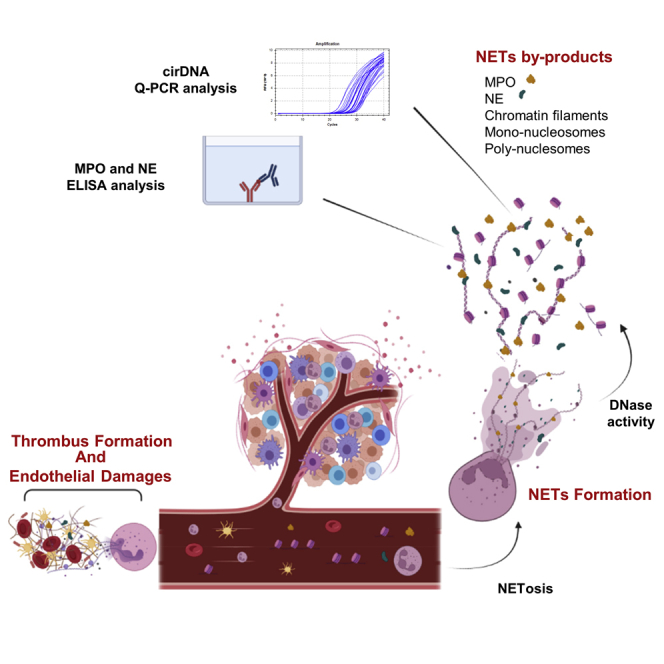
Summary
We postulate that a significant part of circulating DNA (cirDNA) originates in the degradation of neutrophil extracellular traps (NETs). In this study, we examined the plasma level of two markers of NETs (myeloperoxidase (MPO) and neutrophil elastase (NE)), as well as cirDNA levels in 219 patients with a metastatic colorectal cancer (mCRC), and in 114 healthy individuals (HI). We found that in patients with mCRC the content of these analytes was (i) highly correlated, and (ii) all statistically different (p < 0.0001) than in HI (N = 114). These three NETs markers may readily distinguish between patients with mCRC from HI, (0.88, 0.86, 0.84, and 0.95 AUC values for NE, MPO, cirDNA, and NE + MPO + cirDNA, respectively). Concomitant analysis of anti-phospholipid (anti-cardiolipin), NE, MPO, and cirDNA plasma concentrations in patients with mCRC might have value for thrombosis prevention, and suggested that NETosis may be a critical factor in the immunological response/phenomena linked to tumor progression.
Subject areas: Molecular biology, Immunology
Graphical Abstract
Highlights
-
•
NETs markers correlate with cirDNA amounts in patients with mCRC not in healthy subjects
-
•
Quantifying NETs markers and cirDNA could distinguish mCRC from healthy subjects
-
•
Analysis of NETs markers, cirDNA, and aPL may have value for thrombosis prevention
-
•
A strong fraction of cirDNA concentration could be derived from NETs in patients with mCRC
Molecular biology; Immunology
Introduction
Circulating cell-free DNA (cirDNA) is one of the fastest growing and most promising areas in oncology in recent years. CirDNA is defined as extracellular DNA occurring in blood (Bronkhorst et al., 2021). Studies have indicated the great potential of cirDNA for “liquid biopsies” in oncology (Diaz and Bardelli, 2014; Heitzer et al., 2020; Thierry et al., 2016; Wan et al., 2017) and for non-invasive prenatal testing (Dennis Lo and Poon, 2003).
While cirDNA analysis is routinely implemented in cancer theranostics, for example, for the detection of EGFR mutations in lung cancer, it also has significant appeal to researchers and clinicians in many other areas of cancer management care, including the detection of minimal residual disease (Benešová et al., 2019; Parikh et al., 2021; Tie et al., 2016), treatment monitoring (Sefrioui et al., 2021; Thierry et al., 2017b), cancer recurrence surveillance (Parikh et al., 2021; Sefrioui et al., 2021), and even cancer screening (Cohen et al., 2018; Cristiano et al., 2019; Sanchez et al., 2021; Tanos et al., 2020). That said, cirDNA’s biological features and potential functions remain poorly known. Its analytic performance, however, has been greatly improved by critical discoveries regarding its structure/topology (Chandrananda et al., 2015; Jiang et al., 2015; Mouliere et al., 2011) and its origin (nuclear vs mitochondrial (Al Amir Dache et al., 2020; Meddeb et al., 2019a). Recent advances in knowledge acquired through fragmentomics by cirDNA size profile analysis (Chandrananda et al., 2015; Sanchez et al., 2018, 2021; Serpas et al., 2019), methylation (Jensen et al., 2019; Lehmann-Werman et al., 2016; Moss et al., 2018), and nucleosome positioning (Snyder et al., 2016) may contribute to higher capacities in diagnostics or early cancer detection (Cohen et al., 2018; Cristiano et al., 2019; Tanos et al., 2020). Up to now, the impact of malignant cells, tumor microenvironment (TME), and germinal origin on cirDNA release in patients with cancer has not been clearly established. We hypothesize that a significant fraction of cirDNA production derives from the degradation of neutrophil extracellular traps (NETs).
Neutrophils are the most abundant innate immune cells, and offer a first line of immune defense against bacteria, viruses, parasites, yeast, and fungi (Urban et al., 2006; Waisberg et al., 2014; Yipp et al., 2012). The elimination of pathogens by neutrophils involves four distinct mechanisms: phagocytosis, degranulation, cytokines production, and NETosis (Brinkmann, 2018). The latter consists of a release of neutrophil extracellular traps (NETs) which are an extracellular web-like chromatin decorated with cytosolic and bactericidal granules proteins, such as neutrophil elastase (NE) and myeloperoxidase (MPO) (Metzler et al., 2014). While extracellular traps (ETs) formation is prominent in neutrophils, several other types of innate or adaptive immune cells reportedly release (following strong activation signals) chromatin and granular proteins (MPO, NE, …) into the extracellular space, thus forming ETs: macrophages, eosinophils, basophils, mast cells, and lymphocytes (Daniel et al., 2019). NETosis is one of the phenomena by which extracellular DNA may be released into the bloodstream by cell death and active secretion (Al-Khafaji et al., 2016; Papayannopoulos, 2018; Yousefi et al., 2019).
Over the past dozen years, numerous studies have elucidated the role of circulating neutrophils, circulating NETs, and circulating NETs by-product in cancer. It has been demonstrated that various cancer types such as breast, lung, or colorectal cancer exhibit an increase in circulating neutrophil numbers (Gentles et al., 2015; Templeton et al., 2014). There is currently an exponential growth in the literature reporting the emerging role of NETs in tumor progression and metastasis (Erpenbeck and Schön, 2017; Kos and de Visser, 2021; Munir et al., 2021; Nolan and Malanchi, 2020; Yang et al., 2020). CRC is one of the malignant diseases which show the greatest involvement of NETs in tumor progression and metastasis (Kos and de Visser, 2021; Nolan and Malanchi, 2020). It appears that, by sequestering circulating tumor cells in NETs, neutrophils fertilize the pre-metastasis niche. Furthermore, cancers predispose neutrophils to release extracellular DNA traps, which contribute to cancer-associated thrombosis (Wolach et al., 2018).
Interestingly, NETs are found in the same pathological conditions in which high concentrations of cirDNA have been reported, such as autoimmune diseases (Hakkim et al., 2010), inflammatory diseases (Kaplan and Radic, 2012), sepsis (Luo et al., 2014), thrombotic illnesses (Fuchs et al., 2012), and cancer (Daniel et al., 2019; Thierry et al., 2021; Thierry and Roch, 2020). In the case of patients with cancer, we postulate that higher concentrations of circulating neutrophils and their longer lifespan (activated in cancers) could increase the formation of NETs and NETs by-products. These could play a crucial role in cirDNA production, notably in the degradation of the web-like chromatin derived from NETs in blood. In addition to NET’s contribution to the occurrence of thrombosis (Wolach et al., 2018), anti-PL such as aCL were associated with thrombosis in various diseases, in particular with chronic diseases (Abdel-Wahab et al., 2020; Leal Rato et al., 2021). While aPL thrombophilic process or direct association is unclear, it has been suggested that aPL plasma high level may trigger thrombosis in patients with cancer (Gómez-Puerta et al., 2006; Islam, 2020). Thus, concurrent with NET markers and cirDNA, we determined aCL levels in order to investigate their association.
To explore our hypothesis, we will exploit data and plasma samples from patients screened within the UCGI 28 PANIRINOX clinical trial (NCT02980510), which uses cirDNA analysis to determine their RAS and BRAF status. This ongoing clinical study is the first interventional study to use cirDNA as a companion test for selecting patients with mCRC (metastatic colorectal cancer) toward anti-EGFR targeted therapy. Concurrent examination of the conventional NET protein biomarkers (NE and MPO) with the quantification of cirDNA of nuclear and mitochondrial origin is assessed in a large number of patients with mCRC at diagnosis (N = 219). The respective correlation of these circulating biological compounds will be compared with a control cohort constituted of healthy individuals (N = 114).
Results
Concentrations of NETs markers and cirDNA are higher in patients with mCRC than in healthy individuals
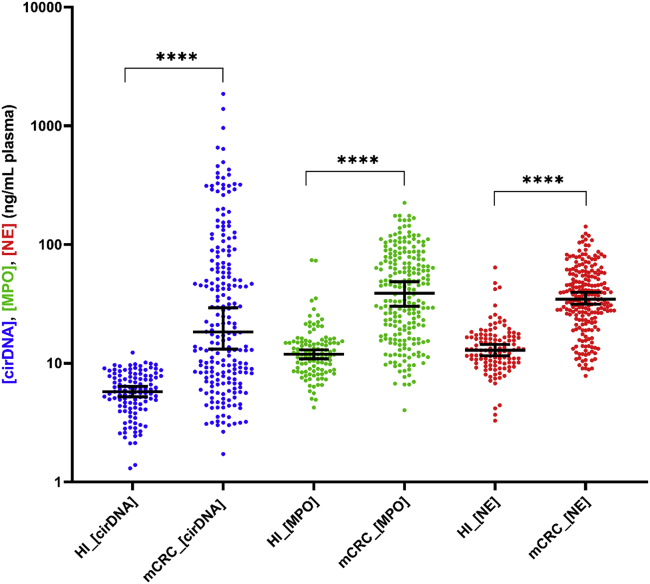
The formation of neutrophil extracellular traps was assessed in plasma samples from 219 patients with mCRC and 114 healthy individuals (Table 1), as estimated by the concentration of NE and MPO. The concentrations of NE and MPO are significantly higher in patients with mCRC than in healthy individuals (Figure 1) (p < 0.0001), with respective median concentrations of 12.90 ng/mL and 11.91 ng/mL in HI and 34.70 ng/mL and 38.90 ng/mL in patients with mCRC. The concentration of cirDNA is significantly higher in patients with mCRC than in HI, with median concentrations of 18.36 ng/mL versus 5.76 ng/mL, respectively (Figure 1) (p < 0.0001). Data revealed the presence of NETs by-products (NE and MPO) in healthy individuals, and their increased presence in patients with mCRC.
Table 1.
Patient characteristic
| Characteristic |
Patients with mCRC |
Healthy Individuals (HI) |
|---|---|---|
| N | 219 | 114 |
| Age, y | ||
| Median (range) | 61 (37–77) | 41 (19–69) |
| Missing data | 96 | |
| Sex | ||
| Male (%) | 73 (58,9) | 59 (51,8) |
| Female (%) | 51 (41,1) | 55 (48,2) |
| missing data | 95 | |
| Location of primary tumor | ||
| Right colon (%) | 34 (27,9) | |
| Left colon (%) | 88 (72,1) | |
| Missing data | 97 | |
| Primary tumor in place | ||
| Yes (%) | 96 (78) | |
| No (%) | 27 (22) | |
| Missing data | 96 | |
| Number of metastatic sites | ||
| Median (range) | 2 (1–4) | |
| 1 (%) | 51 (44,3) | |
| >1 (%) | 64 (55,6) | |
| Missing data | 104 | |
| Limited liver disease | ||
| Yes (%) | 45 (36,3) | |
| No (%) | 79 (63,7) | |
| Missing data | 95 | |
| Leukocytes cell count (G/L) | ||
| Median (range) | 8 (3,93–27,33) | |
| Missing data | 104 | |
| LDH level (U/L) | ||
| Median (range) | 315,5 (148–4502) | |
| Missing data | 123 | |
| CEA level (ng/mL) | ||
| Median (range) | 41,7 (0,70–14034) | |
| Missing data | 107 | |
| Platelet count (G/L) | ||
| Median (range) | 314 (116–849) | |
| Missing data | 152 | |
| Creatinine clearance (mL/min) CKD | ||
| Median (range) | 93,5 (44–120) | |
| Missing data | 155 | |
CEA, carcinoembryonic antigen; LDH, Lactate dehydrogenase; mCRC, metastatic colorectal cancer.
Figure 1.
Comparison of cirDNA, MPO, and NE concentrations (ng/mL of plasma) in patients with mCRC (mCRC) (n = 219) and healthy individuals (HI) (n = 114)
Lines represent median with 95% CI. Mann-Whitney test was performed to compare values for cirDNA, MPO, and NE in patient with mCRC and in HI (∗∗∗∗p < 0.0001). A probability of less than 0.05 was considered statistically significant; ∗∗∗∗p < 0.0001. Each dot represents the values of a single patient or a single healthy individual. CirDNA: circulating cell-free DNA; MPO: myeloperoxidase; NE: neutrophil elastase.
Concentrations of cirDNA and neutrophil extracellular traps markers in patients with mCRC are associated
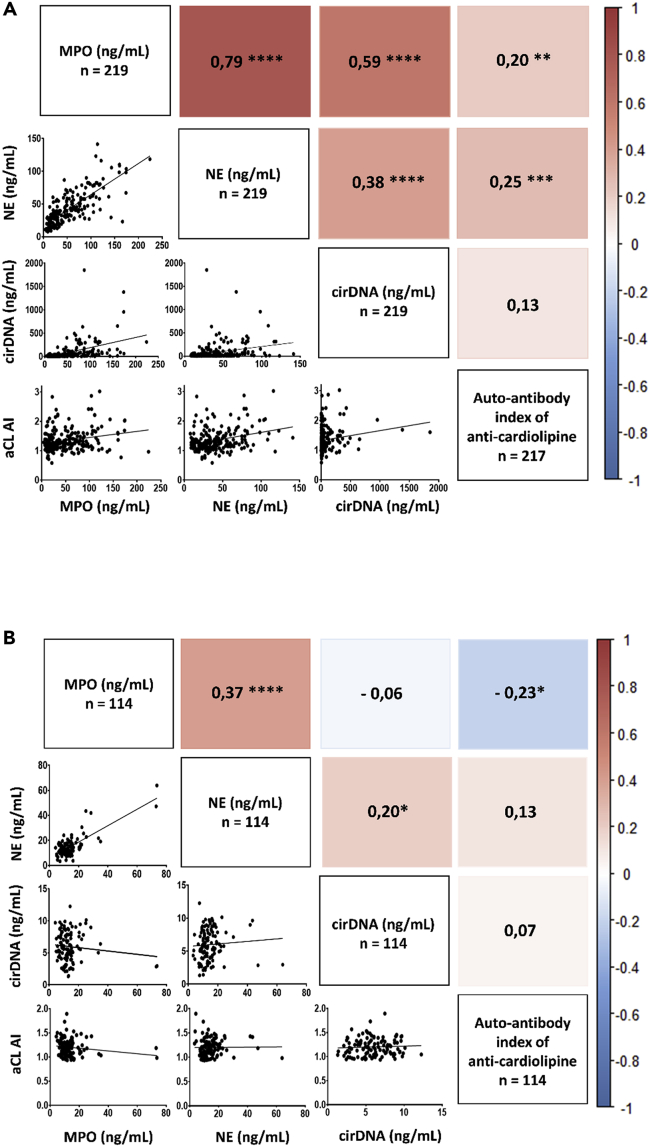
We used a Spearman correlation test to assess the association between cirDNA concentrations and NETs markers. First, data show a significantly positive association between NE and MPO in both patients with mCRC (n = 219) and HI (n = 114), r = 0.79 (p < 0.0001) and r = 0.37 (p < 0.0001), respectively (Figure 2). This positive correlation between NE and MPO shows that NETs are generated in patients with mCRC as well as in healthy individuals. In patients with mCRC, data revealed a significantly positive associations between both MPO and cirDNA concentrations and NE and cirDNA concentrations, r = 0.59 (p < 0.0001) and r = 0.38 (p < 0.0001), respectively (Figure 2A). No or poor association between MPO, NE, and cirDNA concentrations was found in HI (Figure 2B). As observed in our previous work, cirDNA concentrations as determined by targeting KRAS and BRAF wild-type sequences showed a very high correlation, r = 0.97 (p < 0.0001) (Figure S1), confirming earlier observation when validating a KRAS sequence-based Q-PCR system, and using a BRAF sequence-based Q-PCR system as quality control.
Figure 2.
Correlation matrix of cirDNA, MPO and NE concentrations (ng/mL plasma) and the anti-cardiolipin autoantibody index (aCL AI) in mCRC patients (n=219) (A) and in healthy individuals (n=114) (B).
(A and B) Correlation matrix of cirDNA, MPO, and NE concentrations (ng/mL plasma) and the anti-cardiolipin autoantibody index (aCL AI) in patients with mCRC (n=219) (A) and in healthy individuals (n=114) (B). Heatmap manifests the strength of relationship by Spearman’s correlation analysis (red: positive correlation; blue: negative correlation). A probability of less than 0.05 was considered statistically significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗ p < 0.0001. CirDNA, circulating cell-free DNA; MPO, myeloperoxidase; NE, neutrophil elastase; aCL AI, anti-cardiolipin autoantibody index.
To evaluate the influence of strong concentration of cirDNA in patients with mCRC on the correlation between NETs markers and cirDNA concentrations, we dichotomized the mCRC population into subgroups based on their cirDNA concentrations, and compared their NE and MPO values with those of healthy individuals (Figure S2). Given that the median cirDNA concentrations is about 6 ng/mL in HI (Figure 1), we first compared patients with mCRC with cirDNA concentrations below 6 ng/mL to match with cirDNA values of HI. Then, we also dichotomized patients with mCRC into subgroups of cirDNA concentrations over 6 ng/mL. Data reveal that for cirDNA concentrations close to 6 ng/mL, the MPO concentrations are significantly higher in patients with mCRC than in healthy individuals, with median concentrations of 17.76 ng/mL and 11.91 ng/mL (p < 0.0001), respectively (Figure S2A). Similar observations were made for NE concentrations, with median concentrations of 28.18 ng/mL and 12.90 ng/mL (p < 0.001) in patients with mCRC and in HI, respectively (Figure S2B).
In the same way, data show that in patients with mCRC, the higher the concentrations of NETs markers, the higher the cirDNA concentration (Figure S2).
To evaluate whether cirDNA production derives partly from NETs in patients with cancer, we dichotomized the patients with mCRC and the HI cohort by their MPO and NE median values, and then compared the cirDNA concentrations with the concentrations of NETs markers (Figure S3). For patients with mCRC, we observed a significant increase of cirDNA values in patients with MPO and NE concentrations higher than the medians values, as compared to patients with cirDNA concentrations under the median values, 38.9 ng/mL and 34.7 ng/mL (p < 0.0001), respectively (Figures S3A and S3C). Alternatively, when we dichotomized healthy individuals by their MPO median value (11.91 ng/mL), we did not observe a significant variation of cirDNA concentrations between the two groups of HI (Figure S3B). When the HI cohort was dichotomized by the NE median value (12.90 ng/mL), a slightly significant increase of cirDNA concentrations in HI with NE concentrations higher than the median value as compared to HI with NE concentrations below the median value was observed (Figure S3D).
Association between NE, MPO and cirDNA concentrations, and neutrophil and lymphocyte cell counts
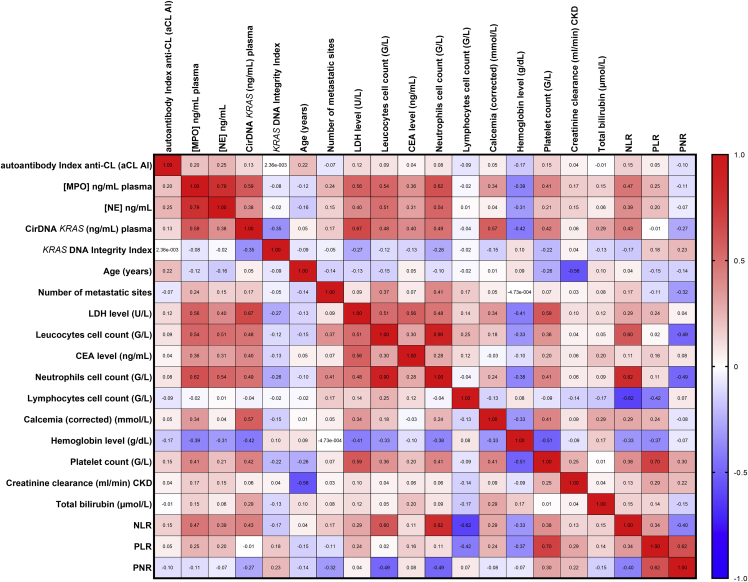
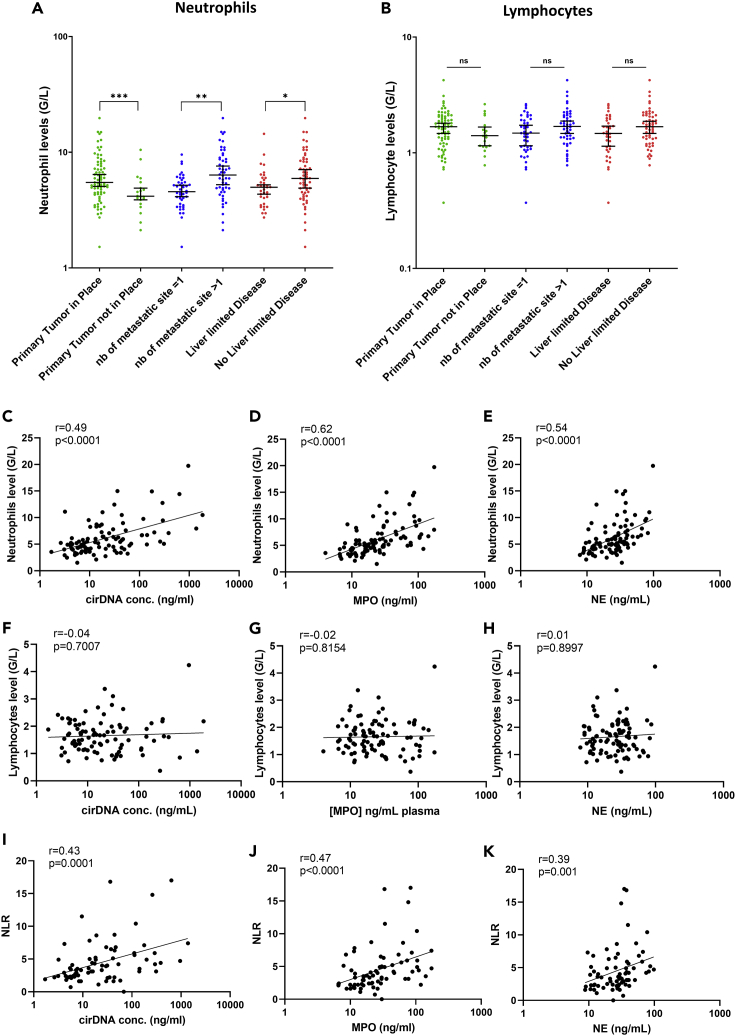
To confirm that NETs markers concentrations and cirDNA concentrations are generated by neutrophils, we interrogated the association between these markers and neutrophil and lymphocyte cell counts in patients with mCRC. Data revealed a very significant association between NE, MPO and cirDNA concentrations, and leukocyte cell counts, with r = 0.51, r = 0.54, and r = 0.48 (p < 0.0001), respectively (Figure 3). Interestingly, there was a significant association between neutrophil cell counts, the absence of surgery on the primary tumor, and the number of metastatic sites (threshold >1) (Figure 4A). Similarly, we found the same results between the leukocytes cell count and the absence of surgery on the primary tumor, and the number of metastatic sites (threshold >1) (Figure S4). Nevertheless, we did not find any association between lymphocyte cell counts and those clinical characteristics (Figure 4B). Similarly, a very significant association was observed between cirDNA, MPO and NE concentrations, and neutrophil cell counts, with r = 0.49, r = 0.62, and r = 0.54 (p < 0.0001), respectively (Figures 4C–4E). Conversely, we did not find any association between lymphocyte cell counts and cirDNA, MPO, and NE concentrations in this cohort (Figures 4F–4H). The neutrophil-to-lymphocyte ratio (NLR) is significantly associated with cirDNA, MPO, and NE concentrations in patients with mCRC (Figures 4I–4K).
Figure 3.
Correlation matrix of cirDNA, MPO, and NE concentrations (ng/mL plasma) with numerous clinical and biological features in patient with mCRC (n = 219)
Heatmap manifests the strength of relationship by Spearman’s correlation analysis (red: positive correlation; blue: negative correlation). CirDNA, circulating cell-free DNA; MPO, myeloperoxidase; NE, neutrophil elastase; aCL AI, anti-cardiolipin autoantibody index; CEA, carcinoembryonic antigen; LDH, Lactate dehydrogenase; NLR, the neutrophil to lymphocyte Ratio; PLR, the platelet to lymphocytes Ratio; PNR, the platelet to neutrophil Ratio.
Figure 4.
Comparison of neutrophils and lymphocytes cell count (G/L) with clinical features and association of neutrophils cell count (G/L) and the neutrophil to lymphocyte ration (NLR) with cirDNA, MPO, and NE concentrations (ng/mL of plasma) in patients with mCRC (n = 219)
(A–K) Lines represent median with 95% CI. Mann-Whitney test was performed to compare (A) neutrophils cell count (G/L) with clinical features, (B) lymphocytes cell count (G/L) with clinical features. Association between (C) neutrophils cell count (G/L) and cirDNA concentrations (ng/mL), (D) neutrophils cell count (G/L) and MPO concentrations (ng/mL), (E) neutrophils cell count (G/L) and NE concentrations (ng/mL), (F) lymphocytes cell count (G/L) and cirDNA concentrations (ng/mL), (G) lymphocytes cell count (G/L) and MPO concentrations (ng/mL), (H) lymphocytes cell count (G/L) and NE concentrations (ng/mL) (I) the NLR and cirDNA concentration (ng/mL), (J) the NLR and MPO concentration (ng/mL), and (K) the NLR and NE concentration (ng/mL). Each dot represents the values of a single patient. A probability of less than 0.05 was considered statistically significant, ∗p < 0.05; ∗∗ p < 0.01; ∗∗∗ p < 0.001. CirDNA, circulating cell-free DNA; MPO, myeloperoxidase; NE, neutrophil elastase; NLR, the neutrophil to lymphocyte Ratio.
Association with other clinical and biological factors
In patients with mCRC, we observed a positive correlation between the NETs markers and NLR, the lactate dehydrogenase level (LDH, U/L), the carcinoembryonic antigen level (CEA, ng/mL), the corrected calcemia level (mmol/L), the platelet count (G/L), and the total bilirubin level (μmol/L) (Figure 3). We observed a negative correlation with the hemoglobin level (g/dL) with NETs markers (Figure 3). Note that, MPO is the marker that correlated most with such others factors. In contrast, in patients with mCRC, poor or no association was detected between NETs markers and the DNA integrity index (DII), age, the number of metastatic sites, and the creatinine clearance (mL/min, CKD) (Figure 3). CirDNA, MPO, and NE globally correlate the same way with respect to those factors.
Anti-cardiolipin autoantibody detection and its association between NE, MPO and cirDNA concentrations in patients with mCRC
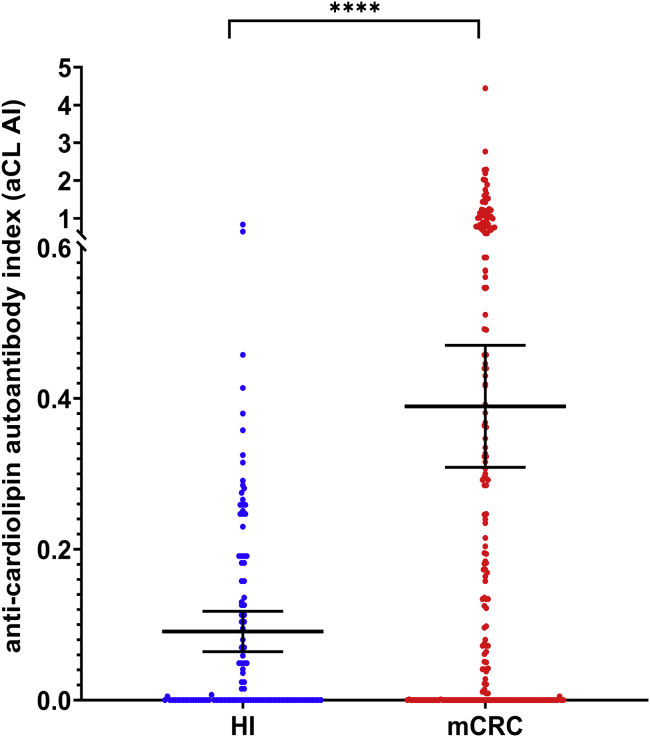
First, the anti-cardiolipin autoantibody index (aCL AI) is significantly higher in patients with mCRC than in HI, with mean concentrations of 0.389 [95%CI, 0.308–0.470] and 0.091 [95% CI, 0.064–0.117], respectively (p < 0.0001) (Figure 5). A statistically significant association was observed between the anti-cardiolipin autoantibody index with MPO and NE (r = 0.20, p < 0.01; and r = 0.25, p < 0.001), respectively; while a slight, non-statistically significant association with cirDNA concentrations was observed in patients with mCRC, (r = 0.13) (Figure 2A). Conversely, we did not observe any correlation between these anti-cardiolipin autoantibodies index and circulating NETs markers in HI (Figure 2B).
Figure 5.
Comparison of the anti-cardiolipin autoantibody index (aCL AI) in patients with mCRC (mCRC) (n = 217) and healthy individuals (HI) (n = 114)
The long horizontal bars indicate the mean, the shorter bars represent the 95% Confidential Interval (95% CI). Each dot indicates the aCL AI of a single patient or a single HI. The Student’s t test was performed to compare means of aCL AI values in patients with mCRC and in HI. A probability of less than 0.05 was considered statistically significant; ∗∗∗p < 0.001.
There are 50.7% of patients with mCRC (110/217) showing an aCL AI value over the mean 95% CI value of the healthy subjects and 62.2% of patients with mCRC (135/217) showing an aCL AI value over the median value of HI. Medians are 0.125 [95% CI, 0.041–0.204] and 0.000 [0.000–0.049] for patients with mCRC and healthy individuals, respectively (p < 0.0001). Moreover, there are 40.6% of patients with mCRC (88/217) showing an aCL AI value over 0.281 which represents the 90% higher aCL AI value of the healthy subjects.
CirDNA, MPO, and NE concentrations could be used to distinguish patients with mCRC from the healthy individuals
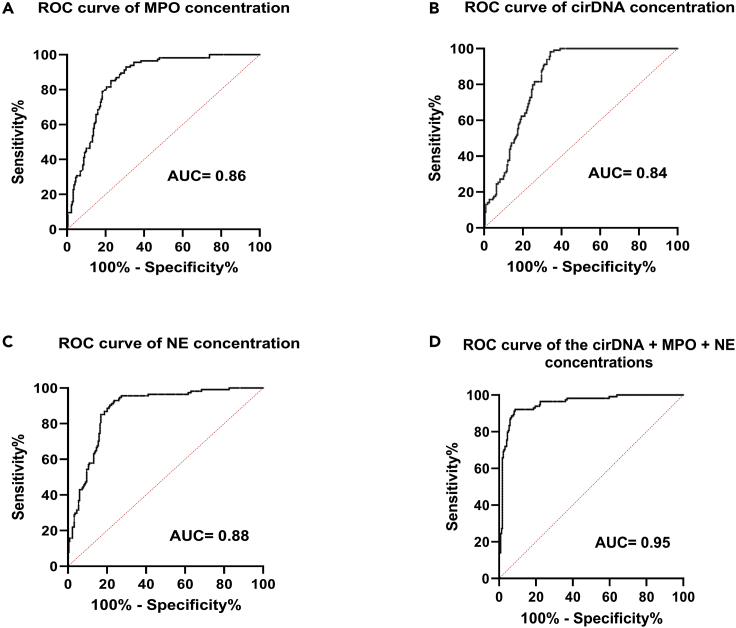
To explore whether cirDNA, MPO, and NE concentrations could be used to distinguish patients with mCRC from the healthy individuals, we interrogated the receiver operating characteristics (ROC) curve analysis for the concentrations of the aforementioned NETs markers. The ROC curves showed an AUC (area under curve) of 0.84 (0.7938–0.8770, 95% CI; confidence interval), 0.86 (0.8213–0.9000, 95% CI), and 0.88 (0.8435–0.9182, 95% CI) for cirDNA, MPO, and NE concentrations, respectively (Figures 6A–6C). When the three NETs marker concentrations were combined (cirDNA, MPO, and NE), the AUC of the ROC curve increased to 0.95 (0.9283–0.9759, 95% CI) (Figure 6D). The concentrations of the combination of these three markers are significantly higher in patients with mCRC than in healthy individuals (p < 0.0001), with respective median concentrations of 29.96 ng/mL in HI and 99.40 ng/mL in patients with mCRC. Similar observations were made when we combined MPO and NE concentrations, producing a new total AUC of the ROC curve of 0.94 (0.9137–0.9676) (Figure S5).
Figure 6.
ROC curves for the cirDNA concentration, the MPO concentration, and the NE concentration between healthy individuals and patients with mCRC
(A–D) ROC curve of (A) the cirDNA concentration (ng/mL), (B) the MPO concentration (ng/mL), (C) the NE concentration (ng/mL), and (D) the addition of three analyte concentrations (cirDNA, MPO, and NE). ROC: receiver operating characteristics; AUC, area under curve; cirDNA, circulating cell-free DNA; MPO, myeloperoxidase; NE, neutrophil elastase
Discussion
For more than two decades, it was postulated that cirDNA originates from cell death (Rostami et al., 2020), in particular from apoptosis and necrosis. This conclusion was based on the results of non-sophisticated methods such as electrophoresis, which revealed a strong 150- to 180-bp electrophoretic band (and weak bands of multiples thereof) (Jahr et al., 2001), highlighting the presence of DNA principally in mononucleosomes, and to a lesser extent in di- or tri-nucleosomes, which is the hallmark of apoptotic DNA cleavage. In addition, necrosis is thought to lead to fragments of high cirDNA fragment size (>10,000 bp) (Jahr et al., 2001; Thierry et al., 2016). Recently, Rostami et al. (2020) provided well-argued evidence that cirDNA release is modulated through a combination of apoptotic and senescent triggers and inhibitors. However, we postulate that short-sized nucleosomal structures could also result from the progressive nuclease degradation of longer cirDNA originating from necrosis, phagocytosis, microparticle-containing DNA, or active release from leukocytes, in particular NETosis. Indeed, chromatin fragments, oligo-nucleosomes, and nucleosomes can be liberated from the degradation process of NETs, and can contribute to the pool of cirDNA and histones. The prominence accorded to apoptosis as the main mechanism of release, therefore, may have to be reconsidered. Paunel-Görgülü et al. have demonstrated that cirDNA amplifies NETosis in an intracellular TLR9-independent manner, and one can speculate that an excess of cirDNA might have a positive feedback loop on NETs formation (Paunel-Görgülü et al., 2017).
Several studies report that tumors release cell-free DNA into the blood stream in quantities proportional to their mass, especially in the case of metastatic colorectal cancer (Bhangu et al., 2017; Thierry et al., 2017b; Wang et al., 2018; Wei et al., 2019; Xu et al., 2020). We may assume that cirDNA content depends upon the release of three cells of origin in a patient with cancer: (i) germline cells, according to cirDNA release in healthy individuals (2–7 ng/mL (Meddeb et al., 2019a; Thierry et al., 2016)); (ii) tumor malignant cells; and (iii) tumor microenvironment (stroma; endothelial and immunological cells) (Papayannopoulos, 2018). The release of wild-type DNA deriving from non-malignant cells may depend upon the quantity and the heterogeneity of the tumor microenvironment, as well as other sources, such as the lymphocytic cells. This might explain the high variation in the mutation allele’s frequency (MAF), as determined by cirDNA analysis. We previously found that MAF varied from 0.03% to 78% in a cohort of patients with mCRC (Mouliere et al., 2013). Thus, MAF is not directly related to tumor aggressiveness (Thierry et al., 2017a), and should not be compared with MAF determined within tumor tissue.
Our data revealed that (i), the concentrations of cirDNA and NETs markers (NE and MPO) are higher in patient with mCRC than in HI; (ii), an association exists between cirDNA concentrations and NETs markers concentrations in an mCRC cohort, and (iii), cirDNA, NE, and MPO concentrations correlate with leukocyte and neutrophil cell counts. A causal link can only be shown if the inhibition of NET formation (i.e. by the addition of DNase I) could lead to a decrease in the amount of cirDNA produced. Although correlation does not mean causation, we speculate that a significant cirDNA fraction derives from NETs formation, which itself is generated by active circulating neutrophils and/or by neutrophils infiltrating the tumor microenvironment. It is likely that NET chromatin filaments are rapidly and dynamically degraded by the very active plasma nucleases, which results in the release of chromatin fragments of high molecular size decreasing down to low molecular size DNA that is associated with mononucleosome/chromatosome chromatin unit (that appeared as the most prominent and the most stabilizing cirDNA structure in blood (Sanchez et al., 2018, 2021; Serpas et al., 2019; Waisberg et al., 2014)). Because MPO and NE are among the most important NET granule constituents, their contents highly correlated in HI and mCRC plasma. Both enzymes are otherwise physiologically independent.
In contrast, an increase in cirDNA concentrations is significantly associated with an increase in NETs conventional markers (NE and MPO) in patients with mCRC, while no such association was observed in healthy individuals. Remarkably, plasma of patient with mCRC with cirDNA concentrations close to the median of HI (6 ng/mL) showed higher concentrations of NE and MPO than HI. Thus, we may assume that cirDNA found in mCRC conditions derives from NETs in a manner unrelated to cirDNA concentrations. Confirming the above observations, ROC experiments revealed that the quantification of NE, MPO, and cirDNA shows a high diagnostic performance. The determination of an optimal test which would combine all NET markers using machine learning assistance is ongoing in our team.
It should be noted that leukocyte and neutrophil cell counts (but not lymphocyte cell count) increased in patients with a primary tumor in place and in patients with two or more metastatic sites. This might suggest that leukocytes and neutrophils—and by extension NETs—are directly implicated in tumor growth or cancer dissemination, as previously suggested (Daniel et al., 2019; Kos and de Visser, 2021; Nolan and Malanchi, 2020; Yang et al., 2020).
Assuming that cirDNA are indirect NETs markers, and are a marker of tumor mass in mCRC, our study suggests the association of increased circulation of NETs markers and disease severity. We may speculate that cirDNA and NETs by-products are markers of the inflammation associated with solid tumors, and more globally with inflammatory diseases. It is possible, therefore, that cirDNA concentrations associated with others NETs circulating markers could provide significant information about cancer severity, cancer prognosis, or treatment guidance, which could constitute a significant advance in cancer along with the advent of immunotherapy and the growing knowledge of tumor immunology.
An association of anti-cardiolipin autoantibodies (aCL) with circulating NETs markers was observed in a significant part of patients with mCRC, but not in HI. ACL is part of the anti-phospholipid antibody family (aPL) characteristic of the anti-phospholipid syndrome (Syrigos et al., 1998; Wichmann et al., 2020), an immune-mediated disorder resulting in pregnancy morbidity and arterial or venous thrombotic events. While aPL is present in 1%–5% of the general population, APS prevalence is 40–50/100,000 subjects (Leal Rato et al., 2021). However, this prevalence can increase to 50% among elderly patients with chronic diseases (Abdel-Wahab et al., 2020; Leal Rato et al., 2021). Several works report higher aPL levels in various hematological and solid tumors (Gómez-Puerta et al., 2006; Islam, 2020), with the percentage of aPL-positive cancer patients varying from 5% to 70%. Because the risk of thrombosis is 4- to 60-fold higher in patients with cancer than in the general population, it has been suggested that an elevated level of aPL might trigger thrombosis in patients with cancer. A meta-analysis revealed that patients with gastrointestinal, genitourinary, and lung cancer are at a higher risk of developing aPL (Gómez-Puerta et al., 2006). While the appearance of aPL such as aCL could be as an essential step in the prevention of thrombosis, the direct or indirect implication of aPL in the thrombophilic process remains unclear. It is clear, however, that neutrophils and NETs contribute to APS pathophysiology (Tambralli et al., 2020). It should also be noted that exacerbated NET formation has been linked to anti-phospholipid syndrome (APS) in numerous auto-immune and non-auto-immune pathologies (such as lupus) which showed elevated levels of aCL (Thierry and Roch, 2020). The clear statistical difference between healthy and mCRC subject study cohorts revealed that a majority or a significant fraction of patients with mCRC at diagnosis showed an auto-production of aCL at a low, moderate, and high level.
In conclusion, our work shows for the first time that a correlation exists between NET markers (MPO and NE) and cirDNA in patients with mCRC, suggesting that cirDNA might appear as a marker of NETs. In addition, this finding redefines existing paradigms of cirDNA release mechanisms, and could suggest that a significant fraction of the cirDNA quantity derives from NETs in cancer patients with mCRC. Moreover, we revealed that concomitant analyses of NETs markers (NE and MPO) and cirDNA also enable the differentiation of patients with mCRC from healthy individuals. This warrants the evaluation of NET by-products analysis in cancer diagnosis, in particular for thrombosis prevention, for patient follow-up, in guiding immunotherapy, or eventually as a means of devising an improved liquid biopsy cancer screening test. Our study is the first to show the close association of aPL with NET markers and cirDNA in patients with cancer, suggesting that the examination of these markers might be useful in preventing thrombosis in patients with cancer. Lastly, our observations contribute to the understanding of the imbalance which cancer can cause between the immunological system and hemostasis. This deepened understanding may prove useful in improving long-term cancer survival rates.
Limitations of the study
Although we showed a positive correlation between cirDNA levels and the levels of NETs markers (NE and MPO), these results should be confirmed on another larger validation cohort. In addition, this study was conducted on patients with mCRC at initial diagnosis, and it would have been interesting to examine this correlation in locally advanced disease, in the course of post-surgery treatment, as well as in other malignancies. Moreover, none of the patients had neutropenia at the time of initial diagnosis; however, the influence of neutropenia at initial diagnosis and during chemotherapy treatment should be evaluated in another study. While we found that the impact of number of neutrophils on the correlation between cirDNA levels with MPO levels is negligible supporting the main observation of this study, the influence of neutrophils numbers during chemotherapy treatment should be evaluated in another study. Finally, a thorough mechanistic study of NETs degradation in blood (as well as in plasma and serum) of patients with cancer and healthy individuals needs to be conducted to confirm the causality between NETs formation and cirDNA production in patients with cancer. Our team is actively working on this last topic.
STAR★Method
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Human ELA2 Capture antibody (Neutrophil Elastase capture antibody) | R&D Systems | Cat #DY9167-05 |
| Biotinylated Mouse Anti-Human ELA2 Detection antibody (Neutrophil Elastase detection antibody) | R&D Systems | Cat #DY9167-05 |
| Rat Anti-Human MPO Capture Antibody | R&D Systems | Cat #DY3174 |
| Biotinylated Goat Anti-Human MPO Detection Antibody | R&D Systems | Cat #DY3174 |
| Human Cardiolipin Total antibody | Boster | Cat #EK7027 |
| Biological samples | ||
| Metastatic colorectal cancer patients blood samples (n = 219) | PANIRINOX study NTC02980510 | N/A |
| Healthy individuals blood samples (n = 114) | Etablissement Français du Sang (EFS) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| iQ SYBR green mix | BIO-RAD | Cat #1708882 |
| Critical commercial assays | ||
| DuoSet Ancillary reagent Kit 2 | R&D Systems | Cat #DY008 |
| Human Cardiolipin Total antibody (IgG, IgM, IgA) ELISA Kit (Direct EIA) | Boster | Cat #EK7027 |
| QIAamp DNA Mini Blood Kit | Qiagen | Cat #51106 |
| Deposited data | ||
| Raw and analyzed data | This paper | N/A |
| Oligonucleotides | ||
| Wild type KRAS sequence : KRAS B1 inv k forward : 5′ CCTTGGGTTTCAAGTTATATG -3′ | Thierry et al., 2014, Nature Medicine | https://doi.org/10.1038/nm.3511 |
| Wild type KRAS sequence: KRAS B2 inv k reverse: 5′ CCCTGACATACTCCCAAGGA -3′ | Thierry et al., 2014, Nature Medicine | https://doi.org/10.1038/nm.3511 |
| Wild type KRAS sequence : KRAS A1 inv k forward: 5′ GCCTGCTGAAAATGACTGA -3′ | Thierry et al., 2014, Nature Medicine | https://doi.org/10.1038/nm.3511 |
| Wild type BRAF sequence: BRAF A1 conv k forward: 5′TTATTGACTCTAAGAGGAAAGATGAA-3′ | Thierry et al., 2014, Nature Medicine | https://doi.org/10.1038/nm.3511 |
| Wild type BRAF sequence: BRAF A2 conv k reverse: 5′ GAGCAAGCATTATGAAGAGTTTAGG -3′ | Thierry et al., 2014, Nature Medicine | https://doi.org/10.1038/nm.3511 |
| Software and algorithms | ||
| PHERAstar control software | BMG LABTECH | https://www.bmglabtech.com/fr/ |
| CFX manager software | BIO-RAD | https://www.bio-rad.com/fr-fr/sku/12004110-cfx-maestro-software?ID=12004110 |
| Prism 8.3.1 software | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alain R. Thierry (alain.thierry@inserm.fr).
Materials availability
This study did not generate newly generated materials or new unique reagents.
Experimental model and subject details
Data recorded during this trial are subject to a computerized treatment at the Unicancer Central Data Center in Montpellier in compliance with the “Loi Informatique et Libertés n° 78-17, 6 January 1978 modified”. The collection of biological samples implemented within the framework of the trial was declared to ANSM in the same time that the request of Clinical Trial Authorization. After the trial, and in case of storage, the storage of the collection of biological samples will be notified to the Minister of Research (and submitted to the CPP to notice if change of purpose of Research)"
Human subjects
In this study, we included 219 mCRC patients (51 females (41.1%) and 73 males (28.9%) (Table 1) from the screening procedure of the ongoing UCGI 28 PANIRINOX study (NCT02980510/EudraCT n°2016-001490-33). We investigated the correlation of MPO concentrations, NE concentrations and the nuclear cell-free DNA (cirDNA) concentrations to demonstrate that a very significant fraction of cirDNA derives from the degradation of neutrophil extracellular traps (NETs). PANIRINOX is the first interventional study to use cirDNA as a companion test for selecting mCRC patients towards anti-EGFR targeted therapy by the IntPlex® method. Briefly: eligible patients were recruited in the PANIRINOX screening procedures at diagnosis; patients accepted for inclusion were male or female, aged between 18 and 75 years old, with a ECOG performance status 0 or 1, a histologically confirmed colo-rectal adenocarcinoma, an untreated synchronous or metachronous metastatic disease deemed unresectable with curative intent; a KRAS (codons 12, 13, 61, 117, 146), NRAS (codons 12, 13, 61) and BRAFV600E wild type (WT) tumor status according to plasma analysis of cirDNA by Intplex technology; a measurable disease according to RECIST version 1.1; and adequate hematologic, hepatic and renal functions. Written, informed consent was obtained from all participants before the screening procedure. However, this work was carried out on an ad hoc cohort study at time of the screening procedure, before the randomization.
We also analyzed 114 healthy individuals (HI) (55 females (48.2%) and 59 males (51.8%) (Table 1) from the Etablissement Français du Sang (EFS), which is Montpellier’s blood transfusion center (Convention EFS-PM N° 21PLER2015-0013). These samples were analyzed (virology, serology, immunology, blood numeration) and ruled out whenever any abnormality was detected.
Samples preparation
Blood from mCRC patients (n = 219) were collected in STRECK tubes (Cell-Free DNA BCT®) and were sent within 24 hours of blood collection at room temperature from the recruiting institutions to our laboratory (IRCM, Institut de Recherche en Cancérologie de Montpellier, U1194 INSERM). Blood tubes were centrifuged for 10 minutes at 1,200 × g at 4 °C within 5 days of blood collection, and the plasma supernatants were immediately centrifuged at 16,000 × g at 4 °C for 10 minutes. Then, plasma samples were stored at −20 °C for several days or used immediately. Total circulating cell-free DNA was extracted from 1 mL of plasma using the QIAamp DNA Mini Blood Kit (Qiagen) in accordance with pre-analytic guidelines (El Messaoudi et al., 2013; Meddeb et al., 2019b) in an elution volume of 130μL. CirDNA extracts were kept at −20 °C until use or used immediately. Blood from healthy individuals (n = 114) was collected in EDTA tubes and was centrifuged for 10 minutes at 1,200 × g at 4 °C within 4 hours of blood collection. Then, plasma supernatants were immediately centrifuged at 16,000 × g at 4 °C for 10 minutes. Finally, plasma samples were stored at −20 °C for several days or used immediately.
Method details
Quantification of cirDNA
Analysis of cirDNA was done by IntPlex®, an allele-specific blocker quantitative PCR (ASB Q-PCR), (Mouliere et al., 2014; Thierry et al., 2014), according to the MIQE guidelines (Bustin, 2010; Bustin et al., 2009). This IntPlex system specifically detect nuclear cirDNA. Q-PCR amplifications were carried out in at least two replicates in a total volume of 25 μL on a CFX96 instrument using the CFX manager software (Bio-Rad). Each PCR reaction was composed of 12.5 μL of IQ Supermix Sybr Green (Bio-Rad), 2.5 μL of DNase-free water (Qiagen) or specific oligoblocker, 2.5 μL of forward and reverse primers (0.3 pmol/mL), and 5 μL of template. Thermal cycling comprised three repeated steps: a hot-start activation step at 95 °C for 3 minutes, followed by 40 cycles of denaturation–amplification at 95 °C for 10 seconds, then at 60°C for 30 seconds. Melting curves were investigated by increasing the temperature from 60°C to 90°C with a plate reading every 0.2°C. Standard curves were performed for each run with a genomic extract of the DiFi cell line at 1.8 ng/μL of DNA. Each PCR run was carried out with no template control and positive control for each primer set. In each single run, were included one standard curve was prepared. Validation of Q-PCR amplification was performed by melt curve differentiation. Quantification of total cirDNA concentration in mCRC patients and HI was obtained by amplifying a 67 bp-length wild-type sequence of the KRAS gene. In addition to routinely performing a standard curve for each primer couple with the PCR system, the accuracy and gene copy number variations were checked by quantifying a WT sequence of the BRAF gene from the amplification of a 105 bp amplicon. This method of quantifying cirDNA has been experimentally (Mouliere et al., 2014) and clinically validated (Thierry et al., 2017a; Thierry et al., 2014), and showed unprecedented specificity and sensitivity, to the point of permitting the detection of a single DNA fragment molecule under Poisson Law distribution (Thierry et al., 2017a). An intra-and inter-experimental reproducibility study shows a 19% and 24% coefficient of variation (Mouliere et al., 2013, 2014) when jointly taking into consideration plasma preparation, cirDNA extraction and Q-PCR measurement.
Myeloperoxidase and neutrophil elastase assay
MPO and NE concentrations were measured using enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's standard protocol (Duoset R&D Systems, DY008, DY3174, and DY9167-05). Briefly, captured antibodies were diluted at the working concentrations in the Reagent Diluent (RD) provided on ancillary reagent kits (DY008) and coated overnight at room temperature (RT) on 96-well microplates with 100 μL per wells. Then, captured antibodies were removed from the microplate, and wells were washed three times with 300 μL of Wash Buffer (WB). Microplates were blocked at RT for 2 hours by adding 300 μL of RD to each well. RD were removed from the microplates, and wells were washed three times with 300 μL of WB. Then, 100μL of negative controls, standards and plasma samples (diluted 1/10) were added to the appropriate wells for one hour at RT. Samples, controls and standards were removed from the microplates, and wells were washed three times with 300 μL of WB. Detection antibodies were diluted at the working concentrations in the RD, and then added by 100 μL per well, for one hour at RT. Detection antibodies were removed from the microplates, and wells were washed three times with 300 μL of WB. Then, 100 μL of Streptavidin-HRP was added to each well and microplates were incubated at RT for 30 minutes. Repeat wash three times. Finally, 100μL per well of substrate solution was added and incubated for 15 minutes, and the Optical Density (O.D) of each well was read immediately at 450 nm with the PHERAstar FS instrument using the PHERAstar control software.
Anti-cardiolipin autoantibody index calculation
The antibody index of total human autoantibodies against cardiolipin (IgG, IgM and IgA) was measured using direct ELISA according to the manufacturer's standard protocol (Boster, EK7027). Briefly, 100 μL of negative controls, positive controls, calibrator and diluted plasma samples (1/21) was dispensed into cardiolipin-coated wells and incubated for 30 minutes at RT. Samples, controls and calibrator were removed from the microplates and the wells were washed three times with 300μL of WB. Then, 100 μL of enzyme conjugate was added in each well for 20 minutes at RT. The washing step was repeated. 100 μL of TMB substrate was dispensed into wells for 10 minutes. Finally, 100 μL of stop solution was added to each well and O.D was immediately read at 450 nm with the PHERAstar FS instrument using the PHERAstar control software. The cut-off value of each plate was calculated as follows: Calibrator O.D × Calibrator Factor (CF) of the kit. The aCL AI is calculated by dividing the O.D value of each sample by cut-off value.
In order to globally evaluate the difference between HI and mCRC subjects, we compared the quantitative data (aCL AI) rather the qualitative data that can be inferred from a positivity threshold determined from a calibrator. In case of the Boster kit, the calibrator corresponds to a diagnosed APLS patient known to have very high level of aCL, and it is consequently not appropriate for studying cancer or pregnancy follow up.
The marketed kits use internal standards whose value has been defined in relation to sera commonly called “Harris standards”. There is a low concordance between the batches of standards: this results in disparities between the kits, depending on whether they have been standardized with a particular batch of Harris standard (the aCL may have varying rates 2 to 3 times, depending on the kit). As with all ELISA techniques, determining the positivity threshold is delicate. Antibody levels in healthy populations are not distributed normally and vary with the source of the antigen used (calibration with each change of antigen lot). Note, a negative result for anti-cardiolipins IgG or IgM indicates that this type of antibody was not present or was present in a too low amount in the blood sample being tested.
In our assay, three negative controls are analyzed in each plate: (1), the blank (no test sample); (2), the kit negative control; and (3), a plasma of a healthy individual. We defined an arbitrary unit of aCL concentration (Anticardiolipin autoantibody index, aCL AI) taking in consideration plate to plate variation and standard batch level: (OD - 1.35 × negative control) / blank. The OD median of healthy individuals is 1.35-fold higher than the kit negative control, irrespective of the plate. The plasma of the healthy individual plasma control is useful as a quality control to check the kit negative control level, and routinely showed an 1.1-fold (+/− 10%) increase as compared to the kit negative control.
Weak or moderately positive results are sometimes observed temporarily in older healthy people without symptoms following infection or following medication. These results are most often of little clinical significance, but should be interpreted in light of other clinical information. This explains the low and moderate aCL AI we found in a small fraction of healthy individuals. Quantification of results is important, since there is a correlation between the rate of aCL (in particular IgG) and the risk of thrombosis. In view of this, low to moderate aCL levels are taken into consideration in contexts of repeated spontaneous miscarriages (Abdel-Wahab et al., 2020).
Quantification and statistical analysis
The Mann-Whitney U test was used for non-parametric data and the Student t-test was used for parametric data. Correlation analysis was performed using the Spearman test (Graph Pad Prism 8.3.1 software). A probability of less than 0.05 was considered to be statistically significant; ∗p <0.05, ∗∗p <0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Additional resources
PANIRINOX clinical trial: NCT02980510
https://clinicaltrials.gov/ct2/show/NCT02980510?term = PANIRINOX&draw = 2&rank = 1
Acknowledgments
The PANIRINOX study is funded by AMGEN. B. Pastor was partially supported by SIRIC Montpellier Cancer Grant INCa_Inserm_DGOS_12553, and AR Thierry by INSERM. PANIRINOX study is sponsored by R&D Unicancer. The sponsor had a role in the collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The funder had no role in the design and conduct of this present study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. We thank our patients and their families for their trust; all the participating physicians and supporting staff; Datacenter Unicancer members (Institut Regional du Cancer de Montpellier) and Marie Bergeaud (R&D Unicancer). The authors thank the excellent technical assistance of C. Sanchez and A. Kudriavtsev (IRCM, Institut de Recherche en Cancérologie de Montpellier, INSERM U1194, Université de Montpellier, Institut régional du Cancer de Montpellier, Montpellier, F-34134298, France) and Cormac Mc Carthy (Mc Carthy Consultant, Montpellier) for English editing (financial compensation). We thank also all CRA and clinical co-investigators, Marie Bergeaud (Unicancer, Paris). We are grateful to K. Billings and Streck corp. for providing the cell-free DNA blood collection tubes. We thank all the patients and healthy donors who participated in this study. We also thank the clinical investigators of the centers who participated in this study.
Author contributions
ART and BP designed the study. BP and ART developed the methodology. BP, CS, AK, EP, and AM did the experiments under the supervision of ART. BP did the statistical analyses. BP and ART analyzed the data and prepared the manuscript. All of the authors (BP, JDA, EP, CS, AK, RT, AM, LM, VP, AA, MY, TM, and ART discussed the results and approved the manuscript.
Declaration of interest
TM reported receiving grants from Amgen SAS; nonfinancial support from Servier and MSD; and personal fees from Merck Serono, Bristol Myers Squibb, Sanofi Genzyme, Sandoz, and Bayer outside the submitted work. AA has reported a consulting/advisory role and/or receiving honoraria from Bristol-Myers Squibb, MSD Oncology, and Servier; and has received research funding from Bayer Pharmaceuticals. ART is a DiaDx stockholder. BP, JDA, EP, CS, AK, LM, RT, VP, MY, and AM declare no competing interests.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103826.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request
-
•
This paper does not report original code.
-
•
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdel-Wahab N., Tayar J.H., Fa’ak F., Sharma G., Lopez-Olivo M.A., Yousif A., Shagroni T., Al-Hawamdeh S., Rojas-Hernandez C.M., Suarez-Almazor M.E. Systematic review of observational studies reporting antiphospholipid antibodies in patients with solid tumors. Blood Adv. 2020;4:1746–1755. doi: 10.1182/bloodadvances.2020001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Amir Dache Z., Otandault A., Tanos R., Pastor B., Meddeb R., Sanchez C., Arena G., Lasorsa L., Bennett A., Grange T., et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020;34:3616–3630. doi: 10.1096/fj.201901917RR. [DOI] [PubMed] [Google Scholar]
- Al-Khafaji A.B., Tohme S., Yazdani H.O., Miller D., Huang H., Tsung A. Superoxide induces neutrophil extracellular trap formation in a TLR-4 and NOX-dependent mechanism. Mol. Med. 2016;22:621–631. doi: 10.2119/molmed.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benešová L., Hálková T., Ptáčková R., Semyakina A., Menclová K., Pudil J., Ryska M., Levý M., Šimša J., Pazdírek F., et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J. Gastroenterol. 2019;25:6939–6948. doi: 10.3748/wjg.v25.i48.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangu J.S., Taghizadeh H., Braunschmid T., Bachleitner-Hofmann T., Mannhalter C. Circulating cell-free DNA in plasma of colorectal cancer patients—a potential biomarker for tumor burden. Surg. Oncol. 2017;26:395–401. doi: 10.1016/j.suronc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Brinkmann V. Neutrophil extracellular traps in the second decade. J. Innate Immun. 2018;10:414–421. doi: 10.1159/000489829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronkhorst A.J., Ungerer V., Diehl F., Anker P., Dor Y., Fleischhacker M., Gahan P.B., Hui L., Holdenrieder S., Thierry A.R. Towards systematic nomenclature for cell-free DNA. Hum. Genet. 2021;140:565–578. doi: 10.1007/s00439-020-02227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. Why the need for qPCR publication guidelines?—the case for MIQE. Methods. 2010;50:217–226. doi: 10.1016/j.ymeth.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chandrananda D., Thorne N.P., Bahlo M. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med. Genomics. 2015;8:29. doi: 10.1186/s12920-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D.C., Jensen S.Ø., Medina J.E., Hruban C., White J.R., et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C., Leppkes M., Muñoz L.E., Schley G., Schett G., Herrmann M. Extracellular DNA traps in inflammation, injury and healing. Nat. Rev. Nephrol. 2019;15:559–575. doi: 10.1038/s41581-019-0163-2. [DOI] [PubMed] [Google Scholar]
- Dennis Lo Y., Poon L.L. The ins and outs of fetal DNA in maternal plasma. Lancet. 2003;361:193–194. doi: 10.1016/S0140-6736(03)12319-7. [DOI] [PubMed] [Google Scholar]
- Diaz L.A., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi S., Rolet F., Mouliere F., Thierry A.R. Circulating cell free DNA: preanalytical considerations. Clinica Chim. Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Erpenbeck L., Schön M.P. Neutrophil extracellular traps: protagonists of cancer progression? Oncogene. 2017;36:2483–2490. doi: 10.1038/onc.2016.406. [DOI] [PubMed] [Google Scholar]
- Fuchs T.A., Kremer Hovinga J.A., Schatzberg D., Wagner D.D., Lämmle B. Circulating DNA and myeloperoxidase indicate disease activity in patients with thrombotic microangiopathies. Blood. 2012;120:1157–1164. doi: 10.1182/blood-2012-02-412197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Puerta J.A., Cervera R., Espinosa G., Aguiló S., Bucciarelli S., Ramos-Casals M., Ingelmo M., Asherson R.A., Font J. Antiphospholipid antibodies associated with malignancies: clinical and pathological characteristics of 120 patients. Semin. Arthritis Rheum. 2006;35:322–332. doi: 10.1016/j.semarthrit.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hakkim A., Furnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V., Herrmann M., Voll R.E., Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. U S A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E., Auinger L., Speicher M.R. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol. Med. 2020;26:519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- Islam Md.A. Antiphospholipid antibodies and antiphospholipid syndrome in cancer: uninvited guests in troubled times. Semin. Cancer Biol. 2020;64:108–113. doi: 10.1016/j.semcancer.2019.07.019. [DOI] [PubMed] [Google Scholar]
- Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.-D., Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- Jensen S.Ø., Øgaard N., Ørntoft M.-B.W., Rasmussen M.H., Bramsen J.B., Kristensen H., Mouritzen P., Madsen M.R., Madsen A.H., Sunesen K.G., et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer—a clinical biomarker discovery and validation study. Clin. Epigenetics. 2019;11:158. doi: 10.1186/s13148-019-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Chan C.W.M., Chan K.C.A., Cheng S.H., Wong J., Wong V.W.-S., Wong G.L.H., Chan S.L., Mok T.S.K., Chan H.L.Y., et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U S A. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.J., Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J. Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos K., de Visser K.E. Neutrophils create a fertile soil for metastasis. Cancer Cell. 2021;39:301–303. doi: 10.1016/j.ccell.2021.01.009. [DOI] [PubMed] [Google Scholar]
- Leal Rato M., Bandeira M., Romão V.C., Aguiar de Sousa D. Neurologic manifestations of the antiphospholipid syndrome—an update. Curr. Neurol. Neurosci. Rep. 2021;21:41. doi: 10.1007/s11910-021-01124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann-Werman R., Neiman D., Zemmour H., Moss J., Magenheim J., Vaknin-Dembinsky A., Rubertsson S., Nellgård B., Blennow K., Zetterberg H., et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U S A. 2016;113:E1826–E1834. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Zhang S., Wang Y., Rahman M., Syk I., Zhang E., Thorlacius H. Proinflammatory role of neutrophil extracellular traps in abdominal sepsis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014;307:L586–L596. doi: 10.1152/ajplung.00365.2013. [DOI] [PubMed] [Google Scholar]
- Meddeb R., Dache Z.A.A., Thezenas S., Otandault A., Tanos R., Pastor B., Sanchez C., Azzi J., Tousch G., Azan S., et al. Quantifying circulating cell-free DNA in humans. Sci. Rep. 2019;9:5220. doi: 10.1038/s41598-019-41593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meddeb R., Pisareva E., Thierry A.R. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin. Chem. 2019;65:623–633. doi: 10.1373/clinchem.2018.298323. [DOI] [PubMed] [Google Scholar]
- Metzler K.D., Goosmann C., Lubojemska A., Zychlinsky A., Papayannopoulos V. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 2014;8:883–896. doi: 10.1016/j.celrep.2014.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018;9:448142. doi: 10.1101/448142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F., El Messaoudi S., Gongora C., Guedj A.-S., Robert B., Del Rio M., Molina F., Lamy P.-J., Lopez-Crapez E., Mathonnet M., et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl. Oncol. 2013;6:319–328. doi: 10.1593/tlo.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F., El Messaoudi S., Pang D., Dritschilo A., Thierry A.R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 2014;8:927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F., Robert B., Arnau Peyrotte E., Del Rio M., Ychou M., Molina F., Gongora C., Thierry A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6:e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir H., Jones J.O., Janowitz T., Hoffmann M., Euler M., Martins C.P., Welsh S.J., Shields J.D. Stromal-driven and Amyloid β-dependent induction of neutrophil extracellular traps modulates tumor growth. Nat. Commun. 2021;12:683. doi: 10.1038/s41467-021-20982-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E., Malanchi I. Neutrophil ‘safety net’ causes cancer cells to metastasize and proliferate. Nature. 2020;583:32–33. doi: 10.1038/d41586-020-01672-3. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 2018;18:134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- Parikh A.R., Van Seventer E.E., Siravegna G., Hartwig A.V., Jaimovich A., He Y., Kanter K., Fish M.G., Fosbenner K.D., Miao B., et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin. Cancer Res, 2021;27:5586–5594. doi: 10.1158/1078-0432.CCR-21-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunel-Görgülü A., Wacker M., El Aita M., Hassan S., Schlachtenberger G., Deppe A., Choi Y.-H., Kuhn E., Mehler T.O., Wahlers T. CfDNA correlates with endothelial damage after cardiac surgery with prolonged cardiopulmonary bypass and amplifies NETosis in an intracellular TLR9-independent manner. Sci. Rep. 2017;7:17421. doi: 10.1038/s41598-017-17561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami A., Lambie M., Yu C.W., Stambolic V., Waldron J.N., Bratman S.V. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 2020;31:107830. doi: 10.1016/j.celrep.2020.107830. [DOI] [PubMed] [Google Scholar]
- Sanchez C., Roch B., Mazard T., Blache P., Dache Z.A.A., Pastor B., Pisareva E., Tanos R., Thierry A.R. Circulating nuclear DNA structural features, origins, and complete size profile revealed by fragmentomics. JCI Insight. 2021;6:e144561. doi: 10.1172/jci.insight.144561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C., Snyder M.W., Tanos R., Shendure J., Thierry A.R. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. NPJ Genomic Med. 2018;3:31. doi: 10.1038/s41525-018-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefrioui D., Beaussire L., Gillibert A., Blanchard F., Toure E., Bazille C., Perdrix A., Ziegler F., Gangloff A., Hassine M., et al. CEA, CA19-9, circulating DNA and circulating tumour cell kinetics in patients treated for metastatic colorectal cancer (mCRC) Br. J. Cancer. 2021;125:725–733. doi: 10.1038/s41416-021-01431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpas L., Chan R.W.Y., Jiang P., Ni M., Sun K., Rashidfarrokhi A., Soni C., Sisirak V., Lee W.-S., Cheng S.H., et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. U S A. 2019;116:641–649. doi: 10.1073/pnas.1815031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M.W., Kircher M., Hill A.J., Daza R.M., Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrigos K.N., Charalampopoulos A., Konstantoulakis M.M., Karayiannakis A., Tsibloulis V., Peveretos P. Autoantibodies against cardiolipin in the serum of patients with colorectal adenocarcinoma:their prognostic significance. Eur. Surg. Res. 1998;30:55–60. doi: 10.1159/000008558. [DOI] [PubMed] [Google Scholar]
- Tambralli A., Gockman K., Knight J.S. NETs in APS: current knowledge and future perspectives. Curr. Rheumatol. Rep. 2020;22:67. doi: 10.1007/s11926-020-00936-1. [DOI] [PubMed] [Google Scholar]
- Tanos R., Tosato G., Otandault A., Al Amir Dache Z., Pique Lasorsa L., Tousch G., El Messaoudi S., Meddeb R., Diab Assaf M., Ychou M., et al. Machine learning-assisted evaluation of circulating DNA quantitative analysis for cancer screening. Adv. Sci. 2020;7:2000486. doi: 10.1002/advs.202000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A.J., McNamara M.G., Šeruga B., Vera-Badillo F.E., Aneja P., Ocaña A., Leibowitz-Amit R., Sonpavde G., Knox J.J., Tran B., et al. Prognostic role of neutrophil-to-lymphocyte Ratio in solid tumors: a systematic review and meta-analysis. JNCI: J. Natl. Cancer Inst. 2014;106:dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- Thierry A.R., El Messaoudi S., Gahan P.B., Anker P., Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A.R., El Messaoudi S., Mollevi C., Raoul J.L., Guimbaud R., Pezet D., Artru P., Assenat E., Borg C., Mathonnet M., et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann. Oncol. 2017;28:2149–2159. doi: 10.1093/annonc/mdx330. [DOI] [PubMed] [Google Scholar]
- Thierry A.R., Mouliere F., El Messaoudi S., Mollevi C., Lopez-Crapez E., Rolet F., Gillet B., Gongora C., Dechelotte P., Robert B., et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014;20:430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- Thierry A.R., Pastor B., Abraham J.-D., Pisareva E., Mazard T. Abstract P11: the elevated level of the main markers of neutrophil extracellular traps in metastatic colorectal cancer plasma highlights the enhanced risk of severe forms of COVID-19 in cancer patients. Clin. Cancer Res. 2021;27:P11. doi: 10.1158/1557-3265.COVID-19-21-P11. [DOI] [Google Scholar]
- Thierry A.R., Pastor B., Jiang Z.-Q., Katsiampoura A.D., Parseghian C., Loree J.M., Overman M.J., Sanchez C., Messaoudi S.E., Ychou M., Kopetz S. Circulating DNA demonstrates convergent evolution and common resistance mechanisms during treatment of colorectal cancer. Clin. Cancer Res. 2017;23:4578–4591. doi: 10.1158/1078-0432.CCR-17-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A.R., Roch B. Neutrophil extracellular traps and by-products play a key role in COVID-19: pathogenesis, risk factors, and therapy. J. Clin. Med. 2020;9:2942. doi: 10.3390/jcm9092942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M., et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C.F., Lourido S., Zychlinsky A. How do microbes evade neutrophil killing? Cell Microbiol. 2006;8:1687–1696. doi: 10.1111/j.1462-5822.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- Waisberg M., Molina-Cruz A., Mizurini D.M., Gera N., Sousa B.C., Ma D., Leal A.C., Gomes T., Kotsyfakis M., Ribeiro J.M.C., et al. Plasmodium falciparum infection induces expression of a mosquito salivary protein (agaphelin) that targets neutrophil function and inhibits thrombosis without impairing hemostasis. PLoS Pathog. 2014;10:e1004338. doi: 10.1371/journal.ppat.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., Pacey S., Baird R., Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang L., Su Y., Yue Z., Xing T., Zhao W., Zhao Q., Duan C., Huang C., Zhang D., et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018;7:3022–3030. doi: 10.1002/cam4.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Zhang Q., Li X., Su W., Li G., Ma T., Gao S., Lou J., Que R., Zheng L., et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol. Cancer Ther. 2019;18:196–203. doi: 10.1158/1535-7163.MCT-17-1298. [DOI] [PubMed] [Google Scholar]
- Wichmann D., Sperhake J.-P., Lütgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schröder A.S., et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolach O., Sellar R.S., Martinod K., Cherpokova D., McConkey M., Chappell R.J., Silver A.J., Adams D., Castellano C.A., Schneider R.K., et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci. Transl. Med. 2018;10:eaan8292. doi: 10.1126/scitranslmed.aan8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu Y., Shen M., Liu M., Wu S., Liang L., Huang F., Zhang C., Guo W., Liu T. Role of circulating free DNA in evaluating clinical tumor burden and predicting survival in Chinese metastatic colorectal cancer patients. BMC Cancer. 2020;20:1006. doi: 10.1186/s12885-020-07516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu Q., Zhang X., Liu X., Zhou B., Chen J., Huang D., Li J., Li H., Chen F., et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583:133–138. doi: 10.1038/s41586-020-2394-6. [DOI] [PubMed] [Google Scholar]
- Yipp B.G., Petri B., Salina D., Jenne C.N., Scott B.N.V., Zbytnuik L.D., Pittman K., Asaduzzaman M., Wu K., Meijndert H.C., et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012;18:1386–1393. doi: 10.1038/nm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Stojkov D., Germic N., Simon D., Wang X., Benarafa C., Simon H.-U. Untangling “NETosis” from NETs. Eur. J. Immunol. 2019;49:221–227. doi: 10.1002/eji.201747053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request
-
•
This paper does not report original code.
-
•
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.