Abstract
Autophagy has been extensively studied and occurs in many biological settings. However, a question remains as to whether ischemia enhances Beclin-1/LC3-II-dependent macroautophagy in vascular endothelial cells, as has been previously thought. Furthermore, the effect of the level of autophagy on cell or skin flap survival still requires elucidation. We created a lethal ischemia model in human umbilical vascular endothelial cells (HUVECs), performed quantitative proteomics and bioinformatics analyses, and verified the autophagic status and effect both in vitro and in vivo. The significantly upregulated proteins encoded by autophagy-related genes (ATGs) included ATG2A, ATG3, ATG4B, ATG5, ATG7, ATG9A, ATG12, ATG16, and ATG101. The significantly enhanced lysosomal proteins were cathepsin B, cathepsin D, lysosome-associated membrane protein 1 (LAMP1), and LAMP2. However, the differentially expressed proteins excluded Beclin-1, microtubule-associated protein light chain 3 (LC3)-I, and LC3-II. Western blot analyses verified that the protein expression levels of Beclin-1, LC3-I, and LC3-II were neither upregulated nor downregulated in ischemia-challenged HUVECs. The autophagic status was not enhanced by rapamycin in ischemic HUVECs but appeared to be inhibited by chloroquine. Our in vivo study on rats showed that a downregulation in autophagic status jeopardized skin flap survival. In conclusion, Ischemia neither enhanced nor inhibited Beclin-1/LC3-II-dependent canonical macroautophagy both in vitro and in vivo, in contradiction to previous studies. An appropriate autophagic homeostasis can minimize cell or skin flap damage.
Keywords: Autophagy, Bioinformatics analysis, Human umbilical vein endothelial cell, Quantitative proteomics study, Skin flap
Introduction
Autophagy aiming to regain post-stress homeostasis is an intracellular self-digestion pathway responsible for the removal of long-lived or malformed proteins and organelles damaged during biosynthesis by lysosomes.1 Macroautophagy, microautophagy, and chaperone-mediated autophagy are the three major autophagy processes involved in lysosomal degradation and extracellular release of cytosolic cargoes under excessive stress.2 Autophagy has been extensively studied in various pathophysiological settings both in vitro and in vivo.
Most researchers have described the activation of autophagy under conditions of ischemia or ischemia/reperfusion (IR). Rogg et al.3 showed that autophagy was upregulated to supply nutrients to endothelial cells during ischemia. Sachdev et al.4 revealed that autophagy occurred in human dermal microvascular endothelial cells in response to hypoxia or serum depletion. Ischemia activates autophagy–lysosome cascades through nitrosative stress, and these actions are pharmacologically inhibited by nitrosative stress antagonists, leading to the suppression of ischemia-induced autophagy–lysosome cascades and the degradation of tight junction proteins.5 Autophagy has been reported to be involved in tissue hypoxia/reoxygenation injury,6 because hypoxia/reoxygenation-induced expression of inducible nitric oxide synthase is associated with the upregulation of autophagy. Furthermore, an inducible nitric oxide synthase-specific inhibitor can abolish IR-induced autophagy.7 IR has been mainly reported to induce marked autophagy through the mammalian target of rapamycin (mTOR) pathway in various settings, such as skin flaps,8 and myocardial hypoxia/reoxygenation injury.6
The effect of autophagy on cell survival remains strongly debatable,9 and most scientists advocate that autophagy has a beneficial role in cell survival. Various autophagic pathways regulate cell homeostasis by supporting the recycling or degradation of macromolecules or organelles, thereby promoting cell survival.10 Li et al.11 reported that autophagy in brain microvascular endothelial cells may have a beneficial effect on the integrity of the blood–brain barrier in IR injuries. During IR, reactive oxygen species (ROS) specifically activate autophagy-related gene (ATG)-encoded protein 7, promoting autophagic flux and formation of microtubule-associated protein light chain 3 (LC3)–II–positive puncta around mitochondria in primary human liver endothelial cells.12 Inhibition of ROS could reduce the autophagic flux in liver endothelial cells during IR and induce necrosis.12 Malat1 serves as a competing endogenous RNA by sequestering miR-26b and upregulating Unc-51-like autophagy-activating kinase 2 expression, promoting the autophagy and survival of brain microvascular endothelial cells under oxygen-glucose deprivation and reoxygenation conditions.13 Xue et al.1 indicated that pituitary adenylate cyclase-activating polypeptide-enhanced autophagy ameliorated cellular damage and diminished necrosis in H2O2-stressed primary hepatocytes. Shi et al.14 proved the protective effect of autophagy by the finding that autophagy suppression by histone deacetylase 9 was associated with endothelial dysfunction. However, under environmental stress or injury, imbalancd autophagic pathways can enhance the degradation of macromolecules or organelles, diminish biosynthetic processes, and cause cell death.9 Zhu et al.7 reported that autophagy was involved in irreversible cell injury and death under extreme conditions. Tang et al.15 further demonstrated a detrimental effect of autophagy on cell survival. They suggested that glycyrrhizic acid could decrease autophagy in coronary artery endothelial cells, and that the protective effect of glycyrrhizic acid against hypoxia/reoxygenation was abolished by autophagy promotion.
The effect of autophagy on tissue survival is also controversial. Upregulated autophagy can supply nutrients, while metabolic switching can improve available substrate utilization. These prosurvival mechanisms may diminish cardiac damage.3 Autophagy in the ischemic brain has been shown to protect brain tissue from damage caused by low oxygen stress.11 Li et al.16 showed that cerebral ischemia-induced autophagy-like injury was regulated by the nuclear factor kappa-B pathway, suggesting a potential treatment for ischemic stroke. ROS-dependent autophagy promotes the survival of liver endothelial cells during liver IR injury, and therapeutic targeting of this signaling pathway may reduce liver damage following transplantation.12 However, it has also been shown that autophagy can contribute to ischemia-induced cell death in the brain.16
Skin flap grafting is a common surgical procedure and frequently results in ischemia or IR injury. Therefore, skin grafts could represent as an ideal model to study the effects of autophagy on tissue survival in vivo. The stimulation of autophagy has been shown to have proangiogenic, antiapoptotic, and antioxidative effects, and to exhibit a prosurvival effect on skin flaps that could be reversed by the autophagy inhibitor 3-methyladenine.17, 18, 19 Wu et al.8 suggested that the prosurvival effect of autophagy may be achieved through the mTOR signaling pathway. Lin et al.20 observed that resveratrol activated autophagy in skin flaps through increased expression of Beclin-1, LC3-II, and cathepsin D, which may be another underlying mechanism for flap survival. These studies indicate that autophagy has a beneficial effect on skin flap survival.
However, no agreement has been reached on the effect of autophagy on skin flap survival. Chen et al.21 revealed that sitagliptin-induced autophagy was detrimental for flap survival and that its inhibition by 3-methyladenine could improve flap survival. Wang et al.22 showed that hypoxia-inducible factor-1α-induced autophagy had a detrimental effect on multiterritory perforator flap survival by augmenting oxidative stress and apoptosis, and that its disadvantageous effect could be reversed by 3-methyladenine. These findings suggest that autophagy has a detrimental effect on skin flap survival.
We aimed to investigate the role of Beclin-1/LC3-II-dependent macroautophagy in human umbilical vascular endothelial cells (HUVECs) under conditions of ischemia, because of the inconsistent effects of autophagy on cell or tissue survival. We established a lethal ischemia model in HUVECs, performed quantitative proteomics and bioinformatics analyses in vitro, and then verified the inactivated autophagy status and its effect on both HUVECs and skin flaps.
Materials and methods
Cell culture
HUVECs were kindly provided by Professor Deqin Yang at Chongqing Medical University.23 HUVECs at 1 × 105 cells/mL (10 mL) and 1 × 104 cells/mL (0.2 mL) were seeded into 100-mm dishes and 96-well culture plates, respectively. The ischemia condition was applied to cells at log phase and 60%–80% confluency, where cells were cultured in ischemia buffer (118 mM NaCl, 24 mM NaHCO3, 20 mM sodium lactate, 16 mM KCl, 2.5 mM CaCl2, 1 mM NaH2PO4, 0.5 mM sodium EDTA, pH 6.8)7 without glucose and fetal bovine serum in a hypoxia incubator (N2:O2:CO2 = 94%:1%:5%) at 37 °C for designated time periods. HUVECs after ischemia treatment were incubated in Dulbecco's modified Eagle's medium supplemented with 1 g/L glucose, 10% fetal bovine serum, 100 units of penicillin and 100 μg of streptomycin in a regular incubator (5% CO2, 37 °C) for 24 h to mimic reperfusion conditions.
Establishment of a lethal ischemia model in HUVECs
In the first round, HUVECs were cultured in ischemia buffer for 0, 3, 6, 9, and 12 h together with a starvation control (ischemia buffer with glucose but without fetal bovine serum) in the hypoxia incubator, followed by a 24-h reperfusion treatment. In the second round, HUVECs were cultured in ischemia buffer for 0, 1, 2, 3, 4, 5, and 6 h together with a starvation control in the hypoxia incubator, prior to the 24-h reperfusion treatment.
Apoptosis analysis
Apoptosis analysis was conducted as previously described.24 Exponentially-growing HUVECs were collected and stained with an Annexin V-FITC/PI Assay Kit (BestBio, Shanghai, China) in accordance with the manufacturer's protocol. The apoptosis detection included three groups: 3-h ischemia group (group I), 3-h ischemia/24-h reperfusion group (group IR), and 3-h normal control group (group NC). The cells were then subjected to flow cytometry analysis using a BD Influx (BD Bioscience, San Jose, CA, USA). The acquired flow cytometry data were analyzed with FlowJo v10.0 software (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). The assay was performed in triplicate under each condition.
Quantitative proteomics study
To determine the autophagic status of HUVECs under conditions of ischemia, ischemic HUVECs were assessed by proteomics analysis using a PTM-BIO (PTM-Biolabs Co. Ltd., Hangzhou, China) protocol. Briefly, HUVECs in the I, IR, and NC groups were collected and sonicated three times on ice using a high-intensity ultrasonic processor (Scientz, Ningbo, China) in lysis buffer (8 M urea, 1% protease inhibitor cocktail) followed by centrifugation at 12,000×g and 4 °C for 10 min. The supernatants were collected, and the protein concentrations were determined with a Bicinchoninic Acid Kit (Beyotime, Shanghai, China) in accordance with the manufacturer's protocol. After trypsin digestion, the peptides were desalted on a Strata X C18 SPE column (Phenomenex, Torrance, CA, USA) and vacuum dried. Then peptides were reconstituted in 0.5 M triethylammonium bicarbonate and processed in accordance with the manufacturer's protocol for the 9-plex tandem mass tag kit (Thermo Fisher Scientific, MA, USA). The tryptic peptides were fractionated by high-pH reverse-phase high-performance liquid chromatography (HPLC) using an Agilent 300Extend C18 column (5-μm particles, 4.6-mm ID, 250-mm length; Agilent Technologies Inc., Santa Clara, CA, USA). The peptides were subjected to mass spectrometry in a Q Exactive Plus Hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific) followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) in a Q Exactive™ Plus (Thermo Fisher Scientific) coupled online to the HPLC system. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for LC during LC-MS/MS using a normalized collision energy of 30 J and ion fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan and 20 LC-MS/MS scans with 15.0 s dynamic exclusion was used. Automatic gain control was employed to prevent overfilling of the Orbitrap. The fixed first mass was set at 100 m/z. Tandem mass spectra were searched against the SwissProt Human database. Trypsin/P was specified as the cleavage enzyme and up to two missing cleavages were allowed. The mass tolerance for precursor ions was set at 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set at 0.02 Da. For the protein quantification method, the LC-MS/MS data were processed using the Mascot search engine (v.2.3.0). The false discovery rate was adjusted to <1% and the peptide ion score was set at ≥ 20.
Bioinformatics analysis
To interpret the proteins that were isolated, fractioned, and purified in the proteomics analysis, gene ontology proteome annotation was performed using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). First, each identified protein ID was converted to a UniProt ID and mapped to the gene ontology ID. For identified proteins that were not annotated by the UniProt-GOA database, InterProScan software (http://www.ebi.ac.uk/interpro/) was used to assign a gene ontology annotation to the protein based on a protein sequence alignment method. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was employed to identify enriched pathways by a two-tailed Fisher's exact test that compared the enrichment of differentially expressed proteins against all identified proteins. The identified pathways were classified into hierarchical categories according to the KEGG website (http://www.genome.jp/kegg/). In the functional enrichment-based clustering analysis, the quantified proteins in this study were initially divided into three quantitative categories according to the following quantification ratios: Q1 (0 < I/NC ratio < 1/1.5), Q2 (1/1.5 < I/NC ratio < 1.5), and Q3 (I/NC ratio > 1.5). A corrected P-value < 0.05 was considered significant. Next, quantitative category-based clustering was performed. All substrate categories obtained after enrichment were collated along with their P-values and then filtered for those categories that were enriched in at least one of the clusters with a P-value < 0.05. The filtered P-value matrix was transformed using the function x = −log10 (P-value). Finally, the x-values were z-transformed for each category. The resulting z-scores were clustered by one-way hierarchical clustering (Euclidean distance, average linkage clustering) in Genesis. Clustering membership was visualized by a heat map using the “heatmap.2” function in the “gplots” R-package (Lucent Technologies Inc., Murray Hill, NJ, USA).
Western blot analysis
Western blot analysis was conducted as previously described25 to verify the differentially expressed proteins in ischemic HUVECs. Briefly, HUVECs after various ischemia/reperfusion treatments were collected, lysed in modified RIPA buffer (Beyotime), centrifuged, and quantified by the bicinchoninic acid method (Beyotime) in accordance with the manufacturer's protocol. After quantification of the protein concentration, equal amounts of protein lysate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis according to established protocols. Proteins were transferred from the gels to PVDF membranes (Millipore Sigma, Burlington, MA, USA) in a sandwich model at 200 mA for 90 min. The membranes were probed and incubated with primary antibodies against Beclin-1 (1:1000; Zen BioScience, Sichuan, China), phospho-AKT (Thr308) (1:500; Zen BioScience), LC3-I/II (1:1000; Zen BioScience), or- actin (1:1000; Zen BioScience) at 4 °C overnight. After washing with phosphate buffered saline with 0.05% Tween-20, all membranes were incubated with horseradish peroxidase-conjugated anti-rabbit/anti-mouse secondary antibodies (1:4000; Abcam, Cambridge, UK) at room temperature for 1 h. The immunoreactive proteins were visualized with a ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA, USA). ImageJ 1.46r software (National Institutes of Health, Bethesda, MD, USA) was used to determine the protein expression levels, which were recorded as the ratio of the target protein relative to -actin.
Gain/loss of function analysis in vitro: cell viability
To assess the effect of the altered autophagic status on HUVECs, cells were treated with rapamycin (MedChemExpress, Monmouth Junction, NJ, USA) or chloroquine phosphate (MedChemExpress). Quantitative cell viability was assessed using a Cell Counting Kit-8 (CCK-8; Beyotime) in accordance with the manufacturer's protocol. Briefly, subconfluent HUVECs were treated during the 3-h ischemia treatment with rapamycin at 0, 10, 50, and 100 nM or chloroquine at 5, 12.5, and 25 μM. After the 3-h ischemia treatment, the CCK-8 reagent was added and the HUVECs were incubated in darkness at 37 °C for 60 min. The absorbance was read at 450 nm. The assay was performed in duplicate.
Autophagic flux detection
To detect the status change of the autophagic flux in ischemia-challenged HUVECs, cells were double-marked with red fluorescent protein (RFP) and green fluorescent protein (GFP) by transduction with Ad-mRFP-GFP-LC3 (Hanbio Biotechnology Co. Ltd., Shanghai, China) in accordance with the manufacturer's protocol. Briefly, HUVECs at log phase and 60%–80% confluency were transduced with recombinant adenovirus encoding the LC3 gene. One million cells were transduced with the purified adenovirus at 1.25 × 1010 pfu/mL and a multiplicity of infection of 50, and lightly agitated on a swing bed incubator for 48 h at 37 °C. Prior to 24-h reperfusion, the cells were exposed to 0, 3, 6, and 12 h nutrient deprivation treatment. The cells were visualized for autophagic flux after reperfusion via a laser scanning confocal microscopy (Leica Microsystems Inc., Mannheim, Germany).
Gain/loss of function analysis in vivo: free flap survival
To verify the effect of the autophagic status change on free flap survival, a rat epigastric axial skin flap model was adopted on Sprague–Dawley rats treated with rapamycin or chloroquine. Briefly, 15 male Sprague–Dawley rats (the Central Animal Laboratory of Chongqing Medical University) weighing 220–280 g were housed under specific pathogen-free conditions and treated in accordance with the National Institute of Health Guidelines. Rats were anesthetized by intraperitoneal injection of sodium pentobarbital (30 mg/kg) to facilitate the establishment of skin flaps. The right superficial epigastric artery- and vein-based axial skin flaps were dissected. Each flap was sized approximately 3 × 8 cm and comprised skin and subcutaneous fascia, including the panniculus carnosus as previously described.25 Rats were randomly allocated to groups I, IR, and NC, and half an hour before the initiation of ischemia, animals received 1 mL intraperitoneal administration of rapamycin (1 mg/kg), chloroquine (10 mg/kg), or sterile saline, respectively (n = 5 per group). Ischemia was induced by clamping the pedicle vessels in the free flaps with double microvascular clamps. The clamps were removed after 3 h, and the flaps were sutured back to their original sites. The diet, mental status, and skin flap survival of the rats were observed and recorded daily. The skin flaps were photographed on postoperative day 7. The survival status of the skin flaps, including skin color, presence of bleeding upon cutting, gross appearance, and surface area regardless of scar tissue coverage, was assessed in a blinded manner as previously described.25 The flap survival area and total flap surface area were evaluated using Image-Pro Plus software (version 5.0; Media Cybernetics LP, Silver Spring, MD, USA). The necrotic ratio was expressed as the percentage of necrotic area relative to the total flap surface area. This in vivo study was approved by the Chongqing Medical University Medical Center Institutional Animal Care and Use Committee (IRB: (2020)044).
Data analysis
Analysis of variance (Tukey test) and/or a non-parametric test (Bonferroni test) were employed to evaluate the differences between groups using SPSS software (Version 19.0; IBM Corp., Armonk, NY, USA). Values of P < 0.05 were considered statistically significant. The original figures were produced using GraphPad Prism 8 software (GraphPad Company, San Diego, CA, USA) and the final figures were assembled using CorelDRAW Graphics Suite X4 (Corel, Ottawa, Ontario, Canada).
Results
Lethal ischemia model in HUVECs
HUVECs were completely disrupted after ≥6-h ischemia followed by 24-h reperfusion on a 0, 3, 6, 9, and 12 h ischemia schedule. HUVECs were completely disrupted after ≥4-h ischemia followed by 24-h reperfusion on a 0, 1, 2, 3, 4, 5, and 6 h ischemia schedule (data not provided). Hence, 3-h ischemia/24-h reperfusion was selected as the optimal lethal ischemia time, which was confirmed by apoptosis detection by flow cytometry (Fig. 1). Apoptosis detection revealed significant differences in apoptotic ratios between the experimental groups and the control group (P < 0.05), but no significant difference between group I and group IR (P > 0.05).
Figure 1.
Flow cytometry analysis. (A) Group NC. (B) Group I. (C) Group IR. There were significant differences between group NC and the experimental groups. However, there was no significant difference between group I and group IR.
Quantitative proteomics study
Quantitative proteomics analyses of the global proteome in I, IR and NC HUVECs were performed by 9-plex tandem mass tag labeling, HPLC fractionation, and LC-MS/MS. A fold change ratio above 1.5 in group comparisons (I/NC, IR/NC, and IR/I) was considered upregulation, while a ratio below 1/1.5 was considered downregulation. In the I/NC ratio comparison, a total of 2050 proteins were upregulated and 1560 proteins were downregulated (Supplementary materials, P < 0.05). The strong negative correlation between protein mass and coverage distribution confirmed extreme peptide quality that met the requirements for quality control (Fig. 2). A heatmap of the Pearson correlation coefficients showed the consistency of the repeated biological measurements. There were strong positive correlations among the specimens in the specific groups (all > 0.60), and negative correlations between the control group and the experimental groups. Boxplots of relative standard deviations indicated intense consistency of repeatability. All data showed very weak fluctuation of relative standard deviations and met the repeatability standard for 75% of relative standard deviations below 20%.
Figure 2.
High credibility and repeatability of data for ischemic HUVECs in a quantitative proteomics study. (A) Protein mass and coverage distribution suggested extremely good peptide quality. (B) Pearson correlation coefficient analysis showed excellent repeatability for the three samples in each group. (C) Analysis of relative standard deviations indicated excellent consistency of repeatability.
Bioinformatics analysis
To assess the autophagic status change in ischemia-challenged HUVECs, a bioinfomatics analysis was performed. Functional enrichment analysis indicated a fold change ratio above 1.5 in the lysosome KEGG pathway (P < 0.05, Fig. 3). Functional enrichment-based clustering analysis suggested that the lysosome KEGG pathway was significantly upregulated in the I/NC group comparison, while the autophagy KEGG pathway was weakly enhanced. The differentially expressed proteins related to autophagic and lysosomal digestion in the I/NC group comparison were specifically analyzed. The significantly upregulated ATG proteins included ATG2A, ATG3, ATG4B, ATG5, ATG7, ATG9A, ATG12, ATG16 (P < 0.001), and ATG101 (P < 0.01). The significantly enhanced lysosomal proteins comprised cathepsin B, cathepsin D, lysosome-associated membrane protein 1 (LAMP1), and LAMP2 (P < 0.001). However, the differentially expressed proteins excluded Beclin-1, LC3-I, and LC3-II.
Figure 3.
Bioinformatics analysis. (A) Functional enrichment analysis indicated enrichment of the lysosome pathway. (B) Functional enrichment-based clustering analysis suggested that the lysosome pathway was strongly enhanced, while autophagy was weakly upregulated. (C) Differentially expressed proteins in group comparisons were assessed. Autophagy-related proteins, such as ATG5, ATG12, ATG16, and ATG101, were significantly upregulated. Lysosome function-related proteins, such as LAMP1, LAMP2, Cathepsin B, and Cathepsin D, were significantly enhanced. However, LC3-I, LC3-II, and Beclin-1 were not activated.
Western blot verification
To verify the expression changes in the critical proteins for autophagy, Western blot analysis was performed in ischemic HUVECs. The protein levels of LC3-II were significantly decreased (P < 0.05) after HUVECs suffered from nutrient deprivation (deprivation of glucose and amino acids) without reperfusion (Fig. 4A–D). In contrast, the protein level of LC3-II was significantly increased (P < 0.05) when HUVECs suffered from starvation (deprivation of amino acids only). The LC3-II protein level returned to normal when HUVECs received reperfusion after nutrient deprivation. The LC3-II protein level was significantly increased (P < 0.05) after the starved HUVECs had access to reperfusion. Interestingly, there were no significant differences in the protein levels of LC3-II/I and Beclin-1 when HUVECs suffered from ischemia (nutrient deprivation plus hypoxia) for 0 and 3 h followed by reperfusion for 24 h. No proteins could be collected when the ischemia time was ≥6 h with a 3-h interval as cells were completely disrupted. To further verify the expression levels of LC3-II/I and Beclin-1, the interval was set at 1 h. No significant differences in the protein levels of LC3-II/I and Beclin-1 were noted when HUVECs suffered from ischemia for 0, 1, 2, and 3 h followed by reperfusion for 24 h. No proteins could be collected when the ischemia time was ≥4 h as the cells were completely disrupted.
Figure 4.
Western blot and CCK-8 analyses. (A, B) HUVECs exposed to nutrient deprivation without reperfusion exhibited LC3-II downregulation but the expression levels of LC3-II returned to normal status after reperfusion. (C, D) HUVECs exposed to various ischemia (nutrient deprivation plus hypoxia) followed by reperfusion verified that LC-II and Beclin-I were neither upregulated nor downregulated. (E) CCK-8 analysis was performed after the addition of rapamycin or chloroquine. There were no significant differences between the control group and the experimental groups, except between the control group and the 25 μΜ chloroquine group (∗, P < 0.05).
Gain/loss of function analysis in vitro
To assess the effect of ischemia on the autophagic level of the HUVECs, a gain/loss of function analysis was conducted. The cell viability of ischemia-challenged HUVECs was not significantly altered by rapamycin (10, 50, and 100 nM) or by lower concentrations of chloroquine (5 or 12.5 μM) (P > 0.05, Fig. 4E). However, it was significantly decreased by 25 μM chloroquine (P < 0.05). These findings suggest that rapamycin does not upregulate the autophagic status of ischemia-challenged HUVECs, while a high dosage of chloroquine could inhibit the autophagic status of ischemic HUVECs.
Autophagic flux detection
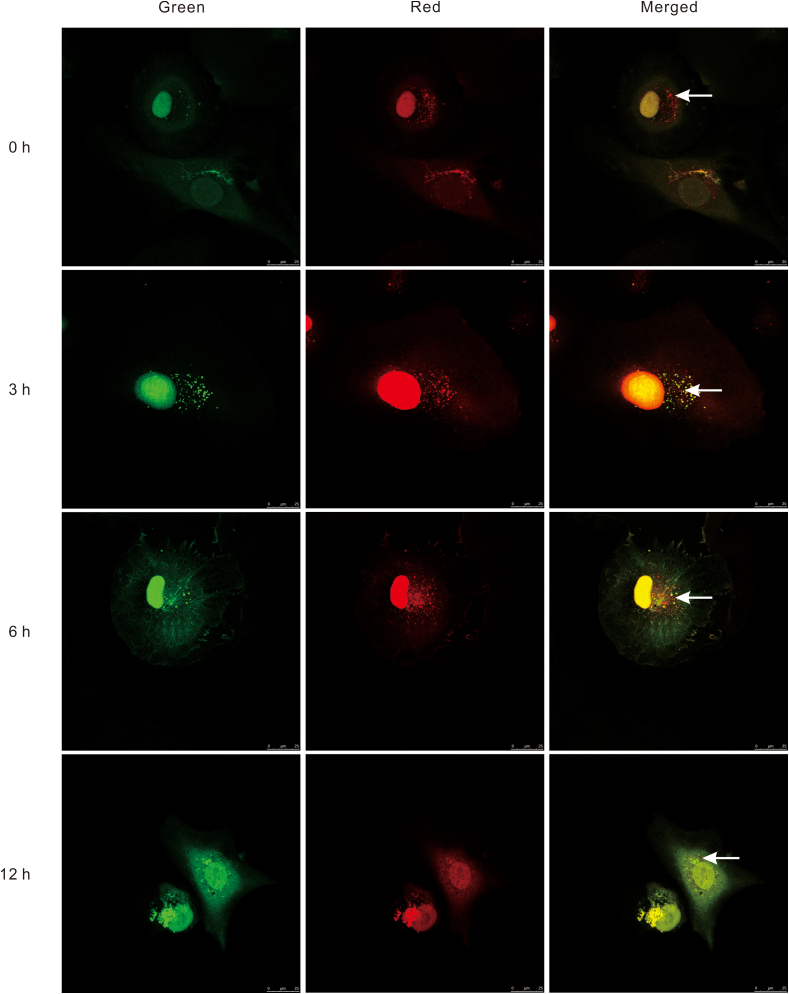
To detect the status change of autophagic flux in ischemia-challenged HUVECs, cells were transduced with Ad-mRFP-GFP-LC3. LC3 was co-marked with RFP and GFP to indicate its presence and status through autophagosome formation. After the autophagosome fused with the lysosome, the green fluorescence became weak and gradually disappeared. Thereafter, only red fluorescence was observed in the autolysosome. The accumulation of red fluorescent puncta in merged images was not obviously enhanced in HUVECs after 0, 3, 6, or 12 h of nutrient deprivation prior to 24-h reperfusion (Fig. 5). The red fluorescence intensity in the 12 h group even showed a decreasing tendency. Some nutrient-deprived HUVECs in the 12 h group showed extreme deformity.
Figure 5.
Autophagic flux in HUVECs transduced with ad-mRFP-GFP-LC3 under nutrient deprivation conditions. LC3 was co-stained with red and green fluorescent labels to indicate its presence and status through autophagosome formation. There was a certain level of macroautophagy in normal HUVECs with 0-h nutrient deprivation/24-h reperfusion. No significant upregulation of autophagic flux was noted in HUVECs with 3- or 6-h nutrient deprivation/24-h reperfusion compared with normal HUVECs. There even appeared to be a tendency toward a decrease in HUVECs with 12-h nutrient deprivation/24-h reperfusion compared with normal HUVECs. White arrows point to the autolysosome.
Gain/loss of function analysis in vivo
To verify the effect of the autophagic status change on free flap survival, a gain/loss of function analysis was performed in Sprague–Dawley rats by utilizing rapamycin and chloroquine, respectively. A 3-h ischemia/7-d reperfusion challenge on the superficial epigastric axial skin flaps (3 cm × 8 cm) showed almost complete survival on postoperative day 7 (Fig. 6). Neither rapamycin nor chloroquine treatment promoted skin flap survival. A larger necrotic area or more severe edema was noted in the chloroquine group. The necrotic ratio in chloroquine group was the highest among the three groups (33.02% ± 5.45%, P < 0.05), while the necrotic ratios of the NC and rapamycin groups were similar (1.92% ± 1.33% and 5.39% ± 2.78%, respectively, both P > 0.05).
Figure 6.
Effect of an autophagy agonist and inhibitor on the flap survival area 7 days after surgery in a rat epigastric axial skin flap model. Rapamycin didn’t promote the skin flap survival. There was no significant difference between the control group and the rapamycin group (P > 0.05). However, chloroquine inhibited the skin flap survival. There were significant differences between the chloroquine group and the control group as well as between the chloroquine group and the rapamycin group (∗∗,P < 0.01).
Discussion
We aimed to verify whether ischemia or IR can upregulate autophagy in HUVECs. Previously reported models of hypoxia, ischemia, IR, and oxygen–glucose deprivation and reoxygenation may not typically represent ischemia or IR, because ischemia consists of hypoxia/anoxia and nutrient deprivation, including deprivation of both glucose and amino acids.26,27 According to our findings, cells were completely disrupted when the ischemia time exceeded 3 h. Thus, 3-h ischemia/24-h reperfusion was selected as the optimal lethal ischemia time. Apoptosis detection revealed significant differences in apoptotic ratios between the experimental groups and the control group, and there was no significant difference between group I and group IR. Thus, an optimal lethal ischemia model was successfully established.
A quantitative analysis of the global proteome in I, IR and NC HUVECs was performed by LC-MS/MS. A strong negative correlation between protein mass and coverage distribution demonstrated excellent peptide quality, consistent with Mertins et al.28 Strong positive correlations between the specimens of individual groups and negative correlations between different groups suggested an extremely high consistency in the biological measurements, which was also confirmed by Bourassa et al.29 The repeatability of the methods was assessed by calculating the percentage of relative standard deviations.30 Our data exhibited very weak fluctuation of relative standard deviations, as evidenced by the finding that 75% of relative standard deviations were below 20%. Therefore, all the data indicated excellent peptide quality and sample repeatability.
In the functional enrichment analysis, a fold change ratio above 1.5 was noted in the lysosome KEGG pathway. This finding indicated the activation of lysosome-related digestion pathways, including macroautophagy, chaperone-mediated autophagy,31 and lysosome-dependent cell death.32 The lysosome KEGG pathway was significantly upregulated in the I/NC group comparison, while the autophagy KEGG pathway was weakly enhanced in the functional enrichment-based clustering analysis. These results indicate that lysosome-dependent cell death may be strongly enhanced, and macroautophagy and chaperone-mediated autophagy may be weakly upregulated. Most previous studies have shown that autophagy, especially macroautophagy, is activated under cell ischemia or IR conditions,33,34 except for one report suggesting that cold ischemia blunted autophagic flux.35 We were eager to determine the activation status and underlying mechanisms of macroautophagy in ischemic HUVECs, and therefore analyzed the differentially expressed proteins related to autophagy and lysosomal digestion in the I/NC group comparison. The significantly upregulated proteins included ATG2A, ATG3, ATG4B, ATG5, ATG7, ATG9A, ATG12, ATG16, ATG101, cathepsin B, cathepsin D, LAMP1, and LAMP2, but excluded Beclin-1, LC3-I, and LC3-II. These findings confirmed that the lysosome digestion pathway was highly activated because lysosome function or dysfunction is shown to be closely related to the expression levels of cathepsin B, cathepsin D, LAMP1, and LAMP2.36,37 The results also indicated that the canonical macroautophagy pathway was not activated, as this pathway is dependent on Beclin-1 and LC3-II.38,39
Western blot analyses were carried out to verify the inactivated status of Beclin-1 and LC3-II. Our study is the first to show that the expression of LC3-II is downregulated under nutrient deprivation (deprivation of glucose and amino acids), and can return to the normal levels after reperfusion. These findings indicate that nutrient deprivation/reperfusion did not activate macroautophagy. This study also demonstrates that ischemia (hypoxia and nutrient deprivation) followed by reperfusion does not enhance the expression levels of LC3-II and Beclin-1. We are the first to propose that the Beclin-1/LC3-II-dependent macroautophagy pathway is neither upregulated nor downregulated under IR conditions, which was verified by Western blot analyses (Fig. 7).
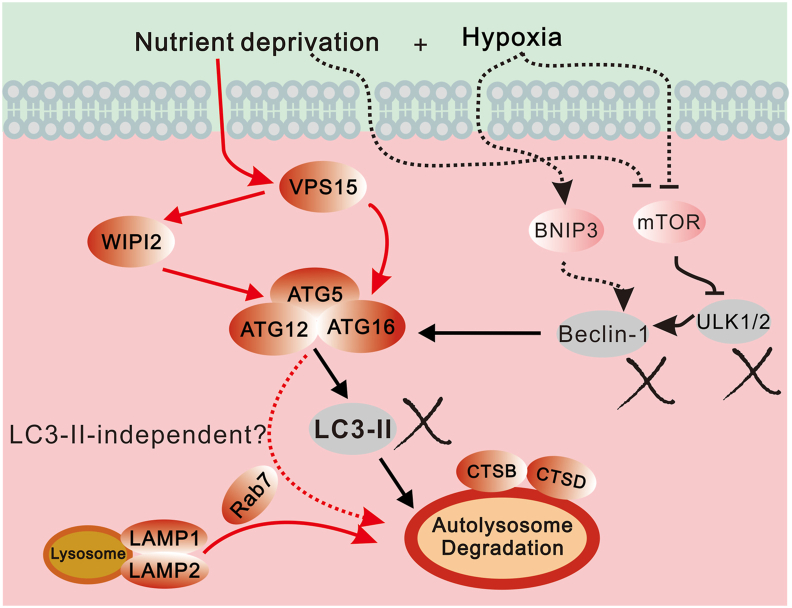
Figure 7.
Schematic graph of inactivation of Beclin-1/LC3-II dependent macroautophagy. This study indicates potential targets of microautophagy or chaperone-mediated autophagy.
A gain/loss of function analysis was conducted to assess the effect of the autophagic level on HUVEC viability. Cell viability was not significantly altered by rapamycin at 10, 50, and 100 nM, which is inconsistent with previous reports that rapamycin treatment could activate autophagy.40,41 However, we believe that our findings are reasonable, because the canonical macroautophagy indicators Beclin-1 and LC3-II were not upregulated in our study. It is interesting to note that cell viability was significantly decreased after treatment with 25 μM chloroquine. This phenomenon can be explained by two reasons. First, chloroquine is a typical inhibitor of macroautophagy,42 and second, chloroquine exhibits its inhibitory function by anchoring the final stage of the macroautophagy pathway involving the fusion of the lysosome and autophagosome.43,44 In contrast, rapamycin targets the upstream of the macroautophagy pathway.45,46
Double fluorescence labeling of LC3 allows the detection of a status change in the autophagic flux in nutrient-deprived HUVECs. LC3 double-tagged with both RFP and GFP indicates the stage from autophagosome initiation to autolysosome formation.47 Subsequently, only red fluorescence is observed in the autolysosome.48 The accumulation of red fluorescent puncta in merged images was clear in normal HUVECs, but was not obviously enhanced in HUVECs treated with nutrient deprivation and reperfusion. These phenomena suggest that normal HUVECs are equipped with a certain level of autophagy that does not change after treatment with nutrient deprivation/reperfusion.
The effect of the autophagic status on free flap survival was verified by utilizing rapamycin and chloroquine. However, the results showed that neither rapamycin nor chloroquine treatment promoted skin flap survival. The necrotic ratios in the chloroquine group were significantly decreased compared with those in the NC group. These findings suggest that neither enhancement nor inhibition of autophagy promotes skin flap survival and that appropriate autophagy could minimize skin flap damage. Thus, it is resonable to expect that autophagic homeostasis plays an important role in cell and skin flap health.49 Disturbance of autophagy can facilitate cell death.50,51
In summary, we demonstrated that ischemia (or IR) neither upregulates nor downregulates autophagy in ischemic HUVECs and free flaps. Disruption of autophagic homeostasis did not improve cell or tissue survival but jeopardized cell or tissue health. Therefore, the Beclin-1/LC3-II-independent noncanonical autophagy pathway, the lysosome-dependent cell death pathway, or other unrecognized signaling pathway may be alternative novel therapeutic targets to prevent cells or tissues from undergoing death.
Author contributions
Fugui Zhang conceived the study and designed experiments. Yaping Ma, Chaofan Li and Tiwei Fu performed experiments, analyzed data and produced figures. Li Song and Fugui Zhang drafted the article. Yan He and Qingsong Ye provided critical input and expertise and revised the article.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81400493), Scientific and Technological Research Program of Chongqing Municipal Education Commission (No. KJQN20200429), and Joint Medical Research Project by Chongqing Health Commission and Natural Science Foundation of Chongqing (No. 2020GDRC008).
Conflict of interests
The authors have no conflict of interests to declare.
Acknowledgements
We would like to appreciate Professor Deqin Yang at Stomatological Hospital of Chongqing Medical University for the kind offer of HUVECs and Dr. Meredith August at Harvard School of Dental Medicine for her excellent help. We thank Alison Sherwin, PhD, and Catherine Perfect, MA (Cantab), from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Data availability statement
Supplementary data to this article can be found online at https://data.mendeley.com/datasets/hjym79tgn6/1.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Qingsong Ye, Email: qingsongye@hotmail.com.
Fugui Zhang, Email: 500290@hospital.cqmu.edu.cn.
References
- 1.Xue Z., Zhang Y., Liu Y., et al. PACAP neuropeptide promotes Hepatocellular Protection via CREB-KLF4 dependent autophagy in mouse liver Ischemia Reperfusion Injury. Theranostics. 2020;10(10):4453–4465. doi: 10.7150/thno.42354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dash S., Aydin Y., Moroz K. Chaperone-mediated autophagy in the liver: good or bad? Cells. 2019;8(11):1308. doi: 10.3390/cells8111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogg E.M., Abplanalp W.T., Bischof C., et al. Analysis of cell type-specific effects of MicroRNA-92a provides novel insights into target regulation and mechanism of action. Circulation. 2018;138(22):2545–2558. doi: 10.1161/CIRCULATIONAHA.118.034598. [DOI] [PubMed] [Google Scholar]
- 4.Sachdev U., Cui X., Hong G., et al. High mobility group box 1 promotes endothelial cell angiogenic behavior in vitro and improves muscle perfusion in vivo in response to ischemic injury. J Vasc Surg. 2012;55(1):180–191. doi: 10.1016/j.jvs.2011.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han F., Chen Y.X., Lu Y.M., et al. Regulation of the ischemia-induced autophagy-lysosome processes by nitrosative stress in endothelial cells. J Pineal Res. 2011;51(1):124–135. doi: 10.1111/j.1600-079X.2011.00869.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Tao T.Q., Song D.D., Liu X.H. Calreticulin ameliorates hypoxia/reoxygenation-induced human microvascular endothelial cell injury by inhibiting autophagy. Shock. 2018;49(1):108–116. doi: 10.1097/SHK.0000000000000905. [DOI] [PubMed] [Google Scholar]
- 7.Zhu T., Yao Q., Wang W., Yao H., Chao J. iNOS induces vascular endothelial cell migration and apoptosis via autophagy in ischemia/reperfusion injury. Cell Physiol Biochem. 2016;38(4):1575–1588. doi: 10.1159/000443098. [DOI] [PubMed] [Google Scholar]
- 8.Wu H., Ding J., Li S., et al. Metformin promotes the survival of random-pattern skin flaps by inducing autophagy via the AMPK-mTOR-TFEB signaling pathway. Int J Biol Sci. 2019;15(2):325–340. doi: 10.7150/ijbs.29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chichger H., Rounds S., Harrington E.O. Endosomes and autophagy: regulators of pulmonary endothelial cell homeostasis in health and disease. Antioxid Redox Signal. 2019;31(13):994–1008. doi: 10.1089/ars.2019.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong W., Liao Y., Qin J.Y., Li W.H., Tang Z.Y. Adverse effects of chemoradiotherapy on invasion and metastasis of tumor cells. Genes Dis. 2020;7(3):351–358. doi: 10.1016/j.gendis.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Gao A., Feng D., et al. Evaluation of the protective potential of brain microvascular endothelial cell autophagy on blood-brain barrier integrity during experimental cerebral ischemia-reperfusion injury. Transl Stroke Res. 2014;5(5):618–626. doi: 10.1007/s12975-014-0354-x. [DOI] [PubMed] [Google Scholar]
- 12.Bhogal R.H., Weston C.J., Velduis S., et al. The reactive oxygen species-mitophagy signaling pathway regulates liver endothelial cell survival during ischemia/reperfusion injury. Liver Transpl. 2018;24(10):1437–1452. doi: 10.1002/lt.25313. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Li J., Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience. 2017;354:1–10. doi: 10.1016/j.neuroscience.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Shi W., Wei X., Wang Z., et al. HDAC9 exacerbates endothelial injury in cerebral ischaemia/reperfusion injury. J Cell Mol Med. 2016;20(6):1139–1149. doi: 10.1111/jcmm.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Q., Cao Y., Xiong W., et al. Glycyrrhizic acid exerts protective effects against hypoxia/reoxygenation-induced human coronary artery endothelial cell damage by regulating mitochondria. Exp Ther Med. 2020;20(1):335–342. doi: 10.3892/etm.2020.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W.L., Yu S.P., Chen D., et al. The regulatory role of NF-kappaB in autophagy-like cell death after focal cerebral ischemia in mice. Neuroscience. 2013;244:16–30. doi: 10.1016/j.neuroscience.2013.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J., Lin R., Li S., et al. Salvianolic acid B promotes the survival of random-pattern skin flaps in rats by inducing autophagy. Front Pharmacol. 2018;9:1178. doi: 10.3389/fphar.2018.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Bao G., ALyafeai E., et al. Betulinic acid enhances the viability of random-pattern skin flaps by activating autophagy. Front Pharmacol. 2019;10:1017. doi: 10.3389/fphar.2019.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R., Chen H., Callow D., et al. Multifaceted effects of astragaloside IV on promotion of random pattern skin flap survival in rats. Am J Transl Res. 2017;9(9):4161–4172. [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J., Lin R., Li S., et al. Protective effects of resveratrol on random-pattern skin flap survival: an experimental study. Am J Transl Res. 2019;11(1):379–392. [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z., Zhang C., Ma H., et al. Detrimental effect of sitagliptin induced autophagy on multiterritory perforator flap survival. Front Pharmacol. 2020;11:951. doi: 10.3389/fphar.2020.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Jin Z., Wang J., et al. Detrimental effect of Hypoxia-inducible factor-1alpha-induced autophagy on multiterritory perforator flap survival in rats. Sci Rep. 2017;7(1):11791. doi: 10.1038/s41598-017-12034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun H., Lei D., Qifang Y., Yuan X., Deqin Y. Effects of concentrated growth factors on the angiogenic properties of dental pulp cells and endothelial cells: an in vitro study. Braz Oral Res. 2018;32:e48. doi: 10.1590/1807-3107bor-2018.vol32.0048. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F., Li Y., Zhang H., et al. Anthelmintic mebendazole enhances cisplatin's effect on suppressing cell proliferation and promotes differentiation of head and neck squamous cell carcinoma (HNSCC) Oncotarget. 2017;8(8):12968–12982. doi: 10.18632/oncotarget.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L., Gao L.N., Wang J., et al. Stromal cell-derived factor-1 α alleviates calcium-sensing receptor activation-mediated ischemia/reperfusion injury by inhibiting caspase-3/caspase-9-induced cell apoptosis in rat free flaps. BioMed Res Int. 2018;2018:8945850. doi: 10.1155/2018/8945850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClung J.M., McCord T.J., Keum S., et al. Skeletal muscle-specific genetic determinants contribute to the differential strain-dependent effects of hindlimb ischemia in mice. Am J Pathol. 2012;180(5):2156–2169. doi: 10.1016/j.ajpath.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodarac A., Saric T., Oberwallner B., et al. Susceptibility of murine induced pluripotent stem cell-derived cardiomyocytes to hypoxia and nutrient deprivation. Stem Cell Res Ther. 2015;6(1):83. doi: 10.1186/s13287-015-0057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mertins P., Tang L.C., Krug K., et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632–1661. doi: 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourassa S., Fournier F., Nehme B., et al. Evaluation of iTRAQ and SWATH-MS for the quantification of proteins associated with insulin resistance in human duodenal biopsy samples. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0125934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jylha A., Nattinen J., Aapola U., et al. Comparison of iTRAQ and SWATH in a clinical study with multiple time points. Clin Proteonomics. 2018;15:24. doi: 10.1186/s12014-018-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tekirdag K., Cuervo A.M. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J Biol Chem. 2018;293(15):5414–5424. doi: 10.1074/jbc.R117.818237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang D., Kang R., Berghe T.V., Vandenabeele P., Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew B., Chennakesavalu M., Sharma M., et al. Autophagy and post-ischemic conditioning in retinal ischemia. Autophagy. 2021;17(6):1479–1499. doi: 10.1080/15548627.2020.1767371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi M.E. Autophagy in kidney disease. Annu Rev Physiol. 2020;82:297–322. doi: 10.1146/annurev-physiol-021119-034658. [DOI] [PubMed] [Google Scholar]
- 35.Guixe-Muntet S., de Mesquita F.C., Vila S., et al. Cross-talk between autophagy and KLF2 determines endothelial cell phenotype and microvascular function in acute liver injury. J Hepatol. 2017;66(1):86–94. doi: 10.1016/j.jhep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 36.Chen S., Zhou C., Yu H., et al. 27-Hydroxycholesterol contributes to lysosomal membrane permeabilization-mediated pyroptosis in Co-cultured SH-SY5Y cells and C6 cells. Front Mol Neurosci. 2019;12:14. doi: 10.3389/fnmol.2019.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M., Pi H., Xi Y., et al. KIF5A-dependent axonal transport deficiency disrupts autophagic flux in trimethyltin chloride-induced neurotoxicity. Autophagy. 2021;17(4):903–924. doi: 10.1080/15548627.2020.1739444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey K., Yu X.W., Steinmetz A., Alberini C.M. Autophagy coupled to translation is required for long-term memory. Autophagy. 2021;17(7):1614–1635. doi: 10.1080/15548627.2020.1775393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B., Xue Q., Guo J., et al. Autophagy induction by the pathogen receptor NECTIN4 and sustained autophagy contribute to peste des petits ruminants virus infectivity. Autophagy. 2020;16(5):842–861. doi: 10.1080/15548627.2019.1643184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian X., Bai Y., Su X., Zhao G., Sun G., Li D. Knockdown of periostin attenuates 5/6 nephrectomy-induced intrarenal renin-angiotensin system activation, fibrosis, and inflammation in rats. J Cell Physiol. 2019;234(12):22857–22873. doi: 10.1002/jcp.28849. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Li X., Jin A. Rapamycin inhibits Nf-KappaB activation by autophagy to reduce catabolism in human chondrocytes. J Invest Surg. 2020;33(9):861–873. doi: 10.1080/08941939.2019.1574321. [DOI] [PubMed] [Google Scholar]
- 42.Cao Q., You X., Xu L., Wang L., Chen Y. PAQR3 suppresses the growth of non-small cell lung cancer cells via modulation of EGFR-mediated autophagy. Autophagy. 2020;16(7):1236–1247. doi: 10.1080/15548627.2019.1659654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Long Z., Ding Y., et al. Dihydroartemisinin ameliorates learning and memory in Alzheimer's disease through promoting autophagosome-lysosome fusion and autolysosomal degradation for Aβ clearance. Front Aging Neurosci. 2020;12:47. doi: 10.3389/fnagi.2020.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., Liu L., Shu Y.Q., et al. Blockade of HCN2 channels provides neuroprotection against ischemic injury via accelerating autophagic degradation in hippocampal neurons. Neurosci Bull. 2020;36(8):875–894. doi: 10.1007/s12264-020-00513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Li Y., Guan Y., et al. TNFAIP8L2/TIPE2 impairs autolysosome reformation via modulating the RAC1-MTORC1 axis. Autophagy. 2021;17(6):1410–1425. doi: 10.1080/15548627.2020.1761748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benito-Cuesta I., Ordonez-Gutierrez L., Wandosell F. AMPK activation does not enhance autophagy in neurons in contrast to MTORC1 inhibition: different impact on β-amyloid clearance. Autophagy. 2020:1–16. doi: 10.1080/15548627.2020.1728095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negoita F., Blomdahl J., Wasserstrom S., et al. PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J Cell Biochem. 2019;120(1):343–356. doi: 10.1002/jcb.27378. [DOI] [PubMed] [Google Scholar]
- 48.Yang H., Shen H., Li J., Guo L.W. SIGMAR1/Sigma-1 receptor ablation impairs autophagosome clearance. Autophagy. 2019;15(9):1539–1557. doi: 10.1080/15548627.2019.1586248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie W., Zheng W., Liu M. BRF1 ameliorates LPS-induced inflammation through autophagy crosstalking with MAPK/ERK signaling. Genes Dis. 2018;5(3):226–234. doi: 10.1016/j.gendis.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pangare M., Makino A. Mitochondrial function in vascular endothelial cell in diabetes. J Smooth Muscle Res. 2012;48(1):1–26. doi: 10.1540/jsmr.48.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharif T., Martell E., Dai C., Singh S.K., Gujar S. Regulation of the proline regulatory axis and autophagy modulates stemness in TP73/p73 deficient cancer stem-like cells. Autophagy. 2019;15(5):934–936. doi: 10.1080/15548627.2019.1586321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary data to this article can be found online at https://data.mendeley.com/datasets/hjym79tgn6/1.