Abstract
Liver cancer presents divergent clinical behaviors. There remain opportunities for molecular markers to improve liver cancer diagnosis and prognosis, especially since tRNA-derived small RNAs (tsRNA) have rarely been studied. In this study, a random forests (RF) diagnostic model was built based upon tsRNA profiling of paired tumor and adjacent normal samples and validated by independent validation (IV). A LASSO model was used to developed a seven-tsRNA-based risk score signature for liver cancer prognosis. Model performance was evaluated by a receiver operating characteristic curve (ROC curve) and Precision-Recall curve (PR curve). The five-tsRNA-based RF diagnosis model had area under the receiver operating characteristic curve (AUROC) 88% and area under the precision–recall curve (AUPR) 87% in the discovery cohort and 87% and 86% in IV-AUROC and IV-AUPR, respectively. The seven-tsRNA-based prognostic model predicts the overall survival of liver cancer patients (Hazard Ratio 2.02, 95% CI 1.36–3.00, P < 0.001), independent of standard clinicopathological prognostic factors. Moreover, the model successfully categorizes patients into high-low risk groups. Diagnostic and prognostic modeling can be reliably utilized in the diagnosis of liver cancer and high-low risk classification of patients based upon tsRNA characterization.
Keywords: Diagnosis, Liver cancer, Prognosis, Random forests, tRNA-derived small RNAs
Introduction
Primary liver cancer, comprised mainly of hepatocellular carcinoma (HCC), remains the sixth most common cancer and the second largest cause of cancer mortality in the world.1 The highest incidence rates of liver cancer alter geographically and parallels with the prevalence of viral hepatitis. China accounts for about 50% of all incidences of liver cancer.2 Chronic hepatitis B virus (HBV) infection is the leading cause of HCC in Asia, in contrast chronic hepatitis C virus (HCV), alcoholic cirrhosis and non-alcoholic steatohepatitis (NASH) are the main causes in the Western world.3 Due to the strong compensatory function of the liver and often undetected early symptoms, liver cancer remains difficult-to-detect until the disease has progressed to an advanced stage.4,5 In China, the five-year survival rate for liver cancer patients is 12.5%, while in the United States, the 2-year survival rate is less than 50% and 5-year survival is only 10%.6 Early identification and timely treatment can significantly improve patient survival rate. Cancer biomarkers can play an important role in the early diagnosis and prognosis of liver cancer.
Transfer RNA (tRNA)-derived small RNA (tsRNA) are a novel regulatory, small non-coding RNA generated from precursor or mature tRNAs and participate in diverse physiological and pathological processes.7, 8, 9 tsRNA is produced by cutting specific sites of tRNA or pre-TRNA and their expression is not consistent with corresponding tRNA levels. This suggests that tsRNAs are not a degradation product, but a precisely regulated non-coding RNA. tsRNAs can be grouped into three distinctive classes, inferring tRNA-derived small RNAs with the characteristic 3’ poly U residues, mature tRNA-derived fragments (tRFs), or tRNA-derived stress-induced RNA (tiRNA). Emerging research has shown that tRNA-derived small RNA (tsRNA) disorders occur in many cancers and that their expression is regulated during cancer development and staging. Furthermore, evidence suggests that tsRNA can influence cellular processes such as cell proliferation, apoptosis, translation inhibition, epigenetic inheritance, and neuronal function, implicating them as a potential biomarker in human diseases.10
However, the presence of tsRNAs in tissue and their diagnostic potential remain unexplored. Here, we demonstrate the presence and expression pattern of tsRNAs within liver cancer tissue, highlighting their potential for cancer diagnosis and prognosis.
Methods
Dataset
The miRNA expression, clinical data, and miRNA-seq binary alignment map (BAM) files were collected from The Cancer Genome Atlas (TCGA-LIHC and TCGA-CHOL) project on 1 September 2020. Among 379 patients diagnosed with liver carcinoma, the median age was 61(IQR, 51–59.14), 66.2% were males, Asians and Caucasian accounted for 42.4% and 50.6%, respectively. A total of 189 (49.8%) individuals were diagnosed with Stage I, 94 (24.8%) with Stage II, 86 (22.6%) with Stage III, and 10 (2.6%) with Stage IV. The training set and internal validation were randomly separated according to the ratio of 3:2. In the independent validation dataset (GSE76903),11 the patient's median age is 50, and males account for 85% (Table 1).
Table 1.
Demographic and clinical characteristic of patients with liver cancer in discovery Corhort1 set (TCGA) and independent validation Corhot2 (GSE76903).
| Cohort | TCGA | GSE76903 |
|---|---|---|
| Num. of patients | 379 | 20 |
| Adjacent Normal | 57 | 20 |
| Primary Tumor | 57 | 20 |
| Age in years, median (IQR) | 61 (51–59.14) | 50 (45.25–60.25) |
| Sex | ||
| Male, count (%) | 251 (66.2) | 17 (85) |
| Race | ||
| Asian | 161 | 20 |
| White | 192 | |
| Black or African American | 15 | |
| American Indian or Alaska Native | 2 | |
| Not Reported | 9 | |
| Treatment Type | ||
| Radiation Therapy | 181 | Not Reported |
| Pharmaceutical Therapy | 198 | |
| Stage | ||
| Stage I | 189 | Not Reported |
| Stage II | 94 | |
| Stage III | 86 | |
| Stage IV | 10 | |
| Primary Diagnosis | ||
| Hepatocellular carcinoma | 331 | 20 |
| Cholangiocarcinoma | 48 | |
| Events | ||
| Death | 130 | Not Reported |
| Alive | 249 | |
| Follow Up time (days, IQR) | 575 (308.5–1106.5) | Not Reported |
Sequencing data analysis
All sequencing data were aligned using sRNAtool to extract tsRNA expression.12 Briefly, all sequencing files were converted to collapsed FASTA format after removal of adapters. The TCGA miRNA-seq BAM files are files that have removed adapters, can be converted directly. Unprocessed FASTQ format reads had adapters trimmed and filtered for ≥16 nucleotides utilizing Cutadpt 2.1.13 Missing values was imputed by MetImp 1.2.14
Statistical analysis
We compared adjacent normal and tumor tissues using paired student t-test. For survival analyses, the Kaplan–Meier method was utilized to analyze the correlation between factors and disease-free survival, and the log-rank test to examine survival curve. Multivariate survival analysis was performed using Cox regression. Diagnostic and prognostic samples, respectively, analyze the correlation between tsRNAs and miRNAs. Then select miRNAs with P < 0.001 through Spearman correlation for further analysis. The Mantel test was used in two matrices correlation test. miRNA pathway analysis was performed using DIANA tools.15 All analyses were performed with SPSS version V26 (IBM, Armonk, NY), Python (version 3.8.5), and R software (version 4.0.2).
Random forests analysis and LASSO regression
Random forests (RF) is a famous and influential statistical classifier that has been well established in biology disciplines. Compared to traditional linear regression, RF has excellent accuracy in classification and determining variable importance.16 We put statistically significant tsRNAs into the RF to select relevant diagnosis biomarkers and built a multi-tsRNA based diagnostic model. Furthermore, we applied an independent validation dataset to test the diagnostic model. “randomForest” package in R software was utilized to conduct RF analysis.
We applied the LASSO regularized linear model to solve the potential overfitting affected by high dimensional ncRNA expression associated with the small number of samples.17 We performed the LASSO Cox regression model to pick the most valuable prognostic tsRNAs associated with liver cancer and constructed a multi-tsRNA-based risk score signature for predicting the disease-free survival of patients. Ten-fold cross-validation was implemented to measure the risk score model. The 'scikit-learn' package in python was used for LASSO regression analysis.
Results
Identifying candidate diagnostic tsRNAs based on miRNA-seq profiling
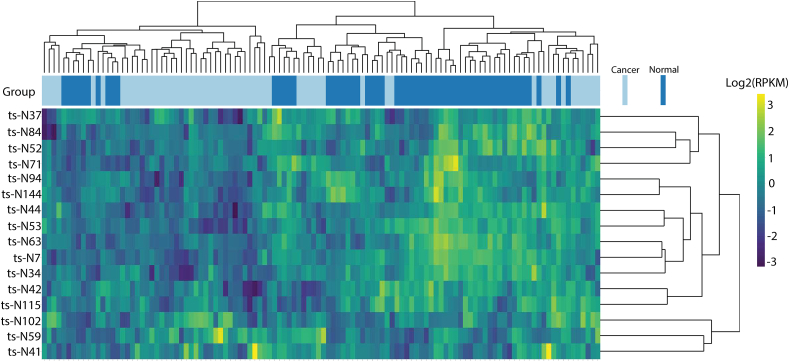
Figure S1 summarizes the workflow for this study and has been described in Methods. 57 paired liver primary tumor and adjacent normal tissues were analyzed by miRNA-seq in the discovery set (TCGA dataset) and differentially expressed tsRNAs in tumors and normal tissue were examined (Fig. 1). Our evaluation revealed 14 tsRNAs whose expression was significantly different by paired student t-test after adjusting for false discovery rate (FDR) (Table 2). Three tsRNAs (ts-N102, ts-N59, and ts-N41) were found to be upregulated in cancer with a foldchange of 2.046, 2.079, and 2.532, respectively; all other tsRNAs were reduced in cancer. In addition, two tsRNAs (ts-N71 and ts-N44) were found to be significantly downregulated in cancer vs normal (both P < 0.05), however these tsRNAs failed to maintain significance when FDR-adjusted (both P = 0.122) and were removed from further analysis. Due to ambiguity in naming conventions across tsRNA databases, we have included both our naming method, as well as Mintbase.
Figure 1.
tsRNA expression analysis of liver cancer diagnosis. Unsupervised hierarchical clustering of all significant tsRNA markers selected for use in the diagnostic model. Each row is tsRNA, and the column is the patient sample.
Table 2.
A list of top 16 tsRNAs that p-value less than 0.05 with tRF sequences when the paired student t-test was evaluated.
| ID | Mintbase ID | tRF Sequences (5′–3′) | P Values | Fold Change (C/N) | FDR |
|---|---|---|---|---|---|
| ts-N7 | NA | GCCCGGATGATCCTCAGTGGTCTGGGGTGCAGGCTTC | 2.91321E-09 | 0.351073565 | 1.19E-07 |
| ts-N63 | tRF-22-RKVP4P9LL | GGGGGTATAGCTCAGTGGTAGA | 2.98038E-07 | 0.420801509 | 6.11E-06 |
| ts-N144 | tRF-18-897PVP04 | TCCTCGTTAGTATAGTGG | 1.58555E-05 | 0.458798884 | 0.000217 |
| ts-N53 | tRF-19-6S7P4PK4 | GGCCGGTTAGCTCAGTTGG | 4.83256E-05 | 0.407564036 | 0.000495 |
| ts-N102 | tRF-18-8R1546D2 | TCCCCAGTACCTCCACCA | 7.6764E-05 | 2.046345387 | 0.000629 |
| ts-N42 | tRF-19-QR18LOJ4 | GCTCCAGTGGCGCAATCGG | 0.000199338 | 0.317559469 | 0.001248 |
| ts-N94 | tRF-18-07QSNHD2 | ACCCTGCTCGCTGCGCCA | 0.000213077 | 0.657910809 | 0.001248 |
| ts-N34 | tRF-20-79MP9P9 M | GTTTCCGTAGTGTAGTGGTC | 0.00031426 | 0.517865005 | 0.001611 |
| ts-N59 | tRF-20-HDK2RSI2 | ATAACCCAGAGGTCGATGGA | 0.000415514 | 2.079287758 | 0.001738 |
| ts-N84 | tRF-28-HJ83RPFQZD0M | ATAGCTCAGTGGTAGAGCATTTGACTGC | 0.000423901 | 0.636436975 | 0.001738 |
| ts-N52 | tRF-25-0P58309NDJ | ACCAGGATGGCCGAGTGGTTAAGGC | 0.002677032 | 0.634413418 | 0.009978 |
| ts-N115 | tRF-17-WSNKP92 | TCTCGCTGGGGCCTCCA | 0.006990814 | 0.775397813 | 0.023885 |
| ts-N37 | tRF-29-RKVP4P9L5FKP | GGGGGTATAGCTCAGTGGTAGAGCATTTG | 0.011555279 | 0.486538973 | 0.036444 |
| ts-N41 | tRF-20-6S7P4PZ3 | GGCCGGTTAGCTCAGTTGGT | 0.013231647 | 2.532122558 | 0.03875 |
| ts-N71 | tRF-20-73VL4YMY | GTGGTTAGTACTCTGCGTTG | 0.045561725 | 0.31115994 | 0.122153 |
| ts-N44 | tRF-24-S3M8309N0Y | GTAGTCGTGGCCGAGTGGTTAAGG | 0.04766928 | 0.711120108 | 0.122153 |
Constructing and validating the multi-tsRNAs based diagnostic model
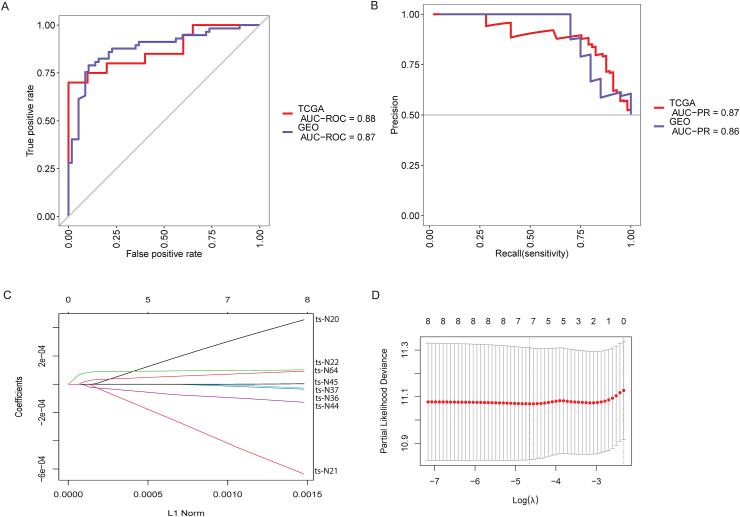
A random forest (RF) model using the 14 statistically significant tsRNAs was then constructed. According to the RF mean decrease of accuracy, five tsRNAs were selected, including up-regulated ts-N102 and down-regulated ts-N7, ts-N94, ts-N84, and ts-N37 in tumor tissue. Out-of-bag (OOB) estimations were applied to evaluate predicted error. We assessed the model performance by a receiver operating characteristic curve (ROC curve) and Precision-Recall curve (PR curve). We found that to differentiate primary tumor from normal, the five-tsRNA-based model has an area under the ROC (AUROC) 88% (Fig. 2A) and area under the precision–recall curve (AUPR) 87% (Fig. 2B). To further test the classification model, we also calculated a diagnostic RF-score for the independent validation dataset (GEO dataset) based on the OOB predicted probabilities. The IV-AUROC and IV-AUPR was 87% and 86%, respectively.
Figure 2.
Random forest diagnostic model and LASSO selection. (A) ROC of the diagnostic prediction model with tsRNA markers in the discovery data (TCGA) and independent validation data sets (GEO dataset). (B) PR curve in the discovery data (TCGA) and independent validation data sets (GEO dataset). (C) LASSO coefficient profiles of the liver-cancer-associated tsRNAs. (D) Seven tsRNAs selected by LASSO Cox regression analysis.
Constructing and validating the multi-tsRNAs based prognostic model
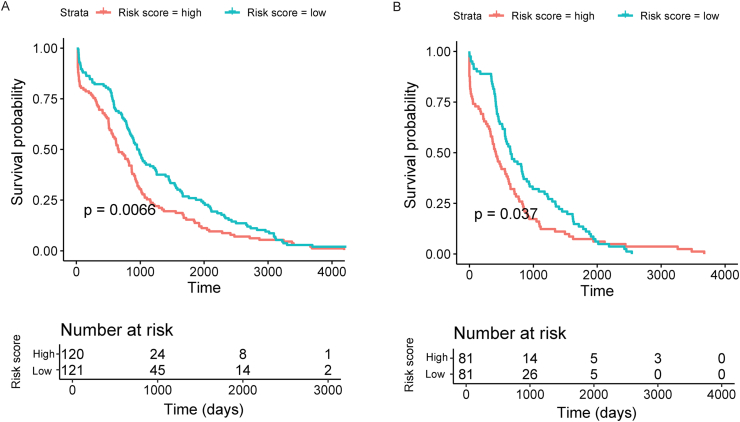
A prognostic model was then built from a TCGA dataset of 379 liver cancer patients (Table 1). Univariate Cox regression analyses of overall survival was utilized and identified 6 significantly associated tsRNAs (Table 3). Two tsRNAs, ts-N20 and ts-N45, trended towards a significance (P < 0.1) with overall survival. A LASSO Cox regression model was adopted to build a prognostic model and included seven of the eight tsRNAs (ts-N20, ts-N21, ts-N22, ts-N36, ts-N37, ts-N44, and ts-N64, Fig. 3). Applying the LASSO Cox regression models, risk score was estimated for each patient based on individualized values of five tsRNAs: risk score = (2.99 × ts-N20) - (4.16 × ts-N21) + (0.96 × ts-22) -(0.132 × ts-N36) - (0.058 × ts-N37) -(0.93 × ts-N44) +(0.7 ∗ ts-N64). Individuals were dichotomized by risk score into low risk (score < 1244.2) or high risk. Classification and survival status were assessed by Log-rank test and low risk individuals were found to have increased survival when compared to high risk in both training (P = 0.0066, Fig. 3A) and validation (P = 0.037, Fig. 3B) groups. Moreover, Figure S3 shows the four stages' survival plot corresponding to the risk model. There are significant differences between the high and low risks in stage II, and stage IV (P < 0.05).
Table 3.
A list of top 8 tsRNAs that p-value less than 0.1 when the univariate Cox models were applied.
| ID | Mintbase ID | tRF Sequences (5′–3′) | P Values | Hazard Ratio | 95% CI |
|---|---|---|---|---|---|
| ts-N20 | tRF-25-395P4PN3FJ | CCTTCGATAGCTCAGCTGGTAGAGC | 0.081739 | 1.3388 | 0.964–1.86 |
| ts-N21 | tRF-23-395P4PN3X | CCTTCGATAGCTCAGCTGGTAGA | 0.008897 | 1.5525 | 1.115–2.163 |
| ts-N22 | tRF-18-YSQSD2D2 | TTCCGGCTCGAAGGACCA | 7.23E-05 | 0.5108 | 0.364–0.716 |
| ts-N36 | tRF-23-HDK2RSI20K | ATAACCCAGAGGTCGATGGATCG | 0.021841 | 1.4735 | 1.056–2.057 |
| ts-N37 | tRF-29-RKVP4P9L5FKP | GGGGGTATAGCTCAGTGGTAGAGCATTTG | 0.016874 | 1.4965 | 1.073–2.087 |
| ts-N44 | tRF-24-S3M8309N0Y | GTAGTCGTGGCCGAGTGGTTAAGG | 0.008527 | 1.5573 | 1.118–2.168 |
| ts-N45 | tRF-30-87R8WP9N1EWJ | TCCCTGGTGGTCTAGTGGTTAGGATTCGGC | 0.085083 | 1.3346 | 0.96–1.855 |
| ts-N64 | tRF-18-8R6546D2 | TCCCCGGCACCTCCACCA | 0.007235 | 0.6342 | 0.453–0.888 |
Figure 3.
Risk score model by the seven-tsRNA-based signature, Kaplan–Meier survival in training (A), internal validation (B). P values calculated by the log-rank test.
A k-fold cross-validation is used to evaluate productive models by partitioning the dataset into k equal subsets. Of each k subset, a single subset is used as validation and the remaining k-1 subsets are designated for training. Cross-validation is then performed k times, with each subset being used once for validation. The k results are then averaged for create a single, overall estimation. The strength of k-fold cross-validation is that the entirety of the dataset is used as both training and validation with each sample being used as validation exactly once. The results of 10-fold cross-validation of this model is summarized in Table S1.
Furthermore, we performed a multivariate Cox regression analysis of the tsRNA-based model with overall survival, the prognostic model remained an independent prognostic factor (Fig. S2. HR 2.02, 95% CI 1.36–3.00, P < 0.001).
tsRNA associated miRNA analysis
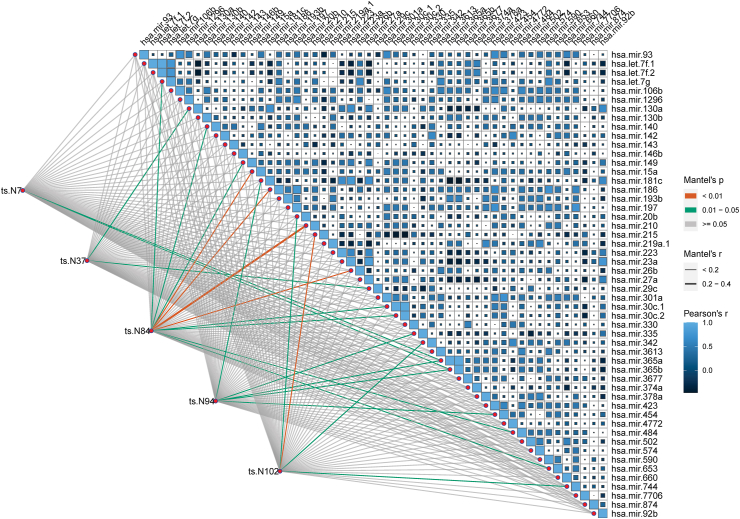
To better understand the functions of tsRNA, we constructed a correlation analysis to find miRNAs that are highly related. Fig. 4 shows miRNAs that are profoundly correlated with five diagnostic tsRNAs. Based on Mantel's P < 0.01, ts-N102 are highly associated with has-mir-215, ts-N84 is highly correlated with hsa-mir-15a, hsa-mir-186, hsa-mir-210 and hsa-mir-26 b. We further performed a pathway analysis on these miRNAs. The analysis results show that hsa-mir-26 b, hsa-mir-15a, hsa-mir-186 are mainly involved in fatty acid synthesis and metabolism, Viral carcinogenesis, and other pathways. Details are showing in Fig. S4. Among prognostic tsRNAs correlated miRNAs, hsa-mir-29b-1, hsa-mir-362, hsa-mir-3653, hsa-mir-425, and hsa-mir-3607 are highly correlated with 3 or more tsRNAs, respectively. Notably, hsa-mir-215 is related to diagnostic biomarker ts-N102 and is associated with prognostic biomarker ts-N64, more details shown in Fig. S3. We further enriched the miRNAs related to prognosis and found that the pathways related to fatty acid synthesis and metabolism. Details in Fig. S4.
Figure 4.
Correlation analysis of diagnostic tsRNA and miRNA. The lower-left corner connection is the correlation between tsRNAs and miRNAs, and the upper-right corner is the correlation analysis between miRNAs and miRNAs. miRNA data down from TCGA-LIHC and TCGA-CHOL project.
Discussion
Although over 90% of the human genome may be involved in transcriptional processes, less than 2% of the whole genome is comprised of protein-coding sequences. Therefore, noncoding RNA (ncRNA) is a vital part of the human transcriptome.18 In recent years, ncRNA, such as miRNA, lncRNA, and snoRNA, has been widely studied in cancer diagnosis and prognosis. However, tsRNA, a novel type of ncRNA, has rarely been investigated, especially in regard to its function in cancer.
In this study, we developed and validated a novel diagnostic and prognostic tool based on tsRNAs for cancer diagnosis which can improve the prediction of patients' overall survival with liver cancer. Our results revealed that the diagnostic tool could successfully distinguish normal and malignant tissue across discovery and validation datasets. The prognostic tool classifies patients into low-risk and high-risk groups with great contrasts in 5-year disease-free survival. Moreover, this suggested classifier can predict the survival of patients as an independent factor and is significantly more reliable than other clinicopathological risk factors.
Since there have been only a limited number of studies involving a small number of tsRNAs, the general and precise mechanisms of action of tsRNAs are not very clear.19 Nevertheless, tsRNA have still been shown to share some characteristics in common with miRNAs in regulating mRNA stability, binding to proteins, and controlling RNA reverse transcription.20 Our research explored correlations between miRNAs and tsRNA to identify candidate regulatory pathways in which tsRNA may participate. Among diagnostic tsRNAs, ts-N102 is significantly up-regulated in cancer tissues, while the highly related hsa-mir-215 is a tumor suppressor in colorectal cancer, and multiple myeloma.21, 22, 23 We infer that ts-N102 is a proto-oncogene and can inhibit the expression of tumor suppressor hsa-mir-215. ts-N84 has a significant correlation with many miRNAs. Among them, hsa-mir-210 has the highest correlation. Studies have shown that hsa-mir-210 promotes venous metastasis in hepatocellular carcinoma when highly expressed, and it participates in hepatic ischemic-reperfusion injury as part of a negative feedback loop with Mothers against decapentaplegic homolog 4 gene (SMAD4).24,25 In regard to the prognostic significance of tsRNA, the regulatory network is more complicated. For instance, let us consider ts-N22 and the related hsa-mir-331 and hsa-mir-33a as examples. ts-N22 is a tsRNA with protective properties (HR = 0.51) in liver cancer, and patients who express ts-N22 have a higher survival rate. The related hsa-mir-331 and hsa-mir-33a are tumor suppressors. Hsa-mir-331 can regulate expression of neuropilin-2 to inhibit glioblastoma cell migration.26 Depletion of hsa-mir-33a is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma (HCC).27 Furthermore, miR-33a-5p can interfere with the cisplatin resistance of HCC cells.28
According to the enrichment analysis results, miRNAs highly related to liver cancer tsRNA are mainly enriched in fatty acid synthesis and metabolism pathways. Researches show that Long-chain acyl-CoA synthetases (ACSLs) effective for activation of the most abundant long-chain fatty acids are usually deregulated in cancer.29 Fatty acid oxidation as a fuel in the metabolic adaptation triggered by β-catenin oncogenic activation in hepatocytes.30 This evidence indicates that tsRNA may be a regulatory factor regulating the fatty acid synthesis and metabolism and potentially as a liver cancer treatment target.
Our study has some limitations. First, in the diagnostic model, we only used one independent validation data set. Second, although a large sample was used in the prognostic model, no suitable independent validation set was found. Third, although we inferred that ts-N37 is a potential tumor suppressor gene, more experiments are needed to verify this, and it also lays the foundation for our future work. Finally, tsRNA fragments are relatively small, and non-specific bands may be generated during PCR amplification, and a more appropriate method needs to be selected.
Author contributions
Youping Deng and Gang Huang had the idea and launched the study. Yi Zuo and Shaoqiu Chen, Ling Hu and Lingling Yan gathered and processed data. Shaoqiu Chen, Yi Zuo Scott Bowler, Emory Zitello drafted the manuscript, and all authors reviewed the manuscript and approved the version for publication.
Conflict of interests
We declare no competing interest.
Funding
This work was also supported by the NIH Grants (No. 5P30GM114737, P20GM103466, U54MD007584 and 2U54MD007601), and Natural Science Foundation of Hubei Province (No. 2019CFB417).
Acknowledgements
The results here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We also thank the Dr. Jin Gu, providers of GSE76903 data in Tsinghua University.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.01.006.
Contributor Information
Gang Huang, Email: huangg@sumhs.edu.cn.
Youping Deng, Email: dengy@hawaii.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Canc. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn K.A., Petrick J.L., London W.T. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19(2):223–238. doi: 10.1016/j.cld.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medavaram S., Zhang Y. Emerging therapies in advanced hepatocellular carcinoma. Exp Hematol Oncol. 2018;7(1):17. doi: 10.1186/s40164-018-0109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsochatzis E., Meyer T., O'Beirne J., Burroughs A.K. Transarterial chemoembolisation is not superior to embolisation alone: the recent European Association for the Study of the Liver (EASL) - European Organisation for Research and Treatment of Cancer (EORTC) guidelines. Eur J Cancer. 2013;49(6):1509–1510. doi: 10.1016/j.ejca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Chong R.J., Abdullah M.S., Hossain M.M., Telisinghe P.U., Chong V.H. Rising incidence of primary liver cancer in Brunei Darussalam. Asian Pac J Cancer Prev. 2013;14(6):3473–3477. doi: 10.7314/apjcp.2013.14.6.3473. [DOI] [PubMed] [Google Scholar]
- 6.Golabi P., Fazel S., Otgonsuren M., Sayiner M., Locklear C.T., Younossi Z.M. Mortality assessment of patients with hepatocellular carcinoma according to underlying disease and treatment modalities. Medicine. 2017;96(9):e5904. doi: 10.1097/MD.0000000000005904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balatti V., Nigita G., Veneziano D., et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114(30):8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Chen L., Gu J., et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q., Ding C., Lang X., Guo G., Chen J., Su X. Small noncoding RNA discovery and profiling with sRNAtools based on high-throughput sequencing. Briefings Bioinformat. 2021;22(1):463–473. doi: 10.1093/bib/bbz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 14.Wei R., Wang J., Su M., et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8(1):663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genuer R., Poggi J.-M. Tuleau-malot CJPrl. Variable selection using random forests. Pattern Recognit Lett. 2010;31(14):2225–2236. [Google Scholar]
- 17.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Stat Soc. 1996;58(1):267–288. [Google Scholar]
- 18.Weng M., Wu D., Yang C., et al. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108–120. doi: 10.1016/j.trsl.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L., Li J., Gong Y., et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18(1):74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin F., Guo Z. Emerging role of a novel small non-coding regulatory RNA: tRNA-derived small RNA. ExRNA. 2019;1(1):39. [Google Scholar]
- 21.Vychytilova-Faltejskova P., Merhautova J., Machackova T., et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogen esis. 2017;6(11):1–14. doi: 10.1038/s41389-017-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Zhang Y., Huang C., Lin S. miR-215-5p is an anticancer gene in multiple myeloma by targeting RUNX1 and deactivating the PI3K/AKT/mTOR pathway. J Cell Biochem. 2020;121(2):1475–1490. doi: 10.1002/jcb.29383. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H., Chen J., Chen J., et al. miR-192/215-5p act as tumor suppressors and link Crohn's disease and colorectal cancer by targeting common metabolic pathways: An integrated informatics analysis and experimental study. J Cell Physiol. 2019;234(11):21060–21075. doi: 10.1002/jcp.28709. [DOI] [PubMed] [Google Scholar]
- 24.Pan W.M., Wang H., Zhang X.F., et al. miR-210 participates in hepatic ischemia reperfusion injury by forming a negative feedback loop with SMAD4. Hepatology. 2020;72(6):2134–2148. doi: 10.1002/hep.31221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji J., Rong Y., Luo C.L., et al. Up-regulation of hsa-miR-210 promotes venous metastasis and predicts poor prognosis in hepatocellular carcinoma. Front Oncol. 2018;8:569. doi: 10.3389/fonc.2018.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epis M.R., Giles K.M., Candy P.A., Webster R.J., Leedman P.J. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J Neuro-oncol. 2014;116(1):67–75. doi: 10.1007/s11060-013-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie R.T., Cong X.L., Zhong X.M., et al. MicroRNA-33a downregulation is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2018;15(4):4571–4577. doi: 10.3892/ol.2018.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng W., Tai Y., Zhao H., et al. Downregulation of miR-33a-5p in hepatocellular carcinoma: a possible mechanism for chemotherapy resistance. Med Sci Monit. 2017;23:1295–1304. doi: 10.12659/MSM.902692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y., Zhou J., Hooi S.C., Jiang Y.M., Lu G.D. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol Lett. 2018;16(2):1390–1396. doi: 10.3892/ol.2018.8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montagner A., Le Cam L., Guillou H.J.G. β-catenin oncogenic activation rewires fatty acid catabolism to fuel hepatocellular carcinoma. Gut. 2019;68(2):183–185. doi: 10.1136/gutjnl-2018-316557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.