Abstract
BACKGROUND
Curative gene therapies for sickle cell disease (SCD) are currently undergoing clinical evaluation. The occurrence of myeloid malignancies in these trials has prompted safety concerns. Individuals with SCD are predisposed to myeloid malignancies, but the underlying causes remain undefined. Clonal hematopoiesis (CH) is a premalignant condition that also confers significant predisposition to myeloid cancers. While it has been speculated that CH may play a role in SCD-associated cancer predisposition, limited data addressing this issue have been reported.
METHODS
Here, we leveraged 74,190 whole-genome sequences to robustly study CH in SCD. Somatic mutation calling methods were used to assess CH in all samples and comparisons between individuals with and without SCD were performed.
RESULTS
While we had sufficient power to detect a greater than 2-fold increased rate of CH, we found no detectable variation in rate or clone properties between individuals affected by SCD and controls. The rate of CH in individuals with SCD was unaltered by hydroxyurea use.
CONCLUSIONS
We did not observe an increased risk for acquiring detectable CH in SCD, at least as measured by whole-genome sequencing. These results should help guide ongoing efforts and further studies that seek to better define the risk factors underlying myeloid malignancy predisposition in SCD and help ensure that curative therapies can be more safely applied.
FUNDING
New York Stem Cell Foundation and the NIH.
Keywords: Hematology
Keywords: Leukemias
Introduction
In addition to the reduced life expectancy from disease complications (1), individuals with sickle cell disease (SCD) are estimated to have an approximately 3- to 10-fold increased lifetime risk for acquiring acute myeloid leukemia (AML) and other myeloid malignancies (2–4). These observations have recently generated profound interest due to a number of reports of myeloid malignancies arising during gene therapy that have halted several ongoing clinical trials (5, 6). In one reported patient, the malignancy was not attributable to insertional mutagenesis, but the malignant clone was noted to harbor RUNX1, KRAS, and PTPN11 mutations (6). While insertional mutagenesis was unlikely the driver of the observed malignancy, it is still possible that other elements of therapy may have contributed to cancer predisposition, such as the conditioning regimen. Analysis of 2 SCD patients with myeloid malignancies in the setting of allogeneic transplantation revealed preexisting hematopoietic clones with TP53 mutations that increased in size following conditioning, resulting in AML and myelodysplastic syndromes (7). It is still unknown what factors underlie the clonal expansion of these mutant cells, but chronic inflammation and functional decline of tissues may contribute.
The baseline rate of clonal hematopoiesis (CH) in SCD patients and thus the overall risk for myeloid malignancies in these individuals attributable to CH remains unclear. We reasoned that the more frequent and earlier onset of myeloid malignancies in people with SCD would be accompanied by the precocious development of CH. Myeloid malignancy predisposition is generally present in the setting of CH with a variant allele frequency (VAF) above 10% (8–12). Here, we have robustly assessed the prevalence of CH in individuals with SCD who were subject to sequencing and variant calling in tandem with other unaffected individuals as part of the National Heart, Lung and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) consortium to interrogate whether SCD may predispose to CH (13).
Results
Prevalence of CH within the SCD population.
To establish the prevalence of CH across human age, whole-genome sequencing (WGS) data from 71,100 individuals unaffected by SCD and 3,090 individuals affected by SCD with ages of 70 years old and below were compiled from 30 distinct cohorts sequenced as part of the TOPMed consortium (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI156060DS1). Previously described methods were employed to identify somatic variants and the presence of clonal expansions indicative of CH (14). In short, somatic mutations in the blood were identified by GATK Mutect2 using a panel of normal samples to correct for genomic loci that were hotspots for sequencing artifacts, followed by filtering of identified loci to those that are reliably classified as preleukemic driver mutations (15). Samples were sequenced to an average depth of 38×, which as we have shown previously, detects almost all clones that exist above 10% VAF, and about half of all clones that exist between VAFs of 5% and 10% (14). Although hematopoietic expansions exist in most individuals, and deep sequencing can retroactively identify early origins of malignancies (1, 7), our analysis is sufficient to reliably detect the more mature clones that are of prognostic value based on analyses done to date (11, 12).
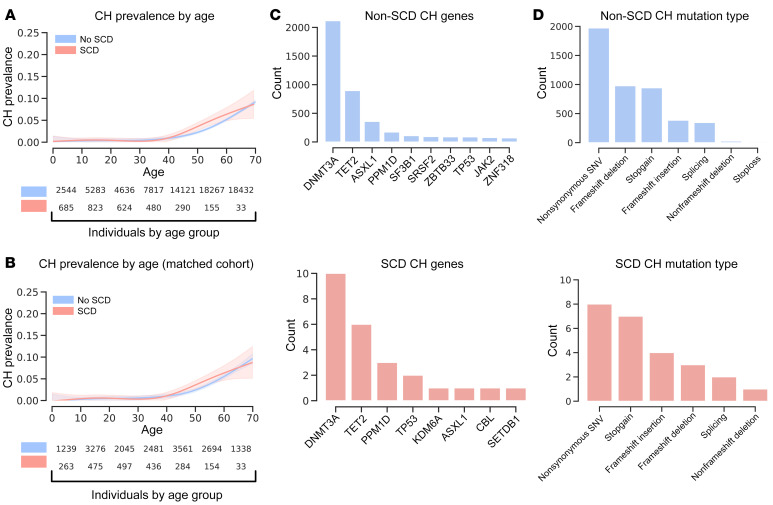
We identified CH in 27 SCD patients and 3,063 unaffected individuals. The prevalence of CH in the SCD cohorts and unaffected individuals strongly correlated with age. There was, however, no significant difference in the prevalence of CH within the SCD cohort when compared to unaffected controls in unadjusted analyses (OR = 1.30, P = 0.20; Figure 1A). Using a generalized linear model to account for covariates that are likely to impact the predicted CH prevalence, including age, age2, sex, study, and 10 principal components (PCs) of genetic ancestry, we found that the prevalence of CH in individuals with SCD was not elevated over unaffected controls (SCD OR = 1.55, P = 0.20; Supplemental Figure 2). As a further sensitivity analysis, we performed a 1:20 case/control matching on age, sex, and the first 10 PCs, as well as excluding several smaller disease-specific cohorts (details in Methods), to identify a group of controls that were as similar as possible to our SCD cases. Even with this constructed set of unaffected controls, the CH prevalence in the SCD cohort was not significantly elevated, and even trended lower (OR = 0.80, P = 0.29; Figure 1B).
Figure 1. Prevalence of CH is similar in unaffected and SCD populations.
(A) A generalized additive model was used here to fit rates of CH within WGS data from a total of 71,100 individuals unaffected by SCD and 3,090 individuals affected by SCD, which indicates no significantly increased prevalence of CH within individuals affected by SCD (OR = 1.30, P = 0.20). (B) Generalized additive model as in A was used to fit rates of CH using a genetically matched cohort without SCD. The matched cohort was created by selecting the 10 most similar individuals by the first 10 PCs for each individual in the SCD cohort. Resampling of individuals without SCD was permitted. SCD samples without sufficient matches were excluded. (C) Genes ranked by variant load across all individuals separated by SCD status into unaffected (blue) and affected (red). (D) Type of genetic change ranked by prevalence in all individuals separated by SCD status into unaffected (blue) and affected (red).
Although our set of controls included individuals with sickle cell trait (2.6% of controls), we do not observe any altered rate of CH in these individuals compared to the other controls (OR = 1.09, P = 0.54; Supplemental Figure 3). Similar subsetting of the SCD cohort to just those individuals with a hemoglobin SS (Hb SS) homozygous genotype, which is the most common SCD genotype, showed no significant difference in prevalence of CH between Hb SS SCD and unaffected individuals (OR = 1.34, P = 0.22; Supplemental Figure 4). Furthermore, separating the different Hb genotypes revealed that Hb Sβ0 thalassemia was closest to a significantly different odds of CH, although it did not reach significance and only 84 individuals had this genotype (OR = 3.81; P = 0.067). Moreover, Hb Sβ+ thalassemia (OR = 1.21; P = 0.85) and Hb SC disease (OR = 1.02; P = 0.97) were both far from significantly different with respect to non-SCD participants. In the absence of explicit annotation of disease severity, the lack of any elevated CH prevalence by genotype, especially for Hb SS which is typically more severe, suggests that there is little impact of disease severity on CH prevalence. Due unfortunately in part to the decreased survival in individuals with SCD, the cohort size diminished with age, but nonetheless with the cohorts available to us, we have 95% power to detect a 2-fold difference and 80% power to detect a 1.75-fold difference in the prevalence of CH in SCD. As the prevalence of CH increases with age, accumulation of additional samples affected by SCD may reveal subtle differences to which we are blind due to the current limited number of individuals.
Tissue microenvironmental and intrinsic conditions have a profound impact on the selective advantages and disadvantages conferred on cells by somatic mutations (16, 17). We therefore expected that while the overall prevalence of CH in SCD may not be elevated, the oncogenic drivers of CH in SCD may notably differ. While categorizing somatic drivers of CH by affected gene and ranking by most prevalent results in slightly different spectra, no genes are significantly more likely to be mutated and represented in expanded clones in the context of SCD (Figure 1C and Supplemental Figure 5). Similarly, classifying somatic mutations by type of change exposes no significant differences in the types of somatic mutations that occur in the context of SCD (Figure 1D and Supplemental Figure 5). By treating the number of CH variants per individual as an ordinal variable and correcting for age in a proportional odds logistic regression, having SCD did not correlate with an increased number of detected mutation-harboring clones (Supplemental Figure 6). The overwhelming majority (>70%) of individuals with CH in our data set had only a single detectable somatic CH mutation, and this did not differ between individuals with SCD and unaffected individuals.
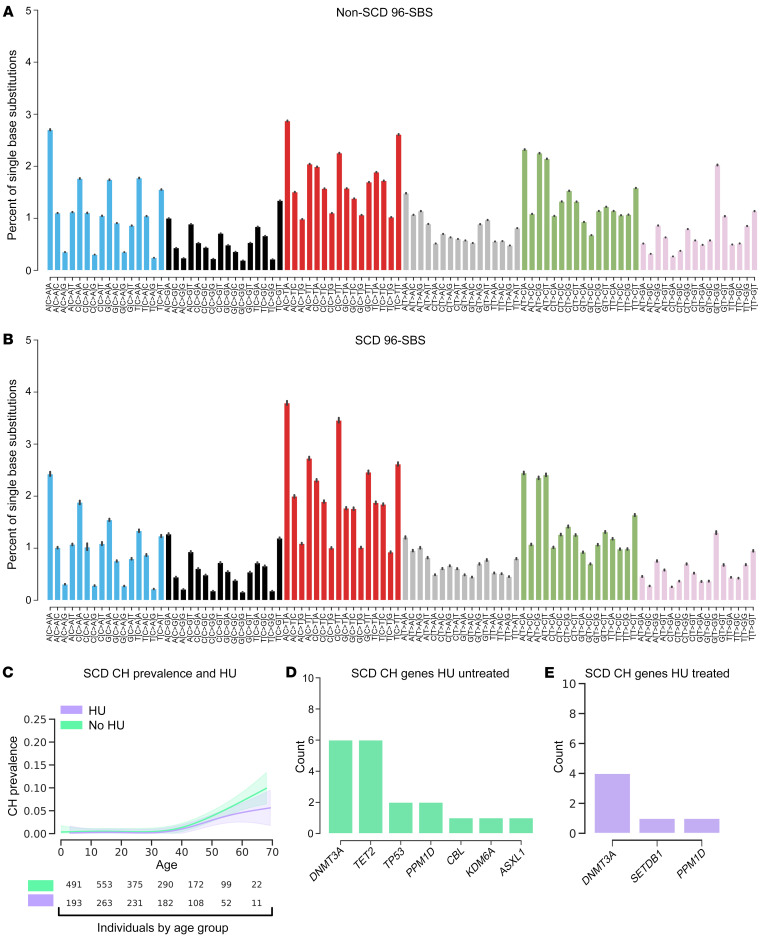
While overall rates of CH may not significantly differ among individuals with or without SCD, mutagenic processes may be altered by specific pathogenic features of SCD. We expect that such mutagenic processes could be reflected as alterations to the relative rates of single-base substitutions (SBSs). Importantly, the trinucleotide SBS mutation spectrum for the unaffected individuals showed no significant difference from those individuals with SCD (cosine similarity of 0.98; Figure 2, A and B). This similarity suggests that at least as seen through SBS, there are no prominent mutagenic processes that are uniquely observed or that predominate in the context of SCD.
Figure 2. Mutation signatures do not vary in SCD and hydroxyurea (HU) treatment does not impact the rate of CH development.
Percentages of total single-base substitutions made up by each possible 96 substitution and trinucleotide context pairs per individual (error is SD across individuals). Signatures are separated into (A) individuals without SCD and (B) individuals with SCD (data represent mean ± SD). (C) A generalized additive model is used here to fit rates of CH within WGS from individuals with SCD separated into never-HU-treated or HU-treated groups. There is no significant difference in the rate of CH in either the HU-treated or untreated groups adjusted for age, age2, sex, study, and the first 10 principal components (OR = 0.58, P = 0.23). (D) Genes ranked by variant load across all individuals with SCD separated by HU treatment status into untreated (green) and (E) treated (purple).
Impact of hydroxyurea treatment on the development of CH in SCD.
Hydroxyurea (HU) has been shown to have a significantly positive clinical impact in individuals with SCD and is thought to primarily influence disease severity by increasing levels of fetal Hb (HbF) (18). Given its cytotoxic effects and evidence from animal models, it has been posited that HU may be mutagenic and potentially carcinogenic, despite demonstrated safety profiles in vivo (19). We therefore reasoned that while the overall SCD cohort may not exhibit an elevated risk of developing CH, those treated with HU may. Treating individuals with SCD categorically as either ever having received HU treatment during their clinical course or not, we found no significant elevation in CH prevalence in the group treated with HU (OR = 0.58, P = 0.23; Figure 2C). We additionally reasoned that while HU treatment may not drive an apparent increased prevalence of CH, it could select for cells with mutations in a different spectrum of genes. As before, we saw no significant difference in the spectrum of mutated genes in the HU-treated group (Figures 2, D and E, and Supplemental Figure 5). There appeared to be a trend toward a reduced rate of CH in the HU-treated individuals with SCD, but there was no significant difference in individuals over 50 years of age (OR = 0.61; P = 0.55). While the data were not available to us, more detailed information such as age, dosage, or duration of treatment would be valuable for additional CH analyses and should be a focus of future studies.
Discussion
Although not well studied, SCD has been associated in multiple studies with predisposition to myeloid malignancies and this has become of acute interest as the cancers observed in ongoing gene therapy trials have halted several current efforts (4). As part of one of these trials being conducted by the company Bluebird bio, 2 participants enrolled have developed myeloid malignancies (5). While this is only a fraction of the individuals enrolled in the study, it may indicate an elevated risk for malignancies in individuals with SCD. This risk may in part be the result of a preexisting elevated myeloid malignancy risk in individuals with SCD. CH is an important factor predisposing to myeloid malignancies and it has been speculated that individuals with SCD may have elevated rates of CH (20). A preliminary study even reported an observed, albeit slight, increase in CH in people with SCD within a pilot cohort in comparison with external controls (21).
To advance our understanding of CH rates in people with SCD, we have taken advantage of the largest genome sequencing study of individuals with SCD to date and do not observe any increased rate of detectable CH compared to other individuals that were sequenced together with the SCD samples. With the size of the SCD cohorts available to us, we are well powered to detect at least a 2-fold increased prevalence of CH within our current cohorts. Additionally, we detected CH using WGS and were unable to detect extremely small clones. The clinical implications of small clones in predisposing to myeloid malignancies is unknown and while small clones have been retroactively identified after expansion in the context of SCD (1, 7), existing literature suggests that only larger clones with VAFs greater than 10% have prognostic impact (8–11). While further studies are needed, especially to better understand the prognostic value of rare mutations, particularly in the setting of conditioning, transplant, or other treatment regimens, our findings are likely to inform clinical decisions and suggest that CH surveillance as part of routine clinical care for SCD patients is no more indicated at the current time than it is in the general population. In the setting of clinical trials testing curative gene therapy or genome editing approaches, CH surveillance may not provide an effective approach to mitigate malignancy risk. The precise underlying factors responsible for this increased risk will require further study, and may include the chronic stress of hemolysis, an inflammatory bone marrow microenvironment, stressful in vitro cell manipulation, and exposure to cytotoxic conditioning regimens. We believe that the rapid reporting of these results will assist in the ongoing search for the causes of myeloid malignancies in SCD and help define ways to reduce this risk, which is particularly relevant to current and future genetic therapies for this disease.
Methods
WGS samples.
Thirty studies from the TOPMed consortium were compiled together into a single data set. The studies were the following: Amish, ARIC, BAGS, BioMe, CARDIA, CFS, CHS, COPDGene, CRA, DHS, FHS, GALAII, GeneSTAR, GenSalt, GOLDN, HCHS_SOL, HyperGEN, JHS, MESA, MLOF, OMG_SCD, REDS-III_Brazil, SAFS, Samoan, SAPPHIRE_asthma, SARP, THRV, VU_AF, walk_PHaSST, and WHI. Together, the studies had 71,100 individuals unaffected by SCD and 3,090 individuals affected by SCD, with a mean of 2,982 individuals per cohort.
Somatic mutation calling.
Somatic mutations were called from WGS samples using GATK-Mutect2 in combination with GATK-Mutect2-PON (14). Analysis was performed using publicly available methods in workflow description language (WDL) on the Broad Institute’s Terra Platform (https://terra.bio/), and BAM files were remapped and harmonized through a unified protocol. Single-nucleotide polymorphisms (SNPs) and short indels were jointly discovered and genotyped across the TOPMed samples using the GotCloud pipeline (https://genome.sph.umich.edu/wiki/GotCloud). An SVM filter was trained to discriminate between true variants and low-quality sites. Sample quality was assessed through pedigree errors, contamination estimates, and concordance between self-reported sex and genotype-inferred sex. Variants were annotated using SnpEff 4.3 (http://pcingola.github.io/SnpEff/). Putative somatic SNPs and short indels were called with GATK Mutect2 (https://software.broadinstitute.org/gatk). In brief, Mutect2 searches for sites where there is evidence for variation and then performs local reassembly. It uses an external reference of recurrent sequencing artefacts termed a “panel of normal samples” to filter out these sites, and calls variants at sites where there is evidence for somatic variation. The panel of normal samples used for our study included 100 randomly selected individuals under the age of 40 years. Absence of a hotspot CH mutation was verified before inclusion in the panel of normal set. An external reference of germline variants was provided to filter out likely germline calls. We deployed this variant calling process on Google Cloud using Cromwell (https://github.com/broadinstitute/cromwell). The caller was run individually for each sample with the same settings. The Cromwell WDL configuration config files for the run conditions found in this manuscript can be found on the Sankaran lab github (https://github.com/sankaranlab/scd-chip). Variants that appeared within a prespecified list of common driver leukemia driver mutations and passed Mutect2 filters were retained and used to classify individuals as having CH of indeterminate potential (CHIP) (14). Passenger mutations were required to pass Mutect2 filtering, and additionally were only permitted to be found within a single individual within the cohort to minimize the risk of including sequencing or library preparation errors.
Sickle trait identification.
Individuals were genotyped and called using the GotCloud/vt pipeline in Freeze 8 of TOPMed (https://genome.sph.umich.edu/wiki/Vt, https://genome.sph.umich.edu/wiki/GotCloud). Bcftools was used to extract the rs334 locus. Any individual carrying a heterozygous call (T/A) at that locus was determined a sickle trait carrier.
CH prevalence modeling.
Somatic variants were retained if they passed Mutect2 filtering, and if they were found within a prespecified list of common leukemia and CHIP driver mutations (14). Other presumably passenger somatic variants that passed Mutect2 filtering were only included in prevalence modeling if they were present within a single individual across all cohorts. All individuals under the age of 70 were included in the study. This cutoff excluded 6 samples affected by SCD, but the limit was chosen to include only those age ranges where there was a sufficient density of samples that a CH prevalence comparison could be made. To derive effect estimates, a binomial logistic regression model was then fit to the data which was annotated for the presence or absence of a clone indicative of CH. Models were then adjusted, where indicated, for age, age2, sex, study, sickle cell genotype (Hb genotype driving the SCD), HU use (samples classified as either never having used HU or having used HU for some period of time during their lifetime), and the first 10 PCs. At all instances in the text, ORs are adjusted for at least age and age2. Graphically, in most cases, generalized additive models were used to represent patterns across age, with shaded regions corresponding to 95% confidence intervals. Model assumptions were checked in all cases. To incorporate HU treatment, individuals were classified as either undergoing HU treatment or not; treatment was not handled as a continuous variable. To build ideally matched control cohorts for individuals with SCD, the first 10 PCs and age were used to select the 20 most closely matching individuals from the entire unaffected cohort for each individual with SCD, using a nearest-neighbor approach on propensity scores, exact matching was performed on sex. For the matching analysis, smaller disease-specific cohorts (DHS, ECLIPSE, VU_AF, CHS, HVH, GOLDN, Mayo_VTE, EOCOPD, IPF, ECLIPSE, ARIC, and FHS) were excluded. Matching quality was evaluated by Love plots, with a threshold of 0.1 for absolute mean differences as well as visual inspection of each covariate against propensity score, separated by SCD diagnosis. Subsequently, weightings from this matching were used in binomial logistic regression analysis, and pair membership was used to ensure estimated effects and variance were cluster robust. This constructed control cohort was then used as a comparison for CH prevalence as matched controls. For all prevalence studies, the full spectrum of ages was included for all analyses.
CH gene ranking.
To rank the most commonly mutated genes, singleton variants were filtered to those that are considered to be indicative of the presence of CH. Genes were then ranked by the number of somatic variants they contained across all cohorts.
Power calculations.
Power calculations were performed using all individuals 70 years of age and younger without SCD as the control population, and using all individuals 70 years of age and younger with SCD as the test population. Power cutoffs were binned at 5% thresholds, and cohort SCD cohort numbers were then used to calculate the maximum bin that powered at a particular fold difference in CH. Across the entire cohort, there was a 4.3% rate of CH, and using all 3,090 individuals with SCD provides at least 95% power to detect a 1.5-fold difference in the rate of CH, requiring a minimum of 1,390 individuals to do so. The probability of type I error (α) used was 0.05. Our SCD cohort was, however, biased toward young individuals, and given that young individuals are typically unlikely to exhibit CH, it is more informative to limit the power calculation to individuals within an older age group that is typically susceptible to developing CH. Within just the range of 40 years of age or older, individuals combined across cohorts without SCD had a 6.78% rate of CH. The experimental population with SCD equal to or above the age of 40 contained 478 individuals. The probability of type I error (α) used was 0.05. In order to detect a 2-fold or 13.56% rate of CH within the experimental SCD population with 95% power, a minimum of 243 individuals are required. To detect a 1.75-fold rate of CH with 80% power, a minimum of 406 individuals are required. We therefore expect that we are at least sufficiently sensitive to detect a 2-fold increase in CH with 95% power and a 1.75-fold increase with 80% power within our entire SCD cohort.
SBS mutation spectra.
To measure relative rates of SBS in each individual, somatic mutations were first binned by individual and annotated by SCD status. Using hg38 as the reference genome, the neighboring upstream and downstream bases were associated with each somatic substitution mutation to define the trinucleotide SBS for each change. SigProfilerMatrixGenerator was then used to concatenate together each of these trinucleotide changes and compile a full matrix containing all individuals used in this study (22). The relative occurrences of these trinucleotide changes were then calculated per individual and plotted across all individuals with standard deviation as the estimation of error.
Data availability.
Individual WGS data for TOPMed whole genomes, individual-level harmonized phenotypes, harmonized germline variant call sets, and the CH somatic variant call sets are available through restricted access via the NCBI’s database of Genotypes and Phenotypes (dbGaP). Accession numbers for these data sets have been described in our prior study (14), except for REDS-III_Brazil (phs001468), OMG_SCD (phs001608), and walk_PHaSST (phs001514). Deidentified variant calls are available from the authors upon request.
Statistics.
Generalized linear models used to compare age-related differences in CH between non-SCD and SCD groups used 95% confidence intervals and significance cutoffs of P less than 0.05. Generalized additive models (GAMs) were used to graphically represent the data of SCD versus no SCD in Figure 1, A and B, and HU treatment versus no HU in Figure 2C. Logistic regression models produced the reported ORs and P values in the manuscript text. To go further, GAMs provide a flexible approach to modeling that accounts for multiple nonlinear effects. All residuals and model parameters were checked and confirmed, and smoothers were checked for overcomplexity. For the logistic regression model outputs reported in the text, P values are a result of Wald testing of the model coefficient.
Study approval.
Written informed consent was obtained from all human participants by each of the studies that contributed to TOPMed with approval of study protocols by ethics committees at participating institutions. All relevant ethics committees approved this study and this work is compliant with all relevant ethical regulations.
Author contributions
LAL, LDC, JSW, AGB, and VGS conceived and designed the study, and wrote the manuscript with input from all authors. LAL, LDC, and JSW analyzed data. YZ directed the Walk-PHasst repository and SMN maintained the Walk-PHaSST database. YZ, SMN, MTG, MEG, AAK, MJT, BC, SK, CLD, ECS, PL, ABCP, CM, APR, GRA, DAW, and PN contributed clinical data or analytic advice. AGB and VGS supervised the study.
Supplementary Material
Acknowledgments
We are grateful to members of the Sankaran laboratory and numerous colleagues for valuable comments and suggestions. This work was supported by the New York Stem Cell Foundation (to VGS), a gift from the Lodish Family to Boston Children’s Hospital (to VGS), the Burroughs Wellcome Fund Career Award (to AGB), and NIH grants R01 DK103794 (to VGS), R01 HL146500 (to VGS), and DP5 OD029586 (to AGB). LAL received support from NIH grants T32 HL066987 and T32 HL007574. VGS is a New York Stem Cell–Robertson Investigator. Molecular data for the TOPMed program was supported by the NHLBI. WGS for “NHLBI TOPMed: DHS” (phs001412) was performed at the Broad Institute Genomics Platform (HHSN268201500014C). WGS for “NHLBI TOPMed: Samoan” (phs000972) was performed at the Northwest Genomics Center (HHSN268201100037C) and the New York Genome Center Genomics (HHSN268201500016C). WGS for “NHLBI TOPMed: walk_PHaSST” (phs001514) was performed at the Baylor Genomics Center (HHSN268201500015C). All other cohorts have been previously analyzed and acknowledged in a prior study on CH (14). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. We thank the investigators of the Walk-PHaSST study and the patients who participated in the study. We also thank the Walk-PHaSST clinical site team (see the Supplemental Acknowledgments for details). Thanks also to the data coordination center team from Rho, Inc.: Nancy Yovetich, Rob Woolson, Jamie Spencer, Christopher Woods, Karen Kesler, Vickie Coble, and Ronald W. Helms. We also thank Jonathan Goldsmith as an NIH program director for this study. Special thanks to the volunteers who participated in the Walk-PHaSST study. This project was funded with federal funds from the NHLBI, NIH, Department of Health and Human Services, under contract HHSN268200617182C. This study is registered at www.clinicaltrials.gov as NCT00492531. Work on the Diabetes Heart Study was supported by NIH grants R01 HL92301, R01 HL67348, R01 NS058700, R01 AR48797, R01 DK071891, and R01 AG058921; the General Clinical Research Center of the Wake Forest University School of Medicine (M01 RR07122 and F32 HL085989); the American Diabetes Association; and a pilot grant from the Claude Pepper Older Americans Independence Center of Wake Forest University Health Sciences (P60 AG10484). Data collection for the Samoan adiposity study was funded by NIH grant R01-HL093093. We thank the Samoan participants of the study and local village authorities. We acknowledge the support of the Samoan Ministry of Health and the Samoa Bureau of Statistics for their support of this research.
Version 1. 01/06/2022
In-Press Preview
Version 2. 02/15/2022
Electronic publication
Footnotes
Conflict of interest: MTG serves as a consultant for Actelion, Bayer Healthcare, Pfizer, Forma, and Fulcrum Therapeutics. GRA is an employee of Regeneron Pharmaceuticals and owns stock and stock options for Regeneron Pharmaceuticals. DAW serves on Novartis steering committee ETB115E2201, is a cofounder of Alerion Biosciences, chief scientific chair of Emerging Therapy Solutions, and serves as an advisor to Geneception, Insertion Site, Bluebird bio, and Beam Therapeutics. PN reports research grants from Amgen, Apple, and Boston Scientific, and is an advisor to Apple, AstraZeneca, Black Stone Life Sciences, Genexwell, Foresite Labs, Novartis, Roche/Genentech, TenSixteen Bio, and Blackstone Life Sciences. AGB serves as an advisor to and has equity in TenSixteen Bio. VGS serves as an advisor to and/or has equity in Novartis, Forma, Cellarity, Ensoma, and Branch Biosciences.
Copyright: © 2022, Liggett et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2022;132(4):e156060.https://doi.org/10.1172/JCI156060.
See the related Commentary at null.
Contributor Information
L. Alexander Liggett, Email: lliggett@broadinstitute.org.
Liam D. Cato, Email: lcato@broadinstitute.org.
Joshua S. Weinstock, Email: jweinstk@umich.edu.
Yingze Zhang, Email: zhanyx@upmc.edu.
S. Mehdi Nouraie, Email: nouraies@upmc.edu.
Mark T. Gladwin, Email: gladwinmt@upmc.edu.
Melanie E. Garrett, Email: melanie.kail@duke.edu.
Allison Ashley-Koch, Email: allison.ashleykoch@duke.edu.
Marilyn J. Telen, Email: telen002@mc.duke.edu.
Brian Custer, Email: bcuster@vitalant.org.
Shannon Kelly, Email: shannon.kelly@ucsf.edu.
Carla L. Dinardo, Email: caludinardo@gmail.com.
Ester C. Sabino, Email: sabinoec@gmail.com.
Paula Loureiro, Email: Paula.loureiro10@gmail.com.
Cláudia Maximo, Email: Clmaximo@uol.com.br.
Alexander P. Reiner, Email: apreiner@u.washington.edu.
Gonçalo R. Abecasis, Email: cdobski@umich.edu.
David A. Williams, Email: DAWilliams@childrens.harvard.edu.
Pradeep Natarajan, Email: PNATARAJAN@mgh.harvard.edu.
Alexander G. Bick, Email: alexander.bick@VUMC.org.
Vijay G. Sankaran, Email: sankaran@broadinstitute.org.
References
- 1.DeBaun MR, et al. Decreased median survival of adults with sickle cell disease after adjusting for left truncation bias: a pooled analysis. Blood. 2019;133(6):615–617. doi: 10.1182/blood-2018-10-880575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, et al. Myeloid neoplasms in the setting of sickle cell disease: an intrinsic association with the underlying condition rather than a coincidence; report of 4 cases and review of the literature. Mod Pathol. 2019;32(12):1712–1726. doi: 10.1038/s41379-019-0325-6. [DOI] [PubMed] [Google Scholar]
- 3.Brunson A, et al. Increased risk of leukemia among sickle cell disease patients in California. Blood. 2017;130(13):1597–1599. doi: 10.1182/blood-2017-05-783233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seminog OO, et al. Risk of individual malignant neoplasms in patients with sickle cell disease: English national record linkage study. J R Soc Med. 2016;109(8):303–309. doi: 10.1177/0141076816651037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Science. Gene Therapy Trials for Sickle Cell Disease Halted After Two Patients Develop Cancer. https://www.sciencemag.org/news/2021/02/gene-therapy-trials-sickle-cell-disease-halted-after-two-patients-develop-cancer Updated February 16, 2021. Accessed May 7, 2021.
- 6.Hsieh MM, et al. Myelodysplastic syndrome unrelated to lentiviral vector in a patient treated with gene therapy for sickle cell disease. Blood Adv. 2020;4(9):2058–2063. doi: 10.1182/bloodadvances.2019001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghannam JY, et al. Baseline TP53 mutations in adults with SCD developing myeloid malignancy following hematopoietic cell transplantation. Blood. 2020;135(14):1185–1188. doi: 10.1182/blood.2019004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman RL, et al. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157–170. doi: 10.1016/j.stem.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465):eaan4673. doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abelson S, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai P, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bick AG et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586(7831):763–768. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaiswal S, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laconi E, et al. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br J Cancer. 2020;122(7):943–952. doi: 10.1038/s41416-019-0721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liggett LA, Sankaran VG. Unraveling hematopoiesis through the lens of genomics. Cell. 2020;182(6):1384–1400. doi: 10.1016/j.cell.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ware RE, et al. Sickle cell disease. Lancet. 2017;390(10091):311–323. doi: 10.1016/S0140-6736(17)30193-9. [DOI] [PubMed] [Google Scholar]
- 19.Ware RE, Dertinger SD. Absence of hydroxyurea-induced mutational effects supports higher utilisation for the treatment of sickle cell anaemia. Br J Haematol. 2021;194(2):252–266. doi: 10.1111/bjh.17323. [DOI] [PubMed] [Google Scholar]
- 20.Jones RJ, DeBaun MR. Leukemia after gene therapy for sickle cell disease: insertional mutagenesis, busulfan, both or neither. Blood. 2021;138(11):942–947. doi: 10.1182/blood.2021011488. [DOI] [PubMed] [Google Scholar]
- 21.Pincez T, et al. Clonal hematopoiesis in sickle cell disease. Blood. 2021;138(21):2148–2152. doi: 10.1182/blood.2021011121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergstrom EN, et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics. 2019;20(1):685. doi: 10.1186/s12864-019-6041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual WGS data for TOPMed whole genomes, individual-level harmonized phenotypes, harmonized germline variant call sets, and the CH somatic variant call sets are available through restricted access via the NCBI’s database of Genotypes and Phenotypes (dbGaP). Accession numbers for these data sets have been described in our prior study (14), except for REDS-III_Brazil (phs001468), OMG_SCD (phs001608), and walk_PHaSST (phs001514). Deidentified variant calls are available from the authors upon request.