Abstract
Members of the gasdermin family contain positively charged N-terminal domains (NTDs) capable of binding phospholipids and assembling membrane pores, and C-terminal domains (CTDs) that bind the NTDs to prevent pore formation in the resting states. The flexible NTD-CTD linker regions of gasdermins are highly variable in length and sequences, which may be attributable to gasdermin recognition by diverse proteases. In addition, protease cleavage within the NTDs is known to inactivate several gasdermin family members. Recognition and cleavage of the gasdermin family members by different proteases share common and distinct features at the protease active sites, as well as exosites recently identified for the inflammatory caspases. Utilization of exosites may strengthen enzyme-substrate interaction, improve efficiency of proteolysis, and enhance substrate selectivity. It remains to be determined if the dual site recognition of gasdermin D (GSDMD) by the inflammatory caspases is employed by other GSDMD-targeting proteases, or is involved in proteolytic processing of other gasdermins. Biochemical and structural approaches will be instrumental in revealing how potential exosites in diverse proteases engage different gasdermin substrates. Different features of gasdermin sequence, structure, expression characteristics, and post-translational modifications may dictate distinct mechanisms of protease-dependent activation or inactivation. Such diverse mechanisms may underlie the divergent physiological and pathological functions of gasdermins, and furnish opportunities for therapeutic targeting of gasdermins in infectious diseases and inflammatory disorders.
Keywords: Gasdermin, Caspase, Exosite, Protease, Cleavage, Pyroptosis
Graphical Abstract
Introduction
The gasdermin family contains six paralogs: GSDMA, GSDMB, GSDMC, GSDMD, GSDME, and DFNB59 (Pejvakin or PJVK) (1, 2). The discovery of GSDMD as an executor molecule for pyroptosis sparked intense interests in the molecular mechanisms underlying the activation and regulation of the gasdermin family members in physiological and pathological conditions (3–5). All gasdermin family members except for PJVK adopt a two-domain structure, with their positively charged N-terminal domains (NTDs) capable of binding phospholipids and assembling membrane pores. Except for GSDMB and PJVK, the negatively charged C-terminal domains (CTDs) of gasdermins bind the NTDs to prevent pore formation in the resting states. By contrast, the PJVK-NTD does not appear to induce cell death when expressed in mammalian cells (6), and the GSDMB-CTD does not appear to prevent its NTD from binding lipids (7). Besides its role as an autoinhibitory domain, the GSDMD-CTD has been reported to function as a recruitment module for the inflammatory caspases (8, 9). Emerging evidence suggests that both NTDs and CTDs adopt similar overall folds and engage in intramolecular domain-domain interaction using largely conserved interfaces (10, 11, 7, 12, 13). Structural studies of human and murine GSDMD and murine GSDMA3 have all revealed similar autoinhibited conformation in which extensive intramolecular domain interfaces are primarily mediated by the α1 helix and β1-β2 loop from the NTDs, which engage a largely hydrophobic pocket in the CTDs. Disruption of the CTD hydrophobic pocket led to disruption of autoinhibition, membrane permeabilization and pyroptosis by the mutant full-length proteins (10, 12).
In contrast to the conservation of the gasdermin NTDs and CTDs, the flexible NTD-CTD linker regions are highly variable in length and sequences, which may be attributable to gasdermin activation by diverse proteases (Table 1) (2). The majority of known gasdermin-processing proteases are either inflammatory or apoptotic caspases. Cleavage of GSDMD by caspases-1, 4, 5, and 11 is an essential step in initiating pyroptosis and is the most well-studied among the gasdermins (8, 9). Prior to the landmark discovery of GSDMD as a pyroptosis executor, it was reported to be a caspase-1 substrate that has similar expression profiles as caspase-1 (14). Current literature reveals that cysteine and serine proteases such as the inflammatory caspases, apoptotic caspases, neutrophil elastase, and granzymes are involved in activation and regulation of different gasdermin family members (3–5, 15–23). The fact that multiple proteases are involved in the activation of gasdermins suggest that diverse mechanisms have evolved to ensure cell death and release of intracellular cytokines and alarmins or danger-associated molecular patterns. These in turn elicit strong inflammatory responses. Here we summarize our current understanding on protease processing of several gasdermin family members, either to activate the cytolysis activities of their NTDs or to suppress such function. The activation mechanisms for other gasdermins such as GSDMA and PJVK remain to be elucidated.
Table 1.
Different gasdermin family members and their cleavage sites by diverse proteases.
| Gasdermin | Protease | Cleavage site (P1 site in bold) | References |
|---|---|---|---|
|
| |||
| GSDMA | unknown | Potential cleavage sites: | N/A |
| hGSDMA: 223-ICNDN-227 | |||
| mGSDMA: 224-ICNDS-228 | |||
| mGSDMA2: 223-ICTDN-227 | |||
| mGSDMA3: 224-ICNDS-228 | |||
|
| |||
| GSDMB | Caspase-1 | 233-EEKD|G-237 (isoform 4 Q8TAX9-4) | (24) |
| Caspases-3, 6, 7 | 88-DNVD|S-92 | (7) | |
| Caspases-1, 3, 4, 6, 7, 8, 9 | Site unknown but may be similar to the above | (26) | |
| Granzyme A | 241-CLGK|S-245 (isoform 4 Q8TAX9-4) | (75) | |
| 226-GKTK|S-230 (isoform 4 Q8TAX9-4) | (75) | ||
|
| |||
| GSDMC | Caspase-8 | 362-LELD|S-366 (human GSDMC) | (76) |
| Caspase-6 | Site unknown but may be similar to the above | (76) | |
|
| |||
| GSDMD | Caspases-1, 4, 5, 11 | 272-FLTD|G-276 (human GSDMD) | (3–5) |
| 273-LLSD|G-277 (mouse GSDMD) | (3–5) | ||
| Caspase-8 | Same sites as above | (18, 20, 21) | |
| Caspases-3, 7 | 84-DAMD|G-88 (human GSDMD) | (15, 20, 21) | |
| 85-DVVD|G-89 (mouse GSDMD) | (15, 20, 21) | ||
| Neutrophil elastase | 265-MMRC|L-269 (human GSDMD) | (19, 32) | |
| 248-RKAV|G-252 (mouse GSDMD) | (19, 32) | ||
| Cathepsin G | 271-LSLL|S-275 (mouse GSDMD) | (22) | |
| 270-HNFL|T-274 (human GSDMD) | (22) | ||
| 3C protease from EV71 | 190-TCLQ|G-194 (human GSDMD) | (37) | |
|
| |||
| GSDME | Caspases-3, 7 | 267-DMPD|A-271 (human GSDME) | (15–17, 20) |
| 267-DMLD|G-271 (mouse GSDME) | (15–17, 20) | ||
| Granzyme B | Same sites as above | (43, 44) | |
| Caspase-3 | 238-DATD|G-242 (coral GSDME) | (40) | |
| 254-DEPD|G-258 (coral GSDME) | (40) | ||
| Caspase-B/caspy2 | 253-SEVD|G-257 (zebrafish GSDMEa) | (16, 41) | |
| 244-FEID|G-248 (zebrafish GSDMEb) | (42) | ||
Regulation of gasdermin B by caspases and granzyme A
Similar to other gasdermins, GSDMB contains an N-terminal domain (NTD) connected to a C-terminal domain (CTD) by a short linker region and is a substrate for caspases (7, 24). The GSDMB N-terminal domain binds phosphoinositides, phosphatidic acid, phosphatidylglycerol, sulfatide, and cardiolipin. What’s perhaps unique about GSDMB is the reported binding of membrane lipids, such as phosphoinositides and sulfatide but not cardiolipin, by the full-length GSDMB in the absence of protease cleavage (7). This may suggest a weak NTD-CTD interaction that allows lipids ready access to the NTD, or the presence of lipid-binding sites that are accessible within the context of the full-length GSDMB. Furthermore, there may be a potential decoupling of lipid-binding and cytolytic activities for GSDMB since its NTD but not the full-length protein has been reported to induce pyroptosis (10). The context and consequences of lipid-binding by the full-length GSDMB in the absence of pore-formation remain to be determined. A likely regulatory mechanism for GSDMB function may be alternative splicing. Among the six GSDMB alternative splice variants, the length and amino acid composition of their interdomain linker regions differ significantly. Such differences may impact differential NTD-CTD association, susceptibility to protease cleavage, lipid-binding, or pore formation (7, 24).
Chao and colleagues reported that recombinant caspases-3, −6, and −7, but not the inflammatory caspases, cleave GSDMB at its N-terminal domain between residues D91 and S92 (88-DNVD|S-92), which may produce a truncated NTD that is incapable of lipid-binding (Table 1). By contrast, Panganiban and colleagues revealed that co-expression of caspase-1 and GSDMB led to cleavage of GSDMB at residue D236 (233-EEKD|G-237, isoform 4 Q8TAX9–4), which led to pyroptosis (24). It remains to be determined whether the overexpression system or the conditions of the in vitro proteolysis contribute to the discrepancy. While studying cytotoxic lymphocyte-mediated immunity, it was revealed that natural killer cells and cytotoxic T lymphocytes target GSDMB-expressing cells through granzyme A-mediated cleavage of GSDMB (23). The cleavage sites are located predominantly at K244 and secondarily at K229 within the interdomain linker. This perforin-granzyme A-dependent cleavage unmasked the pore-forming activity of GSDMB to induce pyroptosis in GSDMB-expressing cells, which contributes to antitumor immunity. In agreement, there is a strong positive correlation between GSDMB expression and patient survival from bladder cancer and melanoma (23). In contrast to GSDMB-mediated killing of mammalian cells, Hansen and colleagues reported that NK cell activation of GSDMB is a host response to infection that kills cytosolic bacteria rather than the infected host cells (25). Side-by-side comparison of liposome-binding by GSDMD and GSDMB revealed that major lipid species found in mammalian cell membranes support robust binding by the former, but much weaker binding by the latter. A possible explanation for this apparent discrepancy is that mitochondria lipid cardiolipin may mis-localize to the plasma membrane in transformed cancer cells. Since GSDMB preferentially binds cardiolipin, this leads to pore-formation and lysis of the plasma membrane in a similar manner as the bacterial membrane. It is therefore important to understand the lipid-binding specificity by GSDMB, which may shed light on its diverse function in both infection and tumor immunity.
Using co-expression system with GSDMB, GSDMD and caspase-4, Chen and colleagues report that the full-length GSDMB promoted caspase-4 cleavage of GSDMD, perhaps through binding of GSDMB N-terminal fragment (residues 1–83) to the CARD of caspase-4 (26). The co-expression system also revealed cleavage of GSDMB in HEK293T cells by caspases-1, 3, 4, 6, 7, 8, and 9 to various extents. The exact site(s) of cleavage were not determined but a truncated NTD fragment appeared to be generated. The physiological context of GSDMB cleavage by the majority of caspases remains to be clarified.
Gasdermin C is activated by caspase-8 cleavage
TNFα has long been known to induce tumour necrosis but the molecular mechanism is poorly understood. One source of the TNFα is the tumor-associated macrophages. In PDL1-expressing breast cancer cells, the expression of GSDMC, but not other gasdermin family members, is upregulated. Upon TNFα treatment, cleavage of GSDMC at residue D365 (362-LELD|S-366) within its CTD by caspase-8 leads to production of a GSDMC fragment that is capable of pore formation (27). This switches from chemotherapy- and TNF/cycloheximide-induced apoptosis to pyroptosis. In vivo, this TNF-dependent tumor necrosis requires nuclear translocation of PD-L1 with phosphorylated Stat3 under hypoxia conditions, which enhances the transcription of the GSDMC gene (27). Paradoxically, tumor necrosis has been correlated with tumour progression and increased resistance to chemotherapy and radiotherapy (28). As a result, high expression of GSDMC correlates with poor survival. Using recombinant proteins, it was also shown that GSDMC can be cleaved by caspase-6 in a similar manner as caspase-8, though the former was not induced by TNFα/cycloheximide treatment (27). As a result, conditions in which caspase-6 is activated in tumor microenvironment may induce cytolysis in GSDMC-expressing cells.
Regulation of Gasdermin D by host and viral proteases
As the best studied member of the gasdermin family, GSDMD was reported to be cleaved within its linker region by activated caspases-1, 4, 5 in human or caspases-1 and 11 in mouse following the stimulation of the canonical or non-canonical inflammasome pathways (3–5). These caspases cleave GSDMD at a short peptide motif 272FLTD|G276 in human and 273LLSD|G277 in mouse. The cleavage generates a ~30 kDa NTD that has intrinsic pore-forming activity, and a ~20 kDa CTD that associates with the NTD and functions as an intramolecular inhibitory domain in the resting state (10). Recent studies suggest that GSDMD can be activated not only by the inflammatory caspases, but also by apoptotic caspase-8 in murine macrophages (18, 20, 21). Caspase-8 was activated upon TGFβ-associated kinase 1 (TAK1) inhibition by Yersinia bacteria protein YopJ or LPS plus TAK1 inhibitor 5Z-7-Oxozeaenol treatment. Activated caspase-8 cleaves GSDMD to generate the pyroptotic p30 fragment that induces pyroptotic cell death and cytokine secretion in murine macrophages, whereas GSDMD deficient cells undergo apoptosis instead. This caspase-8-mediated pyroptosis is dependent on lysosomal membrane-tethered Rag-Ragulator complex, which functions upstream of caspase-8 activation and GSDMD cleavage (29). In vitro cleavage assays revealed that murine GSDMD was directly cleaved by recombinant caspase-8 at the same cleavage motif as the inflammatory caspases 273-LLSD|G-277, even though the processing of GSDMD by caspase-8 was less efficient compared with caspase-1 (18, 20, 21). As such, caspase-8 functions as the protease that drives the activation of both GSDMD through direct cleavage and GSDME through caspase-8 processing of caspases-3 and 7, which may lead to inflammatory cell death. In contrast to what was observed in murine macrophages, human macrophages do not produce IL-1β upon Yersinia infection, and display resistance to pyroptosis upon TAK1 inhibition. This is perhaps due to overexpression of pro-survival proteins such as cellular FLICE-inhibitory protein (cFLIP) and inhibitors of apoptosis (IAPs). Such defects in cell death program upon highly pathogenic bacterial infection may underlie the severe consequences of Yersinia infection in human population, and emphasizes the need to investigate the species-specific cell death programs upon infections (20).
In addition to activation through caspase cleavage, GSDMD has been reported to be cleaved and activated by serine proteases from neutrophils. These proteases have been known to cleave microbial toxins or structural proteins, as well as host cytokines or chemokines, which regulates inflammation upon infections (30, 31). Elastase is a neutrophil serine protease localized in the azurophilic granules in resting neutrophils and is released into cytosol upon pathogen invasion. Recent reports revealed the ability of elastase to cleave GSDMD at the NTD-CTD linker and produce the p30/NTD fragment capable of membrane pore formation and inducing cell death (19, 32). Another GSDMD-activating neutrophil serine protease is cathepsin G (CatG). Burgener and colleagues report that CatG cleaves murine GSDMD at L274, two residues upstream of the caspase cleavage site to generate cytolytic NTD or p30 fragment (22). However, mutation of L274 led to incomplete abrogation of CatG cleavage, suggesting the existence of other less preferred cleavage sites at the NTD-CTD linker. CatG also cleaves human GSDMD and generates a fragment of similar size as the cleaved mouse GSDMD. Together, these findings demonstrate that GSDMD is a substrate for serine proteases such as ELANE and CatG, with preferred and alternative cleavage sites within the GSDMD linker region.
The cleavage of GSDMD by the inflammatory caspases and serine proteases in neutrophils contributes to the formation of neutrophil extracellular traps (NETs) (32, 33), web-like structures of DNA embedded with granule and nuclear proteins that trap and kill bacteria (34). The extrusion of NETs and the concomitant cell death named NETosis is an innate immune defense mechanism often observed during acute inflammation (32, 33, 35). Treatment of neutrophils with elastase inhibitor led to reduced GSDMD processing and NET formation as well as blocked NETosis (32). Infection of neutrophils by gram-negative bacteria induced NETosis that was dependent on caspase-11 and GSDMD (33), in agreement with the important roles of both GSDMD and its activating proteases in NETosis and neutrophil immune defense.
It is perhaps not surprising that cleavage of GSDMD by proteases may also leads to its inactivation. An intriguing example was reported by several groups (15, 20, 21), which showed that both caspase-3 and caspase-7 cleave GSDMD at residue D87 to generate a p43 fragment that inactivates GSDMD. This seems to be a general feature in cells expressing GSDMD and undergoing apoptosis. Depending on the different kinetics for the activation of caspases-3 and −7 by caspase-1, and the cleavage of GSDMD by caspase-1, the former may function as a negative feedback mechanism to dampen excessive inflammation from pyroptotic cell death caused by the latter. Since caspase-3 and caspase-7 are also capable of activating GSDME, caspase-1 appears to function as a crucial “upstream” caspase that regulate the activation of both GSDMD and GSDME. Similarly, caspase-8 also regulates both GSDMD and GSDME through caspases-3 and 7.
GSDMD is inactivated by proteases not only from the host, but also from pathogens as an immune evasion mechanism to counteract pyroptosis. Enterovirus 71 (EV71) is a major causative agent of hand-foot-and-mouth disease (HFMD) in young children and has caused many large-scale epidemics (36). A protease 3C from EV71 cleaves substrate with preference for Gln residue at the P1 site and a small residue at the P1’ site. This protease was found to cleave human GSDMD at residue Q193 within its NTD to inhibit its cytolysis activity (37). Furthermore, inactivation of GSDMD by EV71 3C protease promoted EV71 replication and survival in vivo. It remains to be determined if other pathogens such as SARS-CoV-2, which encodes a 3C-like protease (3CLpro) or main protease (Mpro) with similar substrate specificity, utilizes similar strategy to cleave gasdermins and disable pyroptosis in host cells.
Activation of Gasdermin E by caspases and granzyme B
GSDME, or Deafness, Autosomal dominant 5 (DFNA5), was first discovered in conjunction with a progressive form of hearing impairment and has since been found in a variety of invertebrates and vertebrates as an evolutionarily conserved gene (38). Similar to most gasdermins, the GSDME-NTD and CTD is connected by a flexible linker region (6). Caspases-3 and −7 have been reported to cleave human or mouse GSDME at residue D270 in the linker region to produce a pore-forming NTD that causes secondary necrosis in apoptotic cells (15–17, 20). This was demonstrated as an immune defense mechanism in a recent report that revealed GSDME as a primary mediator of cell death when viral infection of keratinocytes led to suppression of protein synthesis (39). Suppressed translation was sensed by the BCL-2 family members, which led to mitochondria damage, caspase-3 activation and GSDME cleavage. GSDME cleavage is mostly abolished in caspase-3 knockout macrophages and is completely abolished in caspase-3 and caspase-7 double-deficient macrophages (20), suggesting that caspase-3 plays a dominant role in GSDME activation with minor contributions from caspase-7. This is analogous to the more prominent role of caspase-3 in cleaving GSDMD compared with caspase-7 (15).
GSDME is the most ancient gene among the gasdermins and current literature have shed light on its cleavage and activation in fish and invertebrates. Coral GSDME was cleaved by caspase 3 at two sites, 238-DATD|G-242 and 254-DEPD|G-258, to generate two forms of the NTDs that are capable of inducing pyroptosis (40). Human caspases-3 and 7 are also able to cleave coral GSDME into similar sized NTDs. Such cleavage of GSDME in coral mediates cell death upon infection by pathogenic bacteria. Similarly, the zebrafish GSDMEa/DFNA5a is cleaved at 253-SEVD|G-257, perhaps by zebrafish caspy2/caspase-B, a homolog of mammalian caspases-4, 5, and 11, to induce cell death upon bacterial infection (16, 41). The second member of the zebrafish gasdermin GSDMEb/DFNA5b is cleaved at 244-FEID|G-248 by caspy2/caspase-B, to induce pyroptosis and acute kidney injury during sepsis (42).
In addition to caspases, GSDME activation is facilitated by granzyme B, a serine protease similar to the neutrophil elastase. This finding originated from observations that the expression of GSDME led to suppression of tumor growth, and was only partially dependent on caspase-3 (43, 44). It turned out that granzyme B from NK, CD8+, or chimeric antigen receptor (CAR) T cells is delivered through perforin pores to tumor cells such as leukemia, melanoma, breast cancer and colorectal cancer cells. Granzyme B in turn both directly cleaves GSDME at residue D270, and activates caspase-3 which also cleaves GSDME at the same site (43). In addition to the direct and indirect activation of GSDME by granzyme B, the initial GSDME activation may lead to permeabilization of mitochondria by the GSDME-NTD due to engagement with mitochondria membrane lipid cardiolipin. This results in release of cytochrome c and activation of the intrinsic apoptotic pathway and caspase-3 (45). As a result, this propagates a positive feedback loop to cleave and activate additional GSDME. The consummate pyroptosis events may precipitate severe cytokine release syndrome that counteracts the effectiveness of CAR T cell therapy (44). As a result, therapeutic strategies that suppress GSDME-mediate pyroptosis may provide clinical benefits in cancer immunotherapy.
Common and distinct features of gasdermin processing by caspases
Recognition and cleavage of the gasdermin family members by proteases share common and distinct features. The majority of gasdermin-processing proteases are caspases that prefer the P1 site Asp residue. The P1-P4 site residues of caspase substrates have traditionally been the focus of studies in terms of how they fit in the catalytic groove (46–48). These studies revealed the preference for hydrophobic P4 site residues for substrates of the inflammatory caspases, as opposed to acidic P4 site residues for apoptotic caspase-3 substrates. In addition to the caspase active sites, allosteric sites at or near the caspase dimer interfaces were discovered that modulate substrate engagement by caspases-1, 2, 3, 6, 7, 8 and 9 (49–51). A novel exosite was reported for the inflammatory caspases in recent studies of the GSDMD-caspase complexes, and plays a dominant role in the recognition of GSDMD by these caspases (8, 9).
The crystal structures of caspases-1, 4, and 11 in complex with the GSDMD-CTD and the structure of caspase-1 in complex with the full-length GSDMD revealed two interfaces between the enzymes and substrates at the GSDMD linker and GSDMD-CTD, respectively (Figure 1). The absence of contacts between GSDMD-NTD and caspase-1 is consistent with the idea that GSDMD-NTD may be released from the enzyme-substrate complex to oligomerize and assemble membrane pores after the linker cleavage. In the structure of caspase-1 in complex with the full-length GSDMD, the catalytic groove of caspase-1 engages the GSDMD linker in a manner similar to its association with the active site inhibitors such as z-VAD-FMK (52) or the GSDMD-derived peptide inhibitor (53). The conserved P1 site residue D276 in mGSDMD is buried deep in the catalytic groove of caspase-1, in close proximity to the catalytic residues C285 and H237. The P4 site residue L273 forms extensive hydrophobic and van der Waals contacts with caspase-1. Interestingly, the P1’ site residue G277 both anchors the GSDMD linker at the active site, and offers conformational flexibility for a ~90° turn in the linker to exit the caspase-1 catalytic groove and connect to the NTD. This is consistent with the preference for small residues such as Gly/Ala/Ser at the P1’ position reported by previous studies using peptide substrates for caspases (46), and may explain why some of the other Asp residues adjacent to the substrate P1 site Asp are not cleaved by caspases due to the lack of small P1’ residues. Besides P1’, the P1’-P4’ residues have been reported by the Salveson group to influence the recognition of GSDMD by caspases-4, 5, and 11 (54). In fact, grafting the GSDMD P1’-P4’ residues 277-GIDE-280 to pro-IL18 converts this mutant pro-IL18 to an efficient caspase-11 substrate as GSDMD. As such, structural components bordering on the P1-P4 sites in gasdermins may modulate their recognition and cleavage by proteases.
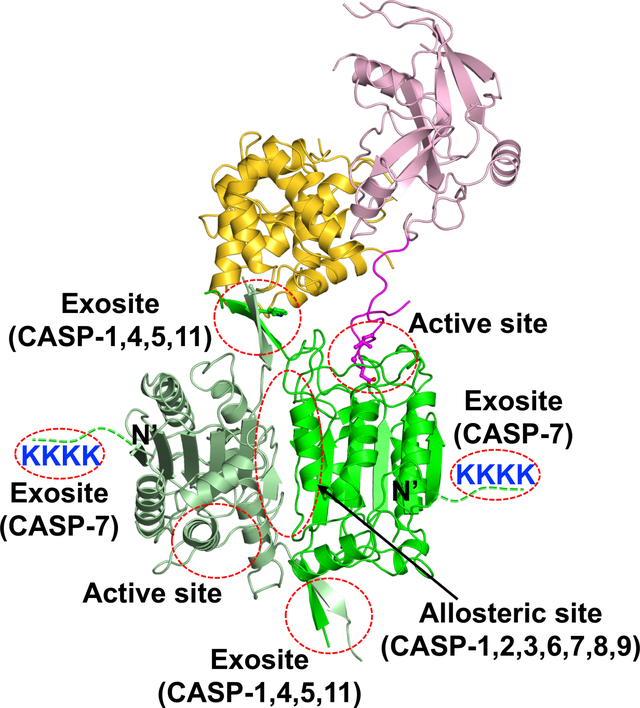
Figure 1.
Exosites from the inflammatory caspases mediate their recognition of GSDMD. Structures of caspase-1 in complex with GSDMD-CTD (A), caspase-4 in complex with GSDMD-CTD (B), caspase-11 in complex with GSDMD-CTD (C), and caspase-1 in complex with the full-length GSDMD (D) are shown in ribbons with the sidechains of the exosite residues displayed. (E) The structure of a dimeric caspase-1 is shown with the active sites, exosites, and allosteric site marked. (F) Sequence alignment adjacent to the active site Cys residues and the exosite residues are shown for different caspases, with the active site Cys in red, and exosite residues from the inflammatory caspases in green and underlined. The equivalent hydrophobic residues from other caspases are in green, and non-hydrophobic resdues are in black.
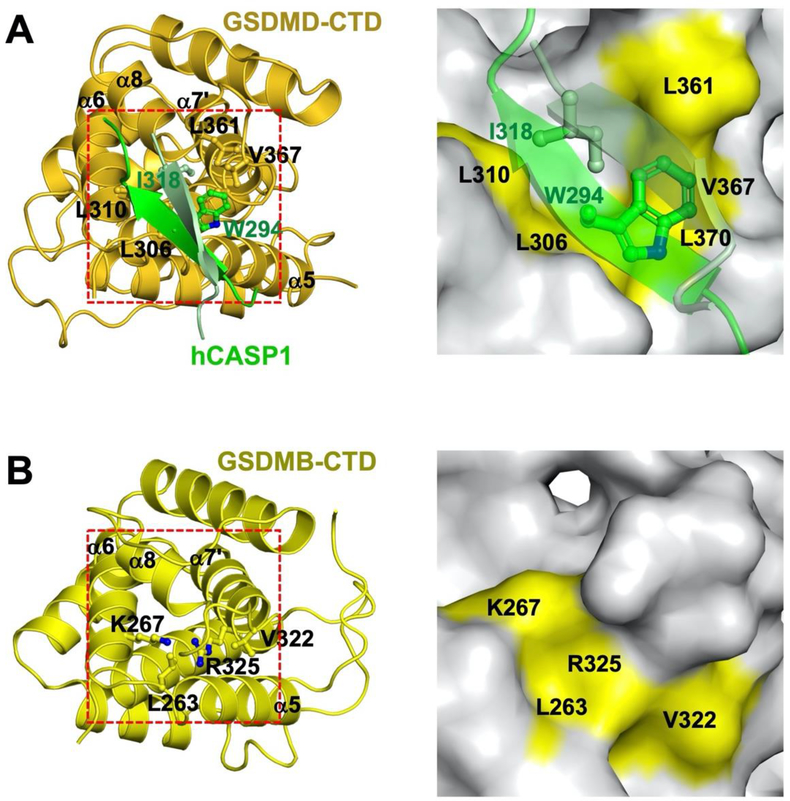
In addition to the caspase active site interface, an exosite ~20 Å away from the catalytic Cys residue was observed between the GSDMD-CTD and an anti-parallel β sheet formed by the L2 and L2’ loops from the dimeric caspases-1, 4, and 11 (8, 9). This interface is centered on caspase-1 residues W294 from L2 and I318 from L2’ loops, or the equivalent residues from caspase-4 (W267 and V291) and caspase-11 (W263 and V287), which dock into a hydrophobic pocket formed by the α5-α6 and α7’-α8 structural elements in GSDMD. The caspase-1 residues W294 and I318 are conserved among the inflammatory caspases, but not in most apoptotic caspases (Figure 1F). The hydrophobic residues in the GSDMD-CTD pocket are conserved between the human and murine GSDMD, but are not conserved in other gasdermins (Figure 2) (9), suggesting that this interface may contribute to the specific recognition of GSDMD but no other gasdermins by the inflammatory caspases. Because this interface is distinct from the active sites of the inflammatory caspases, it is referred to as the “exosite”: structural elements in enzymes that participate in binding of partner proteins and are distinct from the enzyme active sites (55–58). By virtue of its role in mediating caspase-GSDMD binding, the exosite interface endows a novel function for the GSDMD-CTD as a caspase recruitment domain, in addition to its role as an autoinhibition domain. Interestingly, an exosite composed of positively charged residues at the N-terminus of the caspase-7 catalytic domain was previously identified that bind and facilitate the cleavage of its substrate poly(ADP ribose) polymerase 1 (PARP1) (59) (Figure 1). Given that most of the gasdermin CTDs are negatively charged, speculative interaction between the caspase-7 exosite and the gasdermin CTDs may facilitate enzyme-substrate association through electrostatic interactions.
Figure 2.
Structures of the GSDMD-CTD and GSDMB-CTD reveal different pockets at the actual or potential interface with the inflammatory caspase exosites. (A) The left panel shows the structure of the GSDMD-CTD (gold) bound to the caspase-1 exosite (green). The right panel shows the GSDMD-CTD as a surface presentation with the hydrophobic residues binding the caspase exosite marked in yellow. (B) The left panel shows the equivalent view of the GSDMB-CTD. The right panel marks the equivalent GSDMB residues to those in (A).
It is clear that the exosites are the dominant structural determinants for the engagement of GSDMD by the inflammatory caspases. Caspases-1, 4, 11 and GSDMD-CTD or GSDMDs harboring mutated linker regions can still form stable enzyme-substrate complexes (8, 9). It is possible that the initial binding of GSDMD by the inflammatory caspases is through their exosite interfaces, which was referred to as exosite-mediated “priming interaction” (8, 9). This may be followed by the dual-interface engagement between the caspases and the full-length GSDMD, which leads to close proximity of the caspase catalytic residues and the substrate P1 site Asp residues, resulting in the eventual cleavage of the GSDMD linker. The ~20 Å distance between these two interfaces may thus dictate the extent to which the cleavage at the GSDMD linker may occur, as linker residues too close to the CTD may not reach the catalytic groove. The dual site engagement may enhance the overall efficiency of proteolysis through stronger substrate binding at two interfaces. In turn, the engagement of GSDMD by the L2 and L2’ loops from the two protomers may also stabilize the caspase dimers, which are representative of the active enzyme conformation conducive to catalytic activities (8, 60).
Perhaps most importantly, contribution of the dual interface may underlie the more stringent substrate selectivity for the inflammatory caspases compared with apoptotic caspases. In fact, exosites have been previously associated with enhanced substrate specificity for other proteases (61, 62). Conversely, the exosite contribution could be absent or less prominent for promiscuous apoptotic caspases such as caspase-3, which engages substrates such as GSDME primarily through the essential cleavage site sequences. This was demonstrated by the lack of cleavage by caspase-3 and pyroptosis activities when the P4 site residue was mutated in human GSDME (16) or coral GSDME (40), whereas the GSDMD P4 site mutation had minimal impact on its cleavage and pyroptosis as long as the P1 site and the exosite interface is intact (8). Whether this P4 site mutant of GSDME can still be cleaved by caspase-7, which contributes to GSDME-mediated pyroptosis and employ an exosite to recognize its substrate PARP1 (59), remain to be experimentally tested.
Currently, it is not clear whether similar dual site recognition of GSDMD by the inflammatory caspases is employed by other GSDMD-targeting proteases, or are involved in proteolytic processing of other gasdermins. The apoptotic caspase-8 was reported to cleave GSDMD at the same site as the inflammatory caspases to induce pyroptosis, albeit the efficiency of cleavage appeared to be lower than that for caspase-1 (18, 20, 21). Even though the caspase-1 cleavage site at GSDMD matches the signature sequence of a caspase-8 substrate, the exosite hydrophobic residues in caspase-1 are not conserved in caspase-8 (Figure 1F). If caspase-8 indeed employs the same L2 and L2’ loops as an exosite in binding GSDMD, the lack of conserved exosite hydrophobic residues may contribute to weaker association with GSDMD and less efficient cleavage.
Gasdermin processing by non-caspases
The granzymes are a family of death-inducing serine proteases delivered from innate and adaptive immune killer cells to tumor cells or virus-infected cells (63). Among the five human granzymes and eleven mouse granzymes, granzyme A and granzyme B are the most abundant. Granzyme A prefers basic residues R and K at the P1 site, whereas granzyme B cleaves after aspartic acid residues in many of the same substrates as the caspases (63–65). An example is the recent report of granzyme B cleavage of GSDME at the exact site as caspases-3 and 7. It is unclear whether the tertiary structures of gasdermins play any role in their recognition by granzymes, or the cleavage site sequences are sufficient for GSDMB recognition by granzyme A or GSDME by granzyme B. Based on the critical importance of the GSDME P1 and P4 site residues in its cleavage by caspase-3 (16), it is possible that the same sites may play a dominant role in GSDME recognition by granzyme B without the involvement of an exosite. By contrast, exosites have been proposed to mediate electrostatic interactions between granzyme A and its substrates, in light of the known differences in cleavage efficiency between peptide substrates and macromolecular substrates (66, 67). Through analysis of the catalytic groove of granzyme A and modeling of protein substrates and inhibitors, it was suggested that positively charged structural elements outside the granzyme An active site may facilitate substrate access to the enzyme active site. Such structural elements may then constitute exosite(s). This hypothesis remains to be tested with the GSDMB substrate.
Similar to the granzymes, neutrophil elastase and cathepsin G are serine proteases stored in granules and are major components of neutrophil’s microcidal arsenal (68). Cathepsin G and elastase cleave after hydrophobic residues such as Cys, Val, or Leu (69), and are known to activate the IL-1 family members (70). Since cathepsin G cleaves GSDMD at multiple sites (22), it is unlikely that an exosite is involved in limiting the specific sites of cleavage. The elastase cleavage sites in human and murine GSDMDs (C268 vs V251) are not conserved, which is consistent with the known preference of elastase for small hydrophobic amino acids at the P1 site of substrates (69). It remains to be determined if neutrophil elastase utilizes an exosite to recognize GSDMD.
Conclusions and future perspectives
A common theme of gasdermin activation is its trigger by protease cleavage at the flexible and poorly conserved NTD-CTD linker region. These proteases are themselves activated by various signaling pathways or delivered to the cytosol through microbial invasion or perforin from cytotoxic immune cells. It is conceivable that gasdermins may have evolved to sense mislocalized or stimuli-dependent cytosolic proteases as danger signals using their unstructured linker region as easily accessible substrates for such proteases. Emerging evidence suggests that in several cases, the structured regions of gasdermins may provide crucial binding-surface for protease engagement as in exosites for the inflammatory caspases, or perhaps impact the accessibility of enzyme active sites for substrates as suggested for granzyme A. The use of exosites may strengthen enzyme-substrate interactions, improve efficiency of proteolysis, and enhance substrate selectivity. However, exosites may be absent or counterproductive for promiscuous proteases such as caspase-3 or cathepsin G. Furthermore, different structural elements may function as exosites for different substrates. In fact, recent evidence suggests that spatially distinct exosites may be employed by the inflammatory caspases such as caspase-1 to recognize diverse substrates such as GSDMD or pro-IL1β (71). Conceivably unique tertiary structural features of substrates may be recognized by various proteases through different exosites.
Post-cleavage release of the gasdermin NTDs from the CTDs in the presence of membrane lipids allows the NTDs to assemble pores at the plasma membrane, membranes of intracellular organelles, or microbial membranes. The redundancy of the pore-forming function from different gasdermin NTDs suggests crucial importance of the underlying cell death mechanisms in immune defense against infections or tumorigenesis. On the other hand, as the membrane-disrupting function of gasdermin NTDs can have dire consequences for cells expressing gasdermins, multiple mechanisms have evolved to regulate their pore-forming activities. One of such mechanisms is the protease cleavage within the gasdermin NTDs by host or microbial proteases, which may serve to prevent or reverse the membrane-disrupting function of the NTDs. Whether exosites are utilized in such inactivating cleavage events remain to be determined. It should be noted that several gasdermins have been reported to induce cytolysis or liposome leakage in the absence of protease cleavage when autoinhibition from their CTDs were compromised (10, 12). Such pore-forming activities by the mutant full-length gasdermins, though at reduced levels compared with the wildtype NTDs, suggest that even though protease cleavage at the gasdermin NTD-CTD linkers promotes NTD pore formation under physiological conditions, it is not absolutely required for pore assembly.
Recent studies of protease processing of gasdermins suggest a number of future directions going forward. For example, what proteases other than the inflammatory caspases use exosites to engage gasdermins? Do different structural elements in the same protease serve as exosites for different gasdermins? Biochemical and structural approaches will be instrumental in revealing how exosites in diverse proteases engage different gasdermin substrates and their component domains. Such mechanistic understanding will facilitate the development of highly selective inhibitors that target specific exosite interface utilized by each substrate. Are the exosite interface more likely to be located at the CTDs in cases of gasdermin activation by proteases, and at the NTDs in cases of gasdermin inactivation? Most of the protease cleavage events for gasdermins reported thus far activate their pore-forming NTDs, perhaps partially attributed to the technical convenience of assaying membrane disruption events. Are gasdermins besides GSDMD and GSDMB negatively regulated through protease cleavage, particularly by microbial proteases? Are GSDMA or PJVK regulated by protease processing and under what physiological context do they induce cell death? What are the “upstream” signaling events that precipitate the activation of these proteases? Could the expression of different splice variants such as those for GSDMB or the induction of gasdermin expression in different cell types impact their access to and cleavage by proteases? Phosphorylation of GSDME has been reported to suppress its pore-forming activities (45). Could post-translational modifications such as phosphorylation or ubiquitination modulate protease recognition of gasdermins? Unique features of gasdermin sequence, structure, expression characteristics, and post-translational modifications may dictate distinct mechanisms of protease recognition and cleavage. Furthermore, different post-cleavage membrane-binding properties by gasdermin family members are starting to emerge which undoubtedly have important implications for their function in specific cell types or organelles. Such diverse mechanisms may underlie the divergent physiological and pathological functions of gasdermins and may furnish opportunities for therapeutic targeting of gasdermins in infectious diseases and inflammatory disorders. Since the discovery of gasdermins as effectors for pyroptosis a few years ago (3–5), the function of gasdermins has been extended to NETosis (32, 33), secondary necrosis (16, 17), tumor necrosis (23, 27, 43, 44), non-lytic protein secretion (72–74), and lysis of invading microbes (10, 25), etc. Protease recognition and cleavage of gasdermins downstream of diverse signaling pathways underlie many of these activities. It is anticipated that the next few years will witness exciting discovery in the vibrant areas of gasdermin studies.
Highlights.
Flexible linkers connect pore-forming NTDs with autoinhibitory CTDs in gasdermins
Diverse proteases cleave gasdermin linkers or loops to regulate their activities
Exosites strengthen enzyme-substrate interactions and enhance substrate selectivity
Different features of gasdermin regulation by proteases underlie diverse function
Acknowledgements
This work is supported by NIH grants R01GM127609 and P01141360 to T.S.X.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saeki N, Kuwahara Y, Sasaki H, Satoh H, and Shiroishi T (2000) Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mammalian Genome. 11, 718–724 [DOI] [PubMed] [Google Scholar]
- 2.Broz P, Pelegrín P, and Shao F (2020) The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 20, 143–157 [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526, 660–665 [DOI] [PubMed] [Google Scholar]
- 4.Kayagaki N, Stowe IB, Lee BL, and O’Rourke K (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526, 666–671 [DOI] [PubMed] [Google Scholar]
- 5.He W-T, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang Z-H, Zhong C-Q, and Han J (2015) Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Research. 25, 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Beeck KO, Van Camp G, Thys S, Cools N, Callebaut I, Vrijens K, Van Nassauw L, Van Tendeloo VF, Timmermans JP, and Van Laer L (2011) The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. European journal of human genetics : EJHG. 19, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao KL, Kulakova L, and Herzberg O (2017) Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein. Proceedings of the National Academy of Sciences. 114, E1128–E1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, Li Z, Wang Y, Zhao Q, Shao F, and Ding J (2020) Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis. Cell. 180, 941–955.e20 [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Wang C, Yang J, Chen Y, Zhou B, Abbott DW, and Xiao TS (2020) Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage. Immunity. 53, 106–114.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang D-C, and Shao F (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 535, 111–116 [DOI] [PubMed] [Google Scholar]
- 11.Kuang S, Zheng J, Yang H, Li S, Duan S, Shen Y, Ji C, Gan J, Xu X-W, and Li J (2017) Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proceedings of the National Academy of Sciences. 114, 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Wang C, Rathkey JK, Yang J, Dubyak GR, Abbott DW, and Xiao TS (2018) Structures of the Gasdermin D C-Terminal Domains Reveal Mechanisms of Autoinhibition. Structure. 26, 778–784.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Wang C, Yang J, Zhou B, Yang R, Ramachandran R, Abbott DW, and Xiao TS (2019) Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization. Immunity. 10.1016/j.immuni.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agard NJ, Maltby D, and Wells JA (2010) Inflammatory Stimuli Regulate Caspase Substrate Profiles. Molecular & cellular proteomics : MCP. 9, 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taabazuing CY, Okondo MC, and Bachovchin DA (2017) Pyroptosis and Apoptosis Pathways Engage in Bidirectional Crosstalk in Monocytes and Macrophages. Cell Chemical Biology. 24, 507–514.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, and Shao F (2017) Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 547, 99–103 [DOI] [PubMed] [Google Scholar]
- 17.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, and Alnemri ES (2017) Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nature Communications. 8, 14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, Berger SB, Gough PJ, Bertin J, Proulx MM, Goguen JD, Kayagaki N, Fitzgerald KA, and Lien E (2018) Pathogen blockade of TAK1 triggers caspase-8–dependent cleavage of gasdermin D and cell death. Science. 362, 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kambara H, Liu F, Zhang X, Liu P, Bajrami B, Teng Y, Zhao L, Zhou S, Yu H, Zhou W, Silberstein LE, Cheng T, Han M, Xu Y, and Luo HR (2018) Gasdermin D Exerts Anti-inflammatory Effects by Promoting Neutrophil Death. Cell Reports. 22, 2924–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, and Poltorak A (2018) Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. PNAS. 115, E10888–E10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen KW, Demarco B, Heilig R, Shkarina K, Boettcher A, Farady CJ, Pelczar P, and Broz P (2019) Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. The EMBO Journal. 38, e101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgener SS, Leborgne NGF, Snipas SJ, Salvesen GS, Bird PI, and Benarafa C (2019) Cathepsin G Inhibition by Serpinb1 and Serpinb6 Prevents Programmed Necrosis in Neutrophils and Monocytes and Reduces GSDMD-Driven Inflammation. Cell Reports. 27, 3646–3656.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, He H, Wang K, Shi X, Su Y, Wang Y, Li D, Liu W, Zhang Y, Shen L, Han W, Shen L, Ding J, and Shao F (2020) Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 368, eaaz7548. [DOI] [PubMed] [Google Scholar]
- 24.Panganiban RA, Sun M, Dahlin A, Park H-R, Kan M, Himes BE, Mitchel JA, Iribarren C, Jorgenson E, Randell SH, Israel E, Tantisira K, Shore S, Park J-A, Weiss ST, Wu AC, and Lu Q (2018) A functional splice variant associated with decreased asthma risk abolishes the ability of gasdermin B to induce epithelial cell pyroptosis. Journal of Allergy and Clinical Immunology. 142, 1469–1478.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen JM, de Jong MF, Wu Q, Zhang L-S, Heisler DB, Alto LT, and Alto NM (2021) Pathogenic ubiquitination of GSDMB inhibits NK cell bactericidal functions. Cell. 184, 3178–3191.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q, Shi P, Wang Y, Zou D, Wu X, Wang D, Hu Q, Zou Y, Huang Z, Ren J, Lin Z, and Gao X (2019) GSDMB promotes non-canonical pyroptosis by enhancing caspase-4 activity. Journal of Molecular Cell Biology. 11, 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou J, Zhao R, Xia W, Chang C-W, You Y, Hsu J-M, Nie L, Chen Y, Wang Y-C, Liu C, Wang W-J, Wu Y, Ke B, Hsu JL, Huang K, Ye Z, Yang Y, Xia X, Li Y, Li C-W, Shao B, Tainer JA, and Hung M-C (2020) PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 22, 1264–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grivennikov SI, Greten FR, and Karin M (2010) Immunity, Inflammation, and Cancer. Cell. 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Z, Deng W, Bai Y, Miao R, Mei S, Zhang Z, Pan Y, Wang Y, Min R, Deng F, Wu Z, Li W, Chen P, Ma T, Lou X, Lieberman J, and Liu X (2021) The lysosomal Rag-Ragulator complex licenses RIPK1– and caspase-8–mediated pyroptosis by Yersinia. Science. 10.1126/science.abg0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tkalcevic J, Novelli M, Phylactides M, Iredale JP, Segal AW, and Roes J (2000) Impaired Immunity and Enhanced Resistance to Endotoxin in the Absence of Neutrophil Elastase and Cathepsin G. Immunity. 12, 201–210 [DOI] [PubMed] [Google Scholar]
- 31.Clancy DM, Sullivan GP, Moran HBT, Henry CM, Reeves EP, McElvaney NG, Lavelle EC, and Martin SJ (2018) Extracellular Neutrophil Proteases Are Efficient Regulators of IL-1, IL-33, and IL-36 Cytokine Activity but Poor Effectors of Microbial Killing. Cell Reports. 22, 2937–2950 [DOI] [PubMed] [Google Scholar]
- 32.Sollberger G, Choidas A, Burn GL, Habenberger P, Di Lucrezia R, Kordes S, Menninger S, Eickhoff J, Nussbaumer P, Klebl B, Krüger R, Herzig A, and Zychlinsky A (2018) Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Science immunology. 3, eaar6689. [DOI] [PubMed] [Google Scholar]
- 33.Chen KW, Monteleone M, Boucher D, Sollberger G, Ramnath D, Condon ND, von Pein JB, Broz P, Sweet MJ, and Schroder K (2018) Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Science immunology. 3, eaar6676. [DOI] [PubMed] [Google Scholar]
- 34.Brinkmann V (2004) Neutrophil Extracellular Traps Kill Bacteria. Science. 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 35.Kaplan MJ, and Radic M (2012) Neutrophil Extracellular Traps: Double-Edged Swords of Innate Immunity. The Journal of Immunology. 189, 2689–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang P-C, Chen S-C, and Chen K-T (2016) The Current Status of the Disease Caused by Enterovirus 71 Infections: Epidemiology, Pathogenesis, Molecular Epidemiology, and Vaccine Development. International Journal of Environmental Research and Public Health. 13, 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei X, Zhang Z, Xiao X, Qi J, He B, and Wang J (2017) Enterovirus 71 Inhibits Pyroptosis through Cleavage of Gasdermin D. J Virol. 10.1128/JVI.01069-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K, Sumiyama K, Sagai T, and Shiroishi T (2007) Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics. 89, 618–629 [DOI] [PubMed] [Google Scholar]
- 39.Orzalli MH, Prochera A, Payne L, Smith A, Garlick JA, and Kagan JC (2021) Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes Gasdermin-E-dependent pyroptosis in barrier epithelial cells. Immunity. 54, 1447–1462.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang S, Zhou Z, Sun Y, Zhang T, and Sun L (2020) Coral gasdermin triggers pyroptosis. Science Immunology. 10.1126/sciimmunol.abd2591 [DOI] [PubMed] [Google Scholar]
- 41.Li J-Y, Wang Y-Y, Shao T, Fan D-D, Lin A-F, Xiang L-X, and Shao J-Z (2020) The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E–mediated pyroptosis. Journal of Biological Chemistry. 295, 1120–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Gu Z, Hou Q, Chen W, Mu D, Zhang Y, Liu Q, Liu Z, and Yang D (2020) Zebrafish GSDMEb Cleavage-Gated Pyroptosis Drives Septic Acute Kidney Injury In Vivo. J.I 204, 1929–1942 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, Junqueira C, Meza-Sosa KF, Mok TMY, Ansara J, Sengupta S, Yao Y, Wu H, and Lieberman J (2020) Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 579, 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Fang Y, Chen X, Wang Z, Liang X, Zhang T, Liu M, Zhou N, Lv J, Tang K, Xie J, Gao Y, Cheng F, Zhou Y, Zhang Z, Hu Y, Zhang X, Gao Q, Zhang Y, and Huang B (2020) Gasdermin E–mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. Science Immunology. 10.1126/sciimmunol.aax7969 [DOI] [PubMed] [Google Scholar]
- 45.Rogers C, Erkes DA, Nardone A, Aplin AE, Fernandes-Alnemri T, and Alnemri ES (2019) Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun. 10, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stennicke HR, Renatus M, Meldal M, and Salvesen GS (2000) Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 350, 563–568 [PMC free article] [PubMed] [Google Scholar]
- 47.Julien O, and Wells JA (2017) Caspases and their substrates. Cell Death and Differentiation. 24, 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poreba M, Strózyk A, Salvesen GS, and Drag M (2013) Caspase substrates and inhibitors. Cold Spring Harbor Perspectives in Biology. 5, a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy JA, Lam J, Nguyen JT, O’Brien T, and Wells JA (2004) Discovery of an allosteric site in the caspases. Proceedings of the National Academy of Sciences. 101, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman T, Kabaleeswaran V, Jang SB, Antczak C, Djaballah H, Wu H, and Jiang X (2012) A Class of Allosteric Caspase Inhibitors Identified by High-Throughput Screening. Molecular Cell. 47, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tubeleviciute-Aydin A, Beautrait A, Lynham J, Sharma G, Gorelik A, Deny LJ, Soya N, Lukacs GL, Nagar B, Marinier A, and LeBlanc AC (2019) Identification of Allosteric Inhibitors against Active Caspase-6. Scientific Reports. 9, 5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheer JM, Romanowski MJ, and Wells JA (2006) A common allosteric site and mechanism in caspases. Proceedings of the National Academy of Sciences of the United States of America. 103, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Liu Z, Wang C, Yang R, Rathkey JK, Pinkard OW, Shi W, Chen Y, Dubyak GR, Abbott DW, and Xiao TS (2018) Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor. Proceedings of the National Academy of Sciences. 115, 6792–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bibo-Verdugo B, Snipas SJ, Kolt S, Poreba M, and Salvesen GS (2020) Extended subsite profiling of the pyroptosis effector protein gasdermin D reveals a region recognized by inflammatory caspase-11. J. Biol. Chem 10.1074/jbc.RA120.014259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Shavit R, Kahn A, Wilner GD, and Fenton JW (1983) Monocyte chemotaxis: stimulation by specific exosite region in thrombin. Science. 220, 728–731 [DOI] [PubMed] [Google Scholar]
- 56.Rydel TJ, Ravichandran KG, Tulinsky A, Bode W, Huber R, Roitsch C, and Fenton JW (1990) The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 249, 277–280 [DOI] [PubMed] [Google Scholar]
- 57.Steffensen B, Wallon UM, and Overall CM (1995) Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J. Biol. Chem 270, 11555–11566 [DOI] [PubMed] [Google Scholar]
- 58.Kleanthous C, and Walker D (2001) Immunity proteins: enzyme inhibitors that avoid the active site. Trends in Biochemical Sciences. 26, 624–631 [DOI] [PubMed] [Google Scholar]
- 59.Boucher D, Blais V, and Denault J-B (2012) Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. PNAS. 109, 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y (2002) Mechanisms of caspase activation and inhibition during apoptosis. Molecular Cell. 9, 459–470 [DOI] [PubMed] [Google Scholar]
- 61.Fulcher YG, and Van Doren SR (2011) Remote Exosites of the Catalytic Domain of Matrix Metalloproteinase-12 Enhance Elastin Degradation. Biochemistry. 50, 9488–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jabaiah AM, Getz JA, Witkowski WA, Hardy JA, and Daugherty PS (2012) Identifi cation of protease exosite-interacting peptides that enhance substrate cleavage kinetics. Biological Chemistry. 393, 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chowdhury D, and Lieberman J (2008) Death by a Thousand Cuts: Granzyme Pathways of Programmed Cell Death. Annu. Rev. Immunol 26, 389–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Odake S, Kam CM, Narasimhan L, Poe M, Blake JT, Krahenbuhl O, Tschopp J, and Powers JC (1991) Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry. 30, 2217–2227 [DOI] [PubMed] [Google Scholar]
- 65.Waugh SM, Harris JL, Fletterick R, and Craik CS (2000) pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity. nature structural biology. 7, 4. [DOI] [PubMed] [Google Scholar]
- 66.Hink-Schauer C, Estébanez-Perpiñá E, Kurschus FC, Bode W, and Jenne DE (2003) Crystal structure of the apoptosis-inducing human granzyme A dimer. Nat Struct Mol Biol. 10, 535–540 [DOI] [PubMed] [Google Scholar]
- 67.Bell JK, Goetz DH, Mahrus S, Harris JL, Fletterick RJ, and Craik CS (2003) The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity. Nat Struct Mol Biol. 10, 527–534 [DOI] [PubMed] [Google Scholar]
- 68.Pham CTN (2006) Neutrophil serine proteases: specific regulators of inflammation. Nature Reviews Immunology. 6, 541–550 [DOI] [PubMed] [Google Scholar]
- 69.Korkmaz B, Moreau T, and Gauthier F (2008) Neutrophil elastase, proteinase 3 and cathepsin G: Physicochemical properties, activity and physiopathological functions. Biochimie. 90, 227–242 [DOI] [PubMed] [Google Scholar]
- 70.Afonina IS, Müller C, Martin SJ, and Beyaert R (2015) Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity. 42, 991–1004 [DOI] [PubMed] [Google Scholar]
- 71.Devant P, Cao A, and Kagan JC (2021) Evolution-inspired redesign of the LPS receptor caspase-4 into an interleukin-1β–converting enzyme. Science Immunology. 10.1126/sciimmunol.abh3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, and Dubyak GR (2016) Active Caspase-1 Induces Plasma Membrane Pores That Precede Pyroptotic Lysis and Are Blocked by Lanthanides. J.I 197, 1353–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Evavold CL, Ruan J, Tan Y, Xia S, Wu H, and Kagan JC (2018) The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity. 48, 35–44.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karmakar M, Minns M, Greenberg EN, Diaz-Aponte J, Pestonjamasp K, Johnson JL, Rathkey JK, Abbott DW, Wang K, Shao F, Catz SD, Dubyak GR, and Pearlman E (2020) N-GSDMD trafficking to neutrophil organelles facilitates IL-1β release independently of plasma membrane pores and pyroptosis. Nat Commun. 11, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, Wang Y, Li D, Liu W, Zhang Y, Shen L, Han W, Shen L, Ding J, and Shao F (2020) Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 10.1126/science.aaz7548 [DOI] [PubMed] [Google Scholar]
- 76.Hou J, Zhao R, Xia W, Chang C-W, You Y, Hsu J-M, Nie L, Chen Y, Wang Y-C, Liu C, Wang W-J, Wu Y, Ke B, Hsu JL, Huang K, Ye Z, Yang Y, Xia X, Li Y, Li C-W, Shao B, Tainer JA, and Hung M-C (2020) PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nature Cell Biology. 10.1038/s41556-020-0575-z [DOI] [PMC free article] [PubMed] [Google Scholar]