Abstract
Transcriptional regulation underlies many of the growth and developmental processes that shape plants, as well as their adaptation to their environment. Key to transcriptional control are transcription factors, DNA-binding proteins that serve two essential functions: to find the appropriate DNA contact sites in their target genes and to recruit other proteins to execute transcriptional transactions. In recent years, protein structural, genomic, bioinformatic and proteomic analysis has led to new insights into how these central functions are regulated. Here, we review new findings relating to plant transcription factor function, as well as to their role in shaping transcription in the context of chromatin.
INTRODUCTION
Transcription factors are central to all cellular functions; indeed, their activities dictate the contents and identity of each cell. Essentially, transcription factors have two functions - to find the appropriate DNA elements to bind and to recruit additional proteins to that site. Some transcription factors have the additional role of acting as pioneers by accessing closed chromatin and opening it so other transcription factors can bind.
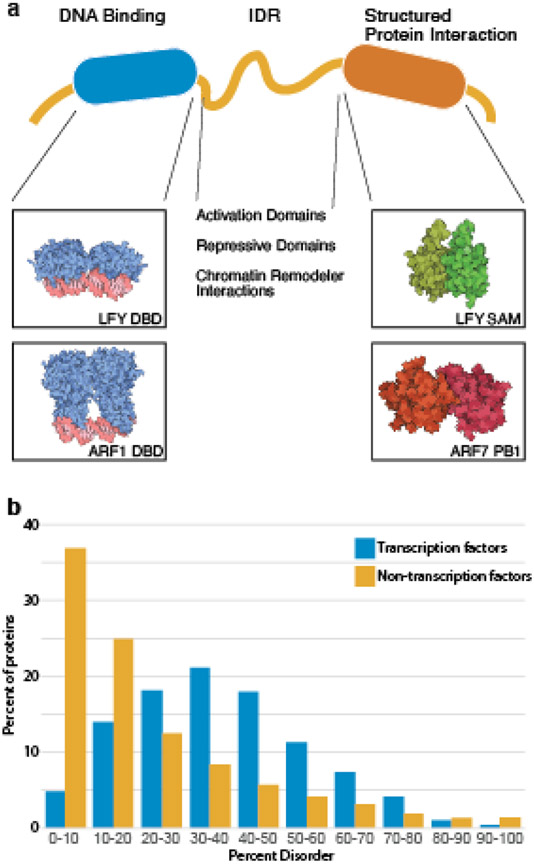
Transcription factors consist of several modular domains that reflect their core functions to find their DNA target and to recruit the appropriate machinery to affect transcription events (Figure 1). Additional domains may act as regulatory modules, for example by interaction with coregulators or proteins that drive post-translational modification of the transcription factor. DNA binding domains are structured domains that facilitate recognition of DNA elements and frequently act as dimers; some have a distinct dimerization motif within the DNA binding domain, such as has been found for AUXIN RESPONSE FACTOR (ARF) proteins ([1-3]) and LFY [4].
Figure 1. Architecture of a transcription factor.
a. Transcription factors consist of multiple modular domains. Structured domains are used for DNA binding and are frequently used for protein interactions. Activation domains, repressive domains, and regions that recruit chromatin remodelers typically reside in intrinsically disordered regions. Protein cartoons created with Illustrate [56] of the LFY DBD (PDB 2VY1) [4], ARF1 DBD (PDB 4LDX) [1], LFY SAM (PDB 4UDE) [57], and ARF7 PB1 (PDB 4NJ7) [58] domains.
b. Analysis of percentage of protein comprised of intrinsically disordered regions from Arabidopsis transcription factors and non-transcription factors on a proteome-wide scale using Metapredict [59] reveals enrichment of intrinsically disordered regions in transcription factors.
Recruitment of transcriptional machinery, such as the Mediator complex, Pol II, or repressor proteins and chromatin regulators, typically occurs in regions of low complexity or intrinsic disorder. Indeed, Arabidopsis transcription factors are enriched for intrinsically disordered regions when compared to the proteome (Figure 1b), suggesting a critical role for protein disorder in the function of transcription factors.
Here, we focus on the two major functions of transcription factors: 1) to find DNA and 2) to find friends, acting as the party planners of the cell by finding the appropriate venue and making the guest list that will ensure specific transcriptional outcomes at certain target loci in response to extrinsic and intrinsic cues.
Getting started in chromatin
Transcription factors direct changes in gene expression that underpin cell fate transitions and response to the environment; this type of ‘reprogramming’ occurs throughout the life of plants. In eukaryotes, transcriptional programs not needed or detrimental at a given developmental stage, tissue or environmental condition are kept in a condensed chromatin state [5,6]. This chromatin state is usually marked by high nucleosome density, presence of linker histones and/or histone modifications (methylation of H3K27 or H3K9) that in turn recruit non histone proteins that further compact chromatin. Since most transcription factors cannot access their cognate binding sites (cis motifs) in the context of a nucleosome [7], this raises the question how new transcriptional programs can be initiated in response to developmental or environmental cues. Studies in animals, initially on the winged helix transcription factor FoxA, uncovered a class of transcription factors termed pioneer transcription factors that can bind to their cis motifs in the context of a nucleosome both in vivo and in vitro [8,9]. This is enabled by the shallow DNA binding domains of pioneer transcription factors that facilitate contact with DNA bases and the sugar-phosphate backbone when the DNA is wrapped around the histone octamer [10]. In addition, in the context of a nucleosome, pioneer transcription factors frequently bind cis motifs that slightly deviate from the consensus, allowing for fewer protein- DNA contacts [11]. The ability to bind nucleosomal DNA with high affinity and in a sequence specific manner in vivo and in vitro is the principal defining property of pioneer transcription factors and uniquely positions them to set in motion the gene expression changes required for cell fate reprogramming.
The plant specific helix-turn-helix transcription factor LEAFY (LFY) was recently shown to be a pioneer transcription factor in Arabidopsis [12,13].(Figure 2). LFY promotes the switch from branch to floral fate in axillary meristems as well as flower patterning. The LFY helix-turn-helix DNA binding domain makes shallow DNA contacts and in vitro LFY binds its cognate binding motif with high affinity in the context of nucleosomes assembled with a native LFY bound region, or when the LFY binding site is combined with a nucleosome positioning sequence [4,12,13] (Figure 2e). Like most pioneer transcription factors, LFY exhibits higher affinity for naked DNA [13]. In root explants, that are can be triggered to synchronously reprogramming to floral fate, the majority of the LFY bound sites are nucleosome occupied immediately prior to LFY binding [13]. Moreover, LFY co-occupies genomic DNA with histones on the basis of sequential ChIP [13]. While LFY can associate with H3K27me3 marked DNA, other types of chromatin condensation block LFY binding, indicating that certain chromatin states can prevent access of pioneer transcription factors to their target sites [12,13]. For some animal pioneer transcription factors H3K9me3 restricts chromatin access [14].
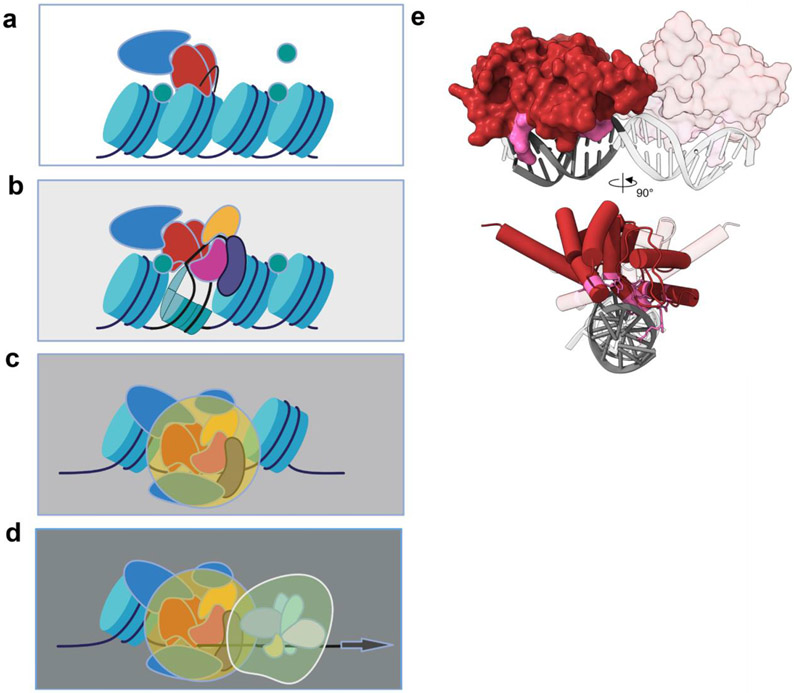
Figure 2. LFY pioneer transcription factor licenses cell fate reprogramming and subsequent activation of transcription at the APETALA1 locus.
a. The LFY dimer (red shapes) binds it cis motif near the dyad (midpoint) of the nucleosome and triggers local chromatin opening via eviction of histone H1 (green circles) and recruitment of SWI/SNF chromatin remodelers (blue shape).
b. Subsequently other transcription factors can access the locus. Their accumulation is either directly activated by LFY (LMI2, yellow) or in response to altered hormone levels and plant age (DELLA proteins plus SQUAMOSA PROTEIN BINDING PROTEINS; purple shape) or photoperiod FLOWERING LOCUS T (together with FD, pink) [23,24,60-63].
c. The assembled transcription factors likely engage with additional chromatin regulators (blue shape) to cooperatively open the AP1 locus.
d. Finally, the general transcriptional machinery (white large shape) is recruited.
e. Shallow DNA contacts with one face of the DNA by the LFY dimer allow co-binding with the histone octamer. Previously published structure of LFY displayed as a dimer (one of the monomers is semi-transparent) binding the pseudo-palindromic recognition site within AP1 (pdb: 2vy1 [4]). DNA is in grey and semi-transparent except for the half-site bound by the opaque monomer on the left. LFY residues which interact with DNA are in pink and displayed in stick style over the cartoon in helix-tube mode in the rotated view below. Visualization using chimera [64].
Interestingly, while LFY binding to closed chromatin is very rapid, target locus chromatin opening and full transcriptional activation occur much later [13]. This temporal separation suggests that the LFY pioneer transcription factor licenses the transcriptional activation for cell fate reprogramming, but does not execute it by itself (Figure 2 a-d). A delay between locus occupancy of pioneer transcription factors and locus opening as defined by ATACseq or similar techniques was also observed for several pioneer transcription factors in animals [15,16]. Decoupling of chromatin binding and transcriptional activation is desirable, as it allows additional tuning of cell fate reprogramming. Indeed, pioneer transcription factors can rapidly open chromatin locally, near their binding site, for example by ejecting linker histone H1 (FoxA and LFY), by altering nucleosome topology or interaction and by recruiting chromatin remodelers [8,13,14,17]. Through this local chromatin opening (enhancer activation), pioneer transcription factors facilitate access of other transcription factors to the DNA. This is a second key property of pioneer transcription factors [8]. The non-pioneer factors cooperatively direct full target locus chromatin opening and robust activation of gene expression with the pioneer factor, in some cases forming transcription hubs [13,19]. Regulation of accumulation or activity of the co-operatively acting non-pioneer transcription factors by specific extrinsic or intrinsic cues can fine-tune cell fate reprogramming by pioneer factors.
Because plants adjust their form and function frequently in response to changing environments, it is likely that pioneer transcription factors play a prominent role in this kingdom. MADS box proteins are good candidates for pioneer transcription factors as they bind chromatin and subsequently enhance accessibility [19]. The Arabidopsis NF-Y component LEC1 was proposed to act as a pioneer transcription factor on the basis of sequence and structural similarity with the mammalian complex; however whether mammalian NF-Y acts as a pioneer transcription factor has not yet been established [21,22].
Transcriptional and cell fate reprogramming in the context of chromatin is also enabled by ‘signal dependent’ transcription factors that are bound to target loci in many different conditions (tissue types developmental stages, intrinsic or extrinsic cues). Their chromatin occupancy may or may not have been the result of initial pioneer transcription factor binding. Signal dependent transcription factors serve both as target locus markers and as scaffolds. In the latter capacity they recruit transcriptional co-activator or co-repressor complexes. One example is ARF5/MONOPTEROS (MP). MP recruits histone deacetylases, which strengthen DNA histone interactions to silence target loci in the absence of the auxin stimulus [22]. In the presence of a local auxin maximum, MP can instead interact with chromatin remodelers that eject or reposition nucleosomes to activate certain target loci [22]. Likewise, the bZIP transcription factor FD binds to the same set of target loci in vegetative and flower producing Arabidopsis but recruits different members of the phosphoethanolamine binding protein (PEBP) family, i.e. TERMINAL FLOWER 1 (TFL1) or FLOWERING LOCUST (FT), depending on the photoperiod or other environmental cues [23-27]. TFL1 acts as a transcriptional co-repressor and FT as a co-activator [23,28].
Finding the right location
Provided that the chromatin context is conducive to transcription factor binding, a next challenge for DNA-binding protein is to home in on its set of unique binding sites. That is: finding the biologically meaningful sites in an ocean of DNA. There is a finite number of DNA-binding domains in transcription factors across the tree of life [29], and in plants, there is likewise a limited number of distinct DNA-binding domains [30]. Systematic surveys of the binding specificity of transcription factors across eukaryotes [31] and in Arabidopsis [32] revealed that DNA-binding domains tend to specifically bind short DNA elements of about 6-10 bases in which specificity is not absolute for some positions. In other words, the effective specificity is in the order of 4-6 effectively unique bases. The crystal structures of a range of DNA-binding proteins in complex with their DNA target have shown the biochemical basis for specificities, and rationalize why only relatively short stretches of non-unique sequence are sufficient for binding [33]. A simple calculation tells that such sites will occur in every 250-4000 bases. In contrast, both DNA-affinity purification (DAP) of Chromatin immunoprecipitation (ChIP), followed by sequencing, showed that transcription factors bind only a subset of the consensus elements in the context of genomes [34]. Therefore, intrinsic DNA-binding specificity of DNA-binding proteins alone is clearly insufficient to explain their DNA binding in the context of a genome, or within a living cell. Other factors must constrain binding. One such constraining factor may be the chemical nature of DNA itself. As a double helix, DNA has a major groove and a minor groove. Accessibility to the (specificity-defining) bases differs between the major and minor groove. As a consequence, the sites for facile base access are spaced by the periodicity of DNA turns.
An interesting question is why transcription factors have not evolved to carry more extensive, more elaborate DNA-binding surfaces, that would extend their DNA target site, and thus increase specificity of protein-DNA interaction. A consideration in this context is that protein-DNA interaction must be viewed as a trade-off between specificity and speed [35]. Transcription factors must scan DNA to bind their target site. Even though this is restricted to the DNA accessible in the context of chromatin for most transcription factors, there must be low affinity for binding to DNA in an unspecific manner, that allows scanning with some speed [36]. Specific interactions at the target locus would then lock the transcription factor in place. A longer, more extensive DNA-binding interface likely also increases the unspecific interactions with DNA, and thus limits the speed by which DNA can be scanned.
Rather than extending the DNA interaction interface within the transcription factor, another strategy that can increase the effective length of a binding site is the binding of DNA by transcription factor dimers. Indeed, many, if not most, transcription factors bind DNA as dimers. These can be homodimers, such as (in plants) is the case for the ARFs [1-3] and LFY [4,37] mentioned earlier. In both cases, proteins dimerize to bind a specific, more extended DNA motif [1,4], and can be part of higher-order interactions, such as MADS protein tetramers [38]. Beyond homodimerization, there are many well-known cases in which transcription factors heterodimerize within the same family (e.g. bHLH factors, bZIP; [39]). Interactions between members of different transcription factor families have been abundantly reported (e.g. [40]), but their contribution to specific DNA binding are less well-understood.In this latter case, the optimal distance between the two binding sites is governed not only by the structure of one, but of two transcription factors, and it will be interesting to see future studies that structurally explore spacing in heterotypic transcription factor dimers, as well as the effects of association of dimerizing transcription factors bound at large distances (such as distal enhancers).
Teaming up with your best friend
Two kinds of dimeric DNA interaction modes are observed: in one (e.g. bHLH factors, LFY and FD), dimerization is a structural requirement for DNA binding a single DNA element [39][4,24]. In the other, two proteins each bind a separate DNA element, and as a complex, they can bind a combination of two DNA elements. In the latter case, when transcription factors dimerize to bind DNA, this has a number of interesting consequences for the mode of DNA binding. First, the effective length of the DNA target site increases by a factor of two. Second, the protein structures will now dictate the orientation and spacing between the individual binding sites that are compatible with protein complex binding (see discussion below). Most importantly, when two proteins bind two DNA elements, there is space for cooperativity in interactions. The affinity of the first transcription factor to bind its target site will be defined by the intrinsic affinity that follows from the binding energies at the protein-DNA interface. While the same energies hold for the second member of the protein dimer, the stability of the entire protein-DNA complex is enhanced by the interaction energies between the two proteins. Thus, affinity of a dimer can be much higher than that of each of the two monomers. This has been illustrated well by an analysis of the Arabidopsis ARF1 and ARF5 proteins [1]. Since the protein-DNA interface could be resolved from a crystal structure, mutants in DNA or protein could be engineered that would allow either a monomer or dimer to bind. Here, it is clear that a dimer binds an order of magnitude better than monomers do. Thus, dimerization offers an instrument to toggle between low (monomer) and high (dimer) affinity DNA binding. In addition, this allows combinatorial inputs into the regulation of gene expression, if the two dimerization partners themselves respond or reflect different inputs. An example of this is the bHLH heterodimer between TMO5 and LHW, where one partner (TMO5) reflects auxin input, and the other (LHW) defines a developmental domain in the root [41].
Within a protein dimer, the structural arrangement of the two monomers relative to one another dictates the topology of DNA element organization and spacing. Given that DNA elements are recognized by interactions of amino acids with both phosphates in the DNA backbone, and by specific interactions with bases, usually on both strands, proteins recognize DNA only in a directional, specific manner. Thus, a head to tail protein dimer will likely recognize a direct repeat, while head-to-head or tail-to-tail dimer will recognize inverted or everted repeats. In the case of ARFs, a symmetric dimer binds an inverted repeat [1-3]. Indeed, the biologically relevant element for auxin-dependent gene expression, an inverted repeat spaced by 7 or 8 bases, can be explained by this mode of protein-DNA binding [3]. However, another repeat of ARF binding sites – a direct repeat spaced by 5 bases- is also very potent in mediating auxin-responsive gene expression [42], but its structural basis is still elusive.
Lastly, structural flexibility of the protein complex, as well as rotational flexibility of the DNA, together define the spacing that is tolerated between two DNA-protein interfaces. For ARF proteins, the optimum is 7-8 bases in the symmetric dimer, but in vitro assays suggest that cooperative binding for Arabidopsis ARF5 extends to shorter and longer distances [1]. While biological relevance is not yet addressed, this could also be a basis for functional distinction among related transcription factors.
An interesting consequence of the protein dimers in protein-DNA interactions is in the constraints it poses for evolution and evolvability of transcriptional regulatory networks. For such interactions, evolution (and selection) can act on protein structures in multiple ways: (1) intrinsic specificity and affinity of monomers for their DNA sites, (2) protein-protein interaction surface position and interaction strength, (3) DNA elements that directly bind to the proteins, (4) length and flexibility of intervening nucleotides. It should be clear that these units are both tunable and therefore, evolvable. Yet at the same time, binding depends on a number of factors, which will also pose constraints on its evolvability. An interesting future question will be how the kinetics of transcription factor binding to complex sites evolves in complex plant genomes.
Party friends
In addition to finding its best friend in the form of another transcription factor with which to dimerize, transcription factors must also recruit a party of friends to drive chromatin remodeling and/or transcriptional events. These interactions are typically driven by fuzzy binding kinetics in low complexity domains [43]. Recruitment of these machines to the appropriate region of DNA is driven by the DNA binding activity of the transcription factor, either to remodel chromatin or to promote gene transcription.
As outlined above, the chromatin state can restrict access of transcription factors to the genomic DNA. On the other hand, the vast majority of the enzymatic activities that alter chromatin do not have inherent DNA binding specificity. As outlined above, the chromatin state can restrict access of transcription factors to the genomic DNA. On the other hand, the vast majority of the enzymatic activities that alter chromatin do not have inherent DNA binding specificity, although exceptions to this rule exist [44]. De novo targeting of chromatin modification and remodeling to the correct genomic location in plants relies strongly on transcription factors (histone acetyltransferases, histone de-acetylases and co-repressor complexes, SWI/SNF chromatin remodelers, histone methyltransferases (H3K27me3)) or on noncoding RNA (DNA methylation, H3K9me2) [5,22,45]. Thus, while chromatin state can restrict transcription factor access to the genomic DNA, most chromatin regulators rely on transcription factors for recruitment. Finally, the act of transcription also alters chromatin through histone loss and recruitment of histone modifying enzymes [46]. In this manner chromatin and transcription factors are mutually dependent on one another.
How transcription factors recruit a diverse set of corepressor and coactivator complexes is not well understood. The transcription factor domains that recruit these complexes typically reside in intrinsically disordered regions that have low conservation. With the exception of the repressor motif of LxLxL that recruits TOPLESS and related transcriptional repressors [47], protein regions that recruit chromatin remodeling and transcriptional machinery are difficult to identify due to their low sequence conservation. Additionally, many of these machineries have been shown to act in transcriptional hubs or condensates, often driven into these assemblies by multivalent interactions in their intrinsically disordered regions.
Protein condensation can act as a friend filter for transcription factors – either by allowing concentration of protein that promotes recruitment of additional proteins to a transcriptional hub or by sequestering transcription factors into inactive bodies [48]. Assemblies of transcription factors that are more stable than individual factor binding has been proposed to drive transcriptional events as transcriptional hubs [43]. Because protein condensation affects the activities of coactivators, corepressors, sequence-specific transcription factors, chromatin modifiers, and RNA Polymerase II by providing a mechanism to concentrate these factors [49], some have proposed protein concentration as the underlying physical basis for formation of transcriptional hubs. Most of these events are thought to promote activity, either by acting as a concentrator of activity or by acting as a mechanism to stabilize the concentrations of the relevant proteins [48].
The examples of transcription factor condensate formation in plants thus far are consistent with condensation acting as an attenuator of their transcriptional activity. Condensation of ARF19 [50] or ELF3 [51] result in sequestration from transcriptional targets. It remains to be seen whether condensation of other plant transcription factors similarly attenuates activity or whether this concentration acts to promote activity as has been seen in other systems.
Transcription factors occupy a pivotal role in orchestration of cell fate and function, and it is becoming increasingly understood how they find and bind their genomic targets. In addition, we are beginning to unravel how their activity is controlled in response to cues in the plant kingdom. Technical advances in imaging, proteomics, and chromatin binding will further this understanding and help address some of the open questions that remain.
OPEN QUESTIONS
Do transcriptional hubs exist in plants? In other systems, transcriptional hubs and/or condensates have been demonstrated for coactivators, corepressors, sequence-specific transcription factors, chromatin modifiers, and RNA Polymerase II [43]. These have not yet been demonstrated in plants; however, given the conservation amongst these core machineries, it seems likely that similar assemblies operate in plant transcriptional control. In Arabidopsis, a large number of genomic regions bound by many transcription factors were recently identified, adding binding peak summits to this excellent resource and filtering out artifact ChIP-seq signal will enable identification of genomic locations bound by many transcription factors [52,53].
Transcriptional pausing in plants? Most eukaryotes have a mechanism to assemble a transcription factor and Pol II on DNA while inhibiting progression of transcription, called promoter pausing. Plants lack the conserved components of this system. Recently, TPL recruitment to ARF proteins has been demonstrated to act in a similar manner by inhibiting full assembly of the Mediator complex [54], resulting in a partially assembled, but paused, complex that has been proposed [55] to act in pausing. Given the widespread recruitment of TPL in various pathways, this mechanism could broadly impact start of transcriptional elongation.
What are the protein domains necessary and sufficient for pioneer activity by transcription factors? How is their binding and activity restricted? Identification of more pioneer transcription factors in plants, followed by functional studies, should help answer these questions and allow harnessing pioneer activity to (re)program cell fate and function to enhance desirable traits.
What are the roles of spatially structured nuclear microenvironments? The nucleus contains many spatially distinct microenvironments, including the nucleolus, nuclear speckles, photobodies, cajal bodies, and dicing bodies. How these microenvironments are formed, what components comprise these bodies, and what roles these and other concentrations of nuclear components play are areas of active study which should reveal their functions in transcription.
Do we know which TFs are activators versus those that are repressors? Transcription factors recruit machinery to drive downstream events – either repression or activation of their gene targets. Because the regions of the protein that recruit these machineries lie in low complexity domains with little sequence conservation, better tools and approaches will be needed to determine whether each transcription factor activates or represses target genes.
Acknowledgements
We thank Ryan Emenecker and Nick Morffy for analyzing IDRs in the Arabidopsis proteome and transcription factors as well as Tomasz Bieluzewski for visualization of the LFY DNA binding domain. This work was supported by the National Institutes of Health R35 GM136338 to LCS, National Science Foundation IOS 1905062 (to DWa), MCB 1916431 (to DWa), and IOS 2112056 (to LCS) and the Netherlands Organization for Scientific Research VICI 865.14.001 and OCENW.KLEIN.027 (to DWe).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. : Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 2014, 156:577–589. [DOI] [PubMed] [Google Scholar]
- **2. Kato H, Mutte SK, Suzuki H, Crespo I, Das S, Radoeva T, Fontana M, Yoshitake Y, Hainiwa E, van den Berg W, et al. : Design principles of a minimal auxin response system. Nat Plants 2020, 6:473–482.32415296 The authors use the minimal Marchantia auxin response system to derive a simple model for auxin-dependent gene expression, in which auxin-sensitive A- and auxin-insensitive B-class ARFs compete for binding the same target sites in auxin-responsive promoters.
- **3. Freire-Rios A, Tanaka K, Crespo I, van der Wijk E, Sizentsova Y, Levitsky V, Lindhoud S, Fontana M, Hohlbein J, Boer DR, et al. : Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in. Proc Natl Acad Sci U S A 2020, 117:24557–24566.32929017 The authors use bioinformatic analysis, coupled to structural biology and Arabidopsis genetics, to demonstrate the requirements for high-affinity DNA binding by ARF transcription factors and rationalize the basis for enrichment of AuxRE repeats in auxin-responsive promoters.
- 4.Hamès C, Ptchelkine D, Grimm C, Thevenon E, Moyroud E, Gérard F, Martiel J-L, Benlloch R, Parcy F, Müller CW: Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J 2008, 27:2628–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieluszewski T, Xiao J, Yang Y, Wagner D: PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci 2021, 0. [DOI] [PubMed] [Google Scholar]
- 6.Klemm SL, Shipony Z, Greenleaf WJ: Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 2019, 20:207–220. [DOI] [PubMed] [Google Scholar]
- 7.Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. : The interaction landscape between transcription factors and the nucleosome. Nature 2018, 562:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaret KS: Pioneer Transcription Factors Initiating Gene Network Changes. Annual Review of Genetics 2020, 54:367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS: Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 2002, 9:279–289. [DOI] [PubMed] [Google Scholar]
- **10. Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, Harrison MM, Zhu H, Zaret KS: Structural Features of Transcription Factors Associating with Nucleosome Binding. Mol Cell 2019, 75:921–932.e6.31303471 Shows that ‘shallow’ DNA binding domain interaction with regulatory DNA is critical for pioneer transcription factors.
- 11.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS: Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161:555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12. Lai X, Blanc-Mathieu R, GrandVuillemin L, Huang Y, Stigliani A, Lucas J, Thévenon E, Loue-Manifel J, Turchi L, Daher H, et al. : The LEAFY floral regulator displays pioneer transcription factor properties. Mol Plant 2021, 14:829–837.33684542 Showed that LFY is pioneer transcription factor, whose binding is blocked in some chromatin contexts.
- **13. Jin R, Klasfeld S, Zhu Y, Fernandez Garcia M, Xiao J, Han S-K, Konkol A, Wagner D: LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nat Commun 2021, 12:626.33504790 Showed LFY is a pioneer transcription factor, that reprograms cell fate by co-occupying DNA with histones, locally opening chromatin for other transcription factors to cooperatively activation of gene expression.
- 14.Larson ED, Marsh AJ, Harrison MM: Pioneering the developmental frontier. Mol Cell 2021, 81:1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R: Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 2018, 32:96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, Drouin J: Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet 2018, 50:259–269. [DOI] [PubMed] [Google Scholar]
- 17.Dodonova SO, Zhu F, Dienemann C, Taipale J, Cramer P: Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020, 580:669–672. [DOI] [PubMed] [Google Scholar]
- **18. Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, Eisen MB: Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 2018, 7. Lattice light sheet microscopy coupled with 4D imaging detects transcription factor hubs for the Zelda pioneer factor in Drosophila
- 19.Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. : Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 2014, 15:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldfield AJ, Henriques T, Kumar D, Burkholder AB, Cinghu S, Paulet D, Bennett BD, Yang P, Scruggs BS, Lavender CA, et al. : NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat Commun 2019, 10:3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaves-Sanjuan A, Gnesutta N, Gobbini A, Martignago D, Bernardini A, Fornara F, Mantovani R, Nardini M: Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J 2021, 105:49–61. [DOI] [PubMed] [Google Scholar]
- 22.Weijers D, Wagner D: Transcriptional Responses to the Auxin Hormone. Annu Rev Plant Biol 2016, 67:539–574. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Klasfeld S, Jeong CW, Jin R, Goto K, Yamaguchi N, Wagner D: TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat Commun 2020, 11:5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taoka K-I, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. : 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476:332–335. [DOI] [PubMed] [Google Scholar]
- 25.Romera-Branchat M, Severing E, Pocard C, Ohr H, Vincent C, Née G, Martinez-Gallegos R, Jang S, Andrés F, Madrigal P, et al. : Functional Divergence of the Arabidopsis Florigen-lnteracting bZIP Transcription Factors FD and FDP. Cell Reports 2020, 32:107966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collani S, Neumann M, Yant L, Schmid M: FT Modulates Genome-Wide DNA-Binding of the bZIP Transcription Factor FD. Plant Physiol 2019, 180:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Klasfeld S, Wagner D: Molecular regulation of plant developmental transitions and plant architecture via PEPB family proteins: an update on mechanism of action. J Exp Bot 2021, 72:2301–2311. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Wang Y, Irish VF: CENTRORADIALIS maintains shoot meristem indeterminacy by antagonizing THORN IDENTITY1 in Citrus. Curr Biol 2021, 31:2261. [DOI] [PubMed] [Google Scholar]
- 29.Charoensawan V, Wilson D, Teichmann SA: Genomic repertoires of DNA-binding transcription factors across the tree of life. Nucleic Acids Res 2010, 38:7364–7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. : Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000, 290:2105–2110. [DOI] [PubMed] [Google Scholar]
- 31.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. : Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R: DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci U S A 2014, 111:2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luscombe NM, Austin SE, Berman HM, Thornton JM: An overview of the structures of protein-DNA complexes. Genome Biol 2000, 1:REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Malley RC, Huang S-SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR: Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 166:1598. [DOI] [PubMed] [Google Scholar]
- 35.Jana T, Brodsky S, Barkai N: Speed-Specificity Trade-Offs in the Transcription Factors Search for Their Genomic Binding Sites. Trends Genet 2021, 37:421–432. [DOI] [PubMed] [Google Scholar]
- 36.Kribelbauer JF, Rastogi C, Bussemaker HJ, Mann RS: Low-Affinity Binding Sites and the Transcription Factor Specificity Paradox in Eukaryotes. Annu Rev Cell Dev Biol 2019, 35:357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thévenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, et al. : A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science 2014, 343:645–648. [DOI] [PubMed] [Google Scholar]
- 38.Puranik S, Acajjaoui S, Conn S, Costa L, Conn V, Vial A, Marcellin R, Melzer R, Brown E, Hart D, et al. : Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 2014, 26:3603–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG: Choose your partners: dimerization in eukaryotic transcription factors. Trends in Biochemical Sciences 2008, 33:220–229. [DOI] [PubMed] [Google Scholar]
- 40.Trigg SA, Garza RM, MacWilliams A, Nery JR, Bartlett A, Castanon R, Goubil A, Feeney J, O’Malley R, Huang S-C, et al. : CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat Methods 2017, 14:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D: A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 2013, 24:426–437. [DOI] [PubMed] [Google Scholar]
- 42.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ: Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9:1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu F, Lionnet T: Transcription Factor Dynamics. Cold Spring Harb Perspect Biol 2021, doi: 10.1101/cshperspect.a040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X, Lu F, Qiu Q, Zhou B, Gu L, Zhang S, Kang Y, Cui X, Ma X, Yao Q, et al. : REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat Genet 2016, 48:694–699. [DOI] [PubMed] [Google Scholar]
- 45.Wendte JM, Schmitz RJ: Specifications of Targeting Heterochromatin Modifications in Plants. Mol Plant 2018, 11:381–387. [DOI] [PubMed] [Google Scholar]
- 46.Li B, Carey M, Workman JL: The role of chromatin during transcription. Cell 2007, 128:707–719. [DOI] [PubMed] [Google Scholar]
- 47.Plant AR, Larrieu A, Causier B: Repressor for hire! The vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol 2021, 231:963–973. [DOI] [PubMed] [Google Scholar]
- 48.Emenecker RJ, Holehouse AS, Strader LC: Biological Phase Separation and Biomolecular Condensates in Plants. Annu Rev Plant Biol 2021, 72:17–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA: A Phase Separation Model for Transcriptional Control. Cell 2017, 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50. Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. : Nucleo-cytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis thaliana. Mol Cell 2019, 76:177–190.e5.31421981 Demonstrated that ARF19 condensation acts to restrict its transcriptional activity.
- **51. Jung J-H, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. : A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585:256–260.32848244 Demonstrated that temperature-regulated ELF3 condensation acts to restrict its transcriptional activity.
- 52.Chèneby J, Ménétrier Z, Mestdagh M, Rosnet T, Douida A, Rhalloussi W, Bergon A, Lopez F, Ballester B: ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments. Nucleic Acids Res 2020, 48:D180–D188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amemiya HM, Kundaje A, Boyle AP: The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci Rep 2019, 9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **54. Leydon AR, Wang W, Gala HP, Gilmour S, Juarez-Solis S, Zahler ML, Zemke JE, Zheng N, Nemhauser JL: Repression by the TOPLESS corepressor requires association with the core mediator complex. Elite 2021, 10. Showed that TPL acts to prevent full Mediator complex formation in a yeast synthetic system.
- 55.Morffy N, Strader LC: Plant promoter-proximal pausing? Nat Plants 2021, 7:862–863. [DOI] [PubMed] [Google Scholar]
- 56.Goodsell DS, Autin L, Olson AJ: Illustrate: Software for Biomolecular Illustration. Structure 2019, 27:1716–1720.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sayou C, Nanao MH, Jamin M, Posé D, Thévenon E, Grégoire L, Tichtinsky G, Denay G, Ott F, Peirats Llobet M, et al. : A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun 2016, 7:11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC: Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A 2014, 111:5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emenecker RJ, Griffith D, Holehouse AS: metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure, [date unknown], doi: 10.1101/2021.05.30.446349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pastore JJ, Limpuangthip A, Yamaguchi N, Wu M-F, Sang Y, Han S-K, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D: LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 2011, 138:3189–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi N, Winter CM, Wu M-F, Kanno Y, Yamaguchi A, Seo M, Wagner D: Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 2014, 344:638–641. [DOI] [PubMed] [Google Scholar]
- 62.Kinoshita A, Vayssières A, Richter R, Sang Q, Roggen A, van Driel AD, Smith RS, Coupland G: Regulation of shoot meristem shape by photoperiodic signaling and phytohormones during floral induction of Arabidopsis. Elite 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J-W, Czech B, Weigel D: miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138:738–749. [DOI] [PubMed] [Google Scholar]
- 64.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE: UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 2018, 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]