Abstract
Neuropeptide Y (NPY) is highly abundant in the brain and involved in various physiological processes related to food intake and anxiety, as well as human diseases such as obesity and cancer. However, the molecular details of the interactions between NPY and its receptors are poorly understood. Here, we report a cryo-electron microscopy structure of the NPY-bound neuropeptide Y1 receptor (Y1R) in complex with Gi1 protein. The NPY C-terminal segment forming the extended conformation binds deep into the Y1R transmembrane core, where the amidated C-terminal residue Y36 of NPY is located at the base of the ligand-binding pocket. Furthermore, the helical region and two N-terminal residues of NPY interact with Y1R extracellular loops, contributing to the high affinity of NPY for Y1R. The structural analysis of NPY-bound Y1R and mutagenesis studies provide molecular insights into the activation mechanism of Y1R upon NPY binding.
Subject terms: Cryoelectron microscopy, G protein-coupled receptors, Neurochemistry
The human neuropeptide Y (NPY) acts through G-protein coupled receptors and is involved in food intake, stress response, anxiety, and memory retention. Here, the authors show that, unlike in other neuropeptides, both the N-terminal and the C-terminal regions of NPY interact with the NPY receptor 1.
Introduction
The human neuropeptide Y (NPY) system comprises three peptide ligands, NPY, peptide YY (PYY), and pancreatic polypeptide (PP), and four functional NPY receptors Y1R, Y2R, Y4R, and Y5R1. These endogenous peptide ligands consisting of 36 amino acids with the amidated C-terminus activate specific NPY receptors generally coupled to Gi or Go protein2. Of these three peptide ligands, NPY, a highly abundant peptide ligand in the brain, can activate all four subtypes of the NPY receptor and is involved in various physiological processes such as food intake, stress response, anxiety, and memory retention3–5. Furthermore, NPY signaling is involved in human diseases such as obesity, mood disorders, and cancers6–8.
The nuclear magnetic resonance (NMR) structure of NPY reveals that its C-terminal segment (13–36) forms an amphipathic α-helix, and the remaining N-terminal part is unstructured and flexible9,10. Previous functional assays using the N-terminal truncation mutants of NPY indicate that the complete N-terminus of NPY is necessary for Gi signaling through Y1R but not through Y2R, which shares approximately 30% sequence identity with Y1R10,11. Similarly, PYY, highly homologous to NPY, binds to Y1R and Y2R in its full-length form; however, PYY (3–36), the N-terminal cleaved form found in the circulation, binds only to Y2R12–14.
NPY receptors belong to the β subgroup of class A G protein-coupled receptors (GPCRs). Among them, Y1R is expressed in the central nervous system (CNS) as well as in the adipose tissue and vascular smooth muscle cells, where it leads to enhanced cell proliferation and the induction of food intake upon activation by NPY15–17. Thus, the Y1R antagonist has been proposed as a potential drug for treating obesity16. Furthermore, Y1R is highly expressed in human primary breast cancer, implying the utility of Y1R as a diagnostic marker for breast cancer18.
The crystal structures of the small molecule antagonist-bound Y1R have been published recently10. Although these structures provide molecular details of Y1R-selective antagonist binding and the overall architecture of an inactive state of Y1R, the molecular mechanism of its activation by binding of the endogenous agonist NPY is still unknown.
Here, we present a single-particle cryo-electron microscopy (cryo-EM) structure of NPY-bound wild-type Y1R with Gi1 protein coupled. The structure reveals that five amino acids at the C-terminus of NPY form an extended conformation and are inserted into a pocket formed by the transmembrane (TM) domain of Y1R. In addition, the helical region and N-terminus of NPY are shown to be involved in Y1R binding. Together with molecular dynamics (MD) simulations and functional analysis of various mutations, our structure provides molecular details of endogenous peptide recognition by Y1R and suggests the activation mechanism of Y1R upon NPY binding.
Results
Structure determination of the NPY–Y1R–Gi1 complex
The published antagonist-bound crystal structure of Y1R was solved using a modified construct involving thermostabilizing mutation, C-terminal truncation, and replacement of intracellular loop 3 (ICL3) with T4 lysozyme10. In our study, we used the wild-type Y1R construct (2–384) with minimal engineering (such as affinity tag) to solve the receptor structure, thereby facilitating molecular analysis of the activation mechanism of the native receptor. Y1R is known to couple Gi1 protein to activate downstream signaling upon NPY binding2. Before purifying the NPY–Y1R–Gi1 complex, we confirmed the functionality of the synthesized NPY by performing bioluminescence resonance energy transfer (BRET) assays, which showed that Y1R specifically recruited Gi1 in response to NPY binding (Supplementary Fig. 1). For the structural study, Y1R and Gi1 heterotrimer were purified separately and mixed in the presence of NPY. The complex was incubated with apyrase to obtain a nucleotide-free Gi1 heterotrimer, and single-chain variable fragment termed scFv16, that specifically recognizes heterotrimeric Gi was added as a stabilizer19 (Supplementary Fig. 2).
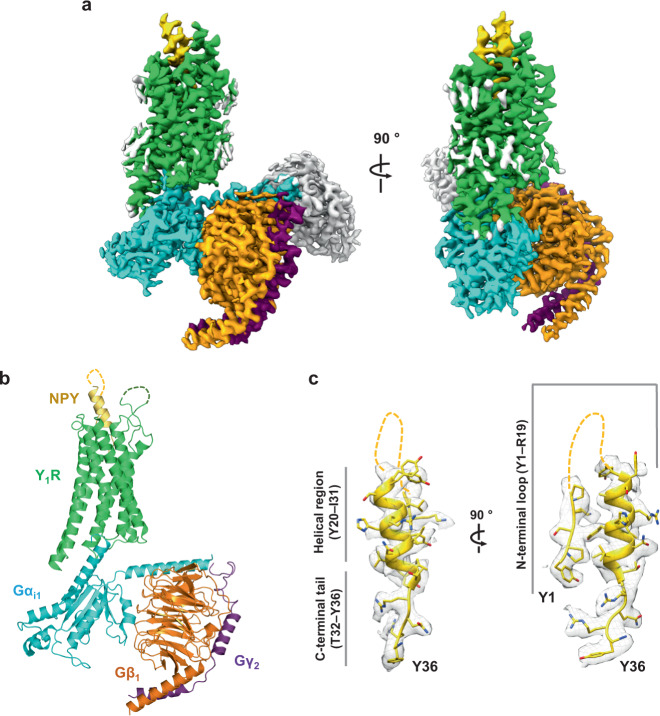
We determined a cryo-EM structure of the NPY–Y1R–Gi1–scFv16 complex at a nominal resolution of 3.2 Å in glyco-diosgenin (GDN) micelles (Supplementary Table 1 and Supplementary Fig. 3). Similar to other structures of nucleotide-free G protein heterotrimers bound to GPCR, the α-helical domain of Gi1 was not modeled in our structure because of its flexibility. Inspection of the cryo-EM map clearly showed the density of NPY protruding into the extracellular region (Fig. 1a). To further focus on the binding of NPY to Y1R, we subtracted the heterotrimeric G protein signal and subjected to local refinement (Supplementary Fig. 3) as used in the cryo-EM analysis of secretin-bound secretin receptor–Gs complex20. The resulting cryo-EM map allowed modeling of the five residues at the N-terminus (1–5) and the C-terminal half of NPY (20–36) (Fig. 1b, c and Supplementary Fig. 4).
Fig. 1. Overall structure of the NPY–Y1R–Gi1–scFv16 complex.
a The cryo-EM map of the NPY–Y1R–Gi1–scFv16 complex is shown. Y1R, NPY, Gαi1, Gβ1, Gγ2, and scFv16 are colored green, yellow, cyan, orange, purple, and gray, respectively. The remaining micelle density is shown in light gray. Details on cryo-EM map generation are described in the “Methods” section. b The structure of the NPY–Y1R–Gi1 complex is shown. scFv16 is included in the final structure but omitted in this figure for clarity. c The sharpened cryo-EM map with the NPY peptide model is shown in two orientations. NPY residues 6–19 are not modeled in this structure and are shown as dashed lines. The three regions of NPY are marked as the N-terminal loop region (Y1–R19), the helical region (Y20–I31), and the C-terminal tail (T32–Y36).
Overall structure of NPY-bound Y1R receptor
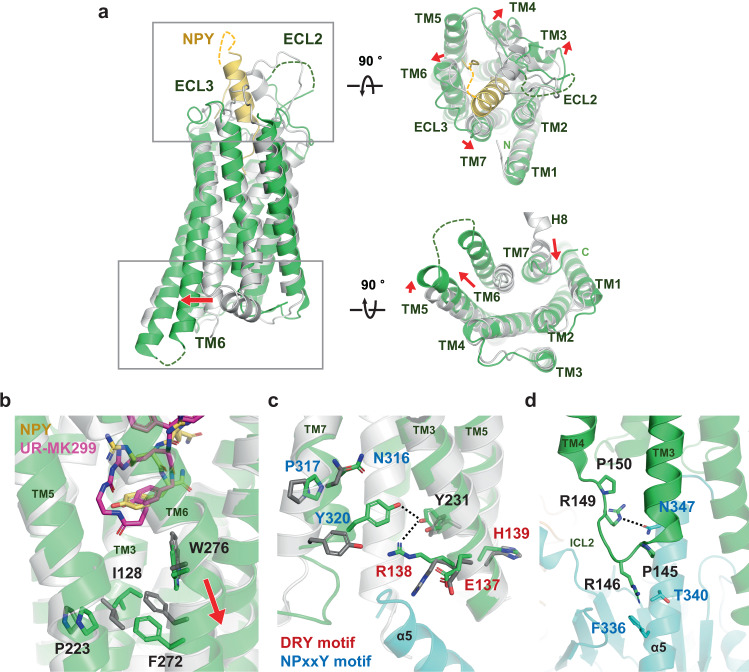
Structural comparison of the NPY-bound and antagonist-bound Y1R reveals distinct conformational changes in the extracellular region, TM core, and cytoplasmic part of the receptor upon activation. As the amidated C-terminus of NPY penetrates deep into the TM core, the extracellular tips of TM3, TM4, TM6, and TM7 slightly move outward by 1.9–2.5 Å in the NPY-bound Y1R structure compared to the antagonist-bound structure, opening up the ligand-binding pocket (Fig. 2a). In fact, the calculated solvent-accessible ligand-binding cavities in the antagonist-bound and NPY-bound structures are ~506 and 730 Å3, respectively (Supplementary Fig. 5). Besides the interaction of the C-terminal tail of NPY with the Y1R TM core, the molecular interaction of the remaining NPY with Y1R was difficult to characterize, as the NPY map was not well resolved (Supplementary Fig. 3). However, we could observe the density of the N-terminal and helical regions of NPY, which are surrounded by extracellular loops (ECLs) and the N-terminal region of Y1R (Supplementary Fig. 6). ECL2, ECL3, and the N-terminal region of Y1R directly interact with NPY to varying degrees. The details of the NPY interaction are discussed later.
Fig. 2. Comparison between NPY-bound and antagonist-bound Y1R structures.
a Structural alignment between NPY (yellow)-bound Y1R (current structure, green) and antagonist-bound Y1R (PDB ID 5ZBQ, light gray, residues 18–32 of Y1R and antagonist are omitted for clarity) clearly shows characteristic TM6 movement as observed in the active structures of class A GPCRs. Views from extracellular (right-upper panel) and cytoplasmic (right-lower panel) sides are shown on the right. Each red arrow represents the movement of the TM helix. b Structural changes at the connector region (P5.50I3.40F6.44 motif) upon NPY binding are shown. The antagonist-bound Y1R (PDB ID 5ZBQ) is colored in light gray and an antagonist, UR-MK299, in magenta. c Structural changes at the D(E)/R/Y(H) (labeled in red) and NPxxY (labeled in blue) motifs are shown. d The binding interface between α5 of Gi1 (in cyan) and ICL2 is shown. Residues participating in the interactions and two Pro residues (P145ICL2 and P150ICL2) in ICL2 are shown as sticks. Dashed lines represent the polar interactions.
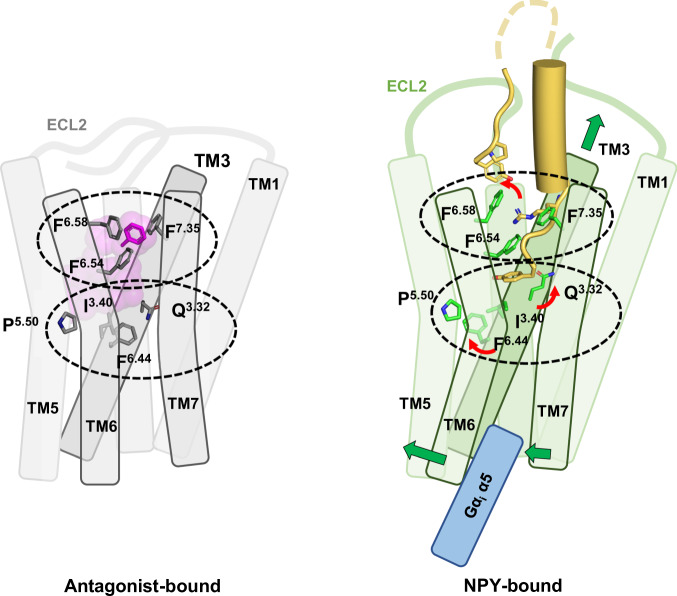
NPY binding results in the TM core of Y1R undergoing conventional conformational changes associated with the activation mechanism of class A GPCRs. Immediately below the C-terminus of NPY peptide, side chains of I1283.40, P2235.50, and F2726.44 (superscripts are the Ballesteros–Weinstein numbers21) in the connector region are repacked to contract the interface of TM3, 5, and 6 (Fig. 2b), resulting in the outward movement of TM6 and inward movement of TM7 at the cytoplasmic part of Y1R (Fig. 2a). In addition, R1383.50 of the (D/E)R(Y/H) motif makes close contact with Y2315.58 and Y3207.53 of the NPxxY motif (Fig. 2c), a well-known key interaction observed in the activated GPCR structures. These conformational changes demonstrate that our structure represents the active conformation of class A GPCRs.
Compared to other Gi1-bound class A GPCRs, such as neurotensin receptor 1 (NTSR1) and μ-opioid receptor (μOR), Y1R has a relatively short ICL2, comprising six amino acids, including two Pro residues (Fig. 2d). In NTSR1 and μOR, ICL2s adopt an α-helix upon activation, interacting with the αN, αN-β1 loop and α5 helix of Gi122,23. In contrast, ICL2 of Y1R remains as a loop in the Gi1-bound state, forming contacts with only the α5 helix of Gi through R146ICL2 and R149ICL2 (Fig. 2d). In addition to ICL2, hydrophobic residues in TM3, TM5, and TM6 (I1423.54, I2345.61, L2385.65, I2616.33, and L2656.37) of Y1R form van der Waals interactions with L348 and L353 in the α5 helix of Gi1 (Supplementary Fig. 7) and R2606.32 of Y1R forms polar interaction with the carboxyl group of F354 in the α5 helix of Gi1 (Supplementary Fig. 7). Most of these interactions are conserved in the hNTSR1-Gi1 and μOR-Gi1 structures (Supplementary Fig. 7). However, when aligning the receptors, the relative position of the α5 helix of Gi1 bound to Y1R slightly differs by ~2 Å displacement of the C-terminus of Gαi1 or ~8° tilt angle of the α5 helix of Gi1 in the NTSR1-bound and μOR-bound Gi1 structures, respectively (Supplementary Fig. 7).
Binding of the C-terminal tail of NPY to Y1R
Our cryo-EM map shows a well-resolved density for the five C-terminal residues of NPY (32–36) (Fig. 1c), forming an extended structure, contrasting the α-helix formation of residues 13–36 in the NMR structure of human NPY (PDB ID 1RON)9. However, helix unwinding at the C-terminal tail of NPY was not unexpected since the previous NMR study of porcine NPY bound to Y1R suggested an extended conformation of the NPY C-tail10. Similarly, the nine amino acids at the C-terminus of orexin-B neuropeptide (OxB) were previously shown to form an extended conformation in the orexin receptor (OX2R)-bound state but form an α-helix in the receptor-free state24,25.
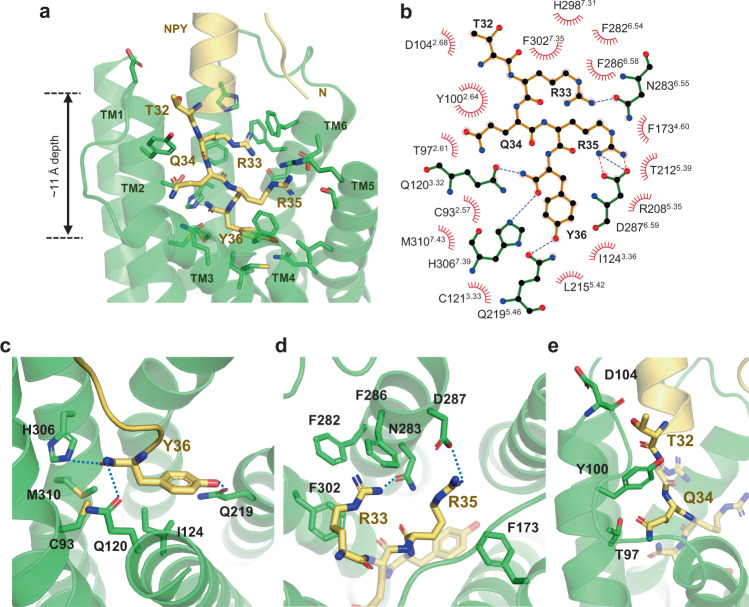
The extended conformation of the NPY C-tail binds to a pocket lined by TM helices 2–7, with a depth of ~11 Å from the top surface of the membrane (Fig. 3a, b). It is well established that the amidation of NPY C-terminal tyrosine is critical for its function; consistently, in this study, non-amidated NPY failed to elicit G protein signaling (Supplementary Fig. 8). At the bottom of the ligand-binding pocket, the C-terminal amide of NPY points toward the side chain of Q1203.32 (Fig. 3c), which has been predicted to interact with the Y36 side chain in the NPY-docked Y1R model10. The importance of Q1203.32 for Gi recruitment and signaling upon NPY treatment was investigated using BRET and calcium signaling assays. As expected, the Q1203.32A mutant exhibited reduced Gαi1 recruitment and an increased EC50 (Supplementary Table 2 and Supplementary Figs. 9, 10, and 11). The C-terminal amide of Y36 is further coordinated by H3067.39 through polar interactions and C932.57 and M3107.43 through van der Waals contacts (Fig. 3c). Furthermore, the ligand-binding pocket of Y1R is highly acidic (Supplementary Fig. 12). The acidic residues of the binding pocket repel the negatively charged C-terminus and favor the amidated C-terminus. A similar acidic patch in the ligand-binding pocket is observed in OX2R, where the amidated C-terminus of OxB binds. In contrast, the ligand-binding pocket is basic in NTSR1 and endothelin B receptor (ETBR), whose peptide agonists have the C-terminal carboxyl groups (Supplementary Fig. 12).
Fig. 3. Binding of NPY C-terminal tail to Y1R.
a The C-terminal tail of NPY (yellow) and Y1R residues that participate in NPY binding are presented as sticks. b Schematic representation of NPY–Y1R interactions analyzed using the LigPlot+ program is shown. The hydrogen bonds and the salt bridges are shown in blue and red dashed lines, respectively. c–e Detailed interactions between C-terminal residues of NPY and Y1R are shown. The polar contacts are shown as blue dashed lines.
The Y36 side chain of NPY forms a hydrogen bond with Q2195.46 through its hydroxyl group and hydrophobic interaction with I1243.36 through its phenyl ring (Fig. 3c). The importance of Q2195.46 and I1243.36 for NPY signaling was demonstrated by 13.5-fold and 3.5-fold reduction in NPY potency in the Q2195.46A and I1243.36A mutants, respectively10. Previous mutational studies of NPY showed that the Y36F mutation had a relatively mild effect on Y1R binding26. In contrast, the Y36A mutation caused a loss of binding to Y1R10,26, suggesting that the phenyl ring of Y36 is more important for NPY signaling. Our structure shows that the phenyl ring of Y36 forms an intramolecular interaction with R35 of NPY (Supplementary Fig. 13), which aids in correctly positioning R35, in addition to hydrophobic interactions with Y1R (Fig. 3a).
R33 and R35 of NPY extensively interact with the Y1R TM core, and alanine scanning mutagenesis of NPY has shown that R33A and R35A mutations of NPY exhibit the most severe defects in Y1R binding10,26. R33 of NPY forms a hydrogen bond with N2836.55 and π–cation interactions with F2866.58 and F3027.35 (Fig. 3d). BRET analysis using the N2836.55A mutant showed a dramatic decrease in Gαi recruitment and reduced NPY potency by 85-fold compared to wild-type Y1R (Supplementary Table 2 and Supplementary Figs. 9, 10, and 11). Previous mutagenesis studies of F2866.58 and F3027.35 showed that substituting of these residues with alanine caused a reduction in NPY potency10,27. R35 forms electrostatic interaction with D2876.59 and van der Waals interaction with F1734.60, both of which have been reported to be essential for NPY signaling10,28.
Q34 and T32 of NPY are oriented opposite to the R35 and R33 side chains, interacting with T972.61, Y1002.64, and the backbone carbonyl of D1042.68 (Fig. 3e). In particular, Y1002.64 is sandwiched between T32 and Q34, forming nonpolar contacts with both (Fig. 3e). Consistent with these structural data, Y1002.64A, a well-known mutation, dramatically decreased downstream NPY signaling10,27,28. In the antagonist-bound structures, none of these residues that interact with T32 and Q34 are involved in antagonist binding.
Altogether, our structural analysis shows that the C-terminal tail of NPY forms extensive interactions with Y1R residues on TM2, 3, 5, and 6. Previous and current mutational studies support the importance of these interacting residues in NPY signaling.
Structural changes in Y1R TM core by binding of NPY C-terminal tail
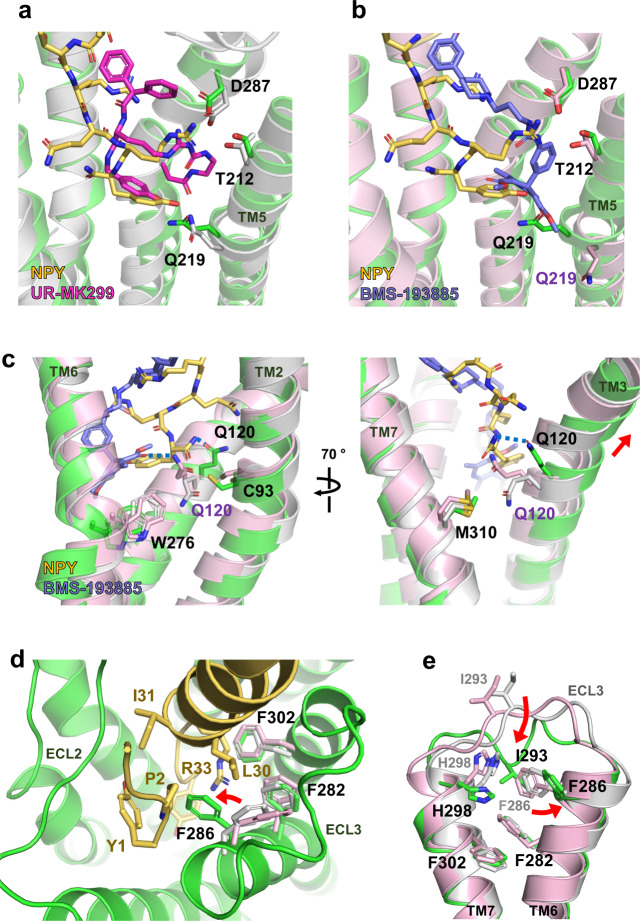
The binding pocket for the NPY C-terminus is where the two antagonists (UR-MK299 and BMS-193885) bind in the previously reported inactive Y1R structures. The structural comparison reveals that the hydroxyphenyl group and guanidine moiety of UR-MK299 exhibit binding modes similar to those of the side chains of Y36 and R35 of NPY, respectively, making polar interactions with Q2195.46 and D2876.59 (Fig. 4a). The BMS-193885-bound inactive Y1R structure shows that D2876.59 of Y1R interacts with the antagonist, whereas Q2195.46 of Y1R is not involved in this antagonist binding, as Q2195.46 is pushed away by the dihydropyridine group (Fig. 4b). In both inactive structures, Q1203.32 makes van der Waals contact with M3107.43, although Q1203.32 rotamers differ in two inactive structures, forming polar interaction with BMS-193885 or van der Waals interaction with W2766.48 (Fig. 4c). In the NPY-bound Y1R structure, the Q1203.32 sidechain adopts an upward-facing rotamer, forming a polar contact with the amidated C-terminus. Also, Q1203.32 no longer interacts with M3107.43 (>7 Å) but with C932.57. Reorganization of interaction network near Q1203.32 by NPY binding stabilizes the conformation of the upward displacement of TM3 (Fig. 4c).
Fig. 4. Comparison of binding mode between NPY and antagonists.
a Ligand-binding site occupied by UR-MK299 (PDB ID 5ZBQ, magenta) and b BMS-193885 (PDB ID 5ZBH, slate) is compared with the binding site for NPY (yellow). The side chains of Q2195.46 are differently oriented in all three structures, whereas D2876.59 and T2125.39 maintain similar interactions. c Q1203.32-mediated interactions in the antagonist-bound and NPY-bound structures are compared. In the NPY-bound active structure of Y1R, TM3 including Q1203.32 shifts upward and Q1203.32 forms a polar interaction with the C-terminal amide of NPY through its upward-facing sidechain. d Upon NPY binding, F2866.58 rotamer is changed to form the interactions with NPY R33, L30, and Y1. e Upon NPY binding, I293ECL3 participates in a hydrophobic interaction network with F2866.58, H2987.31, and F3027.35. UR-MK299-bound Y1R is indicated in light gray, and BMS-193885-bound Y1R is in pink. Red arrows represent positional changes of I293ECL3 and F2866.58 upon NPY binding.
The hydrophobic moieties of the antagonists form hydrophobic networks with I1243.36, I1283.40, F2726.44, W2766.48, and L2796.51 at the bottom of the ligand-binding pocket and with F2826.54, F2866.58, and F3027.35 near the entrance to the binding pocket, to stabilize the inactive Y1R structure. These hydrophobic networks are rearranged in the NPY-bound structure. As mentioned above, at the bottom of the ligand-binding pocket, a rotamer change of I1283.40 and repacking of F2726.44 and W2766.48 occur upon NPY binding (Fig. 2b). The three phenylalanine residues F2826.54, F2866.58, and F3027.35 on TM6 and TM7 form a stable π–π network with phenyl groups present in both antagonists; however, this aromatic network is disrupted in the NPY-bound structure. The phenyl ring of F2866.58 flips to form van der Waals contact with L30 and a π–cation interaction with R33 of NPY (Fig. 4d). F2826.54 and F3027.35 form a new interaction network with H2987.31 and I293ECL3 (Fig. 4e). F2866.58 also interacts with Y1 and P2 of NPY. The NPY N-terminus interactions are discussed in the next section.
Comparison of binding modes between the NPY C-terminal tail and antagonists reveals common interactions of the ligand for Y1R binding and NPY-specific interactions, providing hints for designing novel Y1R ligands that bind to the TM core. In addition, the structural comparison of the TM core between inactive and active states of Y1R suggests the key events of conformational changes during activation by NPY, rearrangement of the hydrophobic network around F2866.58 at the entrance to the ligand-binding pocket, and rearrangement of interaction network around Q1203.32 and I1283.40 in the connector region at the bottom of the ligand-binding pocket.
Binding of the N-terminal and helical regions of NPY to Y1R
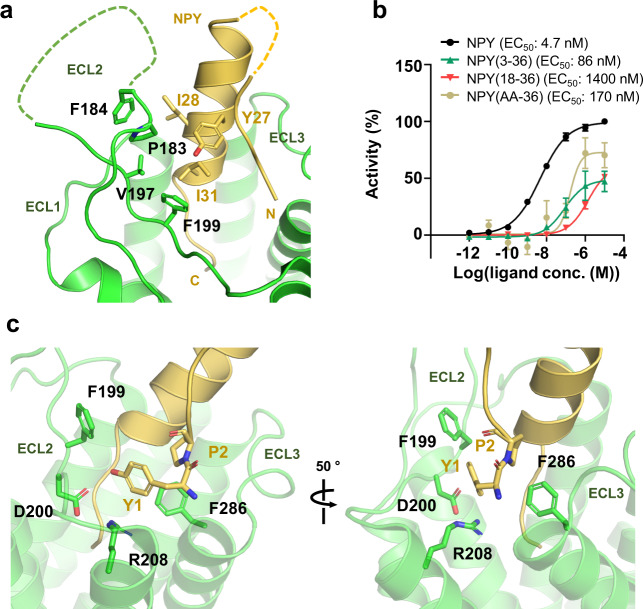
In the OxB-bound OX2R structure, the N-terminus of OxB was not observed despite being necessary for signaling. In our complex structure, we were surprised to observe the density of the helical and the N-terminal loop regions of NPY in addition to the C-terminal tail (Fig. 1c). The NPY residues 20–31 were built as an α-helix based on the continuous cryo-EM density from the C-terminus (Fig. 1c and Supplementary Fig. 4). This α-helical region forms relatively loose interactions with ECLs compared to the C-terminal tail of NPY that forms extensive interactions with the TM core. The cryo-EM map density was weaker toward the N-terminus of the helix, suggesting the flexibility of this region. Three independent 1-μs MD simulations using a model composed of full-length NPY, Y1R (2–339), and Gi1 protein showed that the angle of the helical axis of NPY to the membrane normal varied from 5° to 70° during the simulations (Supplementary Fig. 14). While the C-terminal tail of NPY maintained its binding pose during the simulations, the movement increased toward the N-terminus of the NPY helix, explaining the weak density of the N-terminal end of the NPY helix.
The density of side chains in the ECL2 region spanning residues 185–193 was unclear, but we identified that P183ECL2, F184ECL2, and F199ECL2 interacted with Y27, I28, and I31 of NPY, respectively (Fig. 5a). The F184ECL2A and F199ECL2A mutants reduce the potency of NPY by 38-fold and 2.3-fold, respectively (Supplementary Table 2 and Supplementary Figs. 15, 16, and 17). Although the F202ECL2A mutation exhibited attenuated G protein signaling, F202ECL2 did not interact with NPY. F202ECL2 appears to be important for maintaining the structural integrity of the receptor by forming an aromatic network with F1734.60, Y1764.63, and Y2115.38, as observed in both inactive and active structures (Supplementary Fig. 18).
Fig. 5. Binding of NPY N-terminus to Y1R.
a The sticks show the residues participating in van der Waals interactions between NPY helix and ECL2 of Y1R. Unresolved regions of NPY and ECL2 of Y1R are shown with dashed lines. b The results of Ca2+ assays performed with N-terminally truncated NPY peptides are shown. Removal of two N-terminal residues dramatically reduces NPY potency and efficacy. Symbol and error bar indicate the mean and S.E.M. (standard error of mean) of n = three (NPY(3–36), NPY(18–36), and NPY(AA-36)) or n = 17 (NPY) independent experiments, respectively. Calculated EC50 values (pEC50 ± SEM) are presented on each graph and are summarized in Supplementary Table 2. Source data are provided as a Source Data file. c Van der Waals interactions of Y1 and P2 of NPY with Y1R are shown in two different orientations.
Previously, it was proposed that the complete N-terminus of NPY is necessary for NPY signaling through Y1R10. Consistently, our signaling assays demonstrated the decrease in the potency of NPY(3–36) and NPY(18–36) by 18-fold and 300-fold, respectively, compared to full-length NPY (Fig. 5b), suggesting that the N-terminal residues of NPY are important for Y1R binding. In our structure, NPY residues 1–5 were modeled by fitting these residues into a cryo-EM map (Fig. 1c and Supplementary Fig. 19). Among these five residues, the N-terminal residue Y1 was relatively well defined by the cryo-EM density. Y1 interacts with F199ECL2, D200ECL2, and R2085.35 of Y1R (Fig. 5c), all of which were demonstrated to be important for NPY signaling by previous or current mutational studies27,28 (Supplementary Table 2 and Supplementary Figs. 15, 16, and 17).
The N-terminus of NPY does not enter the TM core like the C-terminus but is exposed to the solvent, suggesting its propensity to interact more dynamically with Y1R. In agreement with our speculation, MD simulations show that the N-terminal region of NPY exhibits relatively higher motions than the C-terminal tail (32–36) during 1-μs simulation. Moreover, the Y1-mediated interactions were broken and reformed in one of the replicates during the simulations (Supplementary Fig. 14). Collectively, our data show that the N-terminal region of NPY interacts dynamically with Y1R. Our model structure represents a possible conformation of the NPY N-terminus, deduced from the cryo-EM map.
Functional role of the N-terminal region of Y1R
Our cryo-EM map reveals a continuous density from the N-terminal end of TM1 toward the NPY ligand (Supplementary Fig. 6), suggesting an interaction between them. Previously, the N-terminal region of Y1R, residues 21–32, was crosslinked to NPY in a photo-crosslinking experiment10. However, the low-resolution map at TM1 and the N-terminal region of Y1R prevented us from unambiguously determining whether the N-terminal region of Y1R directly interacted with NPY. Through signaling assays and structural analysis, it was previously proven that N-terminal residues were critical for peptide ligand binding and activation of other closely related peptide receptors, namely OX2R and ETBR29–32.
To validate the importance of the N-terminal region of Y1R for NPY signaling, we constructed two N-terminal deletion mutants of Y1R, one in which the 25 N-terminal residues were deleted (Y1RΔ25) and the other in which the 31 N-terminal residues were deleted (Y1RΔ31), and performed BRET and Ca2+ assays (Supplementary Table 2 and Supplementary Figs. 20, 21, and 22). Y1RΔ25 behaves like wild-type Y1R in recruiting Gi1 by NPY treatment, whereas Y1RΔ31 showed attenuated response to NPY; this finding suggests that residues 26–31 are involved in NPY binding and thus in Gi1 recruitment. In particular, the hydrophobic residues L26N and F28N in Y1R were important for NPY signaling, as indicated by the 2.8-fold increase in EC50 in the L28NA/F28NA mutant (Supplementary Table 2 and Supplementary Figs. 20, 21, and 22). MD simulations show that although the N-terminal region of Y1R has high mobility, L26N and F28N are within the Cα distance of 9 and 13 Å, respectively, from Y21 in the helical region of NPY (Supplementary Fig. 23). Of note, Y21 of NPY is assumed to be a contact point based on the nearby extra cryo-EM density. In addition, the helical region of NPY was demonstrated to interact with ECL2 during the simulations, suggesting that ECL2 and the N-terminal region of Y1R form dynamic interactions with NPY by reorienting themselves extensively to accommodate the NPY binding (Supplementary Fig. 23). Thus, we hypothesize that the helical region of NPY forms a tripartite interaction with ECL2 and the N-terminal region of Y1R, both are shown to partially cover the ligand-binding pocket in the antagonist-bound Y1R structures.
Discussion
Several structures of the G protein-bound active state of class A GPCRs in complex with endogenous peptide agonists, such as NTS (8–13), OxB, and cholecystokinin-8 (CCK-8), have been reported24,33,34. Commonly, these peptide agonists have a C-terminal region that inserts into the receptor TM core and acts as a “message” domain (Supplementary Fig. 24)35,36. Similarly, in this study, we observed that the five NPY C-terminal residues in the NPY-bound Y1R structure, forming an extended conformation, make extensive contact with residues in the TM core. In addition, our structure shows that the helical region and the N-terminal loop of NPY interact with Y1R, although this interaction is much more dynamic and even transient, as indicated by the weaker density for this region in our cryo-EM map, as well as our MD simulations. This study presents a model candidate containing the five N-terminal residues of NPY, constructed based on our cryo-EM map. The cryo-EM density for Y1 of NPY is relatively well resolved, showing interaction with F199ECL2, D200ECL2, and R2085.35 of Y1R. Despite being a dynamic interaction, the NPY N-terminus is crucial for Gi signaling, as demonstrated by reduced Gi recruitment in the BRET assay and a 18-fold increase in EC50 value in signaling assays after treatment with NPY(3–36). The NPY receptor has two other peptide ligands, PYY and PP. PYY is released in response to nutrient intake along the gut and is highly homologous to NPY with 67% sequence identity; its N-terminus starts with tyrosine, similar to NPY (Supplementary Fig. 25). However, the major circulating form of PYY is the cleaved form PYY(3–36), known to be selective for Y2R12,13,37. Our signaling assays with NPY, PYY, NPY(3–36), and PYY(3–36) also show that Y1R has EC50 values in the nanomolar range for NPY and PYY (4.7 and 6.1 nM, respectively) and 13–18-fold increased EC50 values for NPY(3–36) and PYY(3–36) (86 nM and 77 nM, respectively) (Supplementary Fig. 26), suggesting that PYY would bind Y1R similarly to NPY if its N-terminus remains intact. On the contrary, PP is secreted in the pancreas and has 50% sequence identity with NPY (Supplementary Fig. 25). Reportedly, PP does not bind Y1R at all38. PP has A1 and P34 instead of Y1 and Q34, respectively; thus, the interactions of Y1 and Q34 as shown in our NPY-bound Y1R structure are important for receptor binding. Indeed, our signaling assay shows that Y1R has a 64-fold increased EC50 value for PP compared to NPY (Supplementary Fig. 26). Among four subtypes of the NPY receptor, Y4R was activated in response to PP38–40. Interestingly, Y4R has Glu at the position of 6.58 (E2886.58), instead of Phe as in Y1R (Supplementary Fig. 27), suggesting that Y4R would form a charged interaction network with nearby charged residues (E203ECL2, R2115.35, T2155.39, N2856.55, E2886.58, D2896.59) and basic residues of PP, R33 and R35 (Supplementary Fig. 28). We speculate that this extensive charged interaction network would provide sufficient interaction energy for Y4R to accommodate PP as well as NPY, which should be validated with experimental data.
Unlike the previously predicted NPY binding pose, our structure shows that the C-terminal amide of NPY points toward Q1203.32 of Y1R, and the Y36 side chain interacts with Q2195.46. The importance of Q1203.32 and Q2195.46 for NPY binding and signaling is demonstrated by the approximately 2 and 4-fold increased EC50 values measured using the mutants Q1203.32A and Q2195.46L, respectively. Notably, the Y36 side chain occupies a position similar to the hydroxyphenyl ring of the antagonist UR-MK29910. Thus, in both active and inactive structures, Q2195.46 forms a hydrogen bond with the hydroxyl group of the hydroxyphenyl ring in each ligand. In contrast, Q1203.32 of Y1R forms a hydrogen bond with the C-terminal amide of NPY; however, it is not involved in antagonist binding. Notably, the conserved Gln residue at position 3.32 in OX2R (Q1343.32) forms a hydrogen bond with the peptide agonist. Therefore, it was proposed to be a key residue in facilitating the transition to an active state of OX2R by a rotamer change to its upward-facing extended conformation24. This proposal appears to apply to the Y1R activation mechanism, as a similar rotamer change of Q1203.32 is observed upon activation (Fig. 6).
Fig. 6. Mechanism of Y1R activation by NPY binding.
Schematic figures representing antagonist-bound (left) and NPY-bound (right) Y1R are shown. Two key regions for Y1R activation are indicated by dashed ellipses. The three phenylalanine residues at TM6 and TM7, and Q1203.32, I1283.40, P2235.50, and F2726.44 in the connector region are shown in stick representation. The three phenylalanine residues participate in antagonist binding through π–π interactions, but these interactions are rearranged upon NPY binding. F2866.58 flips and forms a new interaction network with Y1 and R33 of NPY. Since F2866.58 is not conserved in Y2R, F2866.58-mediated interactions are Y1R-selective. Q1203.32, I1283.40 and F2726.44 show rotamer changes upon NPY binding. A series of these changes upon NPY binding pulls TM3 upward and causes outward movement of the cytoplasmic region of TM6.
Comparison of the inactive and active structures suggests the activation mechanism of Y1R upon NPY binding. In the published antagonist-bound structures, the three phenylalanine residues F2826.54, F2866.58, and F3027.35 located near the entrance to the ligand-binding pocket constitute a hydrophobic cluster with the antagonists UR-MK299 and BMS-193885. Upon NPY binding, R33 of NPY is inserted into this Phe network, causing a rotamer change in F2866.58, disrupting the aromatic network. In association with the conformational change of ECL3, a new interaction network including Y1 and R33 of NPY and F2826.54, F2866.58, F3027.35, I293ECL3, and H2987.31 of Y1R is formed, stabilizing the NPY-bound Y1R structure (Fig. 4e). At the bottom of the ligand-binding pocket, Y36 of NPY forms hydrophobic interaction with I1243.36 through the phenyl group and polar interaction with Q1203.32 through the amidated C-terminus, leading to a rotamer change of Q1203.32. This is followed by a rotamer change of I1283.40, which interacts with I1243.36 and repacking of the side chains of P2235.50, F2726.44, and W2766.48. A series of these changes upon NPY binding pulls TM3 upward and causes outward movement of the cytoplasmic region of TM6 (Fig. 6).
One of the key NPY-interacting residues, F2866.58, is not conserved and replaced by valine in Y2R and glutamate in Y4R, both of which cannot form π–cation interaction with R33 as F2866.58 does. The difference in the Phe network may explain the Y1R selectivity of the two antagonists used to determine the inactive Y1R structures; additionally, it suggests that Y1R, Y2R, and Y4R may have different interaction networks with NPY. In addition to F2866.58, Q2195.46, which forms a hydrogen bond with Y36, is replaced with L2275.46 in Y2R, and H2987.31, which forms an interaction network with F2826.54, F2866.58, and F3027.35 is replaced with G3007.31 in Y4R. It should also be noted that F2866.58 is located within a distance of 4–5 Å to Y1 and P2 of NPY. Further details would only be explained by investigating the NPY-bound Y2R and Y4R structures in the future. However, the vicinity of the residue and the importance of the Phe network suggest that F2866.58 may contribute to the difference between Y1R and Y2R in the need for the complete N-terminus to elicit a full response.
Methods
Expression and purification of Y1R
Wild-type human Y1R (2–384) with a FLAG tag at its N-terminus as well as eGFP and a His8 tag at its C-terminus, cleavable by HRV 3C protease, was expressed in Spodoptera frugiperda (Sf9) insect cells using the Bac-to-Bac system (Invitrogen). Cells were harvested 48 h after infection and lysed by repeated dounce homogenization with lysis buffer (20 mM HEPES pH 8.0, 150 mM NaCl, and 1 mM EDTA) supplemented with phenylmethylsulfonyl fluoride (PMSF), benzamidine, and leupeptin. Y1R was extracted from the cell membrane using a solubilization buffer consisting of 20 mM HEPES pH 8.0, 150 mM NaCl, 1% (w/v) n-dodecyl-β-d-maltoside (DDM), 0.1% (w/v) cholesterol hemisuccinate (CHS), PMSF, benzamidine, and leupeptin. After centrifugation, the supernatant was incubated with Ni-NTA resin for 1 h at 4 °C. After column washing with wash buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 20 mM imidazole, 0.05% DDM, and 0.005% CHS), the bound protein was eluted with 300 mM imidazole and subsequently loaded onto anti-FLAG M1 agarose resin (Sigma Aldrich) in the presence of 2 mM CaCl2. After column washing and on-column exchange of detergent from DDM to glyco-diosgenin (GDN), the bound protein was eluted with M1 elution buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 0.01% GDN, 0.1 mg/ml FLAG peptide, and 4 mM EDTA). Heterogeneous glycosylation was removed by PNGase F (NEB), and the eGFP was cleaved by the HRV 3C protease (homemade). Y1R was further purified using a Superdex 200 10/300 gel filtration column (Cytiva) pre-equilibrated with a buffer containing 20 mM HEPES pH 8.0, 150 mM NaCl, and 0.01% GDN. Freshly purified Y1R was used to form a complex with Gi1 heterotrimer.
Purification of the G protein
Human Gαi1 with a His6 tag at its N-terminus was expressed in Escherichia coli (E. coli) Rosetta (DE3) cells. Protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the cells were harvested after incubation at 25 °C overnight. Cells were lysed with Emulsiflex C3 (Avestin), and the cleared lysate after centrifugation was loaded onto the Ni-NTA column. The column was washed with wash buffer (20 mM Tris-HCl pH 8.5, 150 mM NaCl, 30 mM imidazole), and the bound protein was eluted with elution buffer (20 mM Tris-HCl pH 8.5, 150 mM NaCl, and 300 mM imidazole). A His6 tag was cleaved by TEV protease (homemade) treatment at 4 °C overnight. The Hitrap Q column (Cytiva) and a gel filtration column were used for further purification. Purified Gαi1 dissolved in a solution composed of 20 mM Tris-HCl pH 8.5, 50 mM NaCl, 1 mM MgCl2, and 10 μM guanosine diphosphate (GDP) was concentrated, snap frozen in liquid nitrogen, and stored at −80 °C until use.
Human Gβ1 with a His6 tag at its N-terminus and Gγ2 with a C68S mutation were co-expressed in Sf9 insect cells. Cells were harvested 72 h after incubation and lysed with lysis buffer (20 mM Tris-HCl pH 8.5, 0.1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), and protease inhibitors). After centrifugation, the cleared supernatant was loaded onto an Ni-NTA column. The bound protein was eluted with lysis buffer supplemented with 300 mM imidazole after washing with lysis buffer supplemented with 20 mM imidazole. The His6 tag was cleaved with HRV 3C protease, and the Gβγ complex was further purified using a Hitrap Q column.
For G protein heterotrimer formation, purified Gαi1 and Gβγ were mixed in a 1.1:1 molar ratio with excess MgCl2 and GDP. After an hour of incubation on ice, the Gαiβγ complex was purified using a gel filtration column equilibrated with a solution composed of 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 10 μM GDP. The purified Gαiβγ complex was concentrated, snap frozen in liquid nitrogen, and stored at −80 °C until use.
We confirmed that Gαi1 produced from E. coli is still functional using GTP turnover assay (Supplementary Fig. 29). For comparison, we purified Gi1 heterotrimer produced from Sf9 insect cells, as previously described22.
Purification of the scFv16
The scFv16 construct was kindly provided by Dr. Kobilka (Stanford University). Purification of scFv16 was performed by the previously described method with slight modifications19. First, scFv16 with a His6 tag at its C-terminus was expressed in Trichoplusia ni (Hi5) cells. After cell harvesting, the supernatant containing secreted scFv16 was incubated with Ni-NTA resin at 4 °C for 2 h. The resin was washed with buffer (20 mM HEPES pH 7.0, and 150 mM NaCl, 30 mM imidazole), and the bound protein was eluted with the buffer supplemented with an additional 300 mM imidazole. Further purification was performed using a gel filtration column pre-equilibrated with a solution composed of 20 mM HEPES pH 7.0, and 150 mM NaCl. Purified scFv16 was concentrated, snap frozen in liquid nitrogen, and stored at −80 °C until use.
Purification of the NPY–Y1R–Gi1-scFv16 complex
Purified Y1R, Gi1 heterotrimer, and scFv16 were mixed at a molar ratio of 1:1.1:1.2 in the presence of excess NPY (GL Biochem, Shanghai, China) and incubated overnight at 4 °C with apyrase (NEB, MA, USA). The sample was then loaded onto a gel filtration column pre-equilibrated with a solution composed of 20 mM HEPES pH 8.0, 150 mM NaCl, 0.01% GDN, and 2 μM NPY. The purified NPY–Y1R–Gi1–scFv16 complex was concentrated to 10 mg/ml and used for cryo-EM grid preparation.
Cryo-EM grid preparation and data collection
An aliquot (3.5 µl) of purified NPY–Y1R–Gi1–scFv16 complex was applied onto a glow-discharged holey carbon grid (Quantifoil R1.2/1.3, 300 mesh). The grids were blotted for 5 s at 12 °C and 100% humidity and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific, USA) at Center for Macromolecular and Cell imaging of Seoul National University (SNU CMCI). Grids were initially screened with the FEI Glacios (Thermo Fisher Scientific, USA) at SNU CMCI, equipped with a Falcon 4 detector. Images were acquired on a 300-kV Titan Krios (Thermo Fisher Scientific, USA) at Korea Basic Science Institute, equipped with a Falcon 3EC direct electron detector. Movies were recorded in counting mode at a magnification of ×161,850 (corresponding to a calibrated pixel size of 0.865 Å) and a defocus range of −1.25 to −2.75 μm. A total of 4965 movies were collected, each comprising 40 frames, with a total dose of 40 electrons per Å2. A detailed description of the cryo-EM data collection parameters is provided in Supplementary Table 1.
Three-dimensional (3D) reconstruction of NPY–Y1R–Gi1–scFv16 complex
Image stack preprocessing was performed using CryoSPARC v. 3.1 (Structura Biotechnology)41. All movies subjected to beam-induced motion correction using patched-motion correction and contrast transfer function (CTF) parameters for each non-dose-weighted micrograph were determined by patch CTF estimation. After initial particle picking and two-dimensional classification, selected good particles were used for Topaz training42. Topaz picking particles (1,300,000 particles) were extracted and subjected to three rounds of heterogeneous refinement. Further heterogeneity classifications were performed by 3D-variability analysis (3DVA)43, focusing on the NPY–Y1R–Gi1 complex without micelles, Gαi AHD, and scFv16 by masking. Clusters with well-resolved density were obtained and used for final map reconstruction by non-uniform refinement44. Maps for this processing have a global nominal resolution of 3.2 Å, based on gold-standard Fourier shell correlation using the 0.143 criteria. To improve the map quality of NPY–Y1R–Gi1 complex, we performed the local refinement focusing on the NPY–Y1R and Gi1–scFv16 in cryoSPARC v3.2. These local refinements generated the maps at a global nominal resolution of 3.6 Å and 3.1 Å, respectively. Cryo-EM density for ECL2 (176–183, 194–203) and NPY (1–5, 20–36) was clearly observed in the refined map with a mask on NPY–Y1R. These two local refined maps were combined using “vop maximum” command in UCSF chimera to represent and analyze the NPY–Y1R–Gi1 complex45. The combined map is represented in Fig. 1a. The local resolution was determined using the cryoSPARC local resolution estimation. Local sharpening was performed by LocSpiral to trace the Y1R N-terminus (beyond L35N) and ECL2 region (Supplementary Fig. 6)46. No artifacts were observed when compared to the global sharpening map in the other regions.
Model building and refinement
The initial model was obtained by rigid-body-fitting of the structure of inactive Y1R (PDB ID 5ZBH)10 and Gαiβγ and scFv16 from the NTSR1 complex structure (PDB ID 6OS9)22. This initial model was then subjected to iterative rounds of manual rebuilding with COOT and refinement with PHENIX47,48. The geometry of the refined structure was evaluated using the MolProbity49. The final model consisting of NPY, Y1R, Gαi, Gβ, Gγ, and scFv16 was deposited in the PDB with PDB code 7VGX, and the electron density map was deposited in the EMDB with ID EMD-31979. The refinement statistics are presented in Supplementary Table 1. All molecular graphic figures were prepared using the UCSF Chimera, UCSF ChimeraX, and PyMol v2.4.045,50,51.
GTP turnover assay
GTP turnover assay was performed using GTPase-Glo assay kit (Promega)22. Purified Y1R was incubated with NPY for 40 min at room temperature. NPY-bound Y1R (4 μM) was mixed with 1 μM Gi1 heterotrimer (containing Gαi produced from E. coli or Sf9 cells) in an assay buffer consisting of 20 mM HEPES pH 8.0, 150 mM NaCl, 0.03% DDM, 0.003% CHS, 100 μM TCEP, 5 μM GDP, and 2.5 μM GTP. After incubation for 3 h, reconstituted GTPase-Glo reagent was added to the sample and incubated for 30 min at room temperature. Then, detection reagent was mixed and incubated for 10 min at room temperature. Luminescence was measured using a FlexStation 3 multi-mode microplate reader (Molecular Devices) and data were analyzed by GraphPad Prism 9.2.0.
ELISA-based surface expression assay
HEK293T cells were transfected with each expression plasmid of Y1R mutants and wild-type. After 48 h of incubation, 4% paraformaldehyde (PFA, T&I) was treated for fixation and washed with 1× PBS. After 30 min of incubation with blocking solution (2.5% bovine serum albumin, Bovogen), rabbit anti-FLAG antibody (Cell signaling Technology, 1:1000 dilution) was then treated for staining and anti-rabbit HRP antibody (Enzo Life Sciences, 1:1000 dilution) was used for detection. After incubation with HRP antibody, TMB solution (Thermofisher scientific) was added to each well, incubated until blue color was observed. Further reaction was blocked by adding 2 M HCl. The absorbance was detected at 450 nm with FlexStation 3 multi-mode microplate reader. Normalization was carried out by removing TMB substrate solution from the wells and adding Janus Green solution (0.2% w/v, TCL). Further elimination of excess stain was done by washing with milli Q water and adding 0.5 M HCl. Absorbance was read at 595 nm. Normalized expression level of the receptor at the cell surface was calculated by the ratio of the absorbance at 450 and 595 nm (A450/A595). The graphs were plotted using GraphPad Prism 9.2.0.
BRET assay
HEK293T cells were co-transfected with Gαi-Rluc, Gβ, Gγ, and Y1R-eYFP constructs at a 1:1:1:5 ratio. Forty-eight hours of post-transfection, cells were detached with PBS supplemented with 20 mM EDTA and evenly spread into white 96-well microplates (SPL). For ligand-induced conditions, various concentrations of NPY were incubated with each Y1R mutant-transfected group, and Coelenterazine h was added to each well to a final concentration of 5 μM. All BRET data were collected using a Mithras LB940 instrument (Berthold), and graphs were plotted using GraphPad Prism 9.2.0.
Ca2+ signaling assay
For the ligand-induced Ca2+ assay, HEK293T cells were seeded on 96-well black wall/clear bottom microplate (SPL) in Dulbecco’s modified Eagle’s medium (Biowest) containing 10% fetal bovine serum (Biowest) and antibiotic-antimycotic (Gibco) 24 h before plasmid transfection. After transfection with the plasmid containing Y1R constructs, GαΔ6qi4myr52 Gβ, and Gγ in a 3:1:1:1 ratio, followed by 48 h incubation, cells were stained with Cal-520 (AAT Bioquest, Inc.) in assay buffer (HBSS, 0.1% BSA, 20 mM HEPES pH 7.4). After 2 h, Cal-520-stained cells were washed three times with assay buffer. Intracellular Ca2+ influx was measured at Ex/Em = 490/525 nm using the FlexStation 3 multi-mode microplate reader (Molecular Devices). After 30 s of baseline, the ligand was injected to achieve the final concentration. Log (concentration)-response curves, used to estimate EC50, were calculated using GraphPad Prism 9.2.0, by fitting an agonist response curve with a variant slope to the normalized response data.
Calculation of the ligand-binding pocket volume
First, to define the interior of the TM bundle, the receptor structure was aligned along the z-axis by superimposing a pre-aligned GPCR structure from the OPM database53. Thereafter, a 3D grid was constructed with equispaced points covering the receptor structure. The centers of the TM helices for each discrete z-axis value and the lines connecting the neighboring centers were defined as the lateral boundaries. For helices without a defined center point for a given z-axis value, the (x,y) coordinate of the nearest point was used instead. Next, the upper and lower boundaries of the ligand-binding pocket were defined. The z-axis coordinate of the Cα atom of W6.48, a toggle switch residue, was defined as the lower boundary. TM residues closest to the extracellular region were used to define the upper boundary of the pocket volume. Finally, after removing the grid points causing clashes with protein atoms, the cavity volume was calculated from the number of grid points inside the defined boundaries. The python code for calculating the solvent accessible volume of the ligand-binding pocket is available at https://github.com/seoklab/GPCR_binding_cavity_volume_calculation.
Y4R-PP homology modeling
The homology model of PP-bound Y4R was prepared by template-based modeling protocol, GalaxyTBM, using the current NPY-bound Y1R structure as a template54. The initial model was constructed by threading the target sequence on the template structure, followed by energy optimization and additional structure sampling for unreliable local regions. Physics-based optimization method GalaxyRefineComplex refined template-based models55. A scoring function optimized for GPCR structure prediction in Galaxy7TM was used56.
MD simulation
Missing residues were added to the structure model of the NPY–Y1R–Gi1 complex to build an initial model for MD simulation. The AHD domain of Gi1 was added by aligning the previously reported structure57, and the missing residues of Y1R in the N-terminus (25–34), ECL2 (184–193), ICL2 (247–252), and C-terminus (330–339) were built based on the map with a mask on NPY–Y1R at a lower threshold. The rest of the N-terminus (2–24) was extended randomly in a position that did not collide with the existing structure.
This study used the CHARMM36(m) force field for proteins and lipids58–60. The TIP3P water model was utilized along with 0.15 M NaCl solution61. Three independent MD simulations were performed for each system to obtain better sampling and check the convergence. Periodic boundary conditions (PBCs) were employed in all simulations. The van der Waals interactions were smoothly switched off over 10–12 Å by a force-based switching function and the long-range electrostatic interactions were calculated using the particle-mesh Ewald method with a mesh size of ~1 Å62. All simulations were performed using the inputs generated by CHARMM-GUI and GROMACS 2018.6 for both equilibration and production with the LINCS algorithm63–67. The temperature was maintained using a Nosé-Hoover temperature coupling method with a τt of 1 ps68. For pressure coupling (1 bar), the semi-isotropic Parrinello–Rahman method with a τp of 5 ps and compressibility of 4.5 × 10−5 bar−1 was used69. The constant particle number, volume, and temperature (NVT) dynamics were first applied with a 1-fs time step for 250 ps during the equilibration run. Subsequently, the constant particle number, pressure, and temperature (NPT) ensemble was applied with a 1 fs time step (for 2 ns) and with a 2 fs time step (for 18 ns). During the equilibration, positional and dihedral restraint potentials were applied, and their force constants were gradually reduced. The production run was performed with a 4 fs time step using the hydrogen mass repartitioning technique without any restraint potential70. Each system ran about 25 ns/day with 512 CPU cores on NURION in the Korea Institute of Science and Technology Information.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the Creative-Pioneering Researchers Program through Seoul National University (H.-J.C.), the National Research Foundation of Korea funded by the Korean government (NRF-2020R1A2C2003783 and NRF-2019M3E5D6063903 to H.-J.C.), and National Science Foundation, USA (MCB-2111728 to W.I.). This work was also supported by Korea Basic Science Institute grant (2020R1A6C101A183 to H.-J.C.) and the Korea Basic Science Institute under the R&D program (Project No. C140440 to H.-J.C. and H.J.) supervised by the Ministry of Science and ICT. We thank Dr. Brian Kobilka (Stanford University) for providing the scFv16 construct, Chul Won Choi and Jungnam Bae (Seoul National University) for supporting grid preparation, Junsun Park and Dr. Soung-Hun Roh (Seoul National University) for providing materials and methods for grid screening. We also thank Dr. Bum Han Ryu (Institute for Basic Science (IBS), Korea) and Dr. Jin Seok Choi (KAIST Analysis Center for Research Advancement (KARA), Korea) for supporting grid screening at IBS and KARA cryo-EM facilities, respectively, and Global Science experimental Data hub Center (GSDC) at Korea Institute of Science and Technology Information (KISTI) and the data analysis hub, Olaf at the IBS Research Solution Center for computing resources.
Source data
Author contributions
C.P. and J.K. contributed equally to this work. C.P. purified the NPY–Y1R–Gi1–scFv16 complex. J.K. collected and processed cryo-EM data with help from H.J. and I.B. C.P. and J.K. determined and refined the complex structure. S.B.K. performed cell-based assays including surface ELISA, BRET assays, and Ca2+ assays with help from J.K., S.A.K., and T.-Y.Y. Y.K.C. and W.I. performed MD simulations and H.W. and C.S. calculated the volume of ligand-binding pockets and generated a homology model of PP-bound Y4R. H.K. purified Gi1 heterotrimer and scFv16. H.-J.C. conceived and directed the study. C.P., J.K., I.B., and H.-J.C. wrote the manuscript with contributions from all authors.
Peer review
Peer review information
Nature Communications thanks Cheng Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Additional data supporting the findings of this work are available as the Supplementary Information, Supplementary Data and Source Data files. The atomic model has been deposited in the Protein Data Bank under accession code 7VGX and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession code EMD-31979. The structure of inactive Y1R (PDB ID 5ZBH), Gαi1βγ and scFv16 from the NTSR1 complex structure (PDB ID 6OS9) were used as an initial template to build the NPY–Y1R–Gi1 protein model. Structural models used in data analysis were accessed from the Protein Data Bank under the accession codes 5ZBQ (UR-MK299 and Y1R), 6DDE (μOR, Gi), 7L1U (OxB and OX2R), 5GLH (ET1 and ETBR), 7L0Q (NTS and NTSR1), and 7MBX (CCK-8 and CCK1R). Source data are provided with this paper.
Code availability
The python code for calculating the solvent accessible volume of the ligand-binding pocket is available at https://github.com/seoklab/GPCR_binding_cavity_volume_calculation and as a Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chaehee Park, Jinuk Kim.
Change history
2/25/2022
A Correction to this paper has been published: 10.1038/s41467-022-28837-0
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28510-6.
References
- 1.Michel MC, et al. XVI. International Union of Pharmacology recommendations for the nomenclature of neuropeptide Y, peptide YY, and pancreatic polypeptide receptors. Pharm. Rev. 1998;50:143–150. [PubMed] [Google Scholar]
- 2.Herzog H, et al. Cloned human neuropeptide Y receptor couples to two different second messenger systems. Proc. Natl Acad. Sci. USA. 1992;89:5794–5798. doi: 10.1073/pnas.89.13.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno D, Yada T. Arcuate NPY neurons sense and integrate peripheral metabolic signals to control feeding. Neuropeptides. 2012;46:315–319. doi: 10.1016/j.npep.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Tasan RO, et al. The central and basolateral amygdala are critical sites of neuropeptide Y/Y2 receptor-mediated regulation of anxiety and depression. J. Neurosci. 2010;30:6282–6290. doi: 10.1523/JNEUROSCI.0430-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotzsche CR, Woldbye DP. The role of NPY in learning and memory. Neuropeptides. 2016;55:79–89. doi: 10.1016/j.npep.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Vahatalo LH, Ruohonen ST, Ailanen L, Savontaus E. Neuropeptide Y in noradrenergic neurons induces obesity in transgenic mouse models. Neuropeptides. 2016;55:31–37. doi: 10.1016/j.npep.2015.11.088. [DOI] [PubMed] [Google Scholar]
- 7.Farzi A, Hassan AM, Zenz G, Holzer P. Diabesity and mood disorders: Multiple links through the microbiota-gut-brain axis. Mol. Asp. Med. 2019;66:80–93. doi: 10.1016/j.mam.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Bijker MS, Herzog H. The neuropeptide Y system: pathophysiological and therapeutic implications in obesity and cancer. Pharm. Ther. 2011;131:91–113. doi: 10.1016/j.pharmthera.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Monks SA, Karagianis G, Howlett GJ, Norton RS. Solution structure of human neuropeptide Y. J. Biomol. NMR. 1996;8:379–390. doi: 10.1007/BF00228141. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, et al. Structural basis of ligand binding modes at the neuropeptide Y Y(1) receptor. Nature. 2018;556:520–524. doi: 10.1038/s41586-018-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck-Sickinger AG, Jung G. Structure-activity relationships of neuropeptide Y analogues with respect to Y1 and Y2 receptors. Biopolymers. 1995;37:123–142. doi: 10.1002/bip.360370207. [DOI] [PubMed] [Google Scholar]
- 12.Mentlein R, Dahms P, Grandt D, Kruger R. Proteolytic processing of neuropeptide Y and peptide YY by dipeptidyl peptidase IV. Regul. Pept. 1993;49:133–144. doi: 10.1016/0167-0115(93)90435-b. [DOI] [PubMed] [Google Scholar]
- 13.Grandt D, et al. Novel generation of hormone receptor specificity by amino terminal processing of peptide YY. Biochem Biophys. Res. Commun. 1992;186:1299–1306. doi: 10.1016/s0006-291x(05)81547-5. [DOI] [PubMed] [Google Scholar]
- 14.Wieland HA, Willim K, Doods HN. Receptor binding profiles of NPY analogues and fragments in different tissues and cell lines. Peptides. 1995;16:1389–1394. doi: 10.1016/0196-9781(95)02028-4. [DOI] [PubMed] [Google Scholar]
- 15.Zheng H, et al. High-fat intake induced by mu-opioid activation of the nucleus accumbens is inhibited by Y1R-blockade and MC3/4R- stimulation. Brain Res. 2010;1350:131–138. doi: 10.1016/j.brainres.2010.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yulyaningsih E, Zhang L, Herzog H, Sainsbury A. NPY receptors as potential targets for anti-obesity drug development. Br. J. Pharm. 2011;163:1170–1202. doi: 10.1111/j.1476-5381.2011.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhisikar SM, Kokare DM, Nakhate KT, Chopde CT, Subhedar NK. Tolerance to ethanol sedation and withdrawal hyper-excitability is mediated via neuropeptide Y Y1 and Y5 receptors. Life Sci. 2009;85:765–772. doi: 10.1016/j.lfs.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, et al. NPY1R is a novel peripheral blood marker predictive of metastasis and prognosis in breast cancer patients. Oncol. Lett. 2015;9:891–896. doi: 10.3892/ol.2014.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda S, et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 2018;9:3712. doi: 10.1038/s41467-018-06002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong M, et al. Structure and dynamics of the active Gs-coupled human secretin receptor. Nat. Commun. 2020;11:4137. doi: 10.1038/s41467-020-17791-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 22.Kato HE, et al. Conformational transitions of a neurotensin receptor 1-G(i1) complex. Nature. 2019;572:80–85. doi: 10.1038/s41586-019-1337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koehl A, et al. Structure of the µ-opioid receptor-G(i) protein complex. Nature. 2018;558:547–552. doi: 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong C, et al. Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation. Nat. Commun. 2021;12:815. doi: 10.1038/s41467-021-21087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, et al. Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B. Eur. J. Biochem. 1999;266:831–839. doi: 10.1046/j.1432-1327.1999.00911.x. [DOI] [PubMed] [Google Scholar]
- 26.Beck-Sickinger AG, et al. Complete L-alanine scan of neuropeptide Y reveals ligands binding to Y1 and Y2 receptors with distinguished conformations. Eur. J. Biochem. 1994;225:947–958. doi: 10.1111/j.1432-1033.1994.0947b.x. [DOI] [PubMed] [Google Scholar]
- 27.Sautel M, et al. Neuropeptide Y and the nonpeptide antagonist BIBP 3226 share an overlapping binding site at the human Y1 receptor. Mol. Pharm. 1996;50:285–292. [PubMed] [Google Scholar]
- 28.Sjodin P, et al. Re-evaluation of receptor-ligand interactions of the human neuropeptide Y receptor Y1: a site-directed mutagenesis study. Biochem J. 2006;393:161–169. doi: 10.1042/BJ20050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin J, et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol. 2016;23:293–299. doi: 10.1038/nsmb.3183. [DOI] [PubMed] [Google Scholar]
- 30.Rappas M, et al. Comparison of Orexin 1 and Orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J. Med Chem. 2020;63:1528–1543. doi: 10.1021/acs.jmedchem.9b01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takasuka T, Sakurai T, Goto K, Furuichi Y, Watanabe T. Human endothelin receptor ETB. Amino acid sequence requirements for super stable complex formation with its ligand. J. Biol. Chem. 1994;269:7509–7513. [PubMed] [Google Scholar]
- 32.Shihoya W, et al. Crystal structures of human ET(B) receptor provide mechanistic insight into receptor activation and partial activation. Nat. Commun. 2018;9:4711. doi: 10.1038/s41467-018-07094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, et al. Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:258–267. doi: 10.1038/s41594-020-00554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobbs JI, et al. Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity. PLoS Biol. 2021;19:e3001295. doi: 10.1371/journal.pbio.3001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwyzer R. ACTH: a short introductory review. Ann. N. Y Acad. Sci. 1977;297:3–26. doi: 10.1111/j.1749-6632.1977.tb41843.x. [DOI] [PubMed] [Google Scholar]
- 36.Portoghese PS. Bivalent ligands and the message-address concept in the design of selective opioid receptor antagonists. Trends Pharm. Sci. 1989;10:230–235. doi: 10.1016/0165-6147(89)90267-8. [DOI] [PubMed] [Google Scholar]
- 37.Stadlbauer U, Woods SC, Langhans W, Meyer U. PYY3-36: Beyond food intake. Front. Neuroendocrinol. 2015;38:1–11. doi: 10.1016/j.yfrne.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Cabrele C, Wieland HA, Langer M, Stidsen CE, Beck-Sickinger AG. Y-receptor affinity modulation by the design of pancreatic polypeptide/neuropeptide Y chimera led to Y(5)-receptor ligands with picomolar affinity. Peptides. 2001;22:365–378. doi: 10.1016/s0196-9781(01)00339-4. [DOI] [PubMed] [Google Scholar]
- 39.Lundell I, et al. Cloning of a human receptor of the NPY receptor family with high affinity for pancreatic polypeptide and peptide YY. J. Biol. Chem. 1995;270:29123–29128. doi: 10.1074/jbc.270.49.29123. [DOI] [PubMed] [Google Scholar]
- 40.Lin S, et al. Critical role of arcuate Y4 receptors and the melanocortin system in pancreatic polypeptide-induced reduction in food intake in mice. PLoS ONE. 2009;4:e8488. doi: 10.1371/journal.pone.0008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 42.Bepler T, et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punjani A, Fleet DJ. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021;213:107702. doi: 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 44.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Kaur S, et al. Local computational methods to improve the interpretability and analysis of cryo-EM maps. Nat. Commun. 2021;12:1240. doi: 10.1038/s41467-021-21509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams CJ, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrödinger. The PyMOL Molecular Graphics System, Version 2.4.0, Schrödinger LLC. (2020).
- 52.Kostenis E. Potentiation of GPCR-signaling via membrane targeting of G protein alpha subunits. J. Recept Signal Transduct. Res. 2002;22:267–281. doi: 10.1081/rrs-120014601. [DOI] [PubMed] [Google Scholar]
- 53.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 54.Ko J, Park H, Seok C. GalaxyTBM: template-based modeling by building a reliable core and refining unreliable local regions. BMC Bioinform. 2012;13:198. doi: 10.1186/1471-2105-13-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heo L, Lee H, Seok C. GalaxyRefineComplex: refinement of protein-protein complex model structures driven by interface repacking. Sci. Rep. 2016;6:32153. doi: 10.1038/srep32153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee GR, Seok C. Galaxy7TM: flexible GPCR-ligand docking by structure refinement. Nucleic Acids Res. 2016;44:W502–W506. doi: 10.1093/nar/gkw360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang Y, et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klauda JB, Monje V, Kim T, Im W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 60.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 62.Essmann U, et al. A smooth particle Mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 63.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Der Spoel D, et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 65.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 66.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 69.Parrinello M, Rahman A. Polymorphic transitions in single-crystals—a new molecular-dynamics Method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 70.Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Additional data supporting the findings of this work are available as the Supplementary Information, Supplementary Data and Source Data files. The atomic model has been deposited in the Protein Data Bank under accession code 7VGX and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession code EMD-31979. The structure of inactive Y1R (PDB ID 5ZBH), Gαi1βγ and scFv16 from the NTSR1 complex structure (PDB ID 6OS9) were used as an initial template to build the NPY–Y1R–Gi1 protein model. Structural models used in data analysis were accessed from the Protein Data Bank under the accession codes 5ZBQ (UR-MK299 and Y1R), 6DDE (μOR, Gi), 7L1U (OxB and OX2R), 5GLH (ET1 and ETBR), 7L0Q (NTS and NTSR1), and 7MBX (CCK-8 and CCK1R). Source data are provided with this paper.
The python code for calculating the solvent accessible volume of the ligand-binding pocket is available at https://github.com/seoklab/GPCR_binding_cavity_volume_calculation and as a Source Data file.