Abstract
Brain tumors exhibit vast genotypic and phenotypic diversity depending on patient age and anatomical location. Hydrogels hold great promise as 3D in vitro models for studying brain tumor biology and drug screening, yet previous studies were limited to adult glioblastoma cells, and most studies used immortalized cell lines. Here we report a hydrogel platform that supports the proliferation and invasion of patient-derived brain tumor cell cultures (PDCs) isolated from different patient age groups and anatomical locations. Hydrogel stiffness was tuned by varying poly(ethylene-glycol) concentration. Cell adhesive peptide (CGRDS), hyaluronic acid, and MMP-cleavable crosslinkers were incorporated to facilitate cell adhesion and cell-mediated degradation. Three PDC lines were compared including adult glioblastoma cells (aGBM), pediatric glioblastoma cells (pGBM), and diffuse pontine intrinsic glioma (DIPG). A commonly used immortalized adult glioblastoma cell line U87 was included as a control. PDCs displayed stiffness-dependent behavior, with 40 Pa hydrogel promoting faster tumor proliferation and invasion. Adult GBM cells exhibited faster proliferation than pediatric GBM, and DIPG showed slowest proliferation. These results suggest both patient age and tumor location affects brain tumor behaviors. Adult GBM PDCs also exhibited very different cell proliferation and morphology from U87. The hydrogel reported here can provide a useful tool for future studies to better understand how age and anatomical locations impacts brain tumor progression using 3D in vitro models.
Keywords: in vitro 3D cancer models, glioblastoma, diffuse intrinsic pontine glioma, patient-derived brain tumor cell cultures, hydrogels
Graphical Abstract
1. INTRODUCTION
Brain tumors can affect patients of any age and occur in various anatomical locations [1]. Two highly aggressive brain tumors are glioblastoma (GBM), which are found in both adults and children, and diffuse intrinsic pontine glioma (DIPG), a childhood brain tumor. GBMs are the most aggressive and common malignant brain tumors (WHO grade IV) [2]. GBMs typically form in the cerebral hemispheres and can be characterized by various genomic alterations, including isocitrate dehydrogenase mutations, epidermal growth factor receptor (EGFR) amplification, phosphatase and tensin homolog deletion, and p53 mutations [3]. Although pediatric GBMs are histologically indistinguishable from adult GBMs, pediatric GBMs typically exhibit distinct abnormalities, such as K27M and G34V/R gain-of-function mutations in H3F3A, p53 overexpression, lower prevalence of EGFR amplification and overexpression, or platelet-derived growth factor receptor amplification [4–8]. GBM patients typically undergo surgical resection and a combination of radiotherapy and cytotoxic chemotherapy. Despite aggressive treatment regimens, adult and pediatric GBM patients face 5-year survival rates of 5% and 5–15%, respectively [9–11]. DIPG is a brain tumor found exclusively in pediatric patients, representing 10% of all childhood central nervous system tumors [12]. DIPGs are found in the pons, which controls the body’s most vital functions, such as breathing, blood pressure, and heart rate. K27M mutations in H3F3A occur in more than 70% cases of DIPG [13]. Since DIPGs are located in an extremely delicate region in the brain, surgical resection is impossible, and there are currently no FDA-approved chemotherapies for DIPG. Radiotherapy only provides temporary control of the tumor’s growth, and DIPG patients face a 5-year survival rate of less than 1% [14,15].
To better understand disease progression and improve clinical outcomes, there is a critical need for experimental models specifically optimized for various brain tumor types to elucidate tumor biology and screen drug candidates. The current gold standard models for studying brain tumor cell behavior are 2D culture and mouse models. 2D culture is low cost and convenient, but suffers from a few key limitations. First, the stiffness of 2D dishes is on the GPa scale, which is orders of magnitude higher than the stiffness of in vivo tissues. Furthermore, 2D culture fails to recapitulate the three-dimensionality and niche cues of the extracellular matrix. Moreover, 2D culture does not permit studies of tumor cell invasion, a key hallmark of many tumors including GBM and DIPG. On the other end of the spectrum, animal models are arguably the closest approximation of the in vivo tumor niche [16], but are limited by poor scalability and difficulty for mechanistic studies due to complex variables.
To bridge the gap between 2D culture and animal models, recent research has demonstrated that 3D culture can provide an alternative tool to study tumor progression and drug screening [17–19], with 3D culture better preserves tumor cell phenotype than 2D culture [18]. Hydrogels have been employed to engineer 3D in vitro brain tumor models utilizing naturally-derived materials [20–22]. While naturally-derived materials are biomimetic, they are subject to limited tunability and batch-to-batch variability. Unlike naturally-derived polymers, synthetic polymers offer higher degree of tunability and reproducibility [23,24]. Poly(ethylene-glycol) (PEG) is an FDA-approved synthetic polymer widely employed in fabricating hydrogels for tissue engineering applications. PEG hydrogels allows tunable physical and biochemical properties, and have been used for growing various cancer cells in 3D including epithelial ovarian cancer cells [23], melanoma cells [24], and immortalized adult GBM cells [25].
Previous work on engineering 3D in vitro brain tumor models are largely limited to adult GBM tumor cells [20–22, 25–28], yet there is no 3D in vitro brain models designed for brain tumors from different patient age groups and anatomical locations. Furthermore, most previous literature primarily employed immortalized cell lines as model cell types, which differ phenotypically and genotypically from primary tumors [16,29]. To address this critical gap of technology, the goal of this study is to engineer a PEG-based hydrogel platform as 3D brain tumor niche that can support patient-derived brain tumor cell cultures (PDCs) isolated from adult and pediatric brain tumors. Unlike immortalized cell lines, PDCs have been shown to maintain genomic features of the original parental tumor [30]. In the present study, we encapsulated PDCs from different patient age groups (adult vs. pediatric) and anatomical locations (cortex vs. pons).
Based on observations in previous literature [21,22,25,28], we hypothesize that hydrogel stiffness will impact PDC proliferation and invasion in 3D. To determine an optimal hydrogel stiffness, we employed a PEG-based hydrogel platform, which allows tunable stiffness that mimics brain tissues [25]. The stiffness of human brain tissue ranges from 100 to 1000 Pa, and edema in brain can lead to further decrease in tissue stiffness to 45% of normal brain [31,32]. However, how anatomical locations impact brain stiffness remain largely unknown. While one study shows adult GBM led to increasing stiffness up to 26kPa [25], another study reports lower stiffness due to peritumoral edema [32]. Stiffness characterization on DIPG and pediatric GBM is lacking due to tissue scarcity. Given all brain tumor start initially from normal brain tissues, we chose hydrogels with stiffness that covers the full range of normal brain stiffness. Our criteria to identify an optimal hydrogel formulation was based on its ability to support tumor proliferation and invasion in 3D [33,34]. PTDX cells were encapsulated in 3D hydrogels with varying stiffness (40 Pa or 1000 Pa). Cell proliferation and spreading were monitored over 21 days using brightfield microscopy. Using the optimized hydrogel formulation, we further characterize cell proliferation and spreading of all four cell lines using immunostaining and biochemical assays.
2. MATERIALS AND METHODS
2.1. Materials
8-arm PEG (MW ~ 40 kDa) was purchased from JenKem Technology USA (Allen, TX, USA). Linear PEG (MW ~ 1.5 kDa) was purchased from Sigma-Aldrich USA (St. Louis, MO, USA). RGD peptide (CRGDS) and MMP-cleavable peptide (KCGPQGIWGQCK) were purchased from Bio Basic, Inc (Amherst, NY, USA). Sodium hyaluronate (HA) (MW ~ 20–40 kDa) was purchased from Lifecore Biomedical (Chaska, MN, USA). All other reagents were obtained from Fisher Scientific (Pittsburgh, PA, USA) unless otherwise noted.
2.2. Cell culture
Adult GBM D-270 MG (aGBM) cells were provided by the lab of Gerald Grant at Stanford University Medical Center and was derived as previously reported [35,36]. aGBM D-270 MG were expanded in improved minimal essential zinc option medium (Life Technologies) with 10% fetal bovine serum (Gibco, Life Technologies) and 0.1% gentamicin (Life Technologies) at 37°C in 5% CO2 (media change every other day). Pediatric GBM SU-pcGBM-2 (pGBM) and DIPG SU-DIPG-VI (DIPG) cells were provided by the lab of Michelle Monje-Deisseroth at Stanford University. Patient-derived samples were obtained under an IRB-approved protocol and cultures were generated as previously described [37]. pGBM and DIPG cells were expanded in tumor stem medium consisting of Neurobasal (-A) (Life Technologies), B27 (-A) (Life Technologies), human EGF (20 ng/ml) (Shenandoah Biotech, Warwick, PA, USA), human b-FGF (20 ng/ml) (Shenandoah), human PDGF-AA (10 ng/ml) (Shenandoah), human PDGF-BB (10 ng/ml) (Shenandoah), and heparin (2 μg/ml) (StemCell Technologies, Vancouver, BC, Canada) at 37°C in 5% CO2 (media change once a week). U87-MG (U87) cells were obtained from ATCC (Manassas, VA, USA) and expanded in Dulbecco’s minimal essential medium (DMEM, Life Technologies, Carlsbad, CA, USA), supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, Life Technologies), 100 units/ml penicillin (Life Technologies), and 100 μg/ml streptomycin (Life Technologies) at 37°C in 5% CO2 (media change every other day).
2.3. Hydrogel fabrication
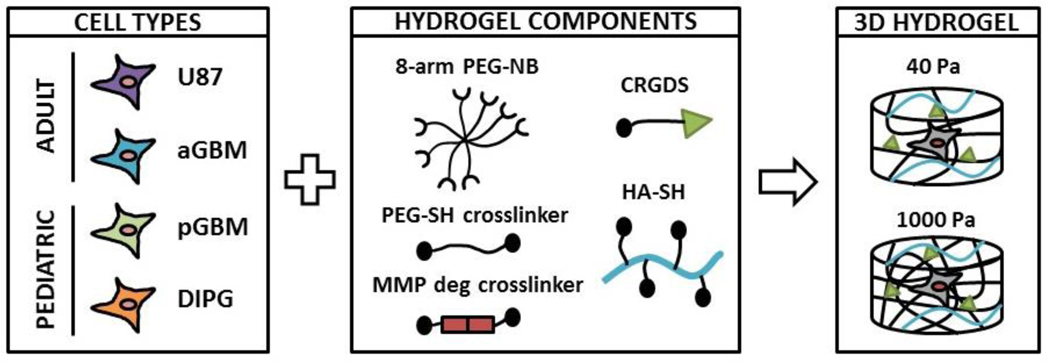
Hydrogels were fabricated using components as shown in Figure 1, which include 8-arm PEG-norbornene (40 kDa) (PEG-NB), linear PEG-dithiol (1.5 kDa) (PEG-SH), and thiolated hyaluronic acid (20 – 40 kDa) (HA-SH). All polymers were synthesized in house as previously reported [38–40]. For 8-arm PEG-NB, 6 arms were used for hydrogel crosslinking, and 2 arms were used for conjugation of biochemical cues. PEG-NB, PEG-SH, and an MMP-cleavable crosslinker (KCGPQGIWGQCK) were mixed at a molar ratio of 2:3:3 to allow crosslinking and cell-mediated degradation. To fabricate 1000 Pa hydrogels, 3% (w/v) PEG was used. To fabricate 40 Pa hydrogels, noncrosslinkable linear PEG (20 kDa) (16% w/v) was added to 3% PEG solution, which will diffuse out after hydrogel formation and further decrease crosslinking density. The hydrogels used in this study were prepared using the same formulation as reported by our lab previously [25, 41]. Specifically, 1000 Pa hydrogel was fabricated as reported in [25] and 40 Pa hydrogel was fabricated as reported in [41]. To support cell adhesion, RGD peptide (CRGDS) was chemically conjugated to PEG-NB at a final concentration of 0.914 mM. To mimic brain ECM content, HA-SH was chemically incorporated at a final concentration of 0.004% (w/v), which was selected based on reported values in human brain tissue [42]. To induce hydrogel formation via thiol-ene photopolymerization, hydrogel components were mixed together in the presence of photoinitiator Igracure D2959 (0.05% w/v, Ciba Specialty Chemicals, Tarrytown, NY, USA). Each hydrogel sample contained 75 μL of hydrogel precursor solution, which was loaded in a cylindrical-shaped mold (3 mm in height, 5 mm in diameter). To induce gelation, hydrogels were exposed to UV light (365 nm, 4mW/cm2) for 5 min at room temperature.
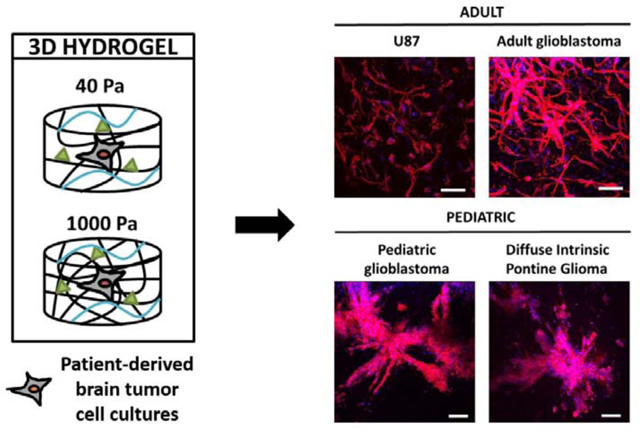
Figure 1. Schematic representation of experimental design.
3 different patient-derived brain tumor cell cultures (PDCs) were encapsulated in 3D hydrogels: aGBM (adult GBM), pGBM (pediatric GBM), and DIPG. An immortalized adult GBM cell line (U87) was also encapsulated as control. Hydrogel stiffness was modulated by tuning the PEG concentration (% w/v).
2.4. Cell encapsulation in 3D hydrogels
For encapsulation in hydrogels, trypsinized cells were resuspended in the hydrogel precursor solution at a final concentration of 0.5M cells/ml. 75 μL of the cell-containing hydrogel solution was pipetted into a cylindrical-shaped mold and UV crosslinked as described above. The samples were then cultured in growth medium as described above at 37°C in 5% CO2.
2.5. Cell viability
Initial cell viability was assessed 2 h after hydrogel encapsulation using Live/Dead Cell Viability Assay kit (Life Technologies). Live/Dead reagent was prepared per manufacturer’s instructions. Hydrogels were immersed in Live/Dead reagent solution for 40 minutes and imaged using a Zeiss fluorescence microscope (Zeiss, Oberkochen, Germany).
2.6. DNA content
Cell proliferation was measured by quantifying the DNA content on Days 1 and 21 using Quant-iT PicoGreen assay (Life Technologies). Briefly, lyophilized hydrogel samples (n = 3) were rehydrated and digested using papain (Worthington Biochemical Corp) at 60°C for 16 h. Samples were then vortexed and centrifuged at 10,000 rpm for 5 min at RT. The supernatant was used to measure DNA content per manufacturer’s instructions.
2.7. Immunohistochemical staining
Hydrogels were fixed in 4% PFA for 1 h at RT and washed in PBS. Samples were dehydrated overnight in 30% sucrose at 4°C and cryopreserved in OCT (Sakura Finetek USA, Torrance, CA, USA) in liquid nitrogen. Samples were then cryosectioned at −20°C (14 μm thickness). To stain for Ki67, samples were first permeabilized in PBS, pH 7.4 containing 0.4% Triton-X 100 for 1 h at RT. Then, samples were blocked in PBS, pH 7.4 containing 2% goat serum, 1% BSA, and 0.1% Triton-X 100 for 1 h at RT. Samples were then incubated overnight with rabbit polyclonal anti-Ki67 antibody (Abcam, Cambridge, UK) at 4°C, followed by secondary antibody (Alexa Fluor 488 goat anti-rabbit, Invitrogen) for 1 hr at RT. Cell nuclei were counterstained with Hoechst dye 33342 (Cell Signaling Technologies, Danvers, MA) for 1 h at RT. Samples were mounted (Vectashield, Vector Laboratories, Burlingame, CA), and images were taken using Zeiss fluorescence microscope (Zeiss, Jena, Germany). Quantification of Ki67 positive and negative nuclei was performed manually using ImageJ software.
2.8. F-actin staining for confocal imaging
Hydrogels were fixed in 4% PFA for 1 h at RT and washed in PBS. Cell permeabilization was performed using 0.1% Triton X-100 in PBS. Samples were blocked in 1% BSA, 0.1% Triton X-100 in PBS overnight at 37°C. F-actin and nuclei were stained using phalloidin-rhodamine (0.5 μg/ml, Sigma) and Hoechst 33342 dye (Cell Signaling Technologies), respectively, for 1 h at 37°C. Samples were washed with PBS and incubated in mounting media (Vector Laboratories) overnight at 4°C. Hydrogels were imaged using Leica SP5 upright laser scanning confocal microscope (Leica, Wetzlar, Germany). Maximal projections were obtained from 200 μm thick Z-stack with 2 μm step size.
2.9. RT-PCR
Gene expression levels of HIF1α were measured at days 1 and 10. Total RNA was isolated from hydrogels (n=3/group) using TRIzol (Life Technologies). cDNA was synthesized using SuperScript III First-Strand Synthesis kit (Life Technologies). RT-PCR was then performed using an Applied Biosystems 7900 Real-Time PCR system (Applied Biosystems, Life Technologies) using Power SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies). Relative expression levels of target genes were determined using the comparative CT method. Target gene expression was first normalized to a housekeeping gene, GAPDH, followed by a second normalization to the gene expression level in the control group (U87 Day 1).
2.10. Statistical analyses
GraphPad Prism (GraphPad Software, San Diego, CA, USA) was used to perform statistical analyses. Unpaired student’s t tests (assuming Gaussian distribution) and two-way analysis of variance (ANOVA) were used to determine statistical significance (p < 0.05). Error was reported as standard deviation unless otherwise noted.
3. RESULTS
3.1. Effects of matrix stiffness on tumor cell proliferation and invasion over time
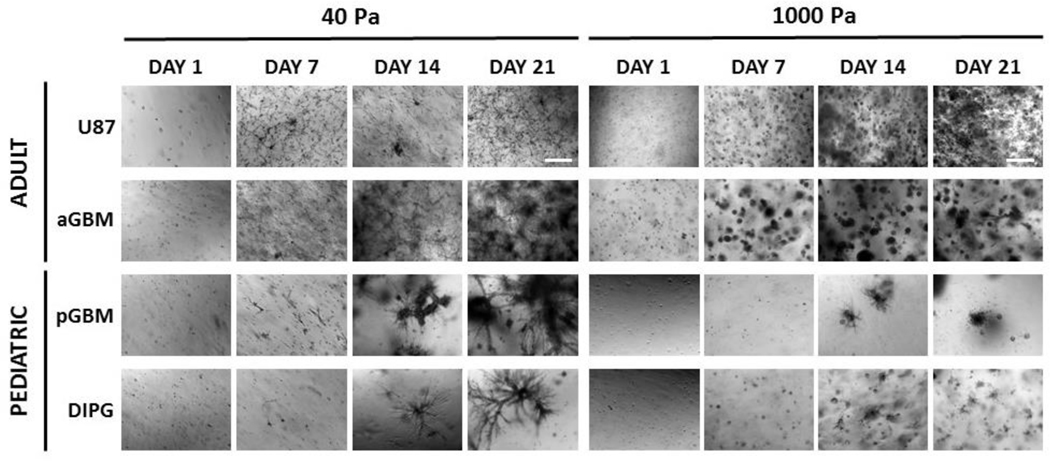
To determine an optimal stiffness that can support PDC cell fates in 3D, hydrogel stiffness was modulated to achieve stiffnesses of 40 Pa and 1000 Pa based on our previous work [25, 41], which fall within the range of edematous and normal brain tissue stiffness [31,32]. The various tumor cell types were homogeneously encapsulated as single cells in 3D hydrogels on day 1 with high cell viability (Figure S1). Using brightfield microscopy, significant differences in proliferation and aggregate formation based on stiffness were observed for the various cell types (Figure 2). aGBM proliferated and formed large cell aggregates in 40 Pa hydrogels, as the cells neared confluence throughout the hydrogel and formed an interconnected cell network. However, in 1000 Pa hydrogels, aGBM cell proliferation was attenuated, and cell aggregates were smaller and remained mostly isolated. In contrast, U87 cell proliferation and aggregate formation was more robust in 1000 Pa hydrogels, as compared to in 40 Pa hydrogels. In 1000 Pa hydrogels, U87 cells proliferated and formed many cell aggregates at a high density throughout the hydrogel by day 21. However, in 40 Pa hydrogels, U87 cells remained largely as isolated single cells, forming few cell aggregates. Similar to aGBM, both pGBM and DIPG cells demonstrated more robust aggregate formation in 40 Pa hydrogels. In 40 Pa hydrogels, pGBM cells formed large interconnected aggregates with long extensions. However, in 1000 Pa hydrogels, pGBM aggregates were smaller and more rounded by day 21. In 40 Pa hydrogels, DIPG cells formed large, highly diffusive cell aggregates with many branch points by day 21, but these aggregates were significantly smaller and isolated in 1000 Pa hydrogels. Furthermore, PDCs exhibited upregulated gene expression of HIF1α at day 10, which were significantly higher compared to U87. In contrast, there was no significant difference in HIF1α expression in U87 from day 1 to 10 (Figure S2).
Figure 2. Effect of matrix stiffness on brain tumor cell proliferation and invasion over time.
Patient-derived brain tumor cell cultures (PDCs) formed larger, more invasive cell aggregates in 40 Pa hydrogels after 21 days in culture. In contrast, U87 cell aggregate formation was more robust in 1000 Pa hydrogels. All images are brightfield images. Dark areas are dense cell aggregates. Scale bar = 500 μm.
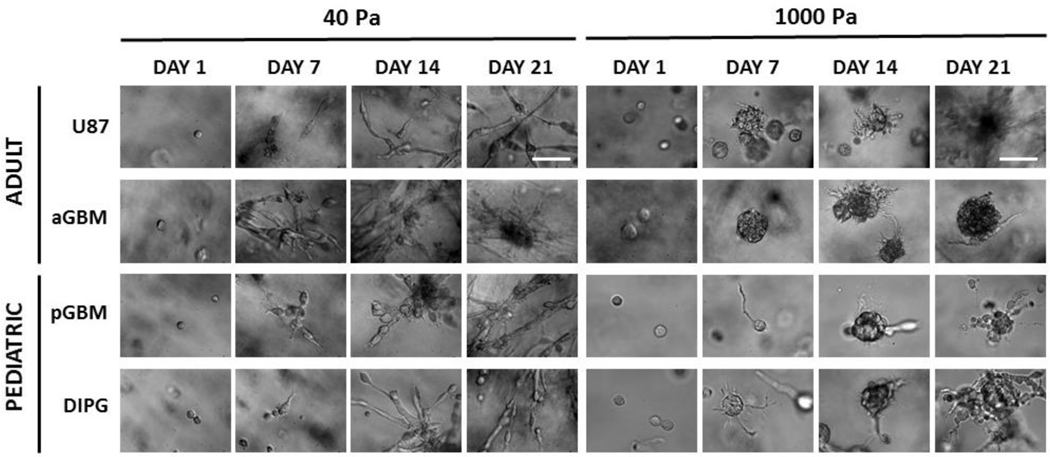
A key hallmark of GBM and DIPG cells is their extreme invasiveness throughout brain [33,43]. To identify an optimal hydrogel stiffness that can support tumor cell invasion, cell spreading and morphology were analyzed, as a measure of cell invasiveness, in 40 Pa and 1000 Pa hydrogels (Figure 3). In 40 Pa hydrogels, aGBM cells were significantly elongated by day 7 and demonstrated considerable spreading by day 21, but remained as dense, isolated aggregates in 1000 Pa hydrogels over 21 days. In contrast, U87 cells were able to spread significantly compared to aGBM by day 21 in the 1000 Pa hydrogel. In the 40 Pa hydrogels, both U87 and aGBM were able to spread to a comparable degree. For pGBM cells, in 40 Pa hydrogels, cells were more bipolar and elongated by day 7 and formed cell aggregates with branches composed of many elongated cells. However, in 1000 Pa hydrogels, pGBM cells formed very long, thin cell protrusions by day 7 and remained as dense cell aggregates with minimal cell protrusions by day 21. In 40 Pa hydrogels, DIPG cells were significantly more elongated throughout the 21 days of culture. In contrast, in 1000 Pa hydrogels, DIPG cells displayed radial cell protrusions by day 7 and formed dense aggregates with short, thin protrusions by Day 21.
Figure 3. Effect of matrix stiffness on brain tumor cell spreading and morphology over time.
Softer hydrogel (40 Pa) supported faster and more elongated cell spreading hydrogels after 21 days in culture. Scale bar = 100 μm.
3.2. Tumor cell proliferation in 3D hydrogels
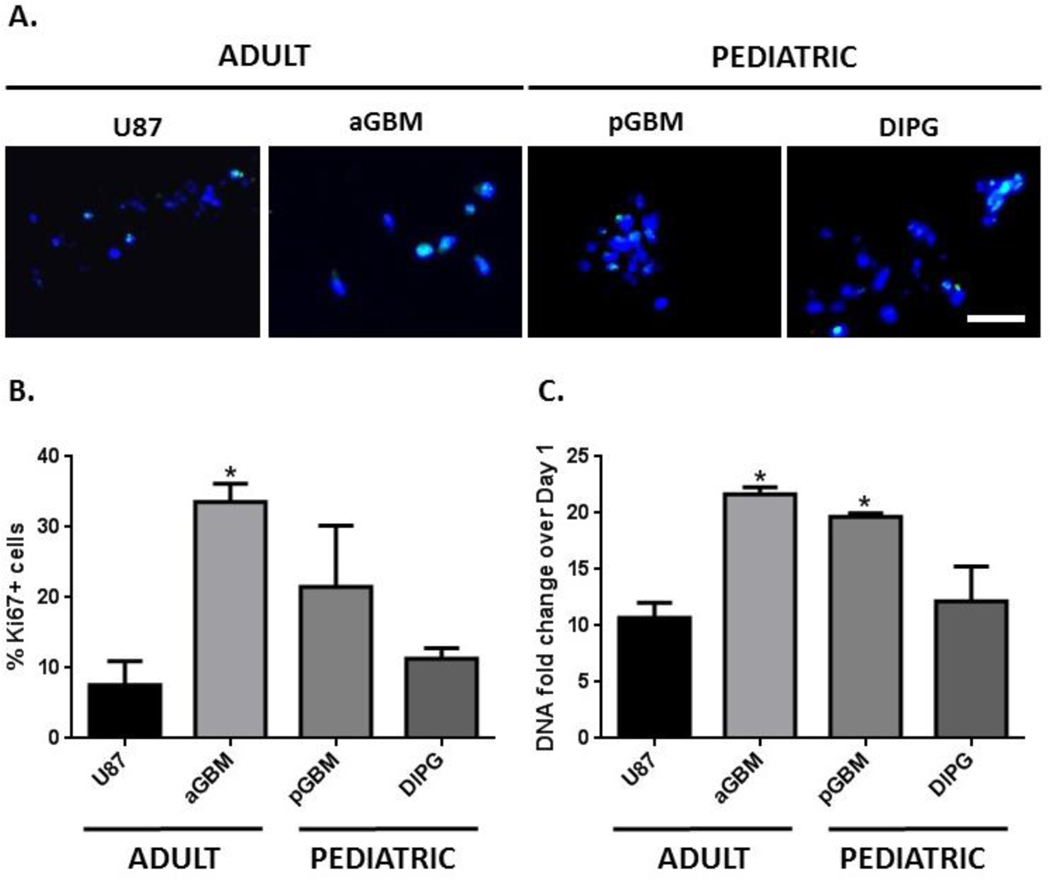
Based on observations of PDC proliferation and spreading, 40 Pa hydrogels were significantly more permissive and conducive to facilitating PDC proliferation and invasion in 3D (Figures 2, 3). Therefore, this hydrogel formulation was selected for further analysis of PDC behavior in 3D. The degree of tumor cell proliferation was quantified in 40 Pa hydrogels using proliferative index (% Ki67+) and fold change in DNA content over Day 1 (Figure 4). aGBM cells had the highest proliferative index (33.6%), followed by pGBM (21.5%), DIPG (11.3%), and U87 (7.60%) (Figures 4A, B). In agreement with the brightfield images and proliferative indices, aGBM cells had the highest fold of cell proliferation (22-fold) after 21 days, followed by pGBM (19-fold), DIPG (12-fold), and U87 (10-fold) (Figure 4C).
Figure 4. Hydrogels supported robust proliferation of brain tumor cells isolated from different patient age groups and anatomical locations.
(A) Ki67+ staining on day 7. Green = Ki67, Blue = nuclei, Scale bar = 50 μm. (B) aGBM cells had the highest percentage of Ki67+ cells (33.6%), followed by pGBM (21.5%), DIPG (11.3%), and U87 (7.60%). Percentage of Ki67+ cells on day 7, *p < 0.05, relative to U87. (C) aGBM cells had the highest fold of cell proliferation after 21 days (22-fold), followed by pGBM (19-fold), DIPG (12-fold), and U87 (10-fold). Day 21 DNA content normalized to day 1, *p < 0.05 relative to U87.
3.3. Tumor cell F-actin cytoskeletal organization in 3D hydrogels
Tumor cell invasion involves numerous cell processes, including organization of the F-actin cytoskeleton [44]. To further analyze the morphology of tumor cells in 40 Pa hydrogels, F-actin staining was performed on cell-laden hydrogels on Day 21 (Figure 5). Confirming our brightfield observations, aGBM cells were able to robustly spread in 40 Pa hydrogels, as seen by extremely elongated cell morphology and brightly stained F-actin-rich stress fibers along the cell membrane. Moreover, aGBM cells were able to organize into cell aggregates bridged together by single cells connected by long cell processes. In contrast, U87 cells remained largely isolated as single elongated cells with diffuse F-actin staining. pGBM and DIPG cells did not display extensive single cell elongation. Instead, pGBM cells formed large, infiltrative cell aggregates composed of cells with smaller bodies at a high cell density. pGBM cells located at the aggregate periphery showed enhanced F-actin staining along the cell membrane. Similarly, DIPG cells organized into densely packed infiltrative aggregates formed from cells with smaller bodies and diffuse F-actin organization.
Figure 5. Hydrogels supported extensive spreading of brain tumor cells isolated from different patient age groups and anatomical locations, as shown by F-actin cytoskeletal staining.
U87 (day 14), aGBM (day 14), pGBM (day 21), and DIPG (day 21). Red = F-actin, blue = nuclei. Scale bar = 100 μm. Z-stack size = 200 μm.
4. DISCUSSION
Brain cancer affects patients of all ages and can occur in various anatomical locations with distinct genotypes and phenotypes based on patient age and tumor type [1]. In the present study, we sought to engineer a 3D hydrogel-based culture system that can support the proliferation and invasion of PDCs isolated from different adult and pediatric brain tumors. Based on previous literature, we hypothesized that hydrogel stiffness may be an important factor that needs to be specifically optimized to facilitate PDC fates in 3D hydrogels [20,22,45]. Our results demonstrate that 1000 Pa hydrogels that previously supported U87 proliferation and spreading [25] did not support similar behavior for PDCs isolated from adult GBM, pediatric GBM, and DIPG. Instead, lowering hydrogel stiffness to 40 Pa significantly enhanced PDC proliferation and spreading after 21 days in culture. Interestingly, although U87 cells were able to spread in 40 Pa hydrogels, cell proliferation was significantly attenuated, as compared to in 1000 Pa hydrogels. In addition, there was significant upregulation in HIF1α expression in the PDCs after day 10 of culture, but not in U87 (Figure S2). This is accompanied by increased sizeof cell aggregate in the PDCs, whereas U87 remained mostly single cells (Figure 2). Increasing in cell aggregation may result in hypoxia within the cell aggregate, which has been reported to induce upregulation of HIF1α [46]. The opposite trends in cell behavior observed for PDC and U87 cells underscore potential differences between immortalized cell lines and PDCs. Furthermore, the various PDCs used in this study displayed significant differences in proliferation and morphology, highlighting differences between tumor cells isolated from different patient age groups and anatomical locations. The hydrogel model developed in this report may provide a valuable 3D in vitro culture platform that may be universally employed for different brain tumors and tailored for other tumor types.
Differences between PDC and U87 cell behavior in 3D hydrogels highlight the need for 3D in vitro models that have been specifically optimized for PDCs. Previous literature has demonstrated that the genomic and gene expression profiles of immortalized cell lines differ greatly compared to primary tumors [29]. In the present study, our results show that immortalized and PDCs behave significantly differently in 3D PEG-based hydrogels with varying stiffness. The distinct trends observed for PDC vs. U87 cell proliferation and invasion in response to changing hydrogel stiffness underscore the importance of optimizing hydrogel stiffness to support PDC fates in 3D. Increasing PEG concentrations stiffen PEG hydrogels and lead to increased crosslinking density and physical restriction of tumor cells. Therefore, in order to proliferate and invade in stiffer matrices, tumor cells must degrade more MMP-degradable crosslinks. In the present study, differences in cell proliferation and invasion found among the various tumor cell types suggests that each cell type may have different expression levels of MMPs for matrix remodeling. Low expression levels of MMPs can lead to more attenuated cell proliferation and invasion in stiffer matrices, which may have been the case of the different PDCs. Decreasing hydrogel stiffness to 40 Pa hydrogels resulted in lower crosslinking density, favoring more robust PDC proliferation and invasion. In contrast, it is possible that for U87 cells potentially through high expression of MMPs facilitated faster cell spreading and invasion. However, in 40 Pa hydrogels, this may have led to over-degradation of hydrogel crosslinks, which can reduce the local biochemical ligand concentration and lead to more attenuated cell proliferation.
Robust PDC proliferation and invasion in 40 Pa hydrogels suggest that this formulation may have potential to be employed universally for different adult and pediatric brain tumors (Figures 2, 3). Further characterization of PDC proliferation and morphology in 40 Pa hydrogels demonstrate that PDCs in 3D hydrogels behave in agreement with expectations based on in vivo reports. First, the proliferative indices (Ki67+) of PDCs in 3D hydrogels fall within the ranges of in vivo values reported in previous literature (Figure 4B). In adult patients with GBM, the mean proliferative index was found to be 19.6%, ranging from 1.9% to 58.3% and with more than 39% of patients with proliferative indices greater than the mean, as reported by Saito et al. from a panel of 43 patients [47]. In our hydrogel, the proliferative index of aGBM cells was 33.6%. For pediatric patients with GBM, the mean proliferative index was 32.1%, ranging from 5.6% to 89.5%, as reported by Pollack et al [48]. pGBM cells cultured in 3D hydrogels had a proliferative index of 21.5%. For DIPG patients, the proliferative index was 12%, ranging from <1% to 40%, as reported by Ballester et al. from a panel of 24 patients [49]. In 3D hydrogel, DIPG cell proliferative index was 11.3%. Furthermore, the trends in proliferative index for the various cell types showed strong correlation with trends in the fold of cell proliferation, suggesting the proliferative index may be a predictor of cell proliferation rates in 3D in vitro.
Furthermore, a key hallmark of GBM and DIPG tumor cells is their extreme ability to invade throughout the dense brain ECM. A significant advantage of using 3D hydrogels as in vitro models is the ability to analyze cell invasion in an intact, 3D microenvironment, which is not feasible using 2D culture and is significantly more challenging using animal models. F-actin confocal staining of PDCs cultured in 40 Pa hydrogels elucidated significant differences in cell shape and F-actin cytoskeletal organization, highlighting the diverse cell morphologies that can arise from tumor cells isolated from different patient age groups and tumor types (Figure 5, Supplementary Videos S1 – S4). aGBM cells were extremely elongated with F-actin-rich stress fibers concentrated at the cell periphery, which resembled GBM cell morphology in in vivo mouse models [50]. In contrast to aGBM cells, pGBM cells were mostly rounded with more intense F-actin staining at the cell periphery and organized into large diffusely infiltrative cell aggregates. Similarly, in mouse models, pediatric GBM cells form poorly demarcated masses with high infiltration of tumor cells into neighboring tissue [51]. Lastly, characteristic of DIPGs in vivo, DIPG cells also had rounded morphology with homogeneous expression of F-actin throughout the cytoplasm and formed diffusely infiltrating cell aggregates [43,52].
Finally, while we demonstrate that PDCs have distinct phenotypic behavior based on patient age and anatomical location, we acknowledge that one limitation to this study is that we only have one patient-derived cells per tumor type. It is possible that differences may also exist among different individual patients within each tumor type. One challenge of studying these rare disease is tissue scarcity. In the future, as more patient-derived cells become available, it would be very helpful to apply the hydrogel model reported in this study to assess multiple patient cell lines for each tumor type.
5. CONCLUSIONS
In summary, here we report a PEG-based hydrogel platform as 3D niche that supports the proliferation and invasion of PDCs isolated from different patient age groups and anatomical locations. Our results demonstrate that PDCs displayed distinct stiffness-dependent behavior when compared to commonly used immortalized cell line, highlighting potential differences between PDC and immortalized cells. Furthermore, among the various PDC types used in this study, significant differences in cell proliferation and morphology in 3D hydrogels underscore the genotypic and phenotypic diversity among different brain tumor types. To the best of our knowledge, this is the first hydrogel platform shown to support robust cell proliferation and invasion of brain tumor cells isolated from different patient age groups and anatomical locations. Such platform may provide a valuable universal culture platform for developing 3D in vitro tumor models for different brain tumors and other cancer types.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Brain tumors exhibit vast genotypic and phenotypic diversity based on patient age and anatomical location. While hydrogels hold great promise as tools for brain tumor research by providing 3D in vitro brain tumor models, previous studies were mostly limited to only adult glioblastoma using immortalized cell lines. Here we report a 3D hydrogel platform that supports robust proliferation and invasion of patient-derived brain tumor cell cultures isolated from adult and pediatric brain tumors from both cortex and brain stem. Our model validates patient-derived brain tumor cell culture exhibited significant different behavior from immortalized cell lines, and both age and anatomical locations impact brain tumor cell fates. The hydrogel reported here can provide a useful tool to better understand how age and anatomical locations impacts brain tumor progression using 3D in vitro models.
ACKNOWLEDGEMENTS
This work was supported by the following grants: NIH R01DE024772 (F.Y.), NIH R01AR074502, Stanford Child Health Research Institute Faculty Scholar Award (F.Y.), Stanford Coulter translational grant, Stanford SPARK program, and Stanford Bio-X IIP program (F. Y.),C.W. would like to thank Stanford Graduate Fellowship and Stanford Interdisciplinary Graduate Fellowship for support. S. S. would like to thank Stanford NIH Biotechnology Training grant for support. The authors also appreciate technical assistance from Stanford Cell Sciences Imaging Facility for confocal imaging and Anthony Behn for mechanical testing.
Footnotes
APPENDIX A. Supplementary Data: Supplementary Figure S1, Figure S2, and Videos S1 – S4.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].A. B. T. Association, Types of Tumors, http://www.abta.org/brain-tumor-information/types-of-tumors/, (accessed August 4, 2016). [Google Scholar]
- [2].A. B. T. Association, Brain Tumor Statistics, (accessed March 22, 2016). [Google Scholar]
- [3].Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM and Hayes DN. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 2010, 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Suri V, Das P, Jain A, Sharma MC, Borkar SA, Suri A, Gupta D. and Sarkar C. Pediatric glioblastomas: a histopathological and molecular genetic study. Neuro-Oncology, 2009, 11, 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ganigi PM, Santosh V, Anandh B, Chandramouli BA and Sastry Kolluri VR. Expression of p53, EGFR, pRb and bcl-2 proteins in pediatric glioblastoma multiforme: a study of 54 patients. Pediatr Neurosurg, 2005, 41, 292–299. [DOI] [PubMed] [Google Scholar]
- [6].Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J. and Zhang Z. The histone H3. 3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev, 2013, 27, 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ and Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science, 2013, 340, 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM and Jabado N. Driver mutations in histone H3. 3 and chromatin remodelling genes in paediatric glioblastoma. Nature, 2012, 482, 226–231. [DOI] [PubMed] [Google Scholar]
- [9].Spostol R, Ertel IJ, Jenkin RDT, Boesel CP, Venes JL, Ortega JA, Evans AE, Waral W. and Hammond D. The effectiveness of chemotherapy for treatment of high grade astrocytoma in children: results of a randomized trial. J Neurooncol, 7, 165–177. [DOI] [PubMed] [Google Scholar]
- [10].Phuphanich S, Edwards MSB, Levin VA, Vestnys PS, Wara WM, Davis RL and Wilson CB. Supratentorial malignant gliomas of childhood: results of treatment with radiation therapy and chemotherapy. J Neurosurg, 1984, 60, 495–499. [DOI] [PubMed] [Google Scholar]
- [11].Sturm D, Bender S, Jones DTW, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, Iavarone A, Aldape K, Brennan CW, Jabado N. and Pfister SM. Paediatric and adult glioblastoma: multiform (epi) genomic culprits emerge. Nat Rev Cancer, 2014, 14, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].D.-F. B. Children’s, Diffuse Intrinsic Pontine Glioma (DIPG), (accessed March 22, 2016). [Google Scholar]
- [13].Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet, 2012, 44, 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hargrave D, Bartels U. and Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol, 2006, 7, 241–248. [DOI] [PubMed] [Google Scholar]
- [15].Jansen MHA, van Vuurden DG, Vandertop WP and Kaspers GJL. Diffuse intrinsic pontine gliomas: a systematic update on clinical trials and biology. Cancer Treat Rev, 2012, 38, 27–35. [DOI] [PubMed] [Google Scholar]
- [16].Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PØ, Miletic H, Thorsen F. and Bjerkvig R. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro-Oncology, 2012, DOI: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Myungjin Lee J, Mhawech-Fauceglia P, Lee N, Cristina Parsanian L, Gail Lin Y, Andrew Gayther S. and Lawrenson K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab Invest, 2013, 93, 528–542. [DOI] [PubMed] [Google Scholar]
- [18].Ridky TW, Chow JM, Wong DJ and Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med, 2010, 16, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatusra T. and Minami H. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep, 2015, 33, 1837–1843. [DOI] [PubMed] [Google Scholar]
- [20].Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS and Weitz DA. Glioma expansion in collagen I matrices: analyzing collagen concentration-dependent growth and motility patterns. Biophys J, 2005, 89, 635–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pedron S. and Harley BA. Impact of the biophysical features of a 3D gelatin microenvironment on glioblastoma malignancy. J Biomed Mater Res A, 2013, 101, 3404–3415. [DOI] [PubMed] [Google Scholar]
- [22].Ananthanarayanan B, Kim Y. and Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials, 2011, 32, 7913–7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA and Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials, 2010, 31, 8494–8506. [DOI] [PubMed] [Google Scholar]
- [24].Singh SP, Schwartz MP, Tokuda EY, Luo Y, Rogers RE, Fujita M, Ahn NG and Anseth KS. A synthetic modular approach for modeling the role of the 3D microenvironment in tumor progression. Sci Rep, 2015, 5, 17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang C, Tong X. and Yang F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Mol Pharm, 2014, 11, 2115–2125. [DOI] [PubMed] [Google Scholar]
- [26].Rape A, Ananthanarayanan B. and Kumar S. Engineering strategies to mimic the glioblastoma microenvironment. Adv Drug Deliv Rev, 2014, 0, 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pedron S, Becka E. and Harley BA. Regulation of glioma cell phenotype in 3D matrices by hyaluronic acid. Biomaterials, 2013, 34, 7408–7417. [DOI] [PubMed] [Google Scholar]
- [28].Pedron S, Becka E. and Harley BA. Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment. Adv Mater, 2015, 27, 1567–1572. [DOI] [PubMed] [Google Scholar]
- [29].Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, Rosenblum M, Mikkelsen T, Zenklusen JC and Fine HA. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res, 2008, 6, 21–30. [DOI] [PubMed] [Google Scholar]
- [30].Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, Yang H, Choi YL, Park WY, Kong DS, Lee JI, Ko YH, Woo HG, Lee J, Kim S. and Nam DH. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep, 2013, 3, 260–273. [DOI] [PubMed] [Google Scholar]
- [31].Engler AJ, Sen S, Sweeney HL and Discher DE. Matrix elasticity directs stem cell lineage specification. Cell, 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- [32].Kuroiwa T, Ueki M, Ichiki H, Kobayashi M, Suemasu H, Taniguchi I. and Okeda R. Time Course of Tissue Elasticity and Fluidity in Vasogenic Brain Edema. in: James H, Marshall L, Raulen H, Baethmann A, Marmarou A, Ito U, Hoff J, Kuroiwa T. and Czernicki Z. (Eds.). Brain Edema X. Springer-Verlag Wien, Vienna, Acta Neurochir Suppl, 1997, vol. 70, pp.87–90. [DOI] [PubMed] [Google Scholar]
- [33].Viapiano MS and Lawler SE. Glioma Invasion: Mechanisms and Therapeutic Challenges. in: Meir GE (Ed.). CNS Cancer: Models, Markers, Prognostic Factors, Targets, and Therapeutic Approaches. Humana Press, Totowa, NJ, 2009, pp. 1219–1252. [Google Scholar]
- [34].Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK and DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev, 2001, 15, 1311–1333. [DOI] [PubMed] [Google Scholar]
- [35].Bigner SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS, Bigner DD. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res, 1990, 50, 8017–8022. [PubMed] [Google Scholar]
- [36].Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA 1992, 89, 2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Venkatesh HS, Johung TB, Caretti V, Noll A, Tang Y, Nagaraja S, Gibson EM, Mount CW, Polepalli J, Mitra SS, Woo PJ, Malenka RC, Vogel H, Bredel M, Mallick P, Monje M. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell, 2015, 161, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN and Anseth KS. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv Mater, 2009, 21, 5005–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shu XZ, Liu Y, Luo Y, Roberts MC and Prestwich GD. Disulfide cross-linked hyaluronan hydrogels. Biomacromolecules, 2002, 3, 1304–1311. [DOI] [PubMed] [Google Scholar]
- [40].Anderson SB, Lin C-C, Kuntzler DV and Anseth KS. The performance of human mesenchymal stem cells encapsulated in cell-degradable polymer-peptide hydrogels. Biomaterials, 2011, 32, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang C, Sinha S, Jiang X, Murphy L, Fitch S, Wilson C, Grant G. and Yang F. Matrix stiffness modulates patient-derived glioblastoma cell fates in 3D hydrogels. Tissue Eng Part A, 2020, ja. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Delpech B, Maingonnat C, Girard N, Chauzy C, Maunoury R, Olivier A, Tayot J. and Creissard P. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumour stroma. Eur J Cancer, 1993, 29, 1012–1017. [DOI] [PubMed] [Google Scholar]
- [43].Caretti V, Bugiani M, Freret M, Schellen P, Jansen M, van Vuurden D, Kaspers G, Fisher PG, Hulleman E, Wesseling P, Vogel H. and Monje M. Subventricular spread of diffuse intrinsic pontine glioma. Acta Neuropathol, 2014, 128, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gardel ML, Schneider IC, Aratyn-Schaus Y. and Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol, 2010, 26, 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ulrich TA, de Juan Pardo EM and Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res, 2009, 69, 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Huang WJ, Chen WW, and Zhang X. Glioblastoma multiforme: Effect of hypoxia and hypoxia inducible factors on therapeutic approaches. Oncol Lett, 2016, 12, 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Saito T, Hama S, Kajiwara Y, Sugiyama K, Yamasaki F, Arifin M-T, Arita K. and Kurisu K. Prognosis of cerebellar glioblastomas: correlation between prognosis and immunoreactivity for epidermal growth factor receptor compared with supratentorial glioblastomas. Anticancer Res, 2006, 26, 1351–1357. [PubMed] [Google Scholar]
- [48].Pollack IF, Hamilton RL, Burnham J, Holmes EJ, Finkelstein SD, Sposto R, Yates AJ, Boyett JM and Finlay JL. Impact of proliferation index on outcome in childhood malignant gliomas: results in a multi-institutional cohort. Neurosurgery, 2002, 50. [DOI] [PubMed] [Google Scholar]
- [49].Ballester LY, Wang Z, Shandilya S, Miettinen M, Burger PC, Eberhart CG, Rodriguez FJ, Raabe E, Nazarian J, Warren K. and Quezado MM. Morphologic characteristics and immunohistochemical profile of diffuse intrinsic pontine gliomas. Am J Surg Pathol, 2013, 37, 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, Huang L, Ratliff M, Karimian Jazi K, Kurz FT, Schmenger T, Lemke D, Gommel M, Pauli M, Liao Y, Haring P, Pusch S, Herl V, Steinhauser C, Krunic D, Jarahian M, Miletic H, Berghoff AS, Griesbeck O, Kalamakis G, Garaschuk O, Preusser M, Weiss S, Liu H, Heiland S, Platten M, Huber PE, Kuner T, von Deimling A, Wick W. and Winkler F. Brain tumour cells interconnect to a functional and resistant network. Nature, 2015, 528, 93–98. [DOI] [PubMed] [Google Scholar]
- [51].Coutinho de Souza P, Mallory S, Smith N, Saunders D, Li X-N, McNall-Knapp RY, Fung K-M and Towner RA. Inhibition of Pediatric Glioblastoma Tumor Growth by the Anti-Cancer Agent OKN-007 in Orthotopic Mouse Xenografts. PloS ONE, 2015, 10, e0134276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Caretti V, Sewing ACP, Lagerweij T, Schellen P, Bugiani M, Jansen MHA, van Vuurden DG, Navis AC, Horsman I, Vandertop WP, Noske DP, Wesseling P, Kaspers GJL, Nazarian J, Vogel H, Hulleman E, Monje M. and Wurdinger T. Human pontine glioma cells can induce murine tumors. Acta Neuropathol, 2014, 127, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.