB132.
Interactions between Gü1303 and the mCatK active site are presented in Figure 4(D) (for details see Table S2). The inhibitor forms a network of hydrogen bonds with the active site residues. The nitrogen atom of the imidate moiety, derived from the warhead nitrile group, is stabilised by two hydrogen bonds to the backbone amide of the catalytic Cys25 and the side chain amide of Gln19 (Figure 4(D)). An analogous interaction pattern was observed for the warhead of an azadipeptide nitrile inhibitor reacted with the protease SmCB1 (PDB: 6YI7)32. The NH of Gly66 acts as a bifurcated hydrogen bond donor for the carbonyl oxygen of the P2 phenylalanine and the noncarbonyl carbamate oxygen in the P3 position of Gü1303. An additional hydrogen bond is formed between the Gly66 oxygen and the carbamate NH of Gü1303. A similar network of Gly66-mediated hydrogen bonds was also reported for a Boc-protected precursor of a reactive activity-based probe for mCatK (PDB: 7NXL)40. Nonpolar interactions of Gü1303 with mCatK are depicted in Figure 4(D). They are absent in the S1 subsite, although the warhead containing the P1 azaalanine residue forms a number of contacts (Supporting information Table S2). At the P2 position, the phenylalanine residue makes nonpolar interactions with Tyr67, Ala134, Leu160, Ala163, and Leu209. The P3 benzyloxycarbonyl capping group forms nonpolar interactions with Asp61 and Tyr67, and the phenyl moiety of this group is stabilised by a T-shaped π-π stacking interaction to the 4-hydroxyphenyl group of Tyr67.
The binding mode of Gü1303 in the active site of iCatK is analogous to that in mCatK, and there is no interaction of the inhibitor with the residual propeptide domain that blocks a part of the primed region of the iCatK active site (Figure 5(B)). High conformational similarity of the inhibitor is indicated by a r.m.s.d. of 0.19 Å, with a certain change in the position of the P3 carbonyl oxygen. Also, Gü1303 forms the same network of hydrogen bonds and nonpolar interactions in the S1 to S3 subsites of iCatK and mCatK (Figure 5(C), Table S2). However, no π-π stacking interaction with Tyr67 residue was observed, due to a slightly different orientation of the benzyloxycarbonyl capping group of Gü1303.
3.3.2. Binding mode of the 3-cyano-3-aza-β-amino acid Gü2602 to mature and zymogen-like cathepsin K
Gü2602 binds to the active site of mCatK in a substrate-like orientation, and the major conformation of the inhibitor occupies the S1 and S2 subsites (Figure 4(A,C)). The cyanohydrazide warhead positioned centrally in the inhibitor molecule forms a covalent isothiosemicarbazide adduct with a thiol group of the catalytic Cys25 (Figure 4(B)). There is a shift in the inhibitor backbone when comparing Gü2602 and Gü1303 that might be the result of nitrogen methylation in the warhead (CO–NH vs. CO–NMe in Gü2602 and Gü1303, respectively) (Figure 4(C)). The CO–NH bond of Gü2602 adopts the Z-configuration, similar to what has been observed for the CO–NMe bond of the enzyme bound inhibitor Gü1303.
The nitrogen atom of the imidate moiety of Gü2602 is strongly stabilised by three hydrogen bonds to the backbone amide of the catalytic Cys25, carbonyl group of Gly23, and the side chain amide of Gln19 (Figure 4(D)). In contrast to Gü1303, there is no hydrogen bonding between Gü2602 and the Gly66 residue. However, a new hydrogen bond is formed between the backbone oxygen of Asn161 and the amide NH of Gü2602. Analysis of the inhibitor-mCatK complexes available in the PDB shows that peptidomimetic inhibitors of mCatK frequently establish hydrogen bonding with Asn161 as well as Gly66 as important interaction determinants. The P1 and P2 residues of Gü2602 are located in the non-primed subsites of the mCatK active site as follows (Figure 4(A,C)). The N-benzyl-N-methylacetamide substructure at P1 occupies the S1 subsite, making contacts with Gly23, Gly64, and Gly65 residues. Its terminal benzyl moiety is oriented out of the S1 subsite and towards the S2 pocket. The P2 Boc-capping group resides in the S2 subsite of mCatK and makes nonpolar interactions with Tyr67, Leu160, and Ala163 (Figure 4(D), Table S2).
In general, Gü2602 binds to the S1 and S2 subsites of iCatK in a manner that is similar to what has been shown for the Gü2602-mCatK complex, with a r.m.s.d. of 1.23 Å; this value is substantially increased compared to Gü1303 in its complexes, with a r.m.s.d. of 0.19 Å (Figure 5(B)). The major conformational change, however, is in the benzyl group of the P1 N-benzyl-N-methylacetamide substructure. It is rotated towards the residual propeptide domain of iCatK and forms new contacts with its Val72p and Gln73p residues (Figure 5(A,B), Table S2). The terminal Boc group accommodates the S2 pocket of iCatK analogously as in mCatK. The network of hydrogen bonds is identical, and the pattern of nonpolar interactions (Tyr67, Asn161, and Ala163 in iCatK) is similar for both Gü2602 complexes (Figure 5(C)).
3.4. Conformational flexibility of cyanohydrazide inhibitors in the cathepsin K active site
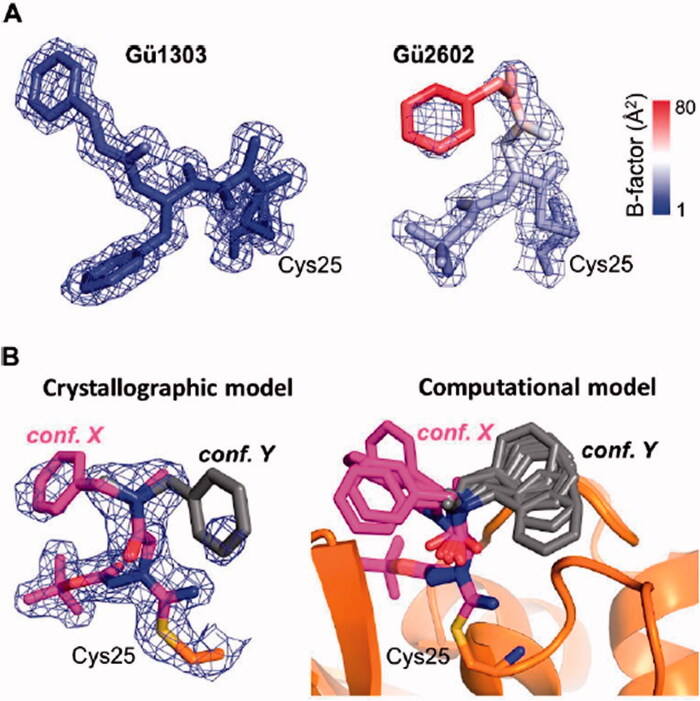
The crystallographic electron density maps used for modelling Gü1303 and Gü2602 in the active site of mCatK were of high quality except for a weak electron density signal of the benzyl group of Gü2602 (Figure 6(A)). This prompted us to analyse the B-factor distribution in both inhibitors. The B-factor values were generally in the low range, but increased values were only found for the P1 N-benzyl-N-methylacetamide part of Gü2602, with the highest value for its benzyl group (Figure 6(A)). An analogous B-factor pattern was also observed for inhibitors in iCatK complexes (Figure S1). This finding indicated an increased flexibility and dynamic disorder of the major crystallographic conformation of the benzyl group of Gü2602, which is located in the non-primed part of the active site and oriented out of the S1 subsite (Figure 5(C)).
Figure 6.
Analysis of conformational flexibility of Gü1303 and Gü2602 inhibitors in the active site of mature cathepsin K. (A) The inhibitors and the side chain of the covalently linked catalytic cysteine residue Cys25 are shown in stick representation. Their 2Fo-Fc electron density maps are contoured at 1 σ and 1.5 σ for Gü1303 and Gü2602, respectively. Structures are coloured according to atomic B-factor values, from blue (low) to red (high). The highest B-factors indicating flexibility are observed for the benzyl moiety of Gü2602. (B) Conformational flexibility of the benzyl moiety of Gü2602. Left panel: Crystallographic model of Gü2602 with the major observed conformation of the benzyl moiety in magenta (conf. X located in the non-primed area) and a predicted alternative minor conformation in grey (conf. Y located in the primed area). The side chain of the covalently linked catalytic Cys25 is in orange; heteroatoms have a standard colour coding (O, red; N, blue; S, yellow). The 2Fo-Fc electron density maps are contoured at 1 σ. Right panel: Conformations of the benzyl moiety were searched by the molecular dynamics/quenching (MD/Q) technique and optimised by the semiempirical quantum mechanical (SQM) method. The lowest-energy conformations up to relative Gibbs “free” energy of 5 kcal/mol are shown for the general orientations X and Y (as defined by crystallography in left panel).
A detailed inspection of electron density maps in the active site of the mCatK-Gü2602 complex provided a further weak electron density signal of poor quality in the primed region of the mCatK active site that can be assigned to Gü2602. Based on this signal, we attempted to model an alternative conformation of the benzyl group of Gü2602, which is oriented towards the S2’ subsite as presented in Figure 6(B) (see conformation Y). For this purpose, we employed molecular modelling to examine conformational space of the flexible N-benzyl-N-methylacetamide substructure of Gü2602. The molecular dynamics/quenching (MD/Q) technique was utilised to generate accessible conformations of this segment in the active site, and semiempirical quantum mechanical optimisation yielded a set of conformations with two general orientations into the non-primed and primed areas (the orientations are marked X and Y, respectively, in Figure 6(B)).
In conclusion, we demonstrated that Gü2602 is more flexible in the enzyme active site in contrast to the rigid ligand Gü1303. The former contains a highly flexible benzyl moiety that is capable of adopting to two different types of conformations that reside in the non-primed part or primed part of the active site.
4. Discussion and conclusions
Cathepsin K (CatK) is a target for the treatment of osteoporosis, arthritis, and bone metastasis, and its potent and selective inhibitors are being intensively pursued as chemotherapeutics6,7,24. In this study, we investigated peptidomimetic inhibitors with a cyanohydrazide warhead, a class of highly efficient inhibitors of CatK that have been recently discovered30,31,39, yet for which the interaction mechanism at the atomic level with CatK is unknown. For two representative cyanohydrazide compounds, Gü1303 and Gü2602, we present crystallographic analysis of the binding mode to mature enzyme mCatK and its zymogen-like activation intermediate iCatK, as well as functional analysis in vitro and in cells.
The crystal structures of the complexes of Gü1303 or Gü2602 with mCatK showed that the inhibitors are bound in a substrate-like orientation, and their cyanohydrazide warhead reacted with a thiol group of the catalytic Cys25, forming a covalent isothiosemicarbazide adduct. The CO–NMe and CO–NH bond (in Gü1303 and Gü2602, respectively) of the warhead adopted a Z-configuration in the mCatK active site. This is in line with our recent analysis of another cyanohydrazide inhibitor (with the azadipeptide nitrile scaffold) in the active site of a cysteine protease32 which also demonstrated that the warhead with the methylated N–N axis provides atropochirality, and the E-configuration of the unbound inhibitor is transformed to a Z-configuration upon binding32. Therefore, such an E- to Z-conformational change in the course of its interaction with CatK is proposed for Gü1303, containing the CO–NMe–NMe portion.
The cyanohydrazide warhead is differently positioned in the inhibitor scaffold of the azadipeptide nitrile Gü1303 and 3-cyano-3-aza-β-amino acid Gü2602. This is reflected in the distribution of the binding subsites that are targeted by the inhibitors. Gü1303 occupies the non-primed subsites S1 to S3, and B-factor distribution showed that Gü1303 is rigid in the mCatK active site. In contrast, Gü2602 primarily occupies the S2 subsite, and the N-benzyl-N-methylacetamide part (especially its benzyl moiety) is highly flexible and has two alternative types of conformations that reside in the non-primed part (the S1 subsite) or primed part (the S2’ subsite) of the active site. The non-primed orientation is the major one observed in the crystal structure, where it might be stabilised by additional contacts, including crystal packing contacts (with a symmetry-related protein molecule) and intramolecular contacts. The primed orientation was clearly demonstrated by molecular modelling using MD/Q and SQM techniques and is further supported by the recent identification of a benzyl group oriented towards the S2’ pocket in the activity-based probe for CatK40. The computational approach showed a set of conformations for both types of orientations, further highlighting the high flexibility of this part of Gü2602 in the enzyme active site.
We hypothesise that the conformational flexibility of the complexed ligand at the boundary of the non-primed and primed subsites provides an entropic advantage contributing to the extraordinary potency of Gü2602 in the low picomolar range. It has been shown that the loss of configurational entropy upon ligand binding contributes significantly unfavourably to the binding free energy65; this entropy loss is decreased for Gü2602 making its binding more favourable. An analysis of active site interactions formed by Gü2602 also identified an important role of hydrogen bonding between the amide NH of the warhead and Asn161 of the S2 subsite, an interaction that is absent in Gü1303. This is supported by analogues of Gü2602 with methylated NH that exhibited a dramatic decrease in inhibitory potency by 3 orders of magnitude39,40.
The differences in the binding mode between Gü2602 and Gü1303 are also reflected in their kinetic behaviour. The slow-binding of Gü1303 is attributed to a conversion from an E- to Z-configuration upon binding that was recently described as the kinetic controlling step for a prototype azadipeptide nitrile with an atropochiral warhead32. In contrast, the 3-cyano-3-aza-β-amino acid scaffold of Gü2602 bears a non-atropochiral warhead, as demonstrated by NMR studies showing the absence of diastereotopic methylene protons39. Hence, the configuration conversion step does not occur, as is reflected by the fast-binding of Gü2602 that resembles analogous kinetics and a reaction mechanism described for carbanitrile inhibitors32.
In addition to mCatK-inhibitor complexes, we also structurally characterised the complexes of Gü2602 and Gü1303 with the activation intermediate iCatK produced during autocatalytic processing of the inactive zymogen to mature enzyme that is associated with the removal of the propeptide. The iCatK structure is described here for the first time. It contains the main catalytic domain, which is non-covalently bound to the residual propeptide domain. Unlike the zymogen structure27,28, the residual propeptide (residues 4p to 83p) of iCatK only partially occluded the active site cleft (mainly the S1’ subsite), and thus the catalytic centre is accessible for ligands. This contrasts to the previously investigated activation intermediate of the cathepsin SmCB1, in which the catalytic centre is still blocked by the residual propeptide41.
iCatK was demonstrated to bind Gü2602 and Gü1303 in an analogous manner to that observed in the mCatK complexes. This indicated that the iCatK active site is competent to bind inhibitory ligands in the available subsites and can be regulated by them. This conclusion is supported by the results on testing Gü2602 and Gü1303 in an in vitro autoactivation assay with the zymogen pCatK. It showed that both inhibitors strongly suppress the formation of mCatK from the zymogen that proceeds as a bimolecular processing reaction catalysed by functional forms of iCatK/mCatK. Furthermore, we provided evidence that Gü2602 and Gü1303 are capable of effectively targeting mCatK as well as the mCatK-generating pathway in the pathophysiologically relevant context of osteosarcoma cells.
In conclusion, our work provides the first crystallographic, computational chemical, and functional insights into the binding mode of the cyanohydrazide inhibitors to the CatK target and will facilitate a further rational design of therapeutics against CatK-mediated pathologies.
Supplementary Material
Acknowledgements
The authors thank Helena Mertlíková-Kaiserová for providing U-2 OS cells. Diffraction data were collected on MX14.1 at the BESSY II electron storage ring operated by the Helmholtz-Zentrum Berlin.
Funding Statement
This work was supported by the project ChemBioDrug CZ.02.1.01/0.0/0.0/16_019/0000729 from ERDF/OPRDE and the institutional project RVO 61388963.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Atomic coordinates and experimental structure factors have been deposited in the Protein Data Bank with accession codes 7QBL, 7QBM, 7QBN, and 7QBO.
References
- 1.Li H, Xiao Z, Quarles LD, Li W.. Osteoporosis: mechanism, molecular target and current status on drug development. Curr Med Chem 2021;28:1489–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa AG, Cusano NE, Silva BC, et al. Cathepsin K: its skeletal actions and role as a therapeutic target in osteoporosis. Nat Rev Rheumatol 2011;7:447–56. [DOI] [PubMed] [Google Scholar]
- 3.Makras P, Delaroudis S, Anastasilakis AD.. Novel therapies for osteoporosis. Metabolism 2015;64:1199–214. [DOI] [PubMed] [Google Scholar]
- 4.Kramer L, Turk D, Turk B.. The future of cysteine cathepsins in disease management. Trends Pharmacol Sci 2017;38:873–98. [DOI] [PubMed] [Google Scholar]
- 5.Drake MT, Clarke BL, Oursler MJ, Khosla S.. Cathepsin K inhibitors for osteoporosis: biology, potential clinical utility, and lessons learned. Endocr Rev 2017;38:325–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brömme D, Lecaille F.. Cathepsin K inhibitors for osteoporosis and potential off-target effects. Expert Opin Investig Drugs 2009;18:585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Wang M, Wang Z, et al. Advances in the discovery of cathepsin K inhibitors on bone resorption. J Enzyme Inhib Med Chem 2018;33:890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejica VM, Mort JS, Laverty S, et al. Increased type II collagen cleavage by cathepsin K and collagenase activities with aging and osteoarthritis in human articular cartilage. Arthritis Res Ther 2012;14:R113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda Y, Kaleta J, Brömme D.. The role of cathepsins in osteoporosis and arthritis: rationale for the design of new therapeutics. Adv Drug Deliv Rev 2005;57:973–93. [DOI] [PubMed] [Google Scholar]
- 10.Aguda AH, Panwar P, Du X, et al. Structural basis of collagen fiber degradation by cathepsin K. Proc Natl Acad Sci USA 2014;111:17474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novinec M, Lenarčič B.. Cathepsin K: a unique collagenolytic cysteine peptidase. Biol Chem 2013;394:1163–79. [DOI] [PubMed] [Google Scholar]
- 12.Novinec M, Kovačič L, Lenarčič B, Baici A.. Conformational flexibility and allosteric regulation of cathepsin K. Biochem J 2010;429:379–89. [DOI] [PubMed] [Google Scholar]
- 13.Cherney MM, Lecaille F, Kienitz M, et al. Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions. J Biol Chem 2011;286:8988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verbovšek U, Van Noorden CJ, Lah TT.. Complexity of cancer protease biology: cathepsin K expression and function in cancer progression. Semin Cancer Biol 2015;35:71–84. [DOI] [PubMed] [Google Scholar]
- 15.Olson OC, Joyce JA.. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer 2015;15:712–29. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Cheng XW, Shi GP, et al. Cathepsin K-mediated Notch1 activation contributes to neovascularization in response to hypoxia. Nat Commun 2014;5:3838. [DOI] [PubMed] [Google Scholar]
- 17.Herroon MK, Rajagurubandara E, Rudy DL, et al. Macrophage cathepsin K promotes prostate tumor progression in bone. Oncogene 2013;32:1580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamolmatyakul S, Chen W, Yang S, et al. IL-1alpha stimulates cathepsin K expression in osteoclasts via the tyrosine kinase-NF-kappaB pathway. J Dent Res 2004;83:791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asagiri M, Hirai T, Kunigami T, et al. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science 2008;319:624–7. [DOI] [PubMed] [Google Scholar]
- 20.Hira VV, Ploegmakers KJ, Grevers F, et al. CD133+ and Nestin + glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J Histochem Cytochem 2015; 63: 481–93. [DOI] [PubMed] [Google Scholar]
- 21.Calio A, Brunelli M, Gobbo S, et al. Cathepsin K: a novel diagnostic and predictive biomarker for renal tumors. Cancers 2021;13:2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leusink FK, Koudounarakis E, Frank MH, et al. Cathepsin K associates with lymph node metastasis and poor prognosis in oral squamous cell carcinoma. BMC Cancer 2018;18:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kos J, Lah TT.. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review). Oncol Rep 1998;5:1349–410. [DOI] [PubMed] [Google Scholar]
- 24.Lecaille F, Brömme D, Lalmanach G.. Biochemical properties and regulation of cathepsin K activity. Biochimie 2008;90:208–26. [DOI] [PubMed] [Google Scholar]
- 25.McQueney MS, Amegadzie BY, D'Alessio K, et al. Autocatalytic activation of human cathepsin K. J Biol Chem 1997;272:13955–60. [DOI] [PubMed] [Google Scholar]
- 26.Linnevers CJ, Mcgrath ME, Armstrong A, et al. Expression of human cathepsin K in Pichia pastoris and preliminary crystallographic studies of an inhibitor complex. Protein Sci 1997;6:919–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivaraman J, Lalumière M, Ménard R, Cygler M.. Crystal structure of wild-type human procathepsin K. Protein Sci 1999;8:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaLonde JM, Zhao B, Janson CA, et al. The crystal structure of human procathepsin K. Biochemistry 1999;38:862–9. [DOI] [PubMed] [Google Scholar]
- 29.Lemaire PA, Huang L, Zhuo Y, et al. Chondroitin sulfate promotes activation of cathepsin K. J Biol Chem 2014;289:21562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Löser R, Frizler M, Schilling K, Gütschow M.. Azadipeptide nitriles: highly potent and proteolytically stable inhibitors of papain-like cysteine proteases. Angew Chem Int Ed Engl 2008;47:4331–4. [DOI] [PubMed] [Google Scholar]
- 31.Frizler M, Lohr F, Furtmann N, et al. Structural optimization of azadipeptide nitriles strongly increases association rates and allows the development of selective cathepsin inhibitors. J Med Chem 2011;54:396–400. [DOI] [PubMed] [Google Scholar]
- 32.Jílková A, Horn M, Fanfrlík J, et al. Azanitrile inhibitors of the SmCB1 protease target are lethal to Schistosoma mansoni: structural and mechanistic insights into chemotype reactivity. ACS Infect Dis 2021;7:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang PY, Wang M, Li L, et al. Design, synthesis and biological evaluation of potent azadipeptide nitrile inhibitors and activity-based probes as promising anti-Trypanosoma brucei agents. Chemistry 2012;18:6528–41. [DOI] [PubMed] [Google Scholar]
- 34.Breidenbach J, Lemke C, Pillaiyar T, et al. Targeting the main protease of SARS-CoV-2: from the establishment of high throughput screening to the design of tailored inhibitors. Angew Chem Int Ed Engl 2021;60:10423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chingle R, Proulx C, Lubell WD.. Azapeptide synthesis methods for expanding side-chain diversity for biomedical applications. Acc Chem Res 2017;50:1541–56. [DOI] [PubMed] [Google Scholar]
- 36.Proulx C, Sabatino D, Hopewell R, et al. Azapeptides and their therapeutic potential. Future Med Chem 2011;3:1139–64. [DOI] [PubMed] [Google Scholar]
- 37.Verhelst SH, Witte MD, Arastu-Kapur S, et al. Novel aza peptide inhibitors and active-site probes of papain-family cysteine proteases. ChemBioChem 2006;7:943–50. [DOI] [PubMed] [Google Scholar]
- 38.Sexton KB, Kato D, Berger AB, et al. Specificity of aza-peptide electrophile activity-based probes of caspases. Cell Death Differ 2007;14:727–32. [DOI] [PubMed] [Google Scholar]
- 39.Schmitz J, Beckmann AM, Dudic A, et al. 3-Cyano-3-aza-β-amino acid derivatives as inhibitors of human cysteine cathepsins. ACS Med Chem Lett 2014;5:1076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemke C, Benýšek J, Brajtenbach D, et al. An activity-based probe for cathepsin K imaging with excellent potency and selectivity. J Med Chem 2021;64:13793–806. [DOI] [PubMed] [Google Scholar]
- 41.Jílková A, Horn M, Řezáčová P, et al. Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch. Structure 2014;22:1786–98. [DOI] [PubMed] [Google Scholar]
- 42.Horn M, Jílková A, Vondrášek J, et al. Mapping the pro-peptide of the Schistosoma mansoni cathepsin B1 drug target: modulation of inhibition by heparin and design of mimetic inhibitors. ACS Chem Biol 2011;6:609–17. [DOI] [PubMed] [Google Scholar]
- 43.Mueller U, Förster R, Hellmig M, et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur Phys J Plus 2015;130:141. [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A.. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 2000;56:1622–4. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011;67:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 2006;62:1002–11. [DOI] [PubMed] [Google Scholar]
- 48.Long F, Nicholls RA, Emsley P, et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr D Struct Biol 2017;73:112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 2007;35:W375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salentin S, Schreiber S, Haupt VJ, et al. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 2015;43:W443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Case DA, Babin V, Berryman JT, et al. AMBER 14. San Francisco (USA): University of California San Francisco; 2014. [Google Scholar]
- 52.Stewart JJ. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model 2007;13:1173–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Řezáč J, Hobza P.. Advanced corrections of hydrogen bonding and dispersion for semiempirical quantum mechanical methods. J Chem Theory Comput 2012;8:141–51. [DOI] [PubMed] [Google Scholar]
- 54.Klamt A, Schuurmann G.. Cosmo - a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J Chem Soc Perk T 2 1993;5:799–805. [Google Scholar]
- 55.Kříž K, Řezáč J.. Reparametrization of the COSMO solvent model for semiempirical methods PM6 and PM7. J Chem Inf Model 2019;59:229–35. [DOI] [PubMed] [Google Scholar]
- 56.Řezáč J. Cuby: an integrative framework for computational chemistry. J Comput Chem 2016;37:1230–7. [DOI] [PubMed] [Google Scholar]
- 57.Stewart JJP. Optimization of parameters for semiempirical methods IV: extension of MNDO, AM1, and PM3 to more main group elements. J Mol Model 2004;10:155–64. [DOI] [PubMed] [Google Scholar]
- 58.Kuzmic P, Sideris S, Cregar LM, et al. High-throughput screening of enzyme inhibitors: automatic determination of tight-binding inhibition constants. Anal Biochem 2000;281:62–7. [DOI] [PubMed] [Google Scholar]
- 59.Horn M, Nussbaumerová M, Šanda M, et al. Hemoglobin digestion in blood-feeding ticks: mapping a multipeptidase pathway by functional proteomics. Chem Biol 2009;16:1053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett AJ, Kembhavi AA, Brown MA, et al. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J 1982;201:189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Billington CJ, Mason P, Magny MC, Mort JS.. The slow-binding inhibition of cathepsin K by its propeptide. Biochem Biophys Res Commun 2000;276:924–9. [DOI] [PubMed] [Google Scholar]
- 62.Husmann K, Muff R, Bolander ME, et al. Cathepsins and osteosarcoma: expression analysis identifies cathepsin K as an indicator of metastasis. Mol Carcinog 2008;47:66–73. [DOI] [PubMed] [Google Scholar]
- 63.Ottersbach PA, Schnakenburg G, Gütschow M.. Induction of chirality: experimental evidence of atropisomerism in azapeptides. Chem Commun 2012;48:5772–4. [DOI] [PubMed] [Google Scholar]
- 64.Ottersbach PA, Schnakenburg G, Gütschow M.. Atropisomerism in azadipeptides: evaluation of N1-methylation and thioamide introduction. Tetrahedron Lett 2015;56:4889–91. [Google Scholar]
- 65.Chia-en AC, Chen W, Gilson MK.. Ligand configurational entropy and protein binding. Proc Natl Acad Sci USA 2007;104:1534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and experimental structure factors have been deposited in the Protein Data Bank with accession codes 7QBL, 7QBM, 7QBN, and 7QBO.