Abstract
Streptococcus sanguinis is an oral commensal and an etiological agent of infective endocarditis. Previous studies have identified the SsaACB manganese transporter as essential for endocarditis virulence; however, the significance of SsaACB in the oral environment has never been examined. Here we report that a ΔssaACB deletion mutant of strain SK36 exhibits reduced growth and manganese uptake under acidic conditions. Further studies revealed that these deficits resulted from the decreased activity of TmpA, shown in the accompanying paper to function as a ZIP‐family manganese transporter. Transcriptomic analysis of fermentor‐grown cultures of SK36 WT and ΔssaACB strains identified pH‐dependent changes related to carbon catabolite repression in both strains, though their magnitude was generally greater in the mutant. In strain VMC66, which possesses a MntH transporter, loss of SsaACB did not significantly alter growth or cellular manganese levels under the same conditions. Interestingly, there were only modest differences between SK36 and its ΔssaACB mutant in competition with Streptococcus mutans in vitro and in a murine oral colonization model. Our results suggest that the heterogeneity of the oral environment may provide a rationale for the variety of manganese transporters found in S. sanguinis.
Keywords: carbon catabolite repression, dental caries, endocarditis, manganese, NRAMP protein, ZIP‐family protein
Streptococcus sanguinis is an oral bacterium that competes with Streptococcus mutans, a causative agent of dental caries. We report that growth of S. sanguinis SK36 in acidic media leads to decreased manganese levels and changes in carbon catabolite repression. A knockout mutant of the primary manganese transporter grows poorly in acidic media but shows similar colonization and competition with S. mutans in vivo when compared with wild type.

1. INTRODUCTION
The oral cavity is a complex environment with dynamic microbial communities, fluctuating metabolite availability, and diverse habitats (Aas et al., 2005; Chen et al., 2010; Jakubovics, 2015; Kolenbrander, 2011; Kolenbrander et al., 2010; Lamont et al., 2018; Marsh et al., 2016). Resident microorganisms must compete for space and nutrients within each of these habitats, including the dental biofilm known as plaque. Some species, such as Streptococcus mutans, utilize acid production to carve out their own niche by lowering the pH of the local environment. This allows competition with other less aciduric (acid‐tolerant) species but also results in demineralization of the tooth enamel, or dental caries (Gross et al., 2012; Moye et al., 2014; Nascimento et al., 2009).
One of the species S. mutans competes with is Streptococcus sanguinis. Although S. sanguinis is acidogenic (acid‐producing), it lacks the aciduricity of S. mutans (Bender et al., 1986; Diaz‐Garrido et al., 2020; Sasaki et al., 2018; Svensater et al., 1997). Conversely, S. sanguinis possesses the ability to produce (Garcia‐Mendoza et al., 1993; Kreth et al., 2008) and survive in (Xu et al., 2014) large quantities of hydrogen peroxide (H2O2), which is thought to enhance its ability to compete against S. mutans, which does not produce H2O2 (Kreth et al., 2005). Thus, S. sanguinis is typically found in much greater abundance at healthy oral sites, whereas S. mutans predominates in carious lesions (Belda‐Ferre et al., 2012; Giacaman et al., 2015; Gross et al., 2012).
Oral bacteria possess a variety of mechanisms for tolerating acid stress (Cotter & Hill, 2003; Guan & Liu, 2020; Liu et al., 2015; Papadimitriou et al., 2016; Quivey et al., 2001). The S. sanguinis genome (Xu et al., 2007) encodes multiple systems that are predicted to play a role: F0F1‐ATPases (Kuhnert & Quivey, 2003), an arginine deiminase system (Burne et al., 1989; Curran et al., 1995; Floderus et al., 1990), various chaperones and proteases (Lemos & Burne, 2002; Lemos et al., 2001; Shabayek & Spellerberg, 2017), and superoxide dismutase (Crump et al., 2014; Kim et al., 2005; Wen & Burne, 2004). S. sanguinis also appears to possess an acid tolerance response (ATR), whereby brief exposure to sub‐lethal acid levels protects against a lethal drop in pH (Cotter & Hill, 2003), although it is less effective than that in the related species Streptococcus gordonii (Cheng et al., 2018). Each of these systems likely contributes to the survival of S. sanguinis in acidic environments.
Manganese (Mn) has recently been implicated in acid stress tolerance in Streptococcus agalactiae (Shabayek et al., 2016) and S. mutans (Kajfasz et al., 2020). In both species, loss of a natural resistance‐associated macrophage protein (NRAMP) family manganese transporter, MntH, led to a reduction in growth in acidic conditions. In S. mutans, the loss of the lipoprotein component, SloC, of the ABC manganese transporter, SloABC, did not impact growth in acidic media unless the sloC mutation was combined with the mntH mutation (Kajfasz et al., 2020). Similarly in Streptococcus parasanguinis, which also possesses a mntH gene, it was previously determined that the homologous fimCBA operon was not important for acid tolerance (Chen et al., 2013). Manganese transport has also been examined in S. sanguinis strain SK36, which possesses an ABC transport system encoded by the ssaACB operon that is homologous to sloABC and fimCBA. However, like most other strains of S. sanguinis, SK36 does not possess a mntH gene. The S. sanguinis studies have established the importance of manganese not for acid tolerance, but for virulence in relation to the disease infective endocarditis (IE) (Baker et al., 2019; Crump et al., 2014). S. sanguinis is a frequent cause of this serious illness, which exhibits a 12%–30% mortality rate (Bor et al., 2013; Jamil et al., 2019; Wilson et al., 2021) and for which the only form of prevention—antibiotic prophylaxis—is controversial (Cahill, Baddour, et al., 2017; Cahill, Dayer, et al., 2017). This has led to the investigation of SsaB, the lipoprotein component of the ABC manganese transport system, as a target for endocarditis prevention. Given the association of S. sanguinis with health in the oral cavity, targeting of SsaB would be of greater value if it would not compromise oral competitiveness.
Here we report for the first time the growth of a naturally MntH‐deficient streptococcal strain, S. sanguinis SK36, under reduced‐pH conditions and for the first time in any bacterium, the effect of reduced pH on manganese transport by a newly described ZIP‐family transporter. We then assess the growth of a MntH‐encoding strain of S. sanguinis, VMC66, in reduced pH conditions. Finally, we report the effect of deleting the ABC manganese transporter on the competitive fitness of S. sanguinis against S. mutans in vitro and in vivo.
2. RESULTS
2.1. Growth of S. sanguinis at reduced pH
As a first step in determining whether manganese transport is important for growth at reduced pH in S. sanguinis, we employed a mutant of SK36 in which the ABC manganese transporter SsaACB has been deleted (Baker et al., 2019). We assessed the growth of wild‐type (WT) SK36 and its ΔssaACB single transport‐system mutant in brain heart infusion (BHI) broth at its unadjusted pH (~7.3) and under increasingly acidic conditions (Figure 1a). We have recently demonstrated that this mutant grows significantly less well than WT in a low‐manganese medium, serum, in 6% O2 but similarly in 1% O2 (Puccio, Kunka, et al., 2022). To rule out oxidative stress as a confounding factor, cells were grown in microaerobic conditions (1% O2). At pH 7.3, the culture densities of the two strains were not significantly different from each other. When the pH was lowered to 6.4 and 6.3, the final density of the WT culture was not significantly different from that at pH 7.3, whereas the final density of the ΔssaACB mutant progressively decreased. The final density of both cultures at pH 6.2 was significantly less than at pH 7.3, but the difference was much greater for the ΔssaACB mutant than for WT.
FIGURE 1.
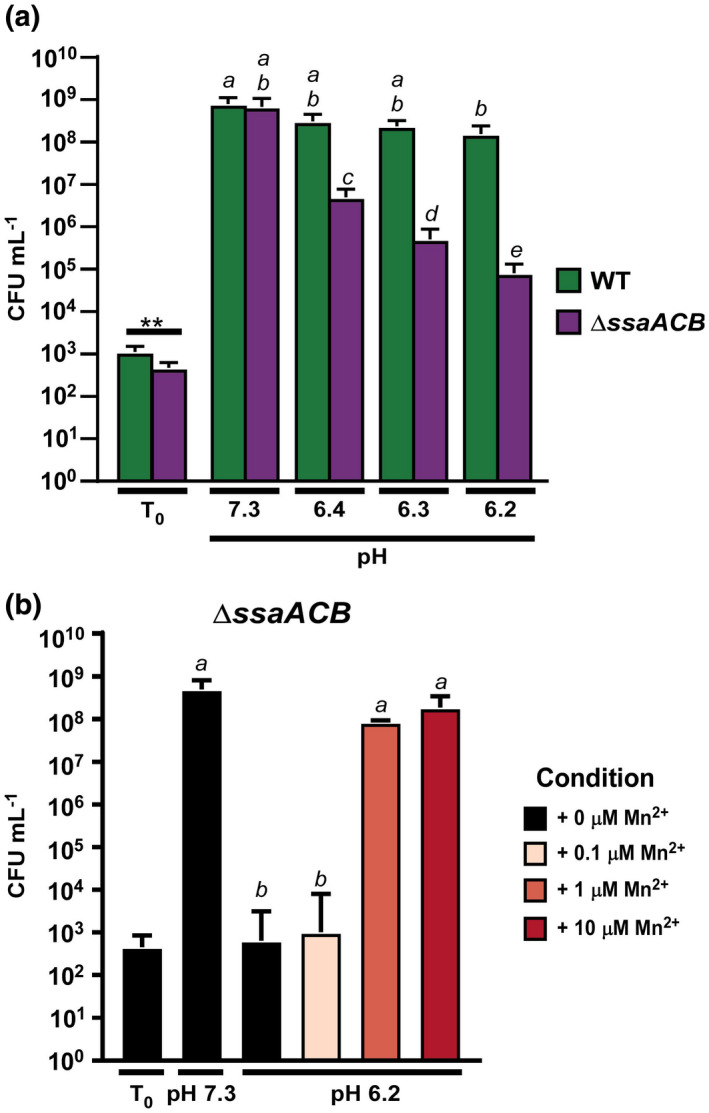
Growth of the Streptococcus sanguinis SK36 WT and a ΔssaACB mutant in reduced‐pH BHI. BHI at different pH levels was preincubated at 1% O2 and inoculated from an overnight culture. Cultures were incubated for 24 hr prior to plating. (a) Growth in BHI at various pH levels was assessed. (b) Exogenous Mn2+ was added to the ΔssaACB mutant at the listed concentrations. Means and standard deviations of at least three replicates are displayed. Significance was determined by unpaired two‐tailed t‐tests for T0 values and one‐way ANOVA with a Tukey multiple comparisons post‐test for T24 values. Bars that share a letter are not significantly different from each other (p > .05)
Although we suspected that the growth arrest of the ΔssaACB mutant at pH 6.2 was due to lower manganese levels in this strain (Murgas et al., 2020), we wanted to test this hypothesis directly. We previously measured the concentration of manganese in BHI from the same supplier as that used in the present study by inductively coupled plasma mass spectrometry (ICP‐MS) and found that it averaged 0.55 µM (Murgas et al., 2020). We added exogenous Mn2+ to pH 6.2 cultures and found that the addition of 1 µM Mn2+ was sufficient to rescue the growth of the ΔssaACB mutant (Figure 1b), suggesting that the reduced manganese levels found in this mutant (Murgas et al., 2020) were contributing to the reduced ability to tolerate low‐pH conditions.
2.2. Contribution of secondary manganese transporters to acid tolerance
The fact that we restored growth by addition of manganese to a strain deleted for the ΔssaACB genes encoding the major manganese transporter (Figure 1b) implies the existence of a second mechanism for manganese uptake, albeit one with lower affinity than the SsaACB transporter. In the accompanying manuscript (Puccio, Kunka, et al., 2022), we report the identification and characterization of a ZIP‐family protein, TmpA, as a secondary manganese transporter present in the majority of 40 fully sequenced S. sanguinis strains examined. Additionally, we noted the presence of a gene encoding a third manganese transporter from the NRAMP family (Nevo & Nelson, 2006), MntH, in four of these strains, including a strain called VMC66. There is no mntH gene, however, in SK36, which was the WT strain used in Figure 1. We hypothesized that each of the three S. sanguinis manganese transporters is at least partly redundant but may have optimal activity under differing conditions. In S. agalactiae, MntH functions best at acidic pH and its gene is expressed highly at pH 5 (Shabayek et al., 2016). The ZIP‐family protein in Escherichia coli was found to function best near neutral pH (Taudte & Grass, 2010) and we have yet to determine a condition where the tmpA gene in S. sanguinis is differentially expressed (Puccio, Kunka, et al., 2022). This is in contrast to SsaB, which was found to be repressed by SsaR, a MntR‐family regulator, in a Mn‐dependent manner, and is therefore expressed most highly under low‐manganese conditions (Crump et al., 2014; Puccio, Kunka, et al., 2022).
To determine whether TmpA contributes to the growth of SK36 WT or ΔssaACB mutant cells at normal or reduced pH, we assessed the growth of ΔtmpA mutants in both backgrounds in BHI pH 7.3 and 6.2 in an atmosphere of 1% O2 (Figure 2a). Growth of the ΔtmpA mutant was not affected at either pH, indicating that SsaACB transports sufficient manganese under both conditions to maintain growth. The double ΔssaACB ΔtmpA mutant grew poorly at both pH 7.3 and 6.2 and its growth was not significantly different from that of its ΔssaACB parent at pH 6.2. These results suggest that when cells are grown at pH 7.3 and 1% O2, an intact tmpA gene is necessary and sufficient to compensate for the deletion of the ΔssaACB genes, allowing for WT levels of growth. The results also suggest, however, that the tmpA gene makes no demonstrable contribution to growth at pH 6.2 under the same microaerophilic (1% O2) condition.
FIGURE 2.
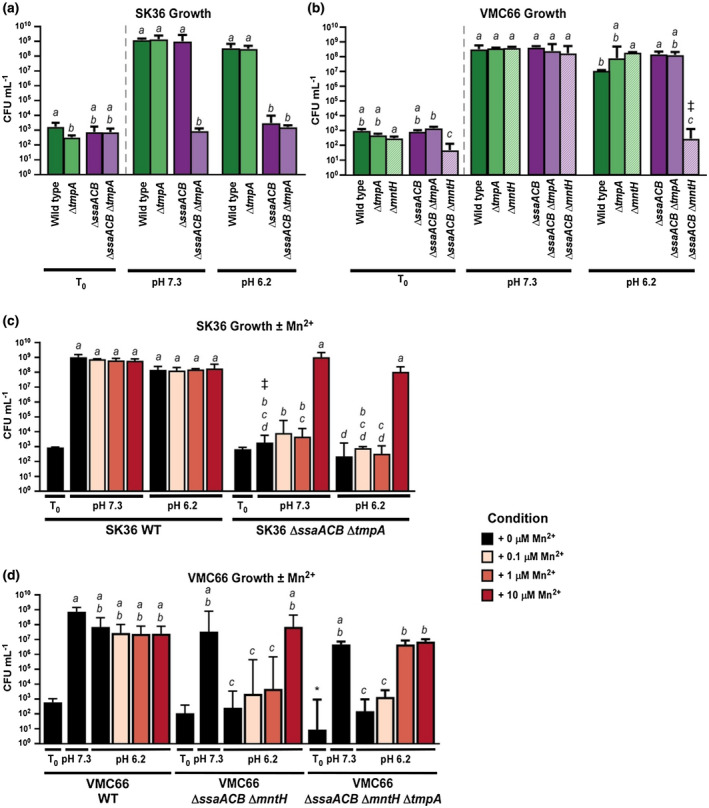
Effect of the loss of manganese transporters on growth of Streptococcus sanguinis SK36 and VMC66 strains in reduced‐pH BHI. Growth of manganese transporter mutants in the SK36 (a) and VMC66 (b) backgrounds was assessed in BHI at pH 7.3 and pH 6.2 in 1% O2 for 24 hr. Strains that grew poorly in (a) & (b), as well as the WT strains, were assessed for growth under the same conditions ± Mn2+ (c and d). Means and standard deviations of at least three replicates are displayed. Significance for T24 values was determined by one‐way ANOVA with a Tukey multiple comparisons post‐test. Bars that share a letter within a chart are not significantly different (p > .05). For (a), (b), & (d), significance for T0 values was determined by a separate one‐way ANOVA. For (c), significance for T0 values was determined by an unpaired t‐test. *p < .01 versus VMC66 WT T0. Bars indicated by ‡ had at least one value that fell below the limit of quantification
In the VMC66 background, we generated mutants deleted for each of the transporters: ΔtmpA, ΔmntH, and ΔssaACB, as well as double mutants lacking SsaACB and one of the secondary transporters: ΔssaACB ΔtmpA, and ΔssaACB ΔmntH. We then assessed their growth in BHI pH 7.3 and pH 6.2 at 1% O2 (Figure 2b). All strains grew identically at pH 7.3 but growth of the ΔssaACB ΔmntH strain was dramatically reduced at pH 6.2.
We also wanted to assess whether the decreased growth of the SK36 ΔssaACB ΔtmpA mutant and the VMC66 ΔssaACB ΔmntH mutant observed in Figure 2a,b was primarily due to decreased manganese levels in these mutants. Before proceeding, we also generated a third VMC66 mutant deleted for all three known manganese transporters. Growth of the SK36 mutant was again poor under both pH conditions. Addition of 10 µM Mn2+ restored growth to WT levels (Figure 2c). Growth of both of the VMC66 mutants was statistically indistinguishably from WT at pH 7.3, although the mean density of triple mutant cultures was more than 2 logs less than that of the WT strain. Addition of 10 µM Mn2+ to the double mutant at pH 6.2 restored growth to the level of VMC66 at pH 7.3, whereas this was not true for the triple mutant (Figure 2d).
We next wanted to determine how the reduced pH affected manganese levels in these strains. In our accompanying study (Puccio, Kunka, et al., 2022), we determined the average manganese levels of the SK36 strains in BHI (pH 7.3) using inductively coupled plasma optical emission spectroscopy (ICP‐OES), which are reproduced in Figure 3a. The levels of manganese were lower in these strains in BHI at pH 6.2 (Figure 3a); so much so that levels in both ΔssaACB and ΔssaACB ΔtmpA fell below the limit of quantification unless exogenous Mn2+ was added. When 10 µM Mn2+ was added to pH 7.3‐grown cells, manganese levels in the ΔssaACB mutant rose to a level at which they were no longer significantly different from WT; however, manganese levels in the ΔssaACB ΔtmpA mutant remained significantly lower than WT (Figure 3b). Interestingly, Mn2+ supplementation of cells grown at pH 6.2 raised manganese levels similarly in all four strains and, as with the unsupplemented cells in the ICP‐OES analysis (Figure 3a) or the cells in the growth study (Figure 2a), the ΔssaACB ΔtmpA mutant was indistinguishable from its ΔssaACB parent strain, and both were significantly lower than WT.
FIGURE 3.

Manganese content of Streptococcus sanguinis SK36 and VMC66 and their respective manganese transporter mutants. Manganese content of transporter mutants in the SK36 (a & b) and VMC66 (c & d) backgrounds in BHI (a & c) or BHI +10 µM Mn2+ (b & d) at pH 7.3 and pH 6.2 was assessed by ICP‐OES. Means and standard deviations of at least three replicates are displayed. Significance was determined by one‐way ANOVA with Tukey multiple comparisons post‐test. Bars that share a letter within a chart are not significantly different (p > .05). Bars indicated by ‡ had values that fell below the lowest standard. Results from pH 7.3 data were originally published in Puccio, Kunka, et al. (2022)
At this point in our study, we made two observations concerning SK36 and its mutant derivatives. First, regardless of the ΔssaACB or ΔtmpA genotype, all cells accumulated significantly less manganese when grown at pH 6.2 than at pH 7.3. This could be explained in various ways, including a general defect in transport at pH 6.2 or reduced bioavailability of Mn2+. Second, when cells are grown at pH 6.2, the ΔssaACB mutant and the ΔssaACB ΔtmpA double mutant are indistinguishable with regard to manganese content (Figure 3a) and growth under microaerophilic conditions (Figure 2a). The simplest explanation for this finding is that TmpA possesses no significant manganese‐transport activity at pH 6.2—a point we will return to. Interestingly, it cannot be concluded that TmpA has no influence at all on metal transport at pH 6.2 because the ΔssaACB ΔtmpA double mutant accumulated less iron than its ΔssaACB parent at this pH (Figure S1). Levels of the biologically relevant metals zinc (Zn) and magnesium (Mg) were not significantly affected by the lack of tmpA in either the WT or ΔssaACB background in BHI ± 10 µM Mn2+ (Figure S1). The ΔtmpA mutation also had no significant effect on any of these metals at pH 7.3 (Puccio, Kunka, et al., 2022). These results suggest that TmpA may have a role in iron uptake or homeostasis at pH 6.2 that was not observed at pH 7.3 (Puccio, Kunka, et al., 2022).
Manganese levels of VMC66 and its single and double mutants in BHI at pH 7.3 were also measured in Puccio, Kunka, et al. (2022) and are shown in Figure 3c,d, alongside measurements made with pH 6.2‐grown cells. Regardless of the pH or whether cultures were supplemented with 10 µM Mn2+ or not, the only strain that accumulated significantly less manganese in the same comparison group was the ΔssaACB ΔmntH mutant. The loss of TmpA did not impact manganese levels in any condition. Levels of iron, zinc, and magnesium were not significantly affected by deletion of the tmpA or mntH gene in either the WT or ΔssaACB background at pH 7.3 (Puccio, Kunka, et al., 2022) or pH 6.2 (Figure S1b,d,f). These results will be examined in relation to our observations of SK36 and its derivatives in the Discussion.
2.3. Effect of pH reduction on SK36 WT and ΔssaACB growth and metal content in a fermentor
We next examined the effects of reduced pH on the metal content and transcriptome of SK36 WT and ΔssaACB when grown in a fermentor (Figure S2). To minimize the impact of oxidative stress, we used the minimum possible airflow (0.03 liters per minute) throughout the experiment. We were unable to turn off the air entirely because cultures without airflow grew poorly (data not shown). Cells were grown at pH 7.4 (pH of human blood) until the OD peaked, and then for 1 hr with a media flow rate of 700 ml/h before collection of the T‐20 min sample for RNA‐seq. Twenty minutes later (T0), the pH was changed to 6.2 by addition of 2 N HCl and maintained at this pH by the addition of 2 N KOH in response to the signal from an indwelling pH probe. A pH of 6.2 was chosen because it did not affect WT (Figure S2a) but led to a decrease in growth rate of the ΔssaACB mutant as evidenced by the drop in OD (Figure S2b). Under constant‐flow conditions, a drop in OD is expected when a culture's doubling rate drops below its dilution rate (Burne & Chen, 1998). Samples at pH 6.2 were removed at T10, T25, and T50 min and RNA was isolated for RNA‐seq analysis.
However, before examination of the RNA‐seq data, we wanted to first establish the effects of reduced pH in the fermentor on cellular levels of not only manganese, but of also iron, zinc, and magnesium. We assessed the metal content of cells at each time point using ICP‐OES (Figure 4). Manganese levels significantly decreased in both strains, further confirming that reduced pH conditions are not conducive to manganese transport. Confirming previous results (Crump et al., 2014; Puccio et al., 2020), the ΔssaACB mutant contained low levels of iron (Figure 4b), further confirming that this system transports both metals. Interestingly, iron levels appeared to increase in both strains after acid addition, although the increase was not significant in SK36 WT. Magnesium increased significantly at T50 only in the ΔssaACB mutant strain. Zinc levels were not affected.
FIGURE 4.

Metal content of fermentor‐grown Streptococcus sanguinis SK36 WT and ΔssaACB mutant cells before and after pH reduction. Fermentor‐grown (a) WT and (b) ΔssaACB cells were collected at each time point and analyzed for cellular metal content using ICP‐OES. Metal concentrations were normalized to protein concentrations. Means and standard deviations of at least three replicates are displayed. Significance for each metal was determined by one‐way ANOVA with Dunnett's multiple comparisons tests with each pH 6.2 time point compared to T‐20 (pH 7.4). *p ≤ .05, *** p ≤ .001
To determine if reduced manganese uptake was the primary cause of the growth deficiency after pH reduction in the fermentor as it appeared to be in tube cultures, we added Mn2+ to a final concentration of 10 µM at T70 in additional fermentor runs (Figure S3). As the WT strain was not deficient in growth at pH 6.2, it was not surprising that Mn2+ addition had no effect (Figure S3a). The addition of Mn2+ led to a recovery in the growth rate of the ΔssaACB mutant as expected (Figure S3c). When metal analysis was performed on these runs, it was observed that manganese levels 10 min after Mn2+ addition (T80) increased ~4.5‐fold in WT (Figure S3b) and ~2.5‐fold in the ΔssaACB mutant (Figure S3d). These results suggest that ΔssaACB mutant cells were still viable at the latest time point (T50), despite the drop in OD, since they were capable of resuming metal uptake and growth 20 min after the addition of Mn2+.
2.4. RNA‐seq analysis of fermentor‐grown SK36 WT and ΔssaACB cells before and after pH reduction
In an attempt to determine the cause of the growth arrest of the ΔssaACB mutant, we examined the transcriptome of fermentor‐grown WT and ΔssaACB cells taken at the same time points as described above. We examined the results of the RNA‐seq analysis using three comparisons: within‐strain comparisons of each pH 6.2 time point to pH 7.4 (T‐20) for (1) WT and (2) the ΔssaACB mutant (Tables S1 and S2), and (3) comparison of the WT and ΔssaACB mutant strains at each time point (Table S3). The number of differentially expressed genes (DEGs; defined as p ≤ .05, |log2 fold change| ≥ 1) for each comparison is listed in Figure 5a. Using principal component analysis (PCA), we were able to demonstrate that each sample grouped with the others from the same time point, indicating that the results were reproducible and had minimal variability (Figure S4a,b). The 95% confidence intervals for the pH 6.2 sample time points of the WT strain overlapped with each other (Figure S4a), whereas those for ΔssaACB did not (Figure S4b). The pH 7.4 samples were well separated from the pH 6.2 samples in both strains. These results suggest that both strains were affected by the drop in pH, but the ΔssaACB mutant experienced a more clearly defined, time‐dependent program of expression changes compared to WT. When all samples were examined together (Figure S4c), the sample groups for both strains overlapped one another at each time point, although the WT T50 group overlapped more with the ΔssaACB T25 samples than those of T50 (Figure S4c). This result again suggests greater separation in expression at each time point in the ΔssaACB mutant compared to WT. The combined results correspond with the volcano plots (Figure S5) and heatmaps (Figure S6). Most of the expression changes occurred at the T50 time point for both strains, although there were fewer changes in WT than the ΔssaACB mutant. When comparing the strains to each other, the T10 and T25 time points were almost identical, whereas T‐20 and T50 time points had more variation (Figure 5a).
FIGURE 5.

Pathway enrichment analysis of the transcriptome of fermentor‐grown Streptococcus sanguinis SK36 WT and ΔssaACB mutant cells after pH reduction. (a) Tallies of DEGs, which are defined as p ≤ .05 and |log2 fold change| ≥ 1. Values in blue indicate the number of genes downregulated at that time point relative to T‐20; red values indicate those that were upregulated. Green values indicate the number of genes that were more highly expressed in WT and purple values indicate the number of genes that were more highly expressed in the ΔssaACB mutant. Pathway enrichment analysis using DAVID functional classification with KEGG annotations of DEGs at T50 compared to T‐20 for (b) WT and (c) ΔssaACB strains
To begin to assess the cause of the growth arrest resulting from addition of acid, we evaluated the expression of stress response genes. As expected, the expression of genes encoding an alkaline stress protein (SSA_RS10520), exinulcease subunit A (uvrA), and all subunits of the F0F1‐Type ATP synthase (Kuhnert & Quivey, 2003) significantly increased in the ΔssaACB mutant. Most other putative stress response genes were decreased or unchanged (Table S4). Notably, expression of the gene encoding superoxide dismutase, sodA, was slightly decreased. Although not unprecedented (Santi et al., 2009), there is a strong connection between SodA and acid stress response in other bacteria (Bruno‐Barcena et al., 2010; Kim et al., 2005). In our previous study (Puccio et al., 2020), we demonstrated that sodA expression is manganese dependent. We thus considered that the unusual decrease in sodA expression might have contributed to the growth arrest of ΔssaACB cells. We therefore tested the growth of a previously generated ΔsodA mutant (Crump et al., 2014) in the fermentor under identical conditions, but growth was not affected (data not shown). Apart from sodA, stress response genes shown in Table S4 had similar expression patterns in both strains (Table S4), although changes were often of slightly lower magnitude in the ΔssaACB mutant.
In examining the KEGG pathways assigned to the DEGs at T50, genes involved in phosphotransferase systems (PTS), biosynthesis of amino acids (specifically arginine and histidine), and oxidative phosphorylation were significantly affected by pH reduction in both strains grown in the fermentor (Figure 5b,c). The most highly enriched pathway for the ΔssaACB mutant was the “Porphyrin/Chlorophyll” pathway but upon examination, these genes encode the cobalamin biosynthetic enzymes (Table S2), which were also significantly downregulated in our recent study of manganese depletion (Puccio et al., 2020). Many of the pathways highlighted by this analysis, such as carbon metabolism and amino acid metabolism, were also affected by manganese reduction in our recent transcriptomic and metabolomic studies (Puccio et al., 2020; Puccio, Misra, et al., 2021).
We observed significant decreases in the expression of sugar transport genes (Table S5). This led us to evaluate whether acidic conditions could also lead to glucose‐independent CcpA‐mediated carbon catabolite repression. We examined expression of all genes found to be within the CcpA‐regulon (Bai et al., 2019) as well as those that we identified that have putative upstream cre sites (Puccio et al., 2020). We found that 75 of the 382 genes with putative cre sites (19.6%) were significantly downregulated in the ΔssaACB mutant at T50, whereas 30 were upregulated (7.9%) (Table S6). In WT, 47 (11.7%) were downregulated and 13 (3.4%) were upregulated. In our manganese‐depletion transcriptomic study (Puccio et al., 2020), we found that 19.9% of genes with putative cre sites were downregulated at T50, although the genes downregulated in the ΔssaACB mutant under both conditions do not match precisely. For example, spxB was significantly upregulated in reduced pH (Table S6) but significantly downregulated in reduced manganese (Puccio et al., 2020). This indicates that although there may be some overlap due to the decrease in manganese levels in both studies, there are also changes that are specific to pH reduction.
As acid stress tolerance has been previously linked to amino acid biosynthesis and transport (Djoko et al., 2017; Guan & Liu, 2020; Quivey et al., 2001; Senouci‐Rezkallah et al., 2011), we evaluated the expression of genes annotated with these functions (Table S7). Many of these genes were significantly affected by acid addition, although more were affected in the ΔssaACB mutant. Of note, aconitate hydratase, citrate synthase, and isocitrate dehydrogenase were significantly upregulated in the ΔssaACB mutant but significantly downregulated in the WT strain at T50 (Table S7).
2.5. Biofilm competition of the S. sanguinis SK36 WT and ΔssaACB strains against S. mutans in vitro
We next assessed whether the lack of the primary manganese transporter, ΔssaACB, would affect growth when in competition with S. mutans, similar to conditions found in the oral cavity. We therefore compared the growth of a spectinomycin (Spc)‐resistant SK36 WT strain, JFP56, to the ΔssaACB mutant in competition with S. mutans (Figure 6). Given that S. mutans often outcompetes WT S. sanguinis if given the opportunity to colonize first (Kreth et al., 2005), we inoculated wells of biofilm media (BM) + 1% sucrose (Loo et al., 2000) with S. sanguinis to stimulate robust biofilm formation and allowed 24 hr of aerobic growth. BM contains 100 µM Mn and 2 mM Mg (Loo et al., 2000). As S. mutans does not grow well in BM, we then refreshed the wells with Tryptone Yeast Extract + 1% sucrose (TY+S) and inoculated with an S. mutans strain containing a plasmid‐encoded erythromycin (Erm) resistance gene. Levels of metals in TY+S were measured and determined to be approximately 0.35 µM manganese, 11 µM iron, and 29 µM zinc. After another 24 hr incubation, we then measured the pH of the media and plated the biofilm cultures. S. mutans growth was unaffected by the presence of S. sanguinis (Figure 6a), whereas both S. sanguinis strains were recovered in significantly lower numbers in the presence of S. mutans (Figure 6b). In fact, only one of six replicates of the ΔssaACB mutant produced a single colony after incubation in the presence of S. mutans. The pH of all culture supernatants after 24 hr growth in TY+S was significantly lower than media alone, with the S. mutans supernatant averaging pH 4.08 and the S. sanguinis WT and ΔssaACB supernatants averaging pH 4.50 and 4.58, respectively (Figure 6c).
FIGURE 6.

Competition of Streptococcus sanguinis SK36 WT and ΔssaACB strains against S. mutans in vitro. S. sanguinis cultures were grown aerobically for 24 hr in BM+S. Media was swapped for TY+S and S. mutans was inoculated. Cultures were incubated an additional 24 hr before media was removed and cells scraped from wells were resuspended in PBS for plating on BHI agar plates with antibiotics for selection. Means and standard deviations of S. mutans (a) and S. sanguinis (b) CFU/ml from three biological replicates with two technical replicates each are shown. The ‡ indicates that 5 of 6 replicates fell below the limit of detection. (c) Means and standard deviation of media pH measurements for each culture are shown. Statistical analysis was performed using one‐way ANOVA with a Tukey multiple comparisons post‐test. Bars with the same letter within a chart are not significantly different (p > .05)
2.6. Contribution of SsaACB to oral colonization and competition in a murine model
Given the results of the in vitro competition assay, we used an in vivo model to compare colonization of the oral cavity and dental biofilms by the SK36 WT and ΔssaACB mutant strains, and once colonization was established, we then compared the ability of each strain to compete against S. mutans under cariogenic conditions. We employed the mouse model described recently by Culp et al. (2020) with mice fed a diet containing 37.5% sucrose. After antibiotic suppression of the oral microbiota, each of two groups of mice received five daily oral inoculations with one of the two S. sanguinis test strains, followed two weeks later by challenge with S. mutans. Oral swabs were taken at weekly intervals, thus providing a measure of colonization within saliva and the oral mucosal pellicle, with subsequent sonication of mandibular molars to assess bacteria recovered from dental biofilms (see timeline of events, Figure 7a). Recoveries of bacteria released from swabs and molar sonicates were determined by strain‐specific qPCR assays of genomic DNA, plus estimates of total recovered bacteria were determined by qPCR assay of the ubiquitous single copy gene, rpsL (30S ribosomal protein S12). Subtraction of strain‐specific recoveries from total recovered bacteria allowed estimations of the population of recovered murine oral commensal bacteria. We observed that both S. sanguinis strains colonized well in competition with native mouse commensals (Figure 7b). Upon introduction of S. mutans, the abundance of the two test strains and the mouse commensals declined and then recovered by the following week. When levels of the two test strains were compared for each time point, we found that the ΔssaACB mutant was recovered from the molars at slightly, yet significantly lower levels than WT 14 days after the introduction of S. mutans (experimental day 28) (Figure 7c). The mouse commensal species present in each animal group were also compared to one another at each time point and were only found to be significantly different on the molars of mice inoculated with the ΔssaACB mutant (Figure 7c). S. mutans levels were slightly but not significantly higher in competition with SK36 WT at day 27 (swab 5) than in competition with the ΔssaACB mutant on the same day (Figure 7b). These results suggest that SsaACB had no impact on oral colonization and was of only minor importance for dental colonization and competition, even in a strain that did not possess the mntH gene. This is in stark contrast to the results obtained from our in vivo endocarditis model showing that the SsaACB transporter is essential for virulence, even in strains possessing TmpA and MntH transporters (Puccio, Kunka, et al., 2022).
FIGURE 7.
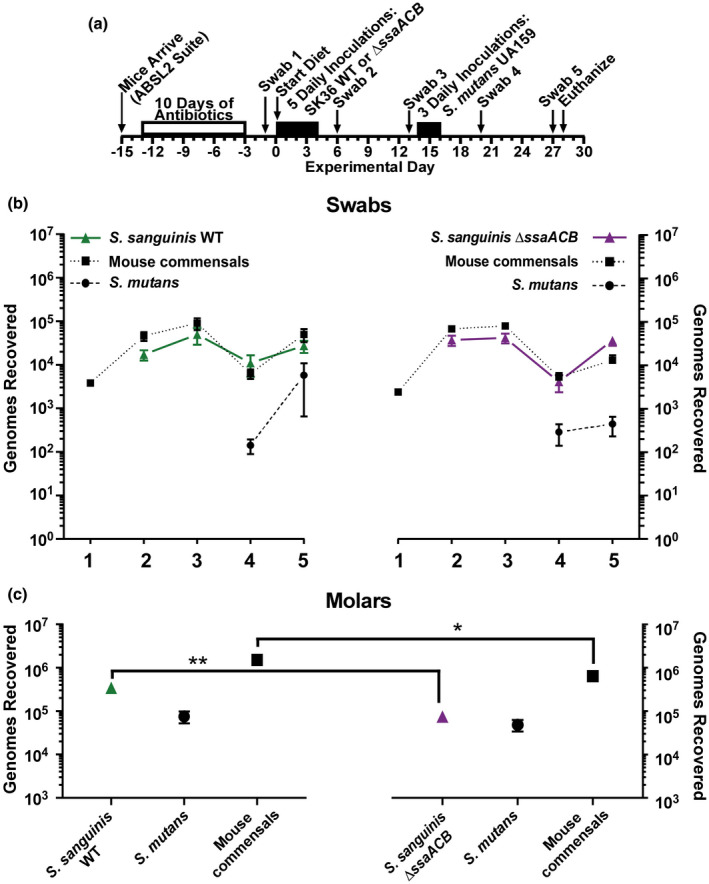
Comparison of oral colonization by Streptococcus sanguinis SK36 WT and ΔssaACB strains, and in subsequent competition with S. mutans UA159 in vivo. (a) Timeline of key events in the experiment. Colonization results for each indicated inoculated S. sanguinis strain (triangles, solid lines), S. mutans UA159 (circles, dashed lines) and mouse oral commensals (squares, dotted lines) from oral swabs 1–5 (b) and from sonicates of mandibular molars (c). S. sanguinis SK36 WT is green (left) and the ΔssaACB mutant is purple (right). Means and standard error (n = 12 mice per cohort) of recovered genomes estimated by qPCR are shown. All samples of each species or group (for commensals) were compared using one‐way ANOVA. Bonferroni's multiple comparisons test was then applied to compare the samples from the WT group to their counterparts in the ΔssaACB group collected at the same time point (*p < .05, **p < .01). There were no significant differences among the swab samples (p > .05). Mice were fed a high‐sucrose powdered diet with sterile drinking water
3. DISCUSSION
Although a relationship between manganese and acid tolerance in streptococci has been suggested previously (Kajfasz et al., 2020; Shabayek & Spellerberg, 2017), it has yet to be fully characterized. Beighton (1982) noted that manganese induced S. mutans to form caries and influenced carbohydrate metabolism. In Streptococcus pneumoniae (Martin‐Galiano et al., 2005) and S. agalactiae (Santi et al., 2009), the orthologs of SsaACB were upregulated in response to acid stress. In S. mutans, expression of the manganese‐dependent regulator SloR was previously linked to ATR (Dunning et al., 2008). Recently, it was appreciated that loss of MntH in S. mutans (Kajfasz et al., 2020) and S. agalactiae (Shabayek et al., 2016) led to a reduction in acid tolerance. Here we report that loss of the high‐affinity manganese ABC transporter in S. sanguinis strain SK36, SsaACB, reduced acid tolerance but did not greatly influence oral colonization or competition with S. mutans. We also characterized the transcriptional changes that accompany the growth arrest of the ΔssaACB mutant observed in reduced pH conditions in vitro and in comparisons with WT.
Initially, we were unsure whether the growth arrest we observed in the ΔssaACB mutant was directly related to manganese, since we have shown previously that the SsaACB system transports both manganese and iron (Crump et al., 2014; Murgas et al., 2020). However, our findings clearly suggest that it is a lack of manganese rather than iron (or any other metal) that is responsible for the growth arrest observed at pH 6.2 in both static broth cultures and the fermentor. First, the ΔssaACB mutant accumulated significantly less manganese than SK36 WT in tube cultures at pH 6.2 (Figure 3), which was not true for iron, zinc, or magnesium (Figure S1). Second, the ΔssaACB mutant accumulated significantly less manganese when the pH of fermentor cultures was reduced from 7.4 to 6.2. In contrast, levels of iron, zinc, and magnesium either remained constant or rose. Third, the addition of 10 µM Mn2+ restored the growth of the ΔssaACB mutant to WT‐like levels in pH 6.2 tube cultures (Figures 1 and 2) and in the fermentor (Figure S3). Furthermore, many transcriptomic responses to pH reduction were similar to those of manganese reduction by EDTA (Puccio, Kunka, et al., 2022).
Although a drop in pH to 6.2 represents a modest decrease, it negatively affected manganese uptake by at least two mechanisms. Figure 3 revealed that at pH 6.2, SK36 WT and the ΔssaACB, ΔtmpA, and ΔssaACB ΔtmpA mutants all accumulated less manganese than they did at pH 7.3. The simplest explanation for this finding may be decreased bioavailability of manganese at this pH, although we have not identified a specific mechanism for this possibility. We also note that some other explanations such as reduced cellular health resulting in a decrease in overall transport are excluded by our finding in the fermentor studies that the shift to pH 6.2 did not result in decreased accumulation of any metal except manganese (Figure 4). As mentioned earlier, growth at pH 6.2 also appears to virtually eliminate the manganese‐transporting activity of TmpA. Figure 3 showed that manganese levels in the ΔssaACB ΔtmpA mutant were indistinguishable from those in the ΔssaACB mutant at pH 6.2. This was also true in the samples with 10 µM Mn2+, indicating that this equivalence is not merely due to the decrease in manganese uptake observed in all of the strains. Our finding that the expression of the tmpA gene increased slightly (though insignificantly) after the shift to pH 6.2 in the fermentor (see Table S8) rules out the possibility that the lack of detectable manganese transport activity at pH 6.2 is due to decreased transcription of the tmpA gene at the lower pH. These results correspond to the activity of an ortholog of TmpA in E. coli, ZupT, which functions best near neutral pH (Taudte & Grass, 2010).
At first glance, VMC66 appears quite different from SK36 with regard to manganese transport at pH 6.2. Notably, the reduced uptake of manganese at pH 6.2 seen with all of the SK36 derivatives is not evident; none of the strains examined took up significantly less manganese at pH 6.2 than at pH 7.3 (Figure 3). Also, deletion of tmpA from the ΔssaACB mutant had no significant effect on growth at either pH. These apparent differences are readily explained by the presence of MntH in VMC66. Figure 3 showed that although the differences do not rise to the level of significance, mean manganese levels in the mntH mutant were lower than in WT cells at pH 6.2, which is not true of even the ΔssaACB mutant, suggesting that MntH is a better transporter of manganese at this pH than SsaACB. We hypothesize that this increased activity is sufficient to compensate for any decrease in manganese transport at this pH due to decreased bioavailability. As for TmpA, we observed no activity at pH 6.2, as in the SK36 background. The triple mutant grew similarly to its parent at pH 6.2, which was expected, but this was also true at 7.3 (Figure 2d). This result is somewhat misleading, however, in that reliable growth of the overnight inoculum of the triple mutant at pH 7.3 often required addition of 15 μM Mn2+, which was more than any other strain, and/or required a longer incubation time, pointing to a hidden growth defect. Taken together, these results strongly support the hypothesis that reductions in growth and manganese levels of the SK36 ΔssaACB mutant at pH 6.2 are due to the reduced function of TmpA (Figure 8). In the VMC66 background, our results suggest that TmpA is also inactive at pH 6.2 and active at pH 7.3, although in a ΔssaACB mutant, MntH fully compensates for the loss of TmpA activity at pH 6.2 and can compensate for loss of the tmpA gene at pH 7.3.
FIGURE 8.
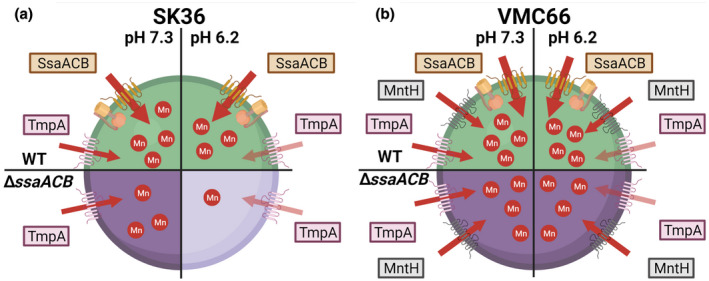
Summary of the role of manganese transporters in the growth of Streptococcus sanguinis in reduced pH conditions. Diagrams of S. sanguinis WT (a) SK36 and (b) VMC66 strains at pH 7.3 (left) and pH 6.2 (right) with their known manganese transporters depicted. In both WT cells (top), all transporters are present, so although TmpA function is reduced at pH 6.2, manganese enters the cell in sufficient quantities to support growth. In SK36 ΔssaACB mutant cells (bottom), manganese uptake solely via TmpA is insufficient to meet cellular needs, leading to reduced growth. In VMC66 ΔssaACB mutant cells (bottom), MntH compensates for the reduced function of TmpA at pH 6.2, resulting in sufficient manganese levels to support growth
The ability of manganese to rescue the growth of mutants lacking all known manganese transporters (SsaACB, TmpA, and in the case of VMC66, MntH; Figure 2) implies the existence of at least one additional manganese transporter in each strain. We have no data concerning the identity or nature of the alternate transporter in either strain, although given the concentrations of manganese required to support growth of the mutant strains compared to that of the WT strains, it seems unlikely that manganese uptake via these transporters is biologically relevant in WT strains.
Due to the short time frame of our transcriptional analysis (50 min), we believe that the changes observed are an acid shock response, as opposed to acid adaptation (Quivey et al., 2001). However, acid shock responses have typically employed a pH of 3.0 to 4.4 (Hamilton & Svensäter, 1998; Martin‐Galiano et al., 2005) so given that the growth rate of WT is unchanged and that we used a modest reduction in pH (6.2), it is likely a relatively mild response. When comparing the reduced‐pH RNA‐seq results in each strain directly to the other, few DEGs were observed and those that were seen occurred only at the T‐20 and T50 time points (Figure 5a). Despite these results, we observed a drastic difference in the fermentor growth rate between WT and ΔssaACB after acid addition (Figure S2). Thus, we hypothesize that either subtle changes between strains have a large combined effect or post‐transcriptional changes not captured by transcriptomics that may be influencing growth. It is also possible that both explanations are correct. One reason for the changes being small could be that WT and ΔssaACB are both experiencing two, typically separate stresses—acid stress and manganese depletion—although SK36 is experiencing both to a lesser degree. Examination of the manganese levels of WT cells in unsupplemented batch cultures at pH 7.3 versus 6.2 (Figure 3a) and fermentor cultures before and after acid addition (Figure 4a) suggests that even with SsaACB intact, WT cells take up less manganese at pH 6.2 than 7.3. This explanation is also supported by the increase in expression of ssaB, ssaC, and ssaA (SSA_RS01450‐01460) after acid addition in the fermentor (Table S1). However, it is also clear that both strains exhibit expression changes expected in response to reduced pH, as discussed previously. It would not be surprising if the effects of the two stresses were additive or synergistic with regard to growth inhibition. Thus, expression changes would be spread over two stress responses rather than one, further diminishing the importance or magnitude of any one expression difference.
We recently reported that manganese depletion led to induction of carbon catabolite repression (CCR) in S. sanguinis (Puccio et al., 2020; Puccio, Misra, et al., 2021). Given that both addition of acid and EDTA (Puccio, Misra, et al., 2021) led to reduced manganese levels in the fermentor (Figure 4), we hypothesize that the impact on glycolysis is similar to what we observed in our previous study. Additionally, expression of genes encoding glycolytic enzymes responded similarly in both studies (Table S2), indicating that the regulatory mechanisms controlling the expression of these genes may have a manganese‐related aspect. Thus, we hypothesize that there is likely an accumulation of fructose‐1,6‐bisphosphate (FBP) occurring in cells growing at reduced pH due to reduced activity of Fba, Fbp, or both enzymes (Puccio, Misra, et al., 2021). This may be the cause behind the changes in expression we observed in the CcpA regulon (Table S6). As noted by Radin et al. (2016), glycolytic enzymes may have a higher demand for manganese than other enzymes and thus in manganese‐deplete conditions, the cells may shift to amino acid metabolism as a source of energy. This, along with the ability of some amino acids and their intermediates, such as alpha‐ketoglutarate, to act as reactive oxygen species scavengers (Fedotcheva et al., 2006; Reinoso et al., 2012), could explain why we observed significant increases in expression of many amino acid biosynthetic and transport genes (Table S7). However, the impact of PTS, CcpA, and central carbon metabolism on gene expression are complex, and it is likely that multiple factors impact the expression of CCR‐responsive genes.
We were also interested in assessing putative metal transporters (Table S8), comparing their levels of expression to the levels of their cognate metals in fermentor‐grown cells (Figure 4). Beginning with manganese, Figure 4 showed that its level dropped in WT and ΔssaACB mutant cells upon shift to pH 6.2 and continued to drop at each time point thereafter. However, the ssaACB genes that encode the chief manganese transporter in SK36 WT showed an increase in transcript levels at each time point, which is consistent with expectations if the decrease in manganese levels was due to a decrease in manganese transporting activity of SsaACB and TmpA, and transcript levels reflect decreased activity of the manganese‐dependent repressor, SsaR (Crump et al., 2014; Murgas et al., 2020). There was no other putative manganese transporter or transport system that shares this pattern of regulation (Table S8).
In contrast to manganese, mean iron levels increased in both strains in the fermentor after pH reduction, although this increase was not significant in WT cells (Figure 4). Also, before the shift in pH, iron levels were much higher in SK36 than in the ΔssaACB mutant. The latter result is consistent with the expectation that the SsaACB system is the main transporter of iron in BHI at its normal pH. The modest increase in iron levels in SK36 at pH 6.2 could be due to increased ssaACB expression levels, but is it? In batch cultures, neither the ΔssaACB mutant nor the ΔtmpA mutant showed reduced iron levels at pH 6.2, but the double ΔssaACB ΔtmpA mutant accumulated significantly less iron. This would suggest that both transporters possess iron‐transport activity at pH 6.2. The finding of a significant increase in iron levels at the 25 min time point in the ΔssaACB mutant could reflect the increased bioavailability of iron that is known to occur under acidic conditions (Ilbert & Bonnefoy, 2013). As for the other putative iron transporters in Table S8, most were expressed at low levels at pH 7.3 (relative to ssaACB), and at much higher levels at T50 than at the other time points. This pattern bears no relationship, either positive or negative, to iron levels, in contrast to expectation for proteins possessing appreciable iron uptake activity or whose genes are downregulated in response to iron, respectively. This was not the case with zinc and magnesium. Zinc levels remained constant in both strains. Every putative zinc uptake transporter was expressed more highly at T50 than at the T10 or T25 time points, whereas the gene encoding the CzdD zinc and cadmium exporter (Martin & Giedroc, 2016; Nies, 1992) displayed the opposite pattern. Mean levels of cellular magnesium increased at each time point in both strains, although only the change in the ΔssaACB mutant was significant. This could be due to expression changes in genes such as SSA_RS03540 (Table S8), which encodes a putative magnesium‐transport protein, CorA (Kehres et al., 1998). This gene was expressed more highly in WT cells than in the ΔssaACB mutant at pH 7.4, but its expression increased more in the mutant, which correlates with magnesium levels in both strains. Further studies will be required to determine which if any of these proteins are responsible for the cellular metal levels observed in these studies and the role of pH in their regulation.
Acid is an important component of the competition between S. sanguinis and S. mutans in vitro (Kreth et al., 2005). Although not significant, the difference in the mean levels of SK36 WT and ΔssaACB mutant growth in direct competition with S. mutans in in vitro biofilms was appreciable, especially given that the ΔssaACB mutant was only recovered in one out of six replicates. Our in vivo study yielded no significant differences in recoveries from oral swabs, a measure of colonization of the pellicle overlaying oral mucosal epithelia in addition to any cells within adsorbed saliva (Culp et al., 2020), and only slight, yet significant differences between the SK36 WT and ΔssaACB mutant strains in colonization of dental biofilms when in competition with S. mutans. It was recently shown using the same mouse model that clinical isolates of streptococci from dental plaque of caries‐free individuals (i.e., Streptococcus mitis, Streptococcus gordonii, Streptococcus A12), which in vitro were highly competitive against S. mutans, poorly colonized oral mucosal and dental biofilms, whereas two strains of S. sanguinis suppressed initial mucosal colonization by S. mutans and persisted in both biofilms, even as S. mutans recovered to levels observed in a control group of mice challenged with S. mutans alone (Culp et al., 2020). Moreover, both strains of S. sanguinis promoted dental colonization of mouse commensals to levels equivalent or higher than S. mutans, similar to the SK36 WT and ΔssaACB mutant strains in the current study. Additionally, oral colonization of SK36 WT and its ΔssaACB mutant quickly recovered after being initially suppressed when challenged with S. mutans, suggesting that persistent colonization of mucosal biofilms may serve as a reservoir for colonization of dental biofilms. In clinical studies, S. sanguinis was shown to precede and delay colonization by S. mutans of newly acquired teeth in children (Caufield et al., 2000) and to persist in cavitated lesions in which S. mutans accounted for up to 55% of the microbiota (Gross et al., 2012). Collective evidence therefore indicates S. sanguinis is well adapted to the oral cavity and dental biofilms in which it copes with innate immune factors in saliva, interacts with other oral commensals either competitively or cooperatively, and importantly can coexist with S. mutans under cariogenic conditions. Although the SsaACB transport system appears to be universally conserved in S. sanguinis strains, its importance was not demonstrable under in vivo conditions. Future studies testing knockout mutants of multiple manganese transporters in several strains may further uncover the role of manganese and each transporter family in S. sanguinis in the oral cavity.
We propose a few possible explanations for the in vivo results. One is that SsaACB was not necessary because oral biofilms on epithelial and dental surfaces contained sufficient manganese to allow comparable growth of the WT and ΔssaACB strains using only the TmpA transporter over the course of this experiment. Salivary proteins in humans and rodents are known to be multifunctional, binding bacteria and calcium (Lamkin & Oppenheim, 1993; Wong & Bennick, 1980). It is thus possible that salivary proteins in addition to colonizing commensals may contribute to provide sufficient levels of manganese within biofilms on epithelial and dental surfaces. As described above from the work of Radin et al. (2016), a shift to amino acid metabolism as a source of energy may have lessened the demand for manganese under conditions of manganese depletion. It has been shown that interactions of oral commensal species and S. mutans with surface‐associated salivary constituents induces a more protease‐active phenotype capable of more effectively degrading salivary proteins and mucin glycoproteins (Kindblom et al., 2012; Wickström et al., 2013), thus potentially providing a reliable source of amino acids for uptake and metabolism. Another possibility is that perhaps there is some aspect of the oral cavity environment that reduces the activity or expression of SsaACB. In this case, SsaACB would not be capable of making a strong contribution, so the WT strain would compete only slightly better than the mutant. Although this explanation seems inherently less plausible, we cannot rule it out at present. If true, it could account for the existence of TmpA in S. sanguinis, which is otherwise inexplicable. Moreover, although we have not yet tested the effect of MntH in an oral model, the fact that it seems to be most active under exactly the conditions in which TmpA is least active (Figure 8) provides a rationale for its presence in some strains.
In conclusion, these results indicate that SsaACB made no more than a minor contribution to competitiveness in this model, even though the only other transporter, TmpA, has now been found to be inactive at reduced pH. These results may guide future studies. For example, there has been much interest concerning the development of probiotic bacteria for promotion of oral health (Culp et al., 2020). Given the results of the studies employing the endocarditis model in the accompanying paper (Puccio, Kunka, et al., 2022) in combination with the murine studies reported here, and the work with the ΔmntH mutant of VMC66, one course of action would be to begin with a promising mntH‐containing S. sanguinis strain or candidate probiotic species such as Streptococcus sp. A12 (Lee et al., 2019), then delete ssaACB. This mutant could be tested in a similar manner to that described here and in the accompanying paper. Assuming favorable results, further development would likely require assessment of colonization ability in the human oral cavity. In conclusion, these results and those of the accompanying study (Puccio, Kunka, et al., 2022) enhance our understanding of the role of manganese in S. sanguinis and highlight the functional and phylogenetic diversity of transporters devoted to uptake of this essential nutrient.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains
Strains and plasmids used in this study are listed in Table 1. All strains were grown in overnight cultures from single‐use aliquots of cryopreserved cells, diluted 1,000‐fold in BHI media (Beckinson Dickinson). Mutant strains were incubated with the appropriate antibiotics: kanamycin (Kan; Sigma‐Aldrich) 500 µg/ml, erythromycin (Erm; Sigma‐Aldrich), 10 µg/ml; spectinomycin (Spc; Sigma‐Aldrich) 200 µg/ml, and chloramphenicol (Cm; Fisher) 5 µg/ml. The pre‐cultures of the SK36 ΔssaACB ΔtmpA and VMC66 ΔssaACB ΔmntH and ΔssaACB ΔmntH ΔtmpA required exogenous Mn2+ for growth (15 µM for the triple mutant; 10 µM for the others). The cultures were then incubated at 37°C for 16–20 hr (or up to 24 hr for the triple mutant) with the atmospheric condition set to 1% O2 (9.5% H2, 9.5% CO2, and 80% N2) or 6% O2 (7% H2, 7% CO2, and 80% N2) using a programmable Anoxomat™ Mark II jar‐filling system (AIG, Inc).
TABLE 1.
Strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| SK36 | Human oral plaque isolate | Mogens Killan (Aarhus University); Xu et al. (2007) |
| JFP56 | ΔSSA_0169::aad9, derived from SK36 | Turner et al. (2009) |
| JFP169 a | ΔssaACB::aphA‐3, derived from SK36 | Baker et al. (2019) |
| JFP173 b | ΔssaACB::tetM, derived from SK36 | Baker et al. (2019) |
| JFP226 | ΔtmpA::aphA‐3, derived from SK36 | Puccio, Kunka, et al. (2022) |
| JFP227 c | ΔssaACB::tetM ΔtmpA::aphA‐3, unintentional mutation: SSA_1414‐W139*; derived from SK36 | Puccio, Kunka, et al. (2022) |
| JFP377 | ΔssaACB::tetM ΔtmpA::aphA‐3, clean version, derived from SK36 | Puccio, Kunka, et al. (2022) |
| VMC66 | Human endocarditis isolate | Kitten et al. (2012) |
| JFP313 | ΔssaACB::aphA‐3, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| JFP317 | ΔtmpA::pSerm, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| JFP320 | ΔssaACB::aphA‐3 ΔtmpA::pSerm, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| JFP367 | ΔmntH::tetM, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| JFP370 | ΔssaACB::cat ΔmntH::tetM, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| JFP397 | ΔssaACB::cat ΔmntH::tetM ΔtmpA:: pSerm, derived from VMC66 | Puccio, Kunka, et al. (2022) |
| UA159 | Wild‐type S. mutans | ATCC 700610 |
| Plasmid | Description | References |
| pVMTeal | Plasmid encoding ErmR for use in S. mutans | Vickerman et al. (2015); Zhu et al. (2017) |
4.2. Tube growth studies
The pH of BHI was modified by addition of 6 N HCl prior to autoclaving. Each tube was pre‐incubated at 37°C in 1% O2 in an Anoxomat jar overnight and then inoculated with a 10–6‐fold dilution of the overnight pre‐culture. The inoculated tubes were returned to incubate at 37°C in 1% O2. In some growth studies, MnSO4 (Alfa Aesar; Puratronic™, >99.999% pure) was added to culture tubes immediately prior to inoculation. To determine CFUs, samples were sonicated for 90 s using an ultrasonic homogenizer (Biologics, Inc) to disrupt chains. Cultures were diluted in PBS and plated on BHI agar (Beckinson Dickinson) plates using an Eddy Jet 2 spiral plater (Neutec Group, Inc.). Plates were incubated for 24 hr at 37°C under anaerobic conditions with a palladium catalyst included in the jars.
4.3. Metal analysis
For metal analysis of cells growing batch cultures, 2 tubes of 38 ml of BHI per strain and condition were pre‐incubated at 37°C overnight. For some samples, Mn2+ was added to 10 µM immediately prior to inoculation. Pre‐cultures (3 ml) were added to each tube. For pH 7.3 samples, pre‐cultures were added directly. For pH 6.2 samples, pre‐cultures were centrifuged and resuspended in pH 6.2 BHI prior to inoculation. Cultures were grown to mid‐logarithmic phase and centrifuged at 3,740 g for 10 min at 4°C. All strains grew under the conditions of this assay; however, some of the mutants grew slower than the WT strains. For fermentor samples, aliquots of 40 ml were collected at each time point and then centrifuged as described above.
The collected cell pellet was then washed twice with cold cPBS (PBS treated with Chelex‐100 resin (Bio‐Rad) for 2 hr, then filter sterilized and supplemented with EDTA to 1 mM). The pellet was then divided for subsequent acid digestion or protein concentration determination. Trace metal grade (TMG) nitric acid (15%) (Fisher Chemical) was added to one portion of the pellet. The pellet was digested using an Anton Paar microwave digestion system using a modified Organic B protocol: 120°C for 10 min, 180°C for 20 min, with the maximum temperature set to 180°C. The digested samples were then diluted 3‐fold with Chelex‐treated dH2O. Metal concentrations were determined using an Agilent 5110 inductively coupled plasma‐optical emission spectrometer (ICP‐OES). Concentrations were determined by comparison with a standard curve created with a 10 μg/ml multi‐element standard (CMS‐5; Inorganic Ventures) diluted in 5% TMG nitric acid. Pb (Inorganic Ventures) was used as an internal standard (100 μg/ml). The other portion of the pellet was resuspended in PBS and mechanically lysed using a FastPrep‐24 instrument with Lysing Matrix B tubes (MP Biomedicals) as described previously (Rhodes et al., 2014). Insoluble material was removed by centrifugation. Protein concentrations were determined using a bicinchoninic acid (BCA) Protein Assay Kit (Pierce) as recommended by the manufacturer, with bovine serum albumin as the standard. For metal analysis of TY+S, media was digested with the microwave digestion system as described above and analyzed on an Agilent 8900 ICP‐QQQ‐MS (ICP‐MS).
4.4. Fermentor growth conditions
The fermentor conditions were similar to Puccio and Kitten (2020) with minor modifications. A BIOSTAT® B bioreactor (Sartorius Stedim) with a 1.5‐L capacity UniVessel® glass vessel was used for growth of 800‐ml cultures at 37°C. Cultures were stirred at 250 rpm and pH was maintained by the automated addition of 2 N KOH and 2 N HCl (Fisher Chemical). A 40‐ml overnight pre‐culture of S. sanguinis was grown as described above and centrifuged for 10 min at 3,740g in an Allegra X‐142 centrifuge at 4°C. The supernatant was discarded and the cells were resuspended in BHI prior to inoculation. The air flow was kept at 0.03 lpm for the entire experiment. Once the cells reached their peak OD in static culture, the input flow of BHI was set to 17% (~700 ml/h), and the output flow of waste was set to 34% for the remainder of the experiment. Cells were allowed to acclimate to this media flow rate for 1 hr. The T‐20 sample was aseptically removed for total RNA isolation or metal analysis. The fermentor culture in the vessel was adjusted to pH 6.2 using an in‐dwelling probe at T0. Samples were taken for each post‐treatment time point (T10, T25, T50). In some experiments, MnSO4 (Puratronic™; Alfa Aesar) was added to the carboy (T66) and vessel (T70) at a final concentration of 10 µM and samples were taken for metal analysis at T80.
4.5. RNA isolation
Fermentor samples (2 ml) were added to 4 ml RNAprotect Bacteria Reagent (QIAGEN) and immediately vortexed for 10 s. The samples were then incubated at room temperature for 5–90 min and centrifuged for 10 min at 3,740g at 4°C. The supernatant was discarded and the samples stored at −80°C. RNA isolation and on‐column DNase treatment were completed using the RNeasy Mini Kit and RNase‐Free DNase Kit, respectively (QIAGEN). RNA was eluted in 50 µl RNase‐Free water (QIAGEN). A second DNase treatment was then performed on the samples (Invitrogen).
4.6. RNA sequencing
Total RNA quantity and integrity were determined using a Bioanalyzer (Agilent). All samples passed quality control assessment with RNA Integrity Numbers (RIN) above 8. Two sequential rounds of ribosomal reduction were then performed on all samples using RiboMinus™ Transcriptome Isolation Kit (ThermoFisher). The resulting depleted RNA was assessed using Bioanalyzer (Agilent) to confirm efficient rRNA removal. Stranded RNA‐seq library construction was then performed on the rRNA‐depleted RNA using the Kapa RNA HyperPlus kit for Illumina (Roche) following manufacturer's specifications for library construction and multiplexing. Final Illumina libraries were assessed for quality using an Agilent Bioanalyzer DNA High Sensitivity Assay and qPCR quantification was performed using Kapa Library Quantification kit for Illumina (Roche). Individual libraries were pooled equimolarly and the final pool was sequenced on an Illumina MiSeq with 2 × 75‐bp paired‐end reads. Demultiplexing was performed on the Illumina MiSeq's on‐board computer. The Virginia Commonwealth University Genomics Core Facility completed all RNA‐seq library preparation and sequencing.
4.7. RNA‐seq analysis pipeline
Using Geneious Prime 2021.1.1 (https://www.geneious.com), sequence reads were trimmed using the BBDuk Trimmer prior to mapping to either the SK36 genome or a modified version, in which the ssaACB operon was replaced with the aphA‐3 sequence. Original and new locus tags from the Genbank® annotations are included (Benson et al., 2013). PATRIC annotations (https://patricbrc.org/) (Wattam et al., 2017) are also included. Reads for each post‐treatment sample were compared to the corresponding pre‐treatment (T‐20) sample using DESeq2 (Love et al., 2014) in Geneious to determine log2 fold changes and adjusted p‐values. The same method was used to compare reads from WT to those from the ΔssaACB mutant at each sample time point. Principal component analysis was completed using R (version 4.0.5) and RStudio (version 13.959) with Bioconductor (Bioconductor.com) package pcaExplorer version 2.160 (Marini & Binder, 2019). Volcano plots were generated using R and RStudio with Bioconductor package EnhancedVolcano version 1.8.0 (Blighe et al., 2018). All DEGs were input into the DAVID database (https://david.ncifcrf.gov/summary.jsp) (Dennis et al., 2003). Since the new locus tags are not accepted in KEGG, only genes with original “SSA_” locus tags were included in the analysis. The KEGG_pathway option was chosen for functional annotation clustering. The p‐value shows the significance of pathway enrichment. DAVID pathway figures were generated using an R script (https://github.com/DrBinZhu/DAVID_FIG).
4.8. Biofilm competition assays
S. sanguinis pre‐cultures (described above) were diluted 100‐fold into 2 ml pre‐warmed BM (SAFC Biosciences) + 1% sucrose in 12 well plates. The cultures were grown for 24 hr at 37°C aerobically (~21% O2). The media supernatant was carefully removed and discarded. Warm TY (3% tryptone, 0.1% yeast extract, 0.5% KOH, 1 mM H3PO4) (Fozo & Quivey, 2004) + 1% sucrose was added to 2 ml and S. mutans + pVMTeal pre‐cultures were diluted 100‐fold into the wells. After 24 hr incubation at 37°C, TY+S supernatant was carefully removed and the pH was measured. PBS was added to 1 ml and biofilms were scraped from each well. Each biofilm culture was sonicated for 90 s using an ultrasonic homogenizer. Cultures were diluted in PBS and plated on BHI plates with antibiotics using a spiral plater. Plates were incubated at 37°C for 24 hr at 0% O2.
4.9. Mouse model of oral colonization
The mouse model was described in detail recently by Culp et al. (2020). Briefly, all procedures with solutions and samples were performed under BSL2 conditions and mice were kept under ABSL2 conditions. Inbred 3‐week‐old female SPF BALB/cJ mice (The Jackson Laboratory, Bar Harbor, ME) were placed in pairs in sterile cages. Two days later, mice were given drinking water containing 0.8 mg/ml sulphamethoxazole/0.16 mg/ml trimethoprim for a total of 10 days to suppress indigenous oral bacteria, followed by a 3‐day washout period with sterile drinking water. On the following day (designated experimental day 0), mice were placed on custom diet TD.160810 (Teklad, Madison, WI) containing 37.5% sucrose and without fluoride, and inoculated daily over five days with 50 µl of 1.5% (wt/vol) carboxymethylcellulose in saliva buffer (50 mM KCl, 1.0 mM KPO4, 0.35 mM K2HPO4, 1.0 mM CaCl2 2H2O, 0.1 mM MgCl2 6H2O, pH 6.5) containing approximately 1 × 109 cells of the indicated strain grown to an OD600 between 0.55 and 0.70 in BHI. Two weeks later, mice received three daily inoculations with approximately 1 × 109 cells of S. mutans UA159. Mice were euthanized by CO2 asphyxiation followed by cervical dislocation. The protocol was approved by the Institutional Animal Care and Use Committee at University of Florida (IACUC protocol #201810470). Oral swabs were taken using HydraFlock® 6″ Sterile Micro Ultrafine Flock swabs (Puritan Medical Products, Guilford, ME). Swab tips were vortexed (3 times for 5 s) in 1 ml sterile PBS, the tips removed and 200 µl added of ice‐cold PBS containing approximately 5 × 108 depurinated cells of laboratory strain S. mitis UF2. Tubes were then vortexed 5 s and centrifuged (10,000g, 10 min at 4°C) to pellet‐recovered cells. Cell pellets were then processed for DNA isolation using the DNeasy UltraClean Microbial kit (Qiagen Inc., Germantown, MD) as per manufacturer's instructions. Depurinated cells were devoid of detectable DNA by qPCR and allowed for quantitative recoveries of DNA from test strains and mouse commensals. To measure dental colonization, the left and right halves of each mandible were aseptically extracted and any remaining extraneous soft tissue removed followed by removal of bone approximately 2 mm anterior and posterior to the three molar teeth. Each pair of molars were sonicated on ice in 1 ml sterile PBS, pH 7.4, in siliconized 2 ml microcentrifuge tubes, the molars aseptically removed and approximately 5 × 108 depurinated cells of S. mitis UF2 then added, followed by vortexing for 5 s and centrifugation (10,000g, 10 min at 4°C). Cell pellets were then processed for DNA isolation as described for swabs. Recovered bacterial genomes in DNA samples were assessed by qPCR in 20 µl reactions run in triplicate in a Bio‐Rad CFX96 real‐time PCR instrument using 10 µl SsoAdvanced Universal SYBR®Green Supermix (Bio‐Rad, Hercules, CA), 0.5 µl each primer and 9 µl of DNA diluted in 4 mM Tris‐HCl, pH 8.0. Samples were run for 3 min at 98°C followed by either 34 cycles (rpsL) or 40 cycles (98°C, 15 s; annealing/elongation, 45 s at 70°C for S. mutans, 55°C for rpsL and 64.5°C for S. sanguinis); followed by a melt curve from 65–95°C at 0.5°C increments. Primers were as follows: S. sanguinis (Forward, 5′‐GAGCGAATCATCAAGGATCAAAC‐3′, Reverse, 5′‐CGAGCAATAGCTTT‐CGTAATAGG‐3′), S. mutans (Forward, 5′‐TGGCAAGTCCTGATGGTTTGAC‐3′, Reverse, 5′‐GGAAGCGGAAGCTGTGATGAAC‐3′), rpsL (Forward, 5′‐CCKAAYTCNGCNYTNCGT‐AA‐3′, Reverse, 5′‐CGHACMCCWGGWARGTCYTT‐3′). Primers were used at final concentrations of 0.50 µM (S. mutans), 0.15 µM (S. sanguinis), and 2.5 µM (rpsL). Standard curves were derived from DNA samples isolated from each strain grown to mid‐exponential phase in BHI. S. mutans UA159 was used as standard for rpsL assays. Efficiencies, slopes, and r 2 values for standard curves were greater than 90%, −3.226, and 0.975, respectively. Results were analyzed using the Bio‐Rad CFX Manager program. Under the listed conditions, all assays were specific for the targeted genomes. All work was performed in a BioSafety cabinet under aseptic conditions.
4.10. Data analysis and presentation
Statistical tests were performed in GraphPad Prism 9.0 (graphpad.com) or R (R Core Team, 2018). Significance was determined by statistical tests indicated in the figure legends. p‐values ≤ .05 were considered significant. DESeq2 calculations of RNA‐Seq datasets were completed in Geneious Prime 2021.1 or in the pcaExplorer R package (Marini & Binder, 2019). Confidence intervals (95%) of replicate samples were determined by the pcaExplorer R package.
CONFLICT OF INTEREST
All authors have no competing interests to declare.
AUTHOR CONTRIBUTIONS
TP and TK designed the in vitro experiments and DJC and RAB designed the in vivo experiment. TP and SSA completed the in vitro experiments and ACS, CAL, and ASB completed the in vivo experiment. TP, DJC, RAB, and TK wrote the manuscript. All authors reviewed and approved the manuscript.
ETHICS APPROVAL STATEMENT
The University of Florida IACUC committee approved all animal procedures (IACUC Study #201810470), following the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Supporting information
Fig S1‐S6
Table S1‐S8
ACKNOWLEDGEMENTS
We would like to thank Karina Kunka, Shannon Green, and Jody Turner for their advice and technical assistance.
Puccio, T. , An, S.‐S. , Schultz, A.C. , Lizarraga, C.A. , Bryant, A.S. , Culp, D.J. , et al. (2022) Manganese transport by Streptococcus sanguinis in acidic conditions and its impact on growth in vitro and in vivo. Molecular Microbiology, 117, 375–393. 10.1111/mmi.14854
Funding information
This work was supported by the National Institutes of Health: award no. R01 AI114926 to TK from the National Institute of Allergy and Infectious Diseases; awards no. R01 DE025832 to RAB and no. F31 DE028468 to TP from the National Institute of Dental and Craniofacial Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
DATA AVAILABILITY STATEMENT
The RNA‐seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE174672. The remaining data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aas, J.A. , Paster, B.J. , Stokes, L.N. , Olsen, I. & Dewhirst, F.E. (2005) Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology, 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Shang, M. , Xu, M. , Wu, A. , Sun, L. & Zheng, L. (2019) Transcriptome, phenotypic, and virulence analysis of Streptococcus sanguinis SK36 wild type and its CcpA‐null derivative (ΔCcpA). Frontiers in Cellular and Infection Microbiology, 9, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S.P. , Nulton, T.J. & Kitten, T. (2019) Genomic, phenotypic, and virulence analysis of Streptococcus sanguinis oral and infective‐endocarditis isolates. Infection and Immunity, 87, e00703‐00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton, D. (1982) The influence of manganese on carbohydrate metabolism and caries induction by Streptococcus mutans strain Ingbritt. Caries Research, 16, 189–192. [DOI] [PubMed] [Google Scholar]
- Belda‐Ferre, P. , Alcaraz, L.D. , Cabrera‐Rubio, R. , Romero, H. , Simon‐Soro, A. , Pignatelli, M. et al. (2012) The oral metagenome in health and disease. ISME Journal, 6, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, G.R. , Sutton, S.V.W. & Marquis, R.E. (1986) Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infection and Immunity, 53, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A. , Cavanaugh, M. , Clark, K. , Karsch‐Mizrachi, I. , Lipman, D.J. , Ostell, J. et al. (2013) GenBank. Nucleic Acids Research, 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe, K. , Rana, S. & Lewis, M. (2018) EnhancedVolcano: Publication‐Ready Volcano Plots with Enhanced Colouring and Labeling. GitHub. [Google Scholar]
- Bor, D.H. , Woolhandler, S. , Nardin, R. , Brusch, J. & Himmelstein, D.U. (2013) Infective endocarditis in the U.S., 1998–2009: a nationwide study. PLoS One, 8, e60033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno‐Barcena, J.M. , Azcarate‐Peril, M.A. & Hassan, H.M. (2010) Role of antioxidant enzymes in bacterial resistance to organic acids. Applied and Environment Microbiology, 76, 2747–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne, R.A. & Chen, Y.‐Y.‐M. (1998) The use of continuous flow bioreactors to explore gene expression and physiology of suspended and adherent populations of oral streptococci. Methods in Cell Science, 20, 181–190. [Google Scholar]
- Burne, R.A. , Parsons, D.T. & Marquis, R.E. (1989) Cloning and expression in Escherichia coli of the genes of the arginine deiminase system of Streptococcus sanguis NCTC 10904. Infection and Immunity, 57, 3540–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill, T.J. , Baddour, L.M. , Habib, G. , Hoen, B. , Salaun, E. , Pettersson, G.B. et al. (2017) Challenges in infective endocarditis. Journal of the American College of Cardiology, 69, 325–344. 10.1016/j.jacc.2016.10.066 [DOI] [PubMed] [Google Scholar]
- Cahill, T.J. , Dayer, M. , Prendergast, B. & Thornhill, M. (2017) Do patients at risk of infective endocarditis need antibiotics before dental procedures? BMJ, 358, j3942. 10.1136/bmj.j3942 [DOI] [PubMed] [Google Scholar]
- Caufield, P.W. , Dasanayake, A.P. , Li, Y. , Pan, Y. , Hsu, J. & Hardin, J.M. (2000) Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infection and Immunity, 68, 4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Yu, W.‐H. , Izard, J. , Baranova, O.V. , Lakshmanan, A. & Dewhirst, F.E. (2010) The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database, 2010, baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.Y. , Shieh, H.R. & Chang, Y.C. (2013) The expression of the fim operon is crucial for the survival of Streptococcus parasanguinis FW213 within macrophages but not acid tolerance. PLoS One, 8, e66163. 10.1371/journal.pone.0066163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Redanz, S. , Cullin, N. , Zhou, X. , Xu, X. , Joshi, V. et al. (2018) Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral streptococci. Applied and Environmental Microbiology, 84(2), e01697‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, P.D. & Hill, C. (2003) Surviving the acid test: responses of gram‐positive bacteria to low pH. Microbiology and Molecular Biology Reviews, 67, 429–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump, K.E. , Bainbridge, B. , Brusko, S. , Turner, L.S. , Ge, X. , Stone, V. et al. (2014) The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Molecular Microbiology, 92, 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp, D.J. , Hull, W. , Bremgartner, M.J. , Atherly, T.A. , Christian, K.N. , Killeen, M. et al. (2020) In vivo colonization with candidate oral probiotics attenuates colonization and virulence of Streptococcus mutans . Applied and Environmental Microbiology, 87, e02490‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, T.M. , Lieou, J. & Marquis, R.E. (1995) Arginine deiminase system and acid adaptation of oral streptococci. Applied and Environment Microbiology, 61, 4494–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, G., Jr. , Sherman, B.T. , Hosack, D.A. , Yang, J. , Gao, W. , Lane, H.C. et al. (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biology, 4, P3. [PubMed] [Google Scholar]
- Diaz‐Garrido, N. , Lozano, C.P. , Kreth, J. & Giacaman, R.A. (2020) Competition and caries on enamel of a dual‐species biofilm model of Streptococcus mutans and Streptococcus sanguinis . Applied and Environmental Microbiology, 86, e01262‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko, K.Y. , Phan, M.D. , Peters, K.M. , Walker, M.J. , Schembri, M.A. & McEwan, A.G. (2017) Interplay between tolerance mechanisms to copper and acid stress in Escherichia coli . Proceedings of the National Academy of Sciences of the United States of America, 114, 6818–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, D.W. , McCall, L.W. , Powell, W.F. , Arscott, W.T. , McConocha, E.M. , McClurg, C.J. et al. (2008) SloR modulation of the Streptococcus mutans acid tolerance response involves the GcrR response regulator as an essential intermediary. Microbiology, 154, 1132–1143. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. & Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotcheva, N.I. , Sokolov, A.P. & Kondrashova, M.N. (2006) Nonezymatic formation of succinate in mitochondria under oxidative stress. Free Radical Biology and Medicine, 41, 56–64. [DOI] [PubMed] [Google Scholar]
- Floderus, E. , Linder, L.E. & Sund, M.‐L. (1990) Arginine catabolism by strains of oral streptococci. APMIS: Acta Pathologica Microbiologica Scandinavica, 98, 1045–1052. [DOI] [PubMed] [Google Scholar]
- Fozo, E.M. & Quivey, R.G., Jr. (2004) Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Applied and Environment Microbiology, 70, 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Mendoza, A. , Liebana, J. , Castillo, A.M. , Higuera, A.d.l. & Piedrola, G. (1993) Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. Journal of Medical Microbiology, 39, 434–439. [DOI] [PubMed] [Google Scholar]
- Giacaman, R.A. , Torres, S. , Gomez, Y. , Munoz‐Sandoval, C. & Kreth, J. (2015) Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion‐free and high caries adults. Archives of Oral Biology, 60, 154–159. [DOI] [PubMed] [Google Scholar]
- Gross, E.L. , Beall, C.J. , Kutsch, S.R. , Firestone, N.D. , Leys, E.J. & Griffen, A.L. (2012) Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One, 7, e47722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, N. & Liu, L. (2020) Microbial response to acid stress: mechanisms and applications. Applied Microbiology and Biotechnology, 104, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, I.R. & Svensäter, G. (1998) Acid‐regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiology and Immunology, 13, 292–300. [DOI] [PubMed] [Google Scholar]
- Ilbert, M. & Bonnefoy, V. (2013) Insight into the evolution of the iron oxidation pathways. Biochimica et Biophysica Acta, 1827, 161–175. [DOI] [PubMed] [Google Scholar]
- Jakubovics, N.S. (2015) Saliva as the sole nutritional source in the development of multispecies communities in dental plaque. Microbiology Spectrum, 3, MBP‐0013‐2014. 10.1128/microbiolspec.MBP-0013-2014 [DOI] [PubMed] [Google Scholar]
- Jamil, M. , Sultan, I. , Gleason, T.G. , Navid, F. , Fallert, M.A. , Suffoletto, M.S. et al. (2019) Infective endocarditis: trends, surgical outcomes, and controversies. Journal of Thoracic Disease, 11, 4875–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz, J.K. , Katrak, C. , Ganguly, T. , Vargas, J. , Wright, L. , Peters, Z.T. et al. (2020) Manganese uptake, mediated by SloABC and MntH, is essential for the fitness of Streptococcus mutans . mSphere, 5, e00764‐00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres, D.G. , Lawyer, C.H. & Maguire, M.E. (1998) The CorA magnesium transporter gene family. Microbial & Comparative Genomics, 3, 151–169. [DOI] [PubMed] [Google Scholar]
- Kim, J.S. , Sung, M.H. , Kho, D.H. & Lee, J.K. (2005) Induction of manganese‐containing superoxide dismutase is required for acid tolerance in Vibrio vulnificus . Journal of Bacteriology, 187, 5984–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindblom, C. , Davies, J.R. , Herzberg, M.C. , Svensater, G. & Wickstrom, C. (2012) Salivary proteins promote proteolytic activity in Streptococcus mitis biovar 2 and Streptococcus mutans . Molecular Oral Microbiology, 27, 362–372. [DOI] [PubMed] [Google Scholar]
- Kitten, T. , Munro, C.L. , Zollar, N.Q. , Lee, S.P. & Patel, R.D. (2012) Oral streptococcal bacteremia in hospitalized patients: taxonomic identification and clinical characterization. Journal of Clinical Microbiology, 50, 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P.E. (2011) Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. International Journal of Oral Science, 3, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P.E. , Palmer, R.J., Jr. , Periasamy, S. & Jakubovics, N.S. (2010) Oral multispecies biofilm development and the key role of cell‐cell distance. Nature Reviews Microbiology, 8, 471–480. [DOI] [PubMed] [Google Scholar]
- Kreth, J. , Merritt, J. , Shi, W. & Qi, F. (2005) Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. Journal of Bacteriology, 187, 7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth, J. , Zhang, Y. & Herzberg, M.C. (2008) Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans . Journal of Bacteriology, 190, 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert, W.L. & Quivey, R.G., Jr. (2003) Genetic and biochemical characterization of the F‐ATPase operon from Streptococcus sanguis 10904. Journal of Bacteriology, 185, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkin, M.S. & Oppenheim, F.G. (1993) Structural features of salivary function. Critical Reviews in Oral Biology and Medicine, 4, 251–259. [DOI] [PubMed] [Google Scholar]
- Lamont, R.J. , Koo, H. & Hajishengallis, G. (2018) The oral microbiota: dynamic communities and host interactions. Nature Reviews Microbiology, 16, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , Walker, A.R. , Chakraborty, B. , Kaspar, J.R. , Nascimento, M.M. & Burne, R.A. (2019) Novel probiotic mechanisms of the oral bacterium Streptococcus sp. A12 as explored with functional genomics. Applied and Environment Microbiology, 85, e01335‐01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, J.A. & Burne, R.A. (2002) Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans . Journal of Bacteriology, 184, 6357–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, J.A. , Chen, Y.Y. & Burne, R.A. (2001) Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans . Journal of Bacteriology, 183, 6074–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Tang, H. , Lin, Z. & Xu, P. (2015) Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnology Advances, 33, 1484–1492. [DOI] [PubMed] [Google Scholar]
- Loo, C.Y. , Corliss, D.A. & Ganeshkumar, N. (2000) Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. Journal of Bacteriology, 182, 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, F. & Binder, H. (2019) pcaExplorer: an R/Bioconductor package for interacting with RNA‐seq principal components. BMC Bioinformatics, 20, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, P.D. , Do, T. , Beighton, D. & Devine, D.A. (2016) Influence of saliva on the oral microbiota. Periodontology 2000, 70, 80–92. [DOI] [PubMed] [Google Scholar]
- Martin, J.E. & Giedroc, D.P. (2016) Functional determinants of metal ion transport and selectivity in paralogous cation diffusion facilitator transporters CzcD and MntE in Streptococcus pneumoniae . Journal of Bacteriology, 198, 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Galiano, A.J. , Overweg, K. , Ferrandiz, M.J. , Reuter, M. , Wells, J.M. & de la Campa, A.G. (2005) Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae . Microbiology, 151, 3935–3946. [DOI] [PubMed] [Google Scholar]
- Moye, Z.D. , Zeng, L. & Burne, R.A. (2014) Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans . Journal of Oral Microbiology, 6, 24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgas, C.J. , Green, S.P. , Forney, A.K. , Korba, R.M. , An, S.S. , Kitten, T. et al. (2020) Intracellular metal speciation in Streptococcus sanguinis establishes SsaACB as critical for redox maintenance. ACS Infectious Diseases, 6, 1906–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento, M.M. , Gordan, V.V. , Garvan, C.W. , Browngardt, C.M. & Burne, R.A. (2009) Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiology and Immunology, 24, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo, Y. & Nelson, N. (2006) The NRAMP family of metal‐ion transporters. Biochimica et Biophysica Acta, 1763, 609–620. [DOI] [PubMed] [Google Scholar]
- Nies, D.H. (1992) CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus . Journal of Bacteriology, 174, 8102–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou, K. , Alegria, A. , Bron, P.A. , de Angelis, M. , Gobbetti, M. , Kleerebezem, M. et al. (2016) Stress physiology of lactic acid bacteria. Microbiology and Molecular Biology Reviews, 80, 837–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccio, T. & Kitten, T. (2020) Fermentor growth of Streptococcus sanguinis . protocols.io. [Google Scholar]
- Puccio, T. , Kunka, K.S. , An, S.‐S. & Kitten, T. (2022) Contribution of a ZIP‐family protein to manganese uptake and infective endocarditis virulence in Streptococcus sanguinis. Molecular Microbiology, 117, 353‐374. 10.1111/mmi.14853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccio, T. , Kunka, K.S. , Zhu, B. , Xu, P. & Kitten, T. (2020) Manganese depletion leads to multisystem changes in the transcriptome of the opportunistic pathogen Streptococcus sanguinis . Frontiers in Microbiology, 11, 592615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccio, T. , Misra, B.B. & Kitten, T. (2021) Time‐course analysis of Streptococcus sanguinis after manganese depletion reveals changes in glycolytic and nucleic metabolites. Metabolomics, 17, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey, R.G. , Kuhnert, W.L. & Hahn, K. (2001) Genetics of acid adaptation in oral streptococci. Critical Reviews in Oral Biology & Medicine, 12, 301–314. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2018) R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://cran.r‐project.org/doc/FAQ/R‐FAQ.html#Citing‐R [Google Scholar]
- Radin, J.N. , Kelliher, J.L. , Parraga Solorzano, P.K. & Kehl‐Fie, T.E. (2016) The two‐component system ArlRS and alterations in metabolism enable Staphylococcus aureus to resist calprotectin‐induced manganese starvation. PLoS Path, 12, e1006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinoso, C.A. , Auger, C. , Appanna, V.D. & Vasquez, C.C. (2012) Tellurite‐exposed Escherichia coli exhibits increased intracellular alpha‐ketoglutarate. Biochemical and Biophysical Research Communications, 421, 721–726. [DOI] [PubMed] [Google Scholar]
- Rhodes, D.V. , Crump, K.E. , Makhlynets, O. , Snyder, M. , Ge, X. , Xu, P. et al. (2014) Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis . Journal of Biological Chemistry, 289, 6273–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi, I. , Grifantini, R. , Jiang, S.‐M. , Brettoni, C. , Grandi, G. , Wessels, M.R. et al. (2009) CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. Journal of Bacteriology, 191, 5387–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M. , Kodama, Y. , Shimoyama, Y. , Ishikawa, T. & Kimura, S. (2018) Aciduricity and acid tolerance mechanisms of Streptococcus anginosus . Journal of General and Applied Microbiology, 64, 174–179. [DOI] [PubMed] [Google Scholar]
- Senouci‐Rezkallah, K. , Schmitt, P. & Jobin, M.P. (2011) Amino acids improve acid tolerance and internal pH maintenance in Bacillus cereus ATCC14579 strain. Food Microbiology, 28, 364–372. [DOI] [PubMed] [Google Scholar]
- Shabayek, S. , Bauer, R. , Mauerer, S. , Mizaikoff, B. & Spellerberg, B. (2016) A streptococcal NRAMP homologue is crucial for the survival of Streptococcus agalactiae under low pH conditions. Molecular Microbiology, 100, 589–606. [DOI] [PubMed] [Google Scholar]
- Shabayek, S. & Spellerberg, B. (2017) Acid stress response mechanisms of group B streptococci. Frontiers in Cellular and Infection Microbiology, 7, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensater, G. , Larsson, U.‐B. , Greif, E.C.G. , Cvitkovitch, D.G. & Hamilton, I.R. (1997) Acid tolerance response and survival by oral bacteria. Oral Microbiology and Immunology, 12, 266–273. [DOI] [PubMed] [Google Scholar]
- Taudte, N. & Grass, G. (2010) Point mutations change specificity and kinetics of metal uptake by ZupT from Escherichia coli . BioMetals, 23, 643–656. [DOI] [PubMed] [Google Scholar]
- Turner, L.S. , Das, S. , Kanamoto, T. , Munro, C.L. & Kitten, T. (2009) Development of genetic tools for in vivo virulence analysis of Streptococcus sanguinis . Microbiology, 155, 2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman, M.M. , Mansfield, J.M. , Zhu, M. , Walters, K.S. & Banas, J.A. (2015) Codon‐optimized fluorescent mTFP and mCherry for microscopic visualization and genetic counterselection of streptococci and enterococci. Journal of Microbiol Methods, 116, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam, A.R. , Davis, J.J. , Assaf, R. , Boisvert, S. , Brettin, T. , Bun, C. et al. (2017) Improvements to PATRIC, the all‐bacterial bioinformatics database and analysis resource center. Nucleic Acids Research, 45, D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z.T. & Burne, R.A. (2004) LuxS‐mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. Journal of Bacteriology, 186, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström, C. , Chávez de Paz, L. , Davies, J.R. & Svensäter, G. (2013) Surface‐associated MUC5B mucins promote protease activity in Lactobacillus fermentum biofilms. BMC Oral Health, 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W.R. , Gewitz, M. , Lockhart, P.B. , Bolger, A.F. , DeSimone, D.C. , Kazi, D.S. et al. (2021) Prevention of viridans group streptococcal infective endocarditis: a scientific statement from the American Heart Association. Circulation, 143, e1‐16. [DOI] [PubMed] [Google Scholar]
- Wong, R.S. & Bennick, A. (1980) The primary structure of a salivary calcium‐binding proline‐rich phosphoprotein (protein C), a possible precursor of a related salivary protein A. Journal of Biological Chemistry, 255, 5943–5948. [PubMed] [Google Scholar]
- Xu, P. , Alves, J.M. , Kitten, T. , Brown, A. , Chen, Z. , Ozaki, L.S. et al. (2007) Genome of the opportunistic pathogen Streptococcus sanguinis . Journal of Bacteriology, 189, 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Itzek, A. & Kreth, J. (2014) Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis . Microbiology, 160, 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B. , Ge, X. , Stone, V. , Kong, X. , El‐Rami, F. , Liu, Y. et al. (2017) ciaR impacts biofilm formation by regulating an arginine biosynthesis pathway in Streptococcus sanguinis SK36. Scientific Reports, 7, 17183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S6
Table S1‐S8
Data Availability Statement
The RNA‐seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE174672. The remaining data that support the findings of this study are available from the corresponding author upon reasonable request.


