Abstract
Background:
In the medical imaging domain, deep learning based methods have yet to see widespread clinical adoption, in part due to limited generalization performance across different imaging devices and acquisition protocols. The deviation between estimated brain age and biological age is an established biomarker of brain health and such models may benefit from increased cross-site generalizability.
Purpose:
To develop and evaluate a deep learning based image harmonization method to improve cross-site generalizability of deep learning age prediction.
Study Type:
Retrospective
Population:
8876 subjects from 6 sites. Harmonization models were trained using all subjects. Age prediction models were trained using 2739 subjects from a single site and tested using the remaining 6137 subjects from various other sites.
Field Strength/Sequence:
Brain imaging with magnetization prepared rapid acquisition with gradient echo (MPRAGE) or spoiled gradient echo sequences (SPGR) at 1.5 and 3T
Assessment:
StarGAN v2, was used to perform a canonical mapping from diverse datasets to a reference domain to reduce site-based variation while preserving semantic information. Generalization performance of deep learning age prediction was evaluated using harmonized, histogram matched, and unharmonized data.
Statistical Tests:
Mean absolute error and Pearson correlation between estimated age and biological age quantified the performance of the age prediction model.
Results:
Our results indicated a substantial improvement in age prediction in out-of-sample data, with the overall mean absolute error improving from 15.81 (±0.21) years to 11.86 (±0.11) with histogram matching to 7.21 (±0.22) years with GAN-based harmonization. In the multisite case, across the 5 out of sample sites, mean absolute error improved from 9.78 (±6.69) years to 7.74 (±3.03) years with histogram normalization to 5.32 (±4.07) years with GAN-based harmonization.
Data Conclusion:
While further research is needed, GAN-based medical image harmonization appears to be a promising tool for improving cross-site deep learning generalization.
Keywords: StarGAN, Deep Learning, Harmonization
INTRODUCTION
Deep learning models can produce useful results on a variety of prediction tasks in medical imaging, including segmentation [1], precision diagnostics [2] and prediction of clinical outcome [3]. However, they often perform inconsistently when applied to data obtained under different conditions such as different imaging devices, acquisition protocols, and patient populations [4]. This imaging heterogeneity can diminish the generalizability of prediction models as they also reflect irrelevant or confounding features. In the medical imaging domain, poor generalization presents a major limitation to the widespread clinical adoption of deep learning-based predictors [5, 6].
Restrictions in generalizability are a common characteristic of modeling high dimensional data with many degrees of freedom, which can be overcome partially if sufficiently large and diverse training sets are available [7]. While there has been progress recently arising from efforts to collect very large and diverse training sets through pooling data across multiple studies [8], it is often the case that the desired labeled data (ie. clinical or genomic variables) are available only for a subset of the studies. Importantly, even if a sufficiently large and diverse labelled dataset can be brought together to support extensive training of a machine learning model, continuous advances in imaging protocols as well as in biomarker and clinical measurements will change the characteristics of future data, thereby raising the need for re-acquiring a new training dataset. Therefore, to date, machine learning-based methods have seen limited applicability in the clinic relative to their potential.
Recently, advances in the computer vision community have shown how generative adversarial networks (GANs) [9] can be used to train deep learning models that are robust to adversarial perturbations [10]. For example, in the case of modeling natural variation, Robey et al. [11] used learnable models of variation, to train robust deep learning for factors such as weather conditions in street sign recognition and background color in digit recognition. In the medical imaging community, there have also been encouraging results using the GAN-based approach to model site-based variation in medical images. In particular, CycleGAN [12] has been used to perform unpaired image to image translation by learning a bijective mapping of scans between imaging sites. In this approach, scans retain their original semantic information through enforcement of an identity mapping back to the original data. Examples include studies by Gao et al. [13], who used a CycleGAN based approach for intensity normalization on multi-site T2-FLAIR MRI data, Modanwal et al. [14], who developed a CycleGAN based image harmonization approach for dynamic contrast-enhanced breast MRI scans from two scanners, and Nguyen et al. [15], who investigated the use of CycleGAN to remove scanner effects at the image level between two sites. Nguyen et al. [16] subsequently investigated the use of the StarGAN v2 architecture for multi-site harmonization of neuroimaging data using a single generator discriminator pair. StarGAN v2 [17] (herein referred to as StarGAN) is an emerging method for unpaired image to image translation that has shown promising results, particularly when jointly learning mappings between multiple sites. In addition to these methods, Dewey et al. [18] and Zuo et al. [19] proposed an alternate harmonization approach using T1 and T2 image pairs to disentangle imaging content from imaging “style”. Additionally, some other deep learning based approaches to site harmonization have been proposed, but their usefulness has been limited by factors such as requiring training data to include paired subjects who have been imaged at both sites [20–22], a condition which is very difficult to meet in practice with sufficiently large and continuously updated training samples. To the best of our knowledge, prior work has not demonstrated the effectiveness of GAN-based harmonization in downstream prediction tasks. An example of such a task is brain age estimation from MRI data. Brain age is an established biomarker of overall brain health and the accuracy of estimation is dependent on neuroanatomical patterns that can be obfuscated by imaging variation across sites.
Thus the aim of this study was to use a StarGAN style model, to perform inter-site mapping of MRI brain neuroimaging data and to show: (1) that StarGAN was able to model domain level variation from unpaired data, while preserving semantic information within the data [17], and (2) that StarGAN harmonization improved the accuracy of brain age prediction models.
METHODS
Datasets
This retrospective study represents a pooled set of imaging studies. These studies received oversight and approval from their respective ethics comities as well as a waiver for written informed consent. All subjects included in this work were determined to be healthy according to criteria established by each individual study. Five of the studies were performed using magnetization prepared rapid acquisition with gradient echo (MPRAGE) sequences and one study was performed with the spoiled gradient echo (SPGR) sequence. Three of the studies were performed using 3T scanners and three used 1.5T scanners. See Table 1 for more details.
Table 1:
Description of the data used for age prediction
| TOTAL SUBJECTS = 8876 | Dataset1 (REFERENCE SITE) | Dataset2 | Dataset3 | Dataset4 | Dataset5 | Dataset6 |
|---|---|---|---|---|---|---|
| TOTAL | 2739 | 952 | 446 | 90 | 247 | 4402 |
| ACQUISITION PROTOCOL – FIELD STRENGTH | MPRAGE – 1.5T | MPRAGE – 3T | MPRAGE – 1.5T | SPGR – 1.5T | MPRAGE – 3T | MPRAGE – 3T |
| SCANNER | Siemens Magnetom Avanto | Philips | Siemens Avanto | GE Signa | Siemens Tim Trio | Siemens Skyra (VD13) |
| STUDY | SHIP | BLSA | AIBL | BLSA | PAC | UK Biobank |
| MEAN AGE (AGE RANGE) | 52.55 (21 - 91) | 67.04 (22 - 96) | 72.77 (60 - 92) | 72.77 (56 - 86) | 61.19 (42 - 77) | 63.20 (45 - 80) |
| RESOLUTION (PER AXIAL SLICE) | 256x256 | 256x256 | 240x256 | 256x256 | 256x256 | 256x256 |
| CITATION | [36] | [37] | [38] | [37] | [39] | [40] |
For age prediction with a single out of sample site, we used two large datasets of T1-weighted brain MRI scans that covered a wide range of ages, Dataset1 (n = 2739, mean age = 52.55, std = 9.27) and Dataset2 (n =952, mean age = 67.04, std = 14.29). Dataset1 was used as the canonical reference domain and training dataset for the age prediction model. See Table 1 for more details.
For multisite age prediction, we used six datasets. Five (n = 6137) were used for the out of sample evaluation and Dataset1 was used as the canonical reference domain and training dataset for the age prediction model. See Table 1 for more details.
Preprocessing
The scans were skull-stripped using an automated multi-atlas label fusion method [23], then affinely registered to a common atlas using FMRIB’s Linear Image Registration Tool FLIRT [24]. Finally, the scans underwent a quality control procedure using automatic outlier detection to flag cases for manual verification (cases reviewed by G. E. – 11 years experience).
Histogram Normalization
For an additional baseline, we compare the StarGAN harmonization method to traditional histogram matching that is commonly used for cross-site image normalization. In our experiments, we replicate our age prediction methodology with scans that have been histogram matched using the Nyúl and Udupa method [25, 26] as implemented in this python package [27].
Image Harmonization to a Reference Domain
StarGAN consists of a style encoder, content encoder, generator, and discriminator. Both the style encoder and the content encoder are convolutional neural networks that map a single axial slice from a T1-weight scan to some lower dimensional representation. In the case of the style encoder, the network learns a mapping of slices to an 8 dimensional vector representing the site-based variation of that slice. In the case of the content encoder, the network learns a mapping of slices to a lower dimensional set of convolutional filters representing the anatomical information of the slice. The generator, a generative adversarial network, then takes in both the style and content encodings and produces a harmonized image with the respective style and content of the input encodings [17]. In this way, a harmonized scan is produced that matches the site-based variation of a reference scan while retaining its original anatomical information. The discriminator network is used during optimization to facilitate adversarial training. This network attempts to discriminate which site a sample originates from and if the sample originates from a real scan or is a synthetic image produced by the generator network. The predictions of this network are then used to optimize the generator to produce more realistic results in order to “fool” the discriminator. In order to ensure that the site-based variation is appropriately encoded into the synthetic image, the distance between the style encodings of the original reference image and the synthetic image is minimized. See supplemental 1 for more details.
For our experiments, we use StarGAN to learn the mapping between unpaired axial slices of scans in our reference domain and out-of-sample sites. As opposed to the age prediction experiments, where only slices from the reference domain were used for training, the StarGAN training uses scans from all sites. Slices were used in training if they contained above 1% of non-zero pixels after processing. Slices during training were randomly sampled to avoid a single scan populating all of the slices in a minibatch. We then apply this mapping to harmonize scans from our new site to the reference domain by using the style encoding of a scan in the reference domain and the content encoding of the scan we aim to harmonize. This harmonized data can then be used in our prediction model (Figure 1). For the purposes of this paper and after qualitative assessment, the style encoding was limited to a vector of length 8. See Supplemental Table 1 for more information.
Figure 1:
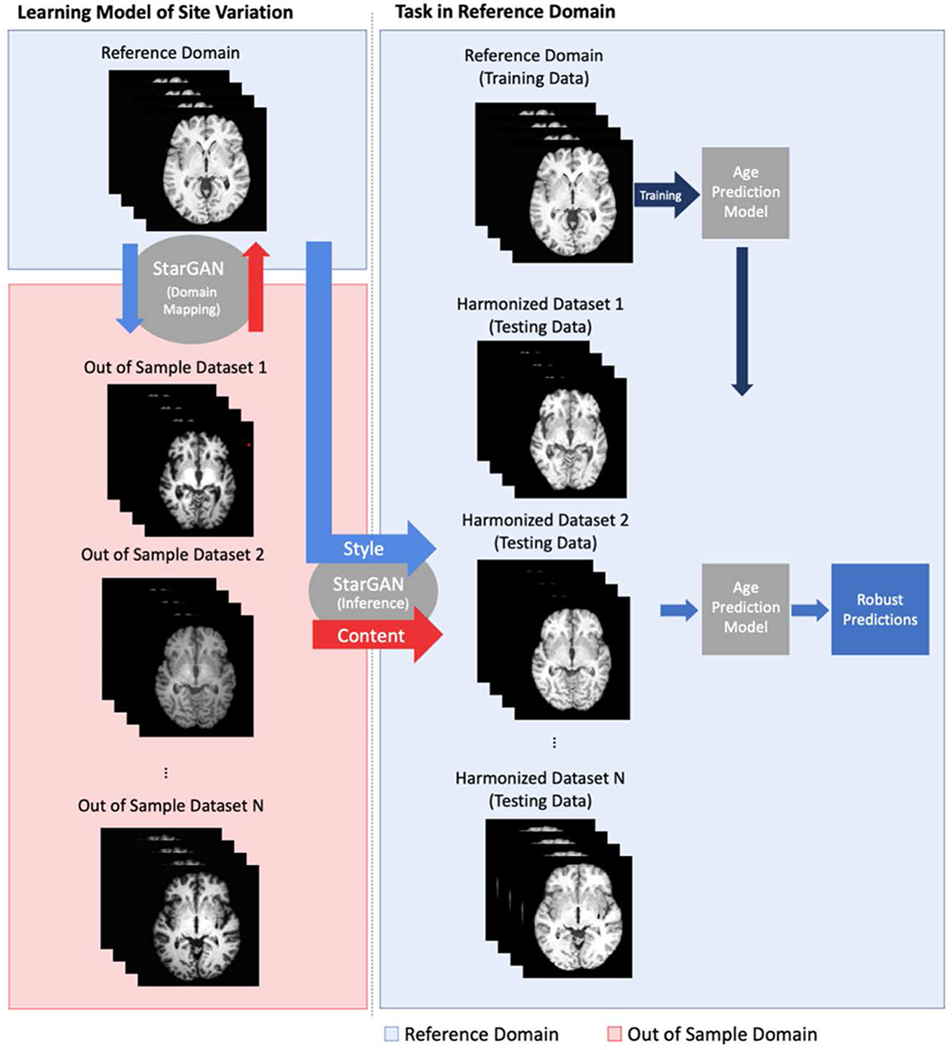
Description of harmonization and prediction workflow. A multimodal mapping is learned between the reference domain and the out-of-sample domains. The harmonized data is obtained using the style encoding from the reference domain and the content encoding of the original image. The age prediction model, trained on data in the reference domain, can then be used on the harmonized data for improved generalization ability.
Pytorch [28] was used to perform all the deep learning experiments in this paper. Models were trained on 2 P100 GPUs with 12GB of vRAM each. Training for the multisite harmonization model took ~20 hours and the multisite age prediction model took 4 hours.
Age Prediction
Age prediction was used to determine the improvement in cross-site prediction generalizability across unharmonized, histogram matched, and GAN harmonized scans. In each of these experiments the prediction model is only trained on Dataset 1 and evaluated on the remaining datasets. In the case of harmonized experiments, the prediction model was trained only on Dataset 1 and evaluated on the histogram or GAN-based transformation of the testing site or sites.
Brain age prediction was modeled after the methodology used in [29]. See Supplementary Figure 1 for a visual depiction of the age prediction methods. Age prediction was performed using a 10-layer ResNet model, which has been shown to perform well on a variety of imaging tasks [30]. Our model used a combination of convolutional and max pooling layers to incrementally reduce the dimensionality of images. After the final max pooling layer, we flattened the output and passed it through a fully connected layer of size 512 with 50% dropout and RELU activation. We attached a single output node with a linear activation, whereby the network could be optimized using mean squared loss. Models were optimized using the Adam optimizer [31] with a learning rate of 3e-4. This learning rate was decreased by a factor of 10 when the training loss remained constant for 5 consecutive epochs. The network was considered to have converged when the training loss was constant for 10 epochs or when the validation loss increased for 5 consecutive epochs. Image augmentation was performed during training with random horizontal flips of images and intensity variation (+/− 5%).
The network was trained using the middle 80 axial slices of each MRI scan in Dataset1, where each slice was treated as an independent training sample. During training, all slices in the training set were randomly shuffled to avoid a single scan populating all the slices in a minibatch. For testing, the median prediction of a scan was used. A baseline in-sample model was trained and evaluated on Dataset1 with 5-fold cross-validation in which training, validation, and testing sets were split at the subject level, such that all slices from a single subject would only be contained in a single set per fold. In the cross-validated experiments, 60% of the total subjects were selected in each fold to be used as the training data to the network. A non-overlapping, 20% of the total subjects are used as the validation set. The mean absolute error and loss on the validation set was evaluated at each epoch in training. These metrics were used to determine when training has been completed and to prevent overfitting. The remaining 20% of subjects are used in the testing set to evaluate the final performance of the fold. This is repeated 5 times until all subjects have been predicted from the test set.
Statistical Analysis
Mean absolute error, Pearson correlation, and Lin’s Concordance Correlation [32] between estimated age and actual biological age were used to quantify the performance of the age prediction models, as biological age was used as the target value during training. All evaluation and statistical analysis was performed using Python.
RESULTS
Capturing Confounding Inter-scanner Variations via StarGAN
Visually, image characteristics became markedly more uniform in terms of grey/white matter contrast, overall and regional intensity, and noise patterns, after canonical mapping via StarGAN. Figure 2 shows that, characterized by the intensity distributions of tissue types, the regional intensities of the mapped image aligned more closely with that of the image from the reference domain than the unmapped image. Additionally, Figure 2 shows that the mean intensity and the standard deviation maps of the mapped image are more similar to the reference domain than those of the unmapped image. Visual characteristics were assessed by 3 neuroradiologists with 13-50 years of experience. See Supplemental Figure 3 for difference maps between harmonized and unharmonized slices from each site.
Figure 2:
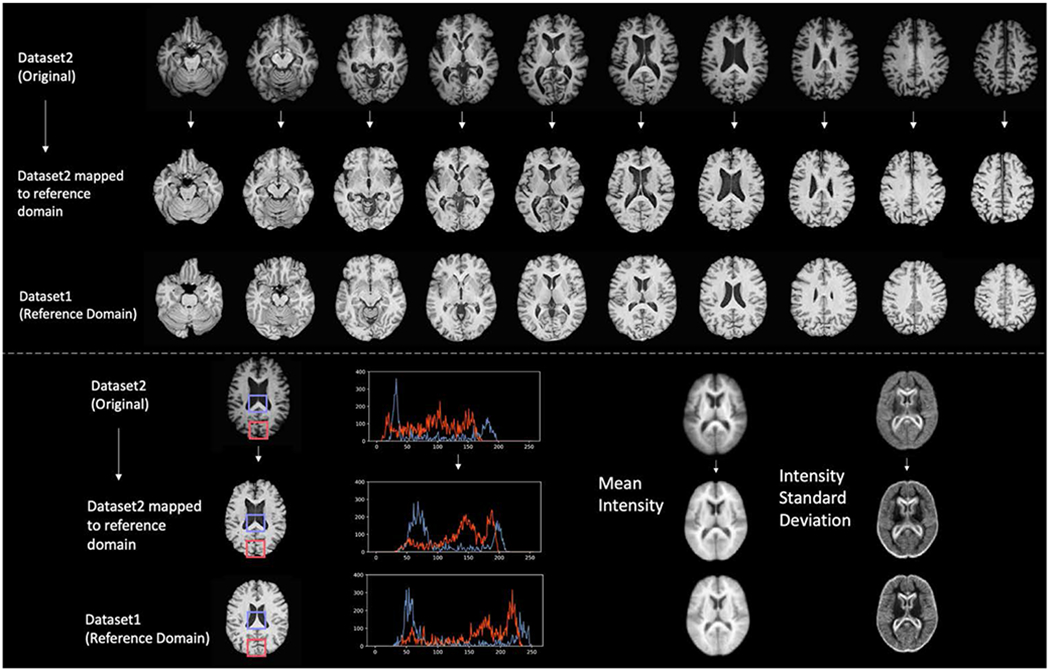
Top: Representation of mapping of axial slices of an example participant from Dataset2 to the reference domain (Dataset 1). Bottom left: Comparison of regional histograms before and after mapping to reference domain. Bottom right: Mean and standard deviation maps across all scans.
Age Prediction
The baseline model (training predictor on Dataset 1 and testing on Dataset 2) resulted in poor generalization ability (Mean Absolute Error (MAE) = 15.81 years, Pearson correlation coefficient = 0.299). We see some improvement using histogram matched scans (MAE = 11.86 years, Pearson correlation coefficient = 0.341).
Using the harmonized data in the single site setting, where Dataset 2 was mapped to the reference domain (Dataset 1, n=2739) and the predictor was trained on the reference dataset (Dataset 1), there was a large improvement in the generalization performance of our predictor (MAE = 7.21 years, Pearson correlation coefficient = 0.779). The results presented in Table 2 are the average of five experiments. For reference, the 5-fold cross-validated accuracy of the age prediction model on Dataset1 was, MAE = 5.24 years and Person correlation coefficient = 0.866.
Table 2:
Age prediction results with model trained on Dataset1 and tested on Dataset2
| HARMONIZED | MAE | PEARSON CORRELATION | CONCORDANCE CORRELATION [32] |
|---|---|---|---|
| No | 15.81 (±0.21) | 0.299 (±0.018) | 0.169 (±0.024) |
| Histogram Matched | 11.86 (± 0.11) | 0.341 (±0.009) | 0.298 (±0.011) |
| GAN | 7.21 (±0.22) | 0.779 (±0.017) | 0.701 (±0.030) |
In the multi-site setting, where 5 datasets were mapped to the reference domain and the predictor was trained on the reference dataset (Dataset 1, n=2739), there was consistent improvement in age prediction performance across all sites in terms of MAE and Pearson correlation (Table 3). With an overall improvement in terms of mean absolute error from 9.78 (±6.69) years to 7.74 (±3.03) years with histogram normalization to 5.32 (±4.07) years with GAN harmonization. The Pearson correlation improved from 0.252 (±0.044) to 0.600 (±0.032) with histogram normalization to 0.870(±0.033) with GAN harmonization. The results are the average of five experiments. See Supplemental Figure 2 for Bland Altman plots of age prediction results broken down by site. See Supplemental Table 2 for an analysis on the proportion of unacceptable predictions (>20% of actual age).
Table 3:
Multisite age prediction results with unharmonized, histogram matched, and GAN harmonized data. The prediction model was only trained with scans from the reference domain (Dataset1). Mean and standard deviation of mean absolute error (MAE), Pearson correlation coefficient (between predicted and actual age), and Lin’s Concordance Correlation (between predicted and actual age) from five runs is shown.
| SITE | HARMONIZED | MAE | PEARSON CORRELATION | CONCORDANCE CORRELATION |
|---|---|---|---|---|
| DATASET2 | No | 14.43 (±8.77) | 0.206 (±0.067) | 0.187 (±0.032) |
| Histogram Matched | 11.52 (±2.62) | 0. 649 (±0.017) | 0.452 (±0.009) | |
| GAN | 7.46 (±5.17) | 0.864 (±0.016) | 0.728 (±0.021) | |
| DATASET3 | No | 14.77 (±6.39) | 0.222 (±0.063) | 0.069 (±0.045) |
| Histogram Matched | 8.74 (±5.34) | 0.493 (±0.043) | 0.258 (±0.021) | |
| GAN | 6.74 (±4.35) | 0.646 (±0.028) | 0.455 (±0.024) | |
| DATASET4 | No | 14.71 (±6.94) | 0.472 (±0.049) | 0.112 (±0.033) |
| Histogram Matched | 11.29 (±3.35) | 0.695 (±0.41) | 0.219 (±0.018) | |
| GAN | 7.42 (±5.07) | 0.666 (±0.042) | 0.516 (±0.032) | |
| DATASET5 | No | 7.94 (±5.16) | 0.334 (±0.092) | 0.174 (±0.057) |
| Histogram Matched | 6.48 (±1.53) | 0.573 (±0.010) | 0.452(±0.008) | |
| GAN | 5.29 (±3.55) | 0.752 (±0.059) | 0.624 (±0.045) | |
| DATASET6 | No | 8.27 (±5.39) | 0.256 (±0.052) | 0.148 (±0.055) |
| Histogram Matched | 6.60 (±1.46) | 0.541 (±0.009) | 0.381 (±0.008) | |
| GAN | 4.67 (±5.54) | 0.756 (±0.037) | 0.627 (±0.036) | |
| ALL | No | 9.78 (±6.69) | 0.252 (±0.044) | 0.149 (±0.032) |
| Histogram Matched | 7.74 (±3.03) | 0.600 (±0.032) | 0.403 (±0.018) | |
| GAN | 5.32 (±4.07) | 0.870 (±0.033) | 0.698 (±0.031) |
DISCUSSION
In this study, we employed a paradigm whereby confounding variation may be removed via canonical mapping across scans acquired at different sites, thereby enabling smaller less diverse datasets to be useful for model construction and to be robust to subsequent variations of image characteristics. Critically, the canonical mapping model derived from unlabeled datasets can continue to evolve, as image acquisition itself evolves, hence allowing for new types of data to be canonically mapped, and therefore retaining the value of classification models derived from relatively limited labeled scans. Such a method has the potential to enable the widespread adoption and standardization of deep learning-based methods, both because it avoids the need for specialized and sophisticated processing of the images, but also because of the good generalization properties achieved via canonical mapping.
StarGAN Harmonization
We examined an image-level harmonization method capable of learning a robust canonical mapping to a reference domain. Our harmonization schema attempted to identify the complex and non-linear imaging variation occurring due to confounding factors, such as scanners, acquisition protocols, and cohorts, while maintaining inter-subject variation related to classification labels. Through harmonization, we observed improvements in image consistency with the reference domain, specifically in terms of grey/white matter contrast, overall intensity, and noise patterns. We found that the use of StarGAN versus prior methods offered benefits in the modeling of multisite data. Instead of modeling pair-wise transformations between sites, StarGAN allowed a joint modeling of site variation, reusing model weights learned across all site transformations. We note that there are various hyperparameters that need to be specified within the harmonization network, in particular, the dimensionality of the style and content encoding. The dimensionality of the style encoding is potentially useful in limiting the amount of variation that can be captured in the harmonization process.
While prior work has shown that GAN-based methods can be powerful for image-level domain adaptation [12], in the medical imaging context the preservation of fine anatomical structure and predictive information is critical, and the downstream effect of harmonization needs to be carefully evaluated. While there is certainly a risk of overfitting and removing non-site variation when using highly non-linear methods, we demonstrated the preservation of this predictive information in neuroimaging data through our experiments on brain age estimation.
Age Prediction
Brain age estimation has become an established biomarker of overall brain, exhibiting overlapping neuroanatomical patterns with a variety of other pathologic processes [29, 33]. Accurate brain age estimation is dependent on fine neuroanatomical patterns that can be obfuscated by imaging variation across sites [34]. Therefore, it is a prime candidate to assess harmonization performance. We demonstrated substantially improved age prediction generalization in five separate sites, following their mapping to the reference domain, in which the predictor was trained. The brain age prediction model was able to perform reasonably well on the out of sample data, indicating that GAN-based harmonization may be a useful tool in multi-site image level harmonization tasks. We note that the generalization performance to the out of sample data is still short of the cross-validated performance on Dataset1, indicating potential for improvements in the harmonization methodology.
We recognize that the age prediction performance, particularly in the harmonized case, can certainly be improved with more specialized prediction networks, optimization techniques, hyperparameter selection, and larger sample sizes. However, our aim was to simply demonstrate improvements with harmonized data with a non-optimized, commonly used network and a reasonable sample size, in view of the scarcity of large, labeled training datasets. We anticipate that future work will incorporate harmonization to a reference domain with finely tuned networks to potentially construct powerful and generalizable imaging predictors. Particularly, in the case of brain age prediction, we also note that the “tightest” fitting model may not be the best at identifying signals of accelerated aging. Highly fit models are incentivized to select features that show consistent aging pattern even in the presence of accelerated aging and may ignore the more general patterns of aging that we are interested in [29].
Limitations
While we have demonstrated a substantial improvement in downstream prediction performance with our particular prediction model, further work will certainly be needed to examine if other prediction methods and tasks will similarly benefit. In addition, while the predictive signal needed for accurate age prediction may be well preserved in the harmonization process, further evaluation on additional prediction tasks is needed to fully evaluate the merits of this harmonization method. Additionally, further work is required to investigate how image level harmonization techniques such as this behave when the reference domain and out of sample domains differ sharply across covariates (such as age, ethnicity, pathology). Some recent work investigating this phenomenon has shown that GAN-based methods can “hallucinate” features in these instances [35]. As it currently stands, it is important to consider group level differences between domains and how that might affect harmonization. Future directions of this work could involve the explicit modeling of such covariates directly within the network.
Conclusion
This work demonstrates the potential for StarGAN based harmonization in multisite T1-weighted MRI brain scans. We show that we can concurrently model and correct for the site effects of multiple scanners while retaining predictive information within scans. While there are certainly limitations in its current formulation, we show a substantial improvement in out-of-sample age prediction performance when using GAN harmonized images.
Supplementary Material
Funding
This work was supported in part by NIH grants RF1AG054409, R01EB022573, R01MH112070, R01MH120482, R01 MH113565, and NIH contract HHSN271201600059C.This work was also supported in part by the Intramural Research Program, National Institute on Aging, NIH, and the Swiss National Science foundation grant 191026. SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. MRI scans in SHIP and SHIP-TREND have been supported by a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania.
Footnotes
Code Availability
The code used in this project will be made available upon publishing.
Data Availability
The data that support the findings of this study are available from their respective institutions. Restrictions apply to the availability of these data, which were used under license for the current study, and are not publicly available. Data may however be available from the authors upon reasonable request and with permission.
References
- 1.Kamnitsas K, et al. , Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical image analysis, 2017. 36: p. 61–78. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, et al. , Landmark-based deep multi-instance learning for brain disease diagnosis. Medical image analysis, 2018. 43: p. 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhagwat N, et al. , Modeling and prediction of clinical symptom trajectories in Alzheimer’s disease using longitudinal data. PLoS computational biology, 2018. 14(9): p. e1006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takao H, Hayashi N, and Ohtomo K, Effect of scanner in longitudinal studies of brain volume changes. J Magn Reson Imaging, 2011. 34(2): p. 438–44. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, et al. , Understanding deep learning (still) requires rethinking generalization. Commun. ACM, 2021. 64(3): p. 107–115. [Google Scholar]
- 6.Zech JR, et al. , Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS medicine, 2018. 15(11): p. e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neyshabur B, et al. Exploring generalization in deep learning. in Advances in neural information processing systems. 2017. [Google Scholar]
- 8.Erus G, Habes M, and Davatzikos C, Machine learning based imaging biomarkers in large scale population studies: A neuroimaging perspective, in Handbook of Medical Image Computing and Computer Assisted Intervention. 2020, Elsevier. p. 379–399. [Google Scholar]
- 9.Goodfellow IJ, et al. Generative Adversarial Networks. arXiv e-prints, 2014. arXiv:1406.2661. [Google Scholar]
- 10.Samangouei P, Kabkab M, and Chellappa R: Defense-GAN Protecting Classifiers Against Adversarial Attacks Using Generative Models. arXiv e-prints, 2018. arXiv:1805.06605. [Google Scholar]
- 11.Robey A, Hassani H, and Pappas GJ Model-Based Robust Deep Learning. arXiv e-prints, 2020. arXiv:2005.10247. [Google Scholar]
- 12.Zhu J-Y, et al. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. arXiv e-prints, 2017. arXiv:1703.10593. [Google Scholar]
- 13.Gao Y, et al. , A Universal Intensity Standardization Method Based on a Many-to-One Weak-Paired Cycle Generative Adversarial Network for Magnetic Resonance Images. IEEE Transactions on Medical Imaging, 2019. 38(9): p. 2059–2069. [DOI] [PubMed] [Google Scholar]
- 14.Modanwal G, et al. , MRI image harmonization using cycle-consistent generative adversarial network. SPIE Medical Imaging. Vol. 11314. 2020: SPIE. [Google Scholar]
- 15.Nguyen H, et al. , Correcting differences in multi-site neuroimaging data using Generative Adversarial Networks. arXiv preprint arXiv:1803.09375, 2018. [Google Scholar]
- 16.Nguyen H, et al. Correcting differences in multi-site neuroimaging data using Generative Adversarial Networks. 2018. arXiv:1803.09375. [Google Scholar]
- 17.Choi Y, et al. StarGAN v2: Diverse Image Synthesis for Multiple Domains. 2019. arXiv:1912.01865. [Google Scholar]
- 18.Dewey B, et al. , A Disentangled Latent Space for Cross-Site MRI Harmonization. 2020. p. 720–729. [Google Scholar]
- 19.Zuo L, et al. Information-based Disentangled Representation Learning for Unsupervised MR Harmonization. 2021. arXiv:2103.13283. [Google Scholar]
- 20.Dewey BE, et al. Deep harmonization of inconsistent MR data for consistent volume segmentation. in International Workshop on Simulation and Synthesis in Medical Imaging. 2018. Springer. [Google Scholar]
- 21.Dewey BE, et al. , DeepHarmony: A deep learning approach to contrast harmonization across scanner changes. Magnetic Resonance Imaging, 2019. 64: p. 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nath V, et al. Inter-scanner harmonization of high angular resolution DW-MRI using null space deep learning. in International Conference on Medical Image Computing and Computer-Assisted Intervention. 2018. Springer. [PMC free article] [PubMed] [Google Scholar]
- 23.Doshi J, et al. , Multi-atlas skull-stripping. Academic radiology, 2013. 20(12): p. 1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkinson M, et al. , Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 2002. 17(2): p. 825–841. [DOI] [PubMed] [Google Scholar]
- 25.Nyúl LG and Udupa JK, On standardizing the MR image intensity scale. Magn Reson Med, 1999. 42(6): p. 1072–81. [DOI] [PubMed] [Google Scholar]
- 26.Shah M, et al. , Evaluating intensity normalization on MRIs of human brain with multiple sclerosis. Med Image Anal, 2011. 15(2): p. 267–82. [DOI] [PubMed] [Google Scholar]
- 27.Reinhold JC, et al. Evaluating the impact of intensity normalization on MR image synthesis. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paszke A, et al. , Automatic differentiation in pytorch. 2017. [Google Scholar]
- 29.Bashyam VM, et al. , MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He K, et al. Deep Residual Learning for Image Recognition. 2015. arXiv:1512.03385. [Google Scholar]
- 31.Kingma DP and Ba J Adam: A Method for Stochastic Optimization. 2014. arXiv:1412.6980. [Google Scholar]
- 32.Lin LI, A concordance correlation coefficient to evaluate reproducibility. Biometrics, 1989. 45(1): p. 255–68. [PubMed] [Google Scholar]
- 33.Jonsson BA, et al. , Brain age prediction using deep learning uncovers associated sequence variants. Nature Communications, 2019. 10(1): p. 5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franke K and Gaser C, Ten Years of BrainAGE as a Neuroimaging Biomarker of Brain Aging: What Insights Have We Gained? Frontiers in Neurology, 2019. 10(789). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen JP, Luck M, and Honari S. Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. 2018. Cham: Springer International Publishing. [Google Scholar]
- 36.Hegenscheid K, et al. Whole-body magnetic resonance imaging of healthy volunteers: pilot study results from the population-based SHIP study. in RöFo-Fortschritte auf dem Gebiet der Röntgenstrahlen und der bildgebenden Verfahren. 2009. © Georg Thieme Verlag KG Stuttgart; · New York. [DOI] [PubMed] [Google Scholar]
- 37.Shock NW, et al. , Normal Human Aging: The Baltimore Longitudinal Study on Aging. 1984. [Google Scholar]
- 38.Ellis KA, et al. , Addressing population aging and Alzheimer’s disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement, 2010. 6(3): p. 291–6. [DOI] [PubMed] [Google Scholar]
- 39.Pomponio R, et al. , Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage, 2020. 208: p. 116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudlow C, et al. , UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. Plos med, 2015. 12(3): p. e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from their respective institutions. Restrictions apply to the availability of these data, which were used under license for the current study, and are not publicly available. Data may however be available from the authors upon reasonable request and with permission.


