Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) system has revolutionized the ability to edit the mammalian genome, providing a platform for the correction of pathogenic mutations and further investigation into gene function. CRISPR reagents can be delivered into the cell as DNA, RNA, or pre-formed ribonucleoproteins (RNPs). RNPs offer numerous advantages over other delivery approaches due to their ability to rapidly target genomic sites and quickly degrade thereafter. Here, we review the production steps and delivery methods for Cas9 RNPs. Additionally, we discuss how RNPs enhance genome and epigenome editing efficiencies, reduce off-target editing activity, and minimize cellular toxicity in clinically relevant mammalian cell types. We include details on a broad range of editing approaches, including novel base and prime editing techniques. Finally, we summarize key challenges for the use of RNPs, and propose future perspectives on the field.
Keywords: CRISPR/Cas9, drug delivery systems, RNP, gene therapy, genome editing, epigenome editing
1. Introduction
The development of genome editing tools has revolutionized the ability to engineer precise changes in mammalian genomes, enabling the correction of detrimental genetic variants and investigation into gene function. Although earlier tools, such as designer zinc finger nucleases (ZNFs) and transcription activator-like effectors (TALENs), enable genome targeting, the recent development of the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) RNA-guided endonuclease (RGEN) system has provided a more efficient, simple, and scalable approach to genome engineering [1–3]. The CRISPR/Cas system was originally recognized to provide prokaryotes with adaptive immunity to invading viruses and plasmids [4–6]. The prokaryotic system relies on complex formation between a Cas endonuclease with a sequence specific CRISPR RNA (crRNA) and structurally integral trans-activating crRNA (tracrRNA), which together enable site-specific cleavage of foreign genetic sequences [4–7]. In mammalian cells, the CRISPR/Cas system has been adapted as a genome editing tool by mediating targeted DNA double-strand breaks (DSBs) [8–14].
Genome editing is most commonly directed by the Streptococcus pyogenes (S. pyogenes) type II CRISPR/Cas nuclease Cas9 and a chimeric single guide RNA (sgRNA), which is formed by the fusion of a crRNA with a tracrRNA [8–11, 15]. The crRNA includes a 20 nucleotide (nt) sequence homologous to a target DNA site, known as a protospacer [15]. Upon Cas9/sgRNA ribonucleoprotein (RNP) complex formation, Cas9 is guided to the sequence-specific protospacer and, in the presence of a downstream trinucleotide 5’-NGG-3’ protospacer adjacent motif (PAM), cleaves the DNA between positions 17 and 18 of the protospacer. In mammalian cells, DSB formation recruits endogenous DNA repair pathways, resulting in multiple editing outcomes. Non-homologous end joining (NHEJ) functions as the primary repair pathway, where DNA blunt ends are repaired in an error-prone manner, often resulting in small insertions or deletions (indels) (Figure 1) [16, 17]. Alternatively, precise genome modifications can be achieved through exploiting the homology-directed repair (HDR) pathway, where an exogenous DNA donor template containing the desired edit is co-introduced with the Cas9 and sgRNA [16]. Inactivating point mutations in one or both of the catalytic RuvC and HNH domains of Cas9 has enabled the development of catalytically impaired Cas9 variants that either nick a single DNA strand (Cas9 nickase, nCas9) or are inactive (dead Cas9, dCas9) [15]. By utilizing these variants, additional genome and epigenome editing outcomes have been achieved beyond traditional NHEJ and HDR editing. For instance, fusion of nCas9 to engineered deaminase enzymes, termed base editors, introduce single base substitutions at target sites [18–21]. Additionally, prime editors mediate precise insertions, deletions, or base-to-base conversions by fusing nCas9 to reverse transcriptase (RT) and simultaneously introducing an RNA template [21–23]. Furthermore, modulation of transcription regulation can be achieved by combining dCas9 with effector domains, such as histone modifying and gene regulating domains [24–27].
Figure 1.
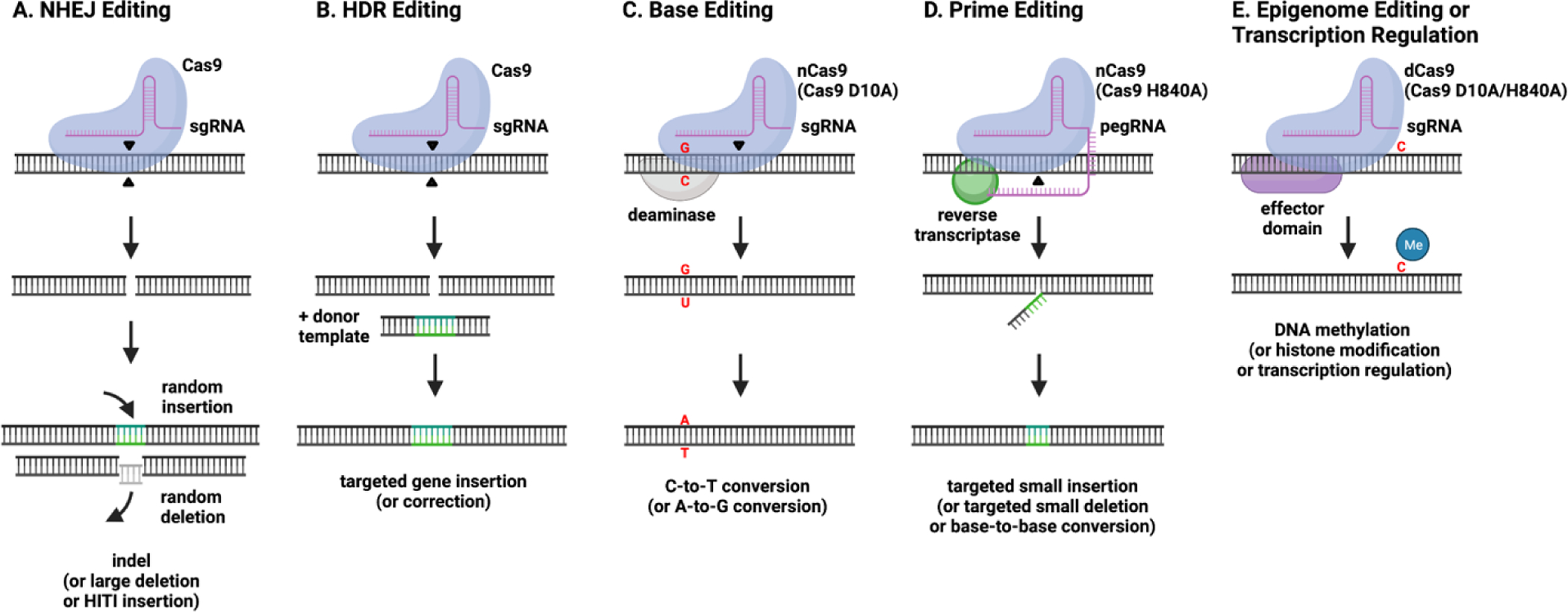
Overview of genome and epigenome editing strategies. A) Non-homologous end joining (NHEJ) is an error prone repair pathway that leads to small insertions or deletions (indels) at the site of Cas9-induced double strand breaks (DSB). When Cas9 is targeted to two genomic sites, large deletions of the intermediate sequence can be generated. Alternatively, targeted insertions can be completed with the addition of a homology-independent targeted integration (HITI) donor template. B) Homology-directed repair (HDR) is a precise DSB repair pathway that utilizes an exogenous DNA donor template with homology arms to achieve targeted gene insertions or corrections. C) Base editing takes advantage of deaminase enzymes to induce C-to-T (cytidine deaminase) or A-to-G (adenosine deaminase) base conversions in a five-base activity window. Cas9 nickase (nCas9; Cas9 D10A) is used to nick a single DNA strand opposite the targeted base in order to facilitate efficient repair. D) Prime editing can achieve targeted insertions, deletions, or base-to-base conversions by the introduction of an extended sgRNA known as a prime editing gRNA (pegRNA) and nCas9 (Cas9 H840A) fused to a reverse transcriptase. E) Epigenome editing and transcription regulation are achieved by guiding effector domains to a targeted DNA site via catalytically inactive dead Cas9 (dCas9; Cas9 D10A/H840A). Effector domains include epigenetic modifying domains, which can alter DNA methylation or histone acetylation, or gene regulatory domains, which directly influence gene expression through interactions with transcriptional machinery.
A prerequisite for successful genome and epigenome editing includes efficient delivery of CRISPR/Cas9 reagents into mammalian cell nuclei. Reagents can be delivered as DNA, RNA, or preassembled RNP complexes between Cas9 protein and sgRNA [28, 29]. However, several drawbacks of DNA and RNA-based methods exist (Table 1). Delivery of plasmids or viral vectors encoding Cas9 and sgRNA can lead to unintended integration into the host genome [30, 31]. When integrated randomly, insertional mutagenesis at critical genomic sites may occur, leading to gene disruption or oncogenesis [30, 32]. Moreover, persistent expression of Cas9 cassettes increases the chance of off-target Cas9 editing activity [33]. Introduction of foreign sequences can also cause a host immune response, limiting the use of nucleic acid approaches for therapeutic applications [34, 35]. RNA approaches, in which Cas9 mRNA and sgRNA are co-introduced into cells, offer a technique that bypasses many of these pitfalls. However, the poor stability of mRNA and the reliance on host translational machinery hinder the efficiency of this approach. To mitigate the problems of DNA and RNA-based methods, CRISPR/Cas reagents can be delivered as pre-formed Cas9/sgRNA RNP complexes.
Table 1.
Advantages and limitations of CRISPR/Cas9 delivery methods.
| Delivery method | Advantages | Limitations |
|---|---|---|
| Viral/Plasmid DNA |
|
|
| Cas9 mRNA and sgRNA |
|
|
| Cas9/sgRNA RNP |
|
|
RNP-mediated genome and epigenome editing holds several advantages over nucleic acid and RNA delivery methods, such as its rapid activity, low off-target editing rates, and minimal cellular toxicity (Table 1). When introduced as an RNP complex, Cas9 does not require cellular transcription or translation steps in order to mediate targeted DSB formation. As such, RNPs cleave DNA more rapidly than other delivery methods, and reach a maximum mutation frequency at 24 hours [36]. Additionally, when delivered as RNPs, Cas9 degrades more quickly as compared to other delivery methods, with minimal protein detected after 24–48 hours [36, 37]. Taken together, these results exhibit that Cas9 RNPs mediate rapid genome editing followed by swift degradation of intracellular Cas9, minimizing the amount of time after precise on-target editing that Cas9 is active within the cell. Due to these characteristics, RNPs can achieve efficient genome editing while minimizing off-target effects and cellular toxicity [36–38].
In addition to commonly used S. pyogenes Cas9, RNPs have successfully been assembled from various Cas9 orthologs, engineered Cas9 variants, and Cas9 fusion proteins [38–43]. Moreover, modified and/or extended sgRNAs, such as those used for prime and epigenome editing, have been delivered with Cas9 as RNP complexes [44]. Together, these additions have expanded the breadth of PAM recognition sequences and targetable genomic sites, increased on-target:off-target editing ratios, and extended genome and epigenome applications that can be achieved using RNPs. A summary of the critical developments in RNP-mediated genome and epigenome editing in mammalian cells is displayed in Figure 2.
Figure 2.
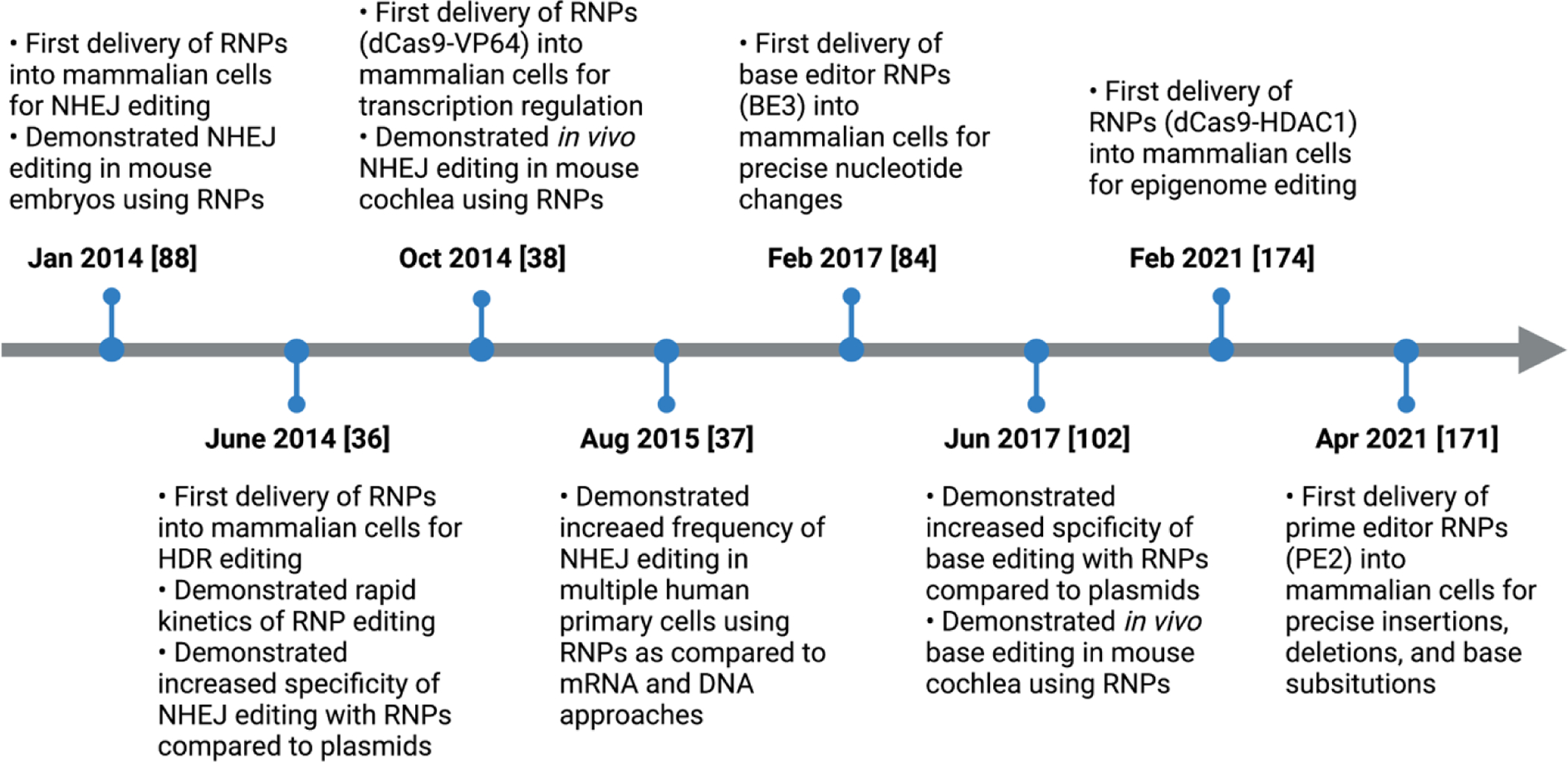
Timeline of critical developments in RNP-mediated genome and epigenome editing in mammalian cells. NHEJ, non-homologous end joining; HDR, homology-directed repair.
In this review, we start by detailing the design of RNPs and methods of intracellular RNP delivery. Next, we describe the numerous editing outcomes that RNPs can facilitate and give examples of editing applications in clinically relevant cell types. Finally, we highlight challenges and future research directions.
2. RNP Production
A ribonucleoprotein is composed of an RNA molecule and an RNA-binding protein. In the context of CRISPR/Cas9-mediated genome and epigenome editing, RNPs form a complex between a Cas9 protein and a site-specific sgRNA. For RNP delivery into target cells, Cas9 and sgRNAs are separately produced and assembled in vitro. Here, we briefly describe the reagents and procedures for RNP production.
2.1. CRISPR-Associated Protein
Numerous Cas9 variants are used to achieve a broad range of CRISPR-mediated editing outcomes. For conventional genome editing, including NHEJ and HDR approaches, the S. pyogenes Cas9 protein flanked by N and C terminal nuclear localization signals (NLS) is used (here on out referred to as WT-Cas9). NLS allows for improved nuclear import of Cas9/sgRNA RNP complexes and higher editing efficiencies [12]. Inactivating point mutations of catalytic RuvC (D10A) or HNH (H840A) domains in Cas9 allows for the generation of single strand nicking nCas9 variants [15]. Single or paired Cas9 nickases have been used to reduce off-target mutagenesis while retaining high on-target editing efficiencies [45–48]. Simultaneous inactivation of RuvC and HNH domains with D10A and H840A mutations, respectively, allows for the generation of a catalytically inactive dCas9 [49]. Perturbations of WT-Cas9, nCas9, and dCas9 have greatly expanded the scope of genome and epigenome editing applications and outcomes [20, 21, 27, 50, 51]. For example, engineered nCas9 fusion proteins have led to the development of novel base and prime editors (protein structures described in sections 4.3 and 4.4, respectively). Moreover, fusion of dCas9 to effector domains, such as transcription regulators or epigenome modifiers, has allowed for the modulation of target gene expression without harmful DSB formation (protein structures described in section 4.5) [26, 27, 52].
WT-Cas9 and some commonly used Cas9 variants are commercially available and can be purchased from various suppliers to use in RNPs. However, Cas9 fusion proteins required for base, prime, and epigenome editing are generally not commercially available. These Cas9 proteins can be produced using E. coli expression systems. Briefly, a plasmid encoding the specific Cas9 variant driven by a T7 promoter is transformed into E. coli [36]. The Cas9 sequence commonly includes an N and C terminal NLS and hemagglutinin (HA) or hexahistidine (6-His) tags for downstream purification [36, 53]. To date, onsite protein production and purification is still a major challenge for labs and represents a barrier for the use of RNPs in research. The commercial availability of these Cas9 variants would greatly enhance the ease and scalability of RNP experimentation.
2.2. sgRNA
sgRNAs are chimeric RNA molecules formed by fusing a site-specific crRNA and tracrRNA [15]. As with some Cas9 nucleases, sgRNAs can be purchased commercially. Chemical modifications to sgRNAs, such as the incorporation of 2’-O-methyl 3’phosphorothioate at the three terminal nucleotides at both the 5’ and 3’ ends, have improved genome editing efficiencies and are now regularly used [54–57]. Some sgRNAs, including those with RNA aptamer extensions used for epigenome editing, may require onsite production. Production of sgRNAs is typically done via in vitro transcription (IVT). First, a transcription template is created with a T7 promoter preceding the sgRNA sequence. Next, the template is transcribed in vitro by T7 RNA polymerase and the sgRNA is purified [53, 58].
2.3. RNP assembly
Complexation of RNPs is completed by mixing Cas9 protein and sgRNA at various molar ratios. The two components are mixed in an aqueous buffer (such as phosphate buffered saline, PBS) and incubated at room temperature for 10–20 minutes [53, 58, 59]. The specific molar ratios may be dependent on various conditions, such as the cell type and editing method, and relevant literature should be consulted. Generally, a molar ratio of 1:1 Cas9 to sgRNA allows for efficient editing. However, in some instances, increased amounts of sgRNA has been shown to overcome its intrinsic instability and generate increased editing efficiencies [59–62].
3. Methods of RNP Delivery
Efficient delivery of RNPs into target cells remains a major barrier in its widespread use. Due to its complex composition and charge properties, RNPs pose particular challenges compared to the delivery of proteins or nucleic acid systems [63]. Cas9 RNP delivery vehicles are classified into three groups: physical approaches, synthetic carriers, and virus-like particles (Figure 3).
Figure 3.
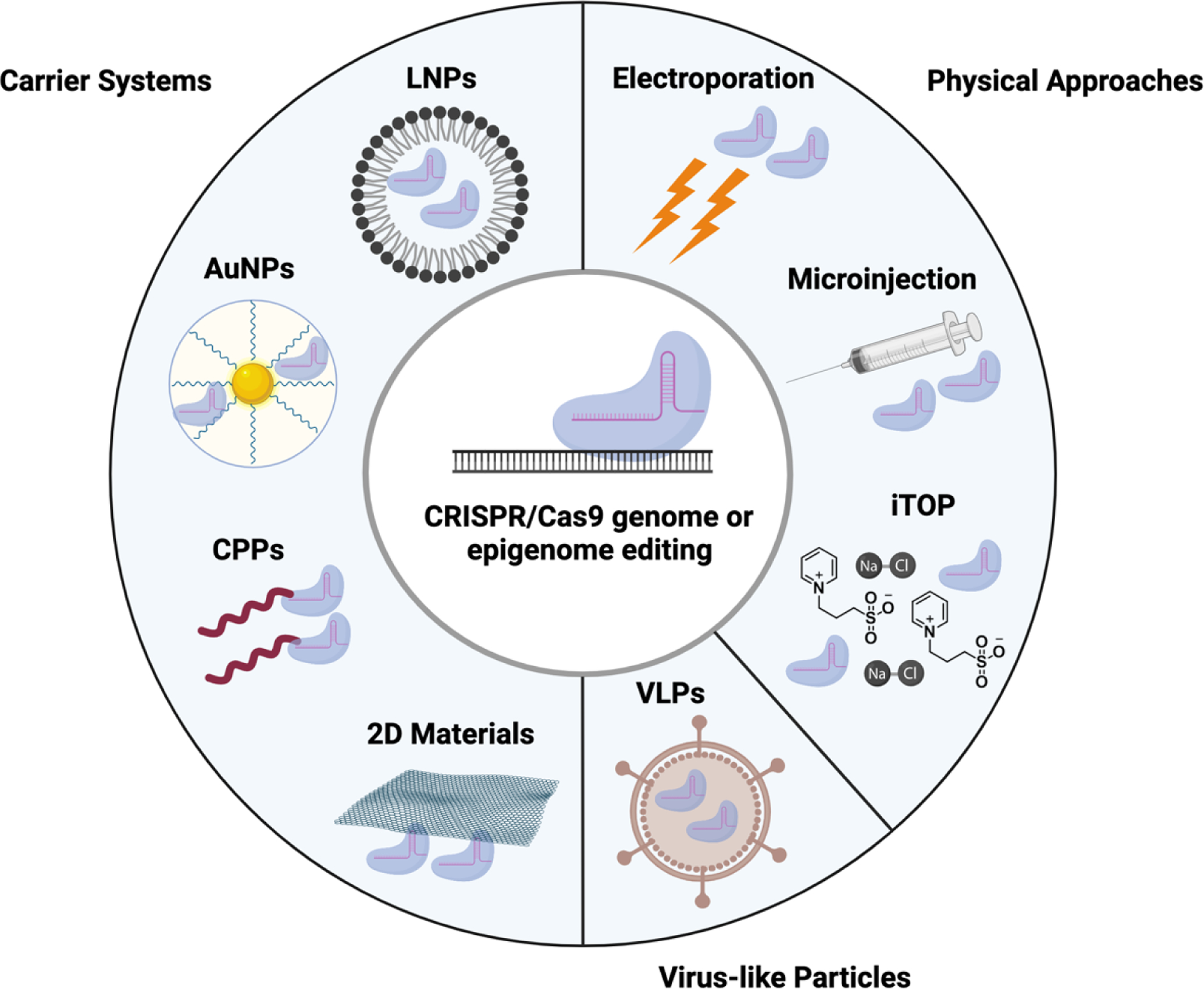
Methods of RNP delivery are categorized into three groups: physical approaches, which include electroporation, microinjection, and iTOP (induced transduction by osmocytosis and propanebetaine), carrier systems, which include LNPs (lipid nanoparticles), AuNPs (gold nanoparticles), CPPs (cell penetrating peptides), and 2D materials, and VLPs (virus-like particles).
3.1. Physical Approaches
3.1.1. Electroporation
Electroporation is one of the most applied techniques for introducing Cas9 RNPs into target cells (Figure 3). A briefly applied electrical pulse creates small pores in the phospholipid bilayer of cellular membranes, temporarily increasing the cellular permeability and allowing for RNP entry. Electroporation efficiently transfects RNPs into various cell types, including but not limited to hematopoietic cells, induced pluripotent stem cells (iPSCs), and zygotes [64–85]. Additionally, electroporation has proven more efficient than other RNP delivery methods, such as lipid-mediated transfection and microinjection (whose mechanisms are explained further on) [79, 86]. For example, RNPs targeting the cystic fibrosis transmembrane receptor (CFTR) gene in iPSCs achieved 89% genome editing when delivered by electroporation, compared to 12% when transfected with Lipofectamine 2000 [79]. In another study, electroporation of mouse zygotes with RNPs and a single strand oligodeoxynucleotide (ssODN) donor template induced higher rates of transgene integration compared to intranuclear microinjection [86]. Although effective, electroporation may cause cellular toxicity due to its transient disruption of the cellular membrane and potential for non-reversible permeabilization [87]. Careful consideration and optimization of voltage and exposure duration is necessary to reduce toxic effects on cells [85].
3.1.2. Microinjection
Microinjection is another physical approach for delivering RNPs directly into cells (Figure 3). In this method, RNPs are injected into a cell’s nucleus or cytoplasm through a glass micropipette or metal syringe, where the amount of RNP can be directly controlled. Due to microinjection operating as a single-cell targeting system, the approach is time consuming and limited in scope. However, microinjection is commonly applied for editing of single-cell systems, such as zygotes, where it has demonstrated efficient delivery [88–91]. For example, in human three pronuclei one-cell embryos, RNP microinjection led to 80% gene knockout at the glucose-6-phosphate dehydrogenase (G6PD) locus [89]. Additionally, microinjection of RNPs into non-human primate zygotes resulted in 100% gene knockout at two separate genetic loci [91]. Although studies have demonstrated safety and efficacy thus far, the process of microinjection may cause cell damage, requiring the need for highly skilled personnel and sophisticated equipment [92].
3.1.3. Induced Transduction by Osmocytosis and Propanebetaine (iTOP)
Induced transduction by osmocytosis and propanebetaine (iTOP) is a method to directly introduce RNPs into cells through the combination of NaCl-mediated hypertonicity and propanebetaine (Figure 3) [93]. To induce RNP entry, iTOP exploits macropinocytosis, a natural active uptake mechanism triggered through hypertonicity. Upon RNP internalization, propanebetaine acts to release RNPs from endolysosomes. This method preserves cell viability and effectively delivers RNPs into fragile, difficult-to-transduce cell types, such as primary human T cells and iPSCs [94]. For example, iTOP-mediated RNP editing in primary human T cells resulted in up to 43% indel formation at the beta-2-microglobulin (B2M) locus, while retaining cellular viability greater than 90% [94]. While iTOP has demonstrated promise for RNP applications, it is not suitable for in vivo RNP editing due to the use of high salt concentrations [92].
3.2. Synthetic Carriers
3.2.1. Lipid Nanoparticles (LNPs)
Lipid nanoparticles (LNPs) are common vehicles for the intracellular delivery of Cas9 RNPs due to their biocompatibility, sustained circulation time, and low toxicity (Figure 3) [95, 96]. Owing in part to their size and surface chemistry, many LNPs enter cells by endocytosis, which allows RNP entry into the cellular cytoplasm and nucleus [97, 98]. LNPs can be formulated from commercially available lipids or synthesized lipids [92]. Commercial reagents that have been developed and used to deliver Cas9 RNPs into cells include Lipofectamine 2000/3000, Lipofectamine RNAiMAX, and Lipofectamine CRISPRMAX [37, 60, 99–104]. For example, Lipofectamine 3000-based delivery of RNPs via subretinal injection in mice achieved local knockout of vascular endothelial growth factor A (VEGFA) [104]. In addition, LNPs formulated with synthetic lipids have been designed by various groups in an effort to increase the encapsulation and delivery efficiency of cargo [60, 105–109]. These include the addition of polymer coatings and targeting ligands, which increase the retention time and aid in the specific targeting and uptake of LNPs. Synthesized LNPs may be formulated with a variety of amine head groups and tails in many variations [105]. Additionally, they can be produced in large batches and are easily modified [106]. Notably, synthesized LNPs have preferentially delivered RNPs into various organs in vivo, including muscle, brain, liver, and lungs [60]. For example, tail vein injection of LNPs packaged with five unique RNPs mediated multiplex gene knockout in mouse lungs [60]. In order to further improve the use of LNPs for RNP packaging and genome and epigenome editing, their stability, specificity, and biocompatibility should be further optimized [110].
3.2.2. Gold Nanoparticles (AuNPs)
Gold nanoparticles (AuNPs) represent another nanoparticle-based delivery system used for RNP editing (Figure 3). RNPs are packaged within AuNPs and are delivered to various cell types to achieve high efficiency of genome and epigenome editing. AuNPs are often utilized due to their low toxicity, ability to package donor DNA, and in vivo editing capabilities [111–113]. For example, cationic arginine AuNPs assembled with Cas9 RNPs generated indels in up to 30% of HeLa cells [112]. In another study, AuNPs packaged with RNPs and donor DNA were injected intramuscularly into a Duchenne muscular dystrophy (DMD) mouse model and achieved gene correction at the pathogenic dystrophin gene [111]. The same group demonstrated a reduction in fragile X syndrome (FXS) symptoms upon intracranial injection of AuNPs packaged with RNPs targeting the overreactive metabotropic glutamate receptor 5 (mGluR5) gene in an FXS mouse model [113]. Some potential challenges of using AuNPs include their aggregation in various mediums, unknown long-term toxicity, and high accumulation in the liver and spleen [114].
3.2.3. Cell Penetrating Peptides (CPPs)
Cell penetrating peptides (CPPs) are short peptides that can mediate the transport of Cas9 RNPs across the cell membrane via passive or active endocytic pathways (Figure 3) [115]. CPPs are advantageous due to their simplicity and convenience [63]. In one study, generation of RNPs containing a Cas9-LMWP (low molecular weight protamine) fusion protein achieved multiplex targeting of immune checkpoint molecules in up to 90% of suspension cancer cells [116]. In another study, RNPs composed of a Cas9 protein fused to a supercharged polypeptide and sgRNAs targeting the clinically relevant CCR5 locus mediated indel formation of 15% in HeLa cells [117]. Although some studies have shown success using CPPs for RNP delivery, the technique usually leads to lower editing efficiency than more established delivery approaches.
3.2.4. 2 Dimensional (2D) Materials
2D materials, such as graphene oxide (GO) and black phosphorous nanosheets (BPs), have been used for the intracellular delivery of RNPs (Figure 3) [118, 119]. Due to their high surface area and ability to shield cargo from enzymatic degradation, large amounts of RNP can be packaged and successfully delivered to target cells [120]. Using GO-polyethylene glycol (PEG)-polyethyleneimine (PEI) nanocarriers, one group demonstrated 33% indel formation at the clinically relevant CXCR-4 locus in a human gastric adenocarcinoma cell line [118]. In another study, BPs loaded with Cas9 RNPs exhibited cytosolic entry via the endocytosis pathway, followed by cargo release upon BPs degradation [119]. Targeting two loci in the MCF-7 breast cancer cell line, BPs-based RNP delivery achieved 27% and 32% genome editing, respectively. Additionally, upon intratumoral injection of BPs in a mouse model, successful in vivo editing was detected. Although 2D materials offer therapeutic potential, some challenges for RNP delivery remain, including the reliable scale up of 2D materials and the potential for in vivo toxicity [120].
3.3. Virus-like Particles
Virus-like particles (VLPs) are being studied for the delivery of RNPs into target cells due to their ease of production and low dosage requirement (Figure 3) [121]. In contrast to the traditional delivery of Cas9 by viral vectors, in which a Cas9 cassette is encoded in the viral genome, VLPs lack a viral genome. Instead pre-formed RNP complexes are packaged in viral capsid proteins that retain their icosahedral or helical capsid structures [121]. A common strategy for packaging RNPs in VLPs is via the interaction of aptamer-modified sgRNA with aptamer-binding proteins (ABPs) fused to viral nucleocapsid proteins [122, 123]. The most prevalent VLPs use lentiviral (LV) capsid proteins for RNP delivery due to their high transduction efficiency [122–125]. For example, LV-based VLPs (LVLPs) packaged with RNPs targeting the interleukin 2 receptor subunit gamma (IL2RG) gene achieved indel formation in 84% of HEK293T cells [123]. In addition to LV particles, VLPs formed with other viral capsid proteins have been studied. For example, VLPs derived from murine leukemia virus demonstrated efficient RNP delivery and genome editing in iPSCs, hematopoietic stem and progenitor cells (HSPCs), and mouse bone-marrow cells [126]. However, one of the major drawbacks of using VLPs is the potential for innate and adaptive immune reactions to the viral particles, making this approach unsuitable for clinical applications [35, 127].
4. Mechanisms of RNP-Mediated Genome and Epigenome Editing
Cas9 RNPs have been employed in various genome and epigenome editing applications. Here, we will discuss the use and advantages of RNPs to facilitate NHEJ, HDR, base, prime, and epigenome editing.
4.1. Non-Homologous End Joining (NHEJ)
Upon RNP delivery and targeted DSB formation, NHEJ functions as the primary DNA repair pathway [16]. NHEJ is considered an error-prone repair mechanism, often resulting in indels at the site of the DSB (Figure 1) [17]. When Cas9 is targeted to a gene of interest, a fraction of indels will result in the introduction of frameshift, nonsense, or missense mutations, resulting in gene knockout. In addition, simultaneous targeting of multiple genomic sites can result in NHEJ-mediated large deletions, inversions, or chromosomal translocations [128]. The NHEJ repair pathway can also be harnessed for transgene knock-in via the homology-independent targeted integration (HITI) method [129, 130].
In mammalian cells, NHEJ repair of Cas9-mediated DSBs has been explored for both research purposes, such as generating gene knockouts for studying protein function, and therapeutic applications, such as silencing of dominant gain-of-function and dominant-negative mutant alleles [101, 131–133]. RNP delivery of Cas9 in mammalian cells was first described in 2014 to generate knockout mice [88]. Since then, countless studies have shown RNP-mediated NHEJ editing in mammalian cells, both in vitro and in vivo.
Head-to-head comparisons of RNPs to other Cas9 delivery systems, such as plasmids, viruses, and mRNA, have demonstrated various advantages of RNPs that make them a promising tool for efficient NHEJ genome editing [36, 37]. For example, one study showed that RNP delivery of Cas9 had up to 13-fold higher on-target:off-target editing ratios compared to plasmid delivery [36]. In addition, RNP delivery was roughly two-fold less toxic than plasmid delivery. Notably, this did not compromise editing efficiencies, as increased indel formation was achieved using RNPs in multiple cell types, including embryonic stem cells (ESCs) and fibroblasts. In another study, RNPs achieved similar on-target editing efficiencies in HEK293T cells compared to plasmids, but up to 28-fold higher on-target:off-target editing ratios [37]. Furthermore, RNP-mediated indel efficiency was higher than plasmid or mRNA approaches in Jurkat T cells, human iPSCs, human ESCs, mouse ESCs, and human cord blood CD34+ cells.
4.2. Homology-Directed Repair (HDR)
HDR functions as an accurate, yet less efficient, DSB repair pathway [134]. For genome editing applications, HDR employs an exogenous DNA donor template to produce precise insertions, deletions, or base substitutions at a target genomic site (Figure 1) [9, 10, 13, 14]. To facilitate HDR editing, donor templates contain a pair of sequences homologous to the genomic DNA flanking the target DSB, which are referred to as homology arms.
During the resolution of DSBs, HDR competes with the more frequent NHEJ pathway [134]. As such, indels are generally more abundant outcomes than the desired HDR edit. Notably, the stage of the cell cycle plays a principal role in determining the relative frequencies of the two DSB repair pathways. NHEJ is active throughout the entire cell cycle, but dominates during the G0, G1 and early S phase [135–137]. In contrast, HDR is limited to late S and G2 phases, as the endogenous HDR system relies on sister chromatids as the primary DNA repair template [135–137]. Harnessing this knowledge, G2/M synchronization of cells with nocodazole led to increased HDR editing in cell lines and primary cells upon delivery of RNPs and an ssODN donor template [138]. Because of its cell cycle characteristics, HDR is largely restricted to actively dividing cells.
HDR can be utilized in mammalian cells to knock-in transgenes, generate disease models, or correct pathogenic mutations to treat monogenic diseases [139, 140]. RNP-mediated HDR editing was initially achieved in mammalian cells in 2014 [36]. Delivery of an ssODN donor template and RNPs targeting the safe-harbor AAVS1 site resulted in 15% HDR editing in the K562 cell line. Soon after, RNP-mediated HDR editing was demonstrated in human primary neonatal fibroblasts and human ESCs [138]. Another study showed 8–11% HDR efficiency upon RNP and ssODN donor template delivery in HEK293T cells, similar to that achieved by optimized plasmid protocols [38]. Since these initial experiments, RNP-mediated HDR editing has been successfully demonstrated in multiple cell types, expanding its use in mammalian genome engineering.
Due to the shared mechanism of Cas9-mediated DSB formation in HDR and NHEJ editing, many of the aforementioned benefits of RNPs apply to HDR editing. For example, RNPs result in DSB formation within 3 hours of delivery, which enables rapid HDR repair [36]. Additionally, when used in HDR editing, the transient nature of RNPs results in decreased cellular toxicity and off-target Cas9 activity compared to plasmid, viral, and mRNA approaches [36, 37].
4.3. Base Editing
Although HDR offers unique possibilities for genome editing applications, its use is inhibited by several pitfalls. For example, Cas9-induced DSBs give rise to variable editing outcomes during HDR, such as undesired indels at target sites, and can lead to harmful p53 activation [10, 141, 142]. Moreover, strategies to correct point mutations using HDR require the introduction of exogenous donor templates and largely remain inefficient, especially in post-mitotic or non-dividing cells. Base editing was designed to circumvent these drawbacks. The system facilitates the precise conversion of one DNA base to another at a target locus without the requirement of potentially harmful DSBs or inefficient HDR mechanisms [20]. The latest versions of base editors contain a catalytically impaired Cas9 nuclease (nCas9; Cas9 D10A) fused to an engineered deaminase enzyme and proteins that modulate DNA repair pathways. Target-specific sgRNAs guide the Cas9 nuclease to a genomic site of interest, where the deaminase enzyme catalyzes a substitution reaction within a five-base activity window spanning protospacer sequences 4–8 (where the PAM sequence is at positions 21–23) (Figure 1) [18, 19, 143–154]. Two classes of base editors exist: cytosine base editors (CBEs), which use cytidine deaminases to convert C-G base pairs to T-A, and adenine base editors (ABEs), which use adenosine deaminases to induce A-T to G-C conversions [18, 19]. As such, base editing strategies can install the four transition mutations (C-to-T, G-to-A, A-to-G, and T-to-C).
Since its development, base editors have been used to correct pathogenic mutations, induce mutagenesis, or knockout genes [19, 76, 155–157]. Its applications have been further expanded by engineering base editors with altered PAM requirements, narrowed activity windows, decreased bystander mutations, reduced DNA and RNA off-target effects, and small molecule dependence [43, 148–154, 158–161].
Initial demonstrations of base editing relied on plasmid transfection for intracellular delivery. However, early studies found problems with expression vectors that reduced base editor transcription and translation efficiency in mammalian cells [146, 147]. Additionally, high levels of guide-dependent and guide-independent DNA and RNA off-target editing using both CBEs and ABEs raised concerns about sustained expression systems [125, 148–154, 162–164]. Due to its transient effects, RNP delivery of base editors has overcome these challenges [84, 102, 125, 152–154, 164]. For example, RNP delivery of the BE4 base editor in HEK293T cells resulted in a 21-fold decrease in guide-independent DNA off-target deamination events compared to plasmid delivery, with similar on-target editing efficiencies [153]. Similarly, RNP delivery of the ABE8e base editor in HEK293T cells showed comparable levels of on-target editing as plasmid delivery, yet achieved a dramatic increase of on-target:off-target editing ratios up to 1,300-fold at nine guide-dependent DNA off-target sites [152]. Moreover, ABE7.10 base editors delivered as RNPs led to no detectable guide-independent RNA off-target activity, whereas plasmid delivery resulted in A-to-G changes at off-target sites in RNA [125].
4.4. Prime Editing
The advancement of genome editing applications requires the ability to make precise insertions, deletions, or point mutations in mammalian genomes. As previously mentioned, Cas9 nucleases can induce site-specific DSBs that, when in the presence of an exogenous DNA donor template, can lead to a fraction of precise genomic changes using HDR [9, 10, 13, 14]. However, HDR editing is accompanied by undesired indels that may compromise genomic integrity and lead to heterogeneous gene edited cell populations. Additionally, HDR editing efficiencies remain low in many cell types. Recently, base editors were developed to install all four transition mutations at target sites without inducing DSBs or requiring a donor template [18, 19]. However, undesired bystander edits and a limitation of target sites due to PAM availability restrict its use in certain instances. To supplement HDR and base editing, and to overcome some of their inherent barriers, the prime editing system was developed [22].
Prime editing can mediate targeted insertions, deletions, and all 12 possible base-to-base conversions, including the eight transversions inaccessible by base editors, without inducing DSBs or requiring the delivery of a donor DNA template (Figure 1) [21]. This ‘search-and-replace’ genome editing technology employs an engineered reverse transcriptase (RT) fused to catalytically impaired Cas9 (nCas9; Cas9 H840A) to copy genetic information from an engineered RNA extension of a guide RNA (termed a prime editing guide RNA; pegRNA) into the target genomic locus.
When initially described, more than 175 edits in human cells were made, including targeted insertions, deletions, and base substitutions at disease-causing variants [22]. Successful prime editing has been established in various human cell lines, mouse primary cortical neurons, induced pluripotent stem cells (iPSCs), organoids, mouse embryos, mouse liver cells, and human primary T cells [22, 165–171].
Prime editors are primarily delivered to cells by DNA or mRNA approaches [21]. However, the large size of prime editors raises challenges for its delivery. For instance, the limiting packaging capacity of viral vectors make them unsuitable in certain applications. Additionally, low efficiency of in vitro synthesis of prime editor mRNA transcripts restricts the scalability of this approach. RNP delivery of prime editors may circumvent these obstacles. To date, we are aware of only one study that has demonstrated prime editing using RNPs in mammalian cells [171]. Here, RNPs were prepared with PE2 editors and specific pegRNAs/sgRNAs and delivered into human cells. Using different pegRNA/sgRNA combinations, RNPs introduced targeted G-to-C substitutions, 3-bp insertions, and 5-bp deletions at efficiencies up to 21% in HEK293T cells and 8% in primary human T cells. Although it is yet to be investigated, prime editing RNPs likely induce fewer off-target edits compared to other delivery methods, owing to its rapid degradation following on-target editing activity [36].
4.5. Transcription Regulation & Epigenome Editing
As the investigation and application of CRISPR/Cas9-mediated genome engineering has blossomed in recent years, parallel exploration of catalytically inactive CRISPR/Cas9 systems (including dCas9) has allowed for the regulation of gene expression [26, 27]. dCas9 (Cas9 D10A/H840A) enables the recruitment of effector domains to target genes, where they can function to modulate transcriptional regulation (Figure 1). Effector domains include gene regulatory domains, which directly influence gene expression through interactions with transcriptional machinery (termed CRISPR activation or CRISPR interference), or epigenetic modifying domains, which alter gene expression following modifications to epigenetic markers, such as histone acetylation and DNA methylation status.
Two broad categories have been described to achieve dCas9-based regulation of gene expression. In the first, dCas9 is directly fused to an effector domain, and the fusion protein is chaperoned to a target gene upon RNP complex formation with a site-specific sgRNA [24, 25, 38, 172–174]. In the second, effector domains are fused to RNA-binding proteins that are recruited to a genomic locus through interactions with an RNA aptamer extension on a sgRNA [25, 175].
While a promising strategy to influence gene expression, epigenome editing has shown evidence of off-target activity [26]. For example, a fusion protein between dCas9 and the catalytic domain of Dnmt3a, a DNA methyltransferase involved in de novo methylation, resulted in global off-target methylation in a novel ESC reporter system [176]. Due to this concern, RNP delivery has been proposed as a potential approach to reduce off-target effects. Additionally, RNPs may be beneficial when transient modifications to gene expression are desired, as strategies employing plasmid or viral vectors confer sustained transcriptional changes.
RNPs were first demonstrated to modulate gene expression in mammalian cells by utilizing the VP64 transcriptional activator [38]. Delivery of RNPs formulated with dCas9-VP64 and sgRNAs targeting neurotrophin-3 (NTF3), a neural growth factor associated with neurodegenerative diseases, resulted in higher than ten-fold gene activation. Compared to plasmid delivery of dCas9-VP64, RNPs achieved rapid, transient transcription activation. Whereas RNPs achieved maximal gene activation 10 hours post-transfection, plasmid delivery reached maximal effects only after 50 hours. Notably, NTF3 transcription returned to near baseline levels 50 hours after RNP delivery, highlighting its transient nature.
Recently, rewriting of epigenetic markers was achieved by RNP delivery of a fusion dCas9-HDAC1 protein and sgRNAs targeting the prominent KRAS oncogene [174]. HDAC1, or histone deacetylase 1, is responsible for the deacetylation of lysine residues on the N-terminal part of the core histone proteins, resulting in gene repression in a context dependent manner. RNP delivery resulted in significant K-Ras silencing up to 9-fold in two cancer cell lines. Efficient epigenome editing led to changes in cancer cell morphology, such as cell shrinkage and loss of cell-cell contact, decreased colony production, and decreased cell viability.
5. Applications of RNP-Mediated Genome and Epigenome Editing in Mammalian Cells
The early illustration of the advantages of RNPs led to a rapid expansion of its use in clinical and research applications (Table 2). Here, we describe the application of RNP-mediated genome and epigenome editing at various target cells and tissues.
Table 2.
Clinical applications of RNP-mediated genome editing.
| Target cells | Editing methods | Clinical application | Reference |
|---|---|---|---|
| T cells | NHEJ | HIV infection treatment | [177] |
| NHEJ, HDR, base editing | CAR-T cell immunotherapy development | [66, 67, 178, 179] | |
| B cells | HDR | Protein secreting plasma cells for protein deficiency treatment (ex. hemophilia B) | [69] |
| HSPCs | NHEJ, HDR, base editing | Hematologic disorder treatment (ex. sickle cell disease) | [41, 73, 74, 76, 180–183] |
| NHEJ, HDR | Immunodeficiency disorder treatment (ex. X-linked severe combined immunodeficiency) | [184–187] | |
| HDR | Metabolic disorder treatment (ex. mucopolysaccharidosis type I) | [188] | |
| iPSCs | HDR | Skin disorder treatment (ex. recessive dystrophic epidermolysis bullosa) | [77] |
| HDR | Hematologic disorder treatment (ex. sickle cell disease) | [78] | |
| HDR | Lung disorder treatment (ex. cystic fibrosis) | [79] | |
| Zygotes | NHEJ, HDR, base editing | Germline gene editing | [80–85, 88, 91] |
| HDR | Germline gene therapy (ex. hypertrophic cardiomyopathy) | [89, 90] | |
| Retinal cells | NHEJ | Ocular disorder treatment (ex. age-related macular degeneration) | [100, 104] |
| Cochlear cells | NHEJ, base editing | Sensorineural hearing loss treatment | [101, 103] |
HPSCs, hematopoietic stem and progenitor cells; iPSCs, induced pluripotent stem cells; NHEJ, non-homologous end joining; HDR, homology directed repair; HIV, human immunodeficiency virus; CAR, chimeric antigen receptor.
5.1. Hematopoietic Cells
Due to their expansive roles, hematopoietic cells represent a promising means for the treatment and prevention of cancers, infectious diseases, autoimmune diseases, blood disorders, and inherited immunodeficiencies [189–192]. These cells can be successfully isolated from diseased or healthy patients in large quantities, allowing for ex vivo editing and subsequent allogeneic or autologous transplantation.
5.1.1. T Cells
Gene edited T cells are being intently explored for cancer immunotherapy and HIV treatment, amongst other applications [193, 194]. Early CRISPR studies in T cells took advantage of viral vectors to mediate intracellular Cas9 delivery. However, low knockout efficiencies were observed, and selection strategies were required to boost edited cell populations [195, 196]. Since then, the introduction of Cas9 RNPs into T cells has dramatically improved editing efficiencies [64, 65, 177–179, 197]. For example, one study showed that both activated and non-activated human T cells routinely achieved greater than 90% knockout efficiencies when electroplated with RNPs [64]. Another study demonstrated efficient multiplex NHEJ editing using RNPs to generate double-knockout T cell populations resistant to HIV infection [177].
Furthermore, the introduction of RNPs into T cells with ssODN or adeno associated virus (AAV) donor templates has led to HDR gene correction and transgene knock-in [66, 197]. For instance, delivery of RNPs targeting T cell receptor alpha chain (TRAC) and AAV donor templates with a CD19-specific chimeric antigen receptor (CAR) led to targeted gene integration resulting in donor-derived non-alloreactive CAR-T cells [66]. An HDR knock-in efficiency of 79% was achieved, exceeding efficiencies in a previous report in which Cas9 mRNA and AAV donor templates were delivered to T cells [198]. Notably, human primary T cells that undergo RNP-based gene knockout or knock-in retain ex vivo expansion potential, vital for the development of cell products at a clinically relevant scale [66, 199].
Base editing has also been employed to create “off-the-shelf” T cells by reducing alloreactivity and disrupting checkpoint inhibitors [67]. To achieve this goal, base editors were designed to mediate multiplex knockout of three genes: TRAC, B-2 microglobulin (B2M), and programmed cell death 1 (PDCD1). Whereas mRNA delivery of first-generation BE4 editors resulted in 22% triple knockout cells, BE4 RNP delivery successfully induced triple knockout in 69% of cells. Interestingly, delivery of codon optimized BE4 mRNA resulted in even higher efficiencies of multiplex knockout, suggesting mRNA delivery may be preferential to RNPs in certain base editing instances.
5.1.2. B Cells
Because some B cells, such as long-lived plasma cells and memory B cells, persist for the duration of an organism’s life, they offer a suitable target for gene therapy. Recently, investigators have achieved efficient Cas9 RNP-mediated genome editing in B cells, offering advantages over other Cas9 delivery methods [68–70, 200]. For example, delivery of RNPs or Cas9 mRNA into human B cells demonstrated similar on-target indel formation (74% and 72%, respectively), while RNPs resulted in greater cell viability compared to mRNA (75% and 59%, respectively) [68]. Additionally, simultaneous RNP-targeting of two genetic loci in B cells led to the successful generation of translocations characteristic of Burkitt Lymphoma, a form of B-cell non-Hodgkin’s lymphoma. In another study, targeted gene knock-in demonstrated the ability to engineer exogenous protein production in plasma cells [69]. This technique may have therapeutic applications in protein deficiency diseases, such as hemophilia B, which is characterized by a deficiency in factor IX (FIX). Delivery of RNPs and an AAV donor template to primary human B cells resulted in up to 20% knock-in of FIX at the CCR5 safe-harbor site and more than 10-fold increase in FIX secretion.
Efficient HDR editing has also been confirmed in primary B cells using non-viral ssODN donor templates [70]. Importantly, RNP editing of B cells has no major impact on the cellular transcriptome and does not induce a detectable type I interferon response, exhibiting the safe nature of gene edited B cells [200].
5.1.3. Myeloid Cells
Myeloid cells are key players in the innate immune system. Due to a restriction of efficient genome editing techniques in primary human and murine myeloid cells, research on innate immunity primarily relies on iPSC-derived myeloid cells or virus-mediated gene delivery in transformed myeloid cell lines [201, 202]. However, using Cas9 RNPs, near population-level genetic knockouts were recently reported, a significant advancement for the study of myeloid cells [71, 72]. One study showed indel formation of up to 90% in primary human monocytes that were edited and subsequently differentiated into either macrophages or dendritic cells [71]. By pooling multiple sgRNAs to the same gene, nearly 100% knockout efficiency was achieved. Another study demonstrated that efficient RNP-mediated indel formation did not cause a selection of subsets of cells or skew macrophage differentiation [72]. Furthermore, edited macrophages retained functionality and the gene editing platform was used to investigate the function of an important myeloid host viral restriction factor.
5.1.4. Hematopoietic Stem and Progenitor Cells (HSPCs)
Hematopoietic stem and progenitor cells (HSPCs) have the unique ability to reconstitute and maintain a functional blood system. As such, HSPCs have been a major target for genome editing applications. Early studies noted that plasmid delivery to HSPCs is extremely toxic, making plasmid-mediated Cas9 delivery an unsuitable method [203]. Additionally, patients undergoing retroviral-mediated gene therapy in HSPCs have developed cancer, revealing potential pitfalls of viral-mediated Cas9 delivery platforms [204, 205]. Therefore, the exploration of Cas9-mediated gene editing in HSPCs has primarily been with RNPs or Cas9 mRNA [73–76, 180–188, 206–210]. It has been exhibited that compared to Cas9 mRNA delivery, RNPs mediate more efficient genome editing, lead to less cellular toxicity, induce fewer global transcriptional changes, and result in reduced off-target genome editing events [73, 74, 211].
For example, one study showed that RNP delivery resulted in 48% on-target indel formation in HPSCs, compared to only 32% with Cas9 mRNA [73]. When delivered with an AAV donor template harboring a GFP transgene, RNPs mediated gene addition in 29% of HSPCs, compared to 15% by Cas9 mRNA. In HDR experiments, RNPs led to no additional toxicity compared to AAV only samples, whereas Cas9 mRNA triggered significant cytotoxicity. Moreover, measurements of off-target indel formation at a highly complementary DNA site showed that RNPs had up to 10-fold higher on-target:off-target editing ratios compared to Cas9 mRNA. Notably, in another experiment, HDR editing mediated by an AAV donor template and Cas9 mRNA delivery invoked greater transcriptional changes compared to RNP delivery, eliciting a distinct viral response and global downregulation of metabolic and cell cycle processes [211]. Another group used simultaneous Cas9 targeting at two genomic target sites to mediate large deletions or inversions [74]. Compared to mRNA delivery, RNPs mediated a higher efficiency of deletion/inversion outcomes. For two pairs of sgRNAs tested, RNPs mediated up to 24%/20% and 23%/11% deletions/inversions, compared to 2%/2% and 10%/7% by Cas9 mRNA. In this study, Cas9 mRNAs led to significant toxicity proportional to the amount of transfected RNA, resulting in impaired clonogenic potential of HSPCs. In contrast, RNPs added no additional toxicity compared to mock-treated controls and retained the ability to differentiate into all progenitors.
RNPs have also been used to mediate other forms of genome editing in HSPCs. For instance, using an NHEJ-based knock-in approach, 21% targeted transgene integration was achieved in repopulating HSPCs upon delivery of RNPs and an AAV HITI donor template [75]. Moreover, RNPs formulated with a high-fidelity Cas9 variant (HiFi Cas9) successfully mediated comparable levels of gene knock-in as wild-type Cas9, while reducing off-target indel formation by 20-fold at a highly complementary site [41].
Base editing strategies utilizing RNPs have also been successfully employed in HSPCs [76, 182]. In one study, base editors were used to disrupt the BCL11A erythroid enhancer, a potent silencer of fetal hemoglobin (HbF) [76]. Successful disruption led to reduced BCL11A expression and increased ɣ-globin levels. In sickle cell disease (SCD) and β-thalassemia patients, where β-globin production is faulty, induction of ɣ-globin can rescue disease phenotype. In HSPCs isolated from SCD patients, RNP delivery of A3A-BE3 editors resulted in a base editing frequency of up to 91%, which increased HbF levels in erythroid progeny from 5% to 32%. Upon sodium metabisulfite treatment, erythroid progeny showed substantially fewer sickled cells after base editing. Additionally, 18% targeted correction of a common β-thalassemia mutation was achieved by RNP delivery of A3A-BE3 in patient HSPCs. Moreover, RNP-mediated base editing was demonstrated in long-term, repopulating HSPCs, with similar editing efficiencies in each hematopoietic lineage. In another study, ABE8e-NRCH base editors were used to correct the pathogenic mutation in SCD [182]. Although RNP delivery of base editors yielded high levels of gene correction, 44% in HSPCs, ABE8e-NRCH mRNA resulted in 80% precise A-to-G conversions. This highlights that, under some circumstances, mRNA delivery may mediate more efficient base conversions than RNPs. In contrast, in mouse HSPCs, RNPs led to more efficient base editing at the SCD mutation. Additionally, guide-dependent DNA off-target editing was substantially lower by RNPs than by mRNA.
5.2. Human Pluripotent Stem Cells (hPSCs)
Human pluripotent stem cells (hPSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), have unlimited self-renewal capacity and, in principle, can differentiate into all specialized cell types. The development of iPSCs has led to significant strides in human disease modeling and regenerative cell therapy. Notably, patient-derived iPSCs can be genetically modified, differentiated into target cell types, and transplanted for disease correction. As such, developing effective genome editing strategies in iPSCs is of paramount importance.
RNP delivery has demonstrated effective HDR gene correction and transgene knock-in at clinically relevant target sites in iPSCs [77–79]. The COL7A1 gene encodes type VII collagen (C7), and homozygous or compound heterozygous frameshift mutations are responsible for recessive dystrophic epidermolysis bullosa (RDEB), a rare genetic skin disorder [212]. In patient derived iPSCs with homozygous frameshift mutations, delivery of a Cas9 plasmid and an ssDNA donor template led to 10% biallelic correction and 40% monoallelic correction in selected clones [77]. When Cas9 was delivered as an RNP, selected clones showed 58% biallelic correction and 42% monoallelic correction. In addition, iPSCs from patients with compound heterozygous frameshift mutations targeted with RNPs and ssODN donor templates demonstrated multiplex gene correction, with 19% biallelic correction of the two mutant alleles and 48% monoallelic correction. Differentiation of gene-corrected iPSCs into skin cells, generation of human skin equivalents (HSEs), and grafting of HSEs onto nude mice demonstrated skin integrity and C7 restoration.
Another group demonstrated efficient transgene knock-in and gene correction in iPSCs and ESCs upon Cas9 RNP and AAV donor template delivery [78]. Targeting occurred at the hemoglobin subunit beta (HBB) and MYD88 loci, involved in SCD and Waldenstrom’s macroglobulinemia, respectively. AAV donor templates harboring a GFP expression cassette and RNP delivery led to a targeted integration frequency of 51% at the HBB locus in iPSCs, and 91% at the HBB locus and 59% at the MYD88 locus in ESCs. HDR editing was precise, with no incorporation of AAV inverted terminal repeats. Karyotype analysis relieved no chromosomal abnormalities and edited iPSCs and ESCs retained their pluripotent nature. Moreover, in an iPSC line with a homozygous Glu6Val SCD-causing mutation, AAV donor templates harboring a SNP with the corrected allele and RNP delivery achieved 63% allele correction, including a majority of biallelic correction. Differentiated erythrocytes demonstrated high levels of gene-corrected adult hemoglobin and virtually eliminated sickle hemoglobin expression.
In another study, Cas9 RNPs were used to correct the most common pathogenic mutation (3-bp deletion; ΔF508) in the cystic fibrosis transmembrane conductance regulator (CFTR) gene associated with cystic fibrosis (CF) [79]. First, iPSCs were introduced with either RNPs or Cas9 plasmid to determine the most efficient platform for DSB formation. In dF508 patient iPSCs, RNPs achieved greater than 80% indel formation at two sgRNA sites neighboring the pathogenic mutation, compared to just 6% at both sides upon plasmid delivery. Next, RNP and ssODN delivery demonstrated greater than 10% correction of the dF508 mutation in patient iPSCs. Subsequent analysis in proximal lung organoids derived from gene edited iPSCs validated phenotypic correction.
5.3. Zygotes
In vivo editing of zygotes is an important tool for studying mammalian embryogenesis and generating genome edited animals for disease modeling. This method could also be used for the germline correction of pathological sequences in humans, although important ethical considerations are required [213–215]. To date, RNP-mediated editing has been demonstrated in mice, human, non-human primate, and bovine zygotes [80–85, 88–91].
5.3.1. Mouse Zygotes
In initial CRISPR experiments to obtain genetically modified mice, Cas9 mRNA was introduced into zygotes by microinjection or electroporation [13, 14, 216, 217]. However, generation of modified mice was restricted by low editing efficiencies and abundant mosaicism. Evidence of mosaicism implied that Cas9-mediated DSB formation occurred after the first genome replication in the mouse embryos. Notably, investigators demonstrated that protein translated from electroporated mRNA was only detected after E0.8 (E: day), indicating that translational machinery is only fully active at this time in mouse zygotes [82]. Because zygotes generally enter the genome replication phase before E0.8, introduction of Cas9 mRNA would lead to Cas9 protein production after the first genome replication, resulting in mosaic outcomes. In contrast, RNPs do not rely on cellular translational machinery, and were thus proposed as a strategy to decrease mosaicism. Indeed, Cas9 delivery by RNPs in mouse zygotes has led to decreased mosaicism [82, 83]. Additionally, RNPs have achieved improved editing efficiencies and greater viability of embryos after genome editing compared to Cas9 mRNA delivery [80–83, 88].
One study analyzed mosaicism in naturally bred mouse zygotes that were gene edited with either Cas9 mRNA or RNPs and transferred into pseudopregnant females. Cas9 mRNA delivery led to twice as many mosaic embryos compared to RNPs [82]. Additionally, RNP-mediated editing of zygotes resulted in a higher occurrence of biallelic knockout compared to Cas9 mRNA (94% and 87%, respectively). Furthermore, RNPs achieved HDR gene correction when electroporated with an ssODN and demonstrated a 1 kb genomic deletion when simultaneously edited with two sgRNAs. In fact, large deletions up to 16.2 kb have been achieved in mouse zygotes using RNPs [80, 83]. Another group demonstrated in situ editing of mouse zygotes using RNPs, referred to as Improved Genome-editing via Oviductal Nucleic Acids Delivery (i-GONAD) [83]. They showed that RNP-mediated genome editing led to mosaicism in 57% of embryos, whereas Cas9 mRNA resulted in 82% mosaic embryos. RNPs also led to increased editing efficiencies, generating indels in 97% of mouse embryos compared to 31% with Cas9 mRNA. Moreover, in situ RNP and ssDNA injection in zygotes mediated HDR knock-in of a reporter cassette. In a different study, mouse zygotes were electroporated with ssODNs and either RNP or Cas9 mRNA, and subsequently transferred to pseudopregnant females [80]. Cas9 mRNA only resulted in HDR editing after six electroporation pulses, which gave rise to HDR editing in 25% of birthed pups. In contrast, RNPs mediated high efficiency HDR editing in zygotes, including 60% of birthed pups after one electroporation pulse and up to 100% with 8 pulses.
RNPs have also been demonstrated to induce targeted point mutations in mouse zygotes using the BE3 base editor [84]. First, BE3 RNPs designed to induce a premature stop codon (pmSTOP) at the dystrophin (DMD) locus were delivered to mouse zygotes. Analysis revealed that 56% of blastocysts carried the precise C-to-T conversion, and another 25% showed evidence of bystander C-to-T conversions, undesired C-to-A conversions, or indels. Similarly, when targeting the tyrosinase (TYR) locus with BE3 RNPs designed to induce a pmSTOP, analysis showed that 69% of blastocysts carried the precise C-to-T conversion, with another 15% carrying various other mutations. Because the authors were attempting to induce targeted mutagenesis and gene knockout, the variation in editing outcomes was tolerable. Finally, zygotes edited at the TYR locus were transplanted into surrogate mothers and offspring were obtained for analysis. Of the analyzed pups, 57% carried the precise pmSTOP mutation, and the remainder carried various other mutations.
5.3.2. Human Zygotes
RNP delivery has also achieved superior genome editing in human zygotes [89, 90]. In human 3PN (three pronuclei) one-cell embryos, investigators demonstrated up to 80% indel formation upon RNP microinjection [89]. Additionally, the efficiency of HDR editing in 3PN zygotes upon introduction of ssODNs and RNPs or Cas9 mRNA was compared. Whereas Cas9 mRNA did not facilitate HDR editing, RNPs mediated HDR editing in 20% of embryos. Furthermore, RNP and ssODN injection into normal 2PN (dual pronuclei) zygotes achieved greater HDR efficiencies of up to 100%. This technique was then used to correct the B-thalassemia disease causing mutation in 2PN zygotes. In another study, RNPs and ssODN donor templates targeting the heterozygous MYBPC3 mutation causing hypertrophic cardiomyopathy were co-injected with sperm into M-phase oocytes by intracytoplasmic sperm injection (ICSI) [90]. A high yield of gene-corrected homozygous embryos with no cytogenetic abnormalities or off-target indels was achieved. Interestingly, genomic analysis revealed that the wild-type oocyte allele, rather than the exogenous ssODN, was used as a template for HDR repair. This suggests that human embryos employ different DNA repair mechanisms than somatic or pluripotent cells and that providing an exogenous donor template may not be required for gene correction of heterozygous human embryos.
5.3.3. Non-Human Primate Zygotes
In marmosets, Cas9 RNP injection into zygotes led to increased editing efficiencies at two genetic loci compared to Cas9 mRNA injection [91]. Whereas Cas9 mRNA delivery to marmoset zygotes led to indel formations in 78% and 80% of embryos, RNP delivery achieved 100% editing of embryos at both loci. Additionally, compared to Cas9 mRNA delivery, RNPs led to greater embryo viability after editing at the two loci. RNPs also mediated a higher efficiency of biallelic editing, achieving 83% and 88% in marmoset embryos, compared to Cas9 mRNA, which resulted in 37% and 46% biallelic editing. Although statistically insignificant, RNP-mediated editing showed a tendency to produce less mosaic embryos than Cas9 mRNA editing. Finally, upon ssODN and RNP delivery into marmoset zygotes, precise HDR editing was achieved in 32% of embryos. In contrast, Cas9 mRNA exhibited no signs of HDR-mediated gene correction.
5.3.4. Bovine Zygotes
RNP delivery into bovine zygotes resulted in efficient indel formation of 92% at the Oct4 encoding gene [85]. Of the edited embryos, 92% exhibited biallelic editing events. Oct4 is required in blastocyst formation, and phenotypic analysis of embryos showed that only one out of 87 gene-corrected embryos reached the blastocyst stage. Furthermore, authors demonstrated that electroporation of RNPs did not affect embryo viability more than mock-electroporated zygotes.
5.4. Retinal Cells
Inherited retinal disorders and multifactorial retinal diseases cause visual impairment in millions of people worldwide. Due to the post-mitotic nature of most retinal cells, HDR efficiencies are low and CRISPR-mediated gene correction strategies have been limited [218]. Furthermore, RNP-mediated DSBs were shown to induce a p53-dependent G1 arrest in retinal pigment epithelium 1 (RPE1) cells, leading to decreased HDR efficiencies [142]. Notably, p53 inhibition increased HDR gene correction rates in RPE1 cells, which should be further tested in in vivo studies to advance retinal gene therapy applications.
Despite limited HDR editing in retinal cells, the use of Cas9 to induce gene or aberrant splice site disruption for clinical applications has shown promising results [219, 220]. AAV vectors represent the primary strategy for Cas9 delivery to retinal cells, and safety and efficacy evaluation in a clinical trial is underway (ClinicalTrials.gov ID: NCT03872479). However, non-viral approaches are being considered to reduce the inflammatory effects of AAV vectors in the retina and to achieve more transient Cas9 activity.
Overexpression of VEGFA in RPE cells leads to choroidal neovascularization (CNV), the major pathologic feature of age-related macular degeneration (AMD). As such, localized gene knockout of VEGFA in RPE cells has emerged as a potential gene therapy strategy. As a non-viral approach, RNPs were evaluated in their ability to mediate VEGFA disruption [100]. RNPs led to indel formation in the human RPE cell line ARPE-19 at higher efficiencies than Cas9 plasmid delivery. Subretinal injection of RNPs in mice gave rise to indels with a frequency of 25% in RPE cells isolated from the injected area. Importantly, CNV reduction was observed in a mouse model due to VEGFA knockout. Additionally, RNPs were demonstrated to be completely degraded in the mouse retina by 72 hours post-injection, confirming their transient nature and potential clinical benefits. Indeed, no off-target effects were detected at predicted sites. Another group also analyzed the ability for RNPs to induce in vivo VEGFA knockout [104]. Here, investigators improved on the previous report by using sgRNAs with chemical modifications at the terminal three nucleotides and an improved transfection reagent. Additionally, subretinal injection of mice with lipoplexes containing Cas9 RNPs and EGFP mRNA allowed for precise indel analysis of transfected RPE cells only. Testing various concentrations of RNPs and EGFP mRNA, investigators showed up to 6% indel formation at VEGFA in transfected cells.
5.5. Cochlear Cells
Sensory cochlear cells in the mammalian inner ear, including hair cells, have emerged as gene therapy targets for sensorineural hearing loss caused by either genetic factors or environmental insults. Silencing of dominant-negative mutant alleles has shown promising results in animal models [221]. Additionally, gene editing strategies designed to stimulate proliferation of inner-ear cells can potentially curb progressive hearing loss after damage. In order to expand the clinical applicability of CRISPR/Cas9 genome editing strategies in sensorineural hearing loss, in vivo Cas9 RNP delivery has been explored [101–103].
Dominant-negative mutations in transmembrane channel-like gene family 1 (TMC1) leads to progressive post-lingual sensorineural hearing loss in humans. One group designed a TMC1 mutant allele specific sgRNA that was demonstrated to selectively knockout the mutant allele 23-fold more efficiently than the wild-type TMC1 allele [101]. To optimize specificity of the genome editing system, off-target editing was compared after Cas9 plasmid or RNP delivery into primary fibroblasts derived from a homozygous TMC1 mutant mouse model. Plasmid delivery of Cas9 resulted in greater off-target editing, and thus RNPs were used for subsequent experiments. RNP injection into the scala media of a heterozygous TMC1 mutant mouse model led to increased survival of inner and outer hair cells, enhanced cochlear function, and preservation of acoustic behavioral reflex. Cochlear tissue was harvested and indel formation in the TMC1 mutant allele was detected in the organ of Corti, spiral ganglion, and spiral ligament. Overall, the RNP-mediated gene therapy system exhibited specific on-target gene editing resulting in a reduction of progressive hearing loss.
Due to the post-mitotic nature of inner ear sensory cells, HDR editing has been insufficient. Thus, when single nucleotide changes have been desired, base editing strategies have been successfully applied [102, 103]. Base editing was first shown in inner ear cells by intracochlear injection of RNPs formulated with the cytosine-base editor BE3 and a sgRNA targeting the VEGFA locus [102]. Following RNP injection, high-throughput DNA sequencing of harvested cochlear tissue demonstrated C-to-T conversions in the editing window at the organ of Corti, stria vascularis, and modiolus. Additionally, no C-to-T conversions or indels were observed at four predicted off-target loci, confirming the clinical applicability of in vivo RNP base editing in the cochlea. In another study, RNP-mediated base editing with BE3 editors was applied to introduce a precise C-to-T mutation in the B-catenin gene [103]. The conversion was hypothesized to reduce B-catenin degradation, leading to increased Wnt signaling that would stimulate proliferation of hair cells and reverse progressive hearing loss after environmental damage. First, using HEK293T cells, RNP delivery of base editors demonstrated more efficient nucleotide substitution than HDR machinery. Then, RNP injection of base editors in mouse cochlea resulted in transdifferentiation of supporting cells into hair cells. Moreover, DNA sequencing confirmed precise C-to-T conversions in the organ of Corti, stria vascularis, and modiolus.
6. Challenges & Future Perspectives
6.1. Temporal and Spatial Regulation of CRISPR/Cas9 Activity
The transient nature of Cas9 RNPs promotes rapid genome and epigenome editing, reduced off-target effects, and decreased cell toxicity. Despite these advantages, RNPs may not be the preferred method of Cas9 delivery for every application. For instance, due to the quick degradation of Cas9 when delivered as an RNP, Cas9 cannot be effectively regulated in a spatial or temporal manner. In contrast, plasmid or viral delivery systems, which rely on persistent expression of Cas9 cassettes, can successfully achieve both transcriptional and post-translational regulation. Consequently, RNPs lack the benefits of inducible CRISPR/Cas9 systems. Such systems may allow for functional perturbations of Cas9 that will facilitate its use in both research and clinical contexts. For example, a plasmid-encoded chemical-inducible CRISPR/Cas9 system employing a split Cas9 architecture gave rise to on-target indel formation or dCas9-VP64-mediated gene activation upon rapamycin-activated dimerization [222]. In another study, a plasmid-encoded chemical-induced Cas9 variant (termed iCas) was employed to repeatedly toggle Cas9 activity on and off and induce multiple gene knock-outs in succession [223]. Other scientists engineered a light activated CRISPR/Cas9 effector (LACE) system with fusion proteins between CRISPR reagents (dCas9 and VP64) and photoinducible dimerization domains [224]. The LACE system was encoded in plasmids and exhibited optogenetic activation of target gene expression that was reversible. Finally, another group devised a split Cas9 (termed paCas9) system that could be optogenetically controlled to induce multiplexed indel formation, HDR when co-transfected with an ssODN, single strand nicking with a paCas9 D10A variant, and gene suppression with an inactivated paCas9 [225]. The aforementioned split Cas9, iCas, LACE, and paCas9 systems utilize post-translational control of Cas9 activity. Therefore, in theory, if constructed using stable Cas9 variants that extend its half-life, RNP delivery and spatio-temporal regulation may be feasible with these inducible systems. However, it is unclear as to whether split Cas9 fragments can form RNP complexes with sgRNAs. In contrast, other inducible techniques often rely on transcriptional modulation, making them exclusively compatible with DNA methods of Cas9 delivery. For example, thermal gene switches were integrated with dCas9 complexes to drive heat-activated dCas9 complex expression and subsequent gene activation and suppression both in vitro and in vivo [226]. Methods to finely tune the control of Cas9 expand the potential applications of genome and epigenome engineering. As of now, RNPs are unable to be regulated in a spatio-temporal fashion and investigations that require Cas9 modulation should use plasmid or viral delivery systems.
The rapid quick degradation of Cas9 when delivered as an RNP makes its use in ex vivo cellular reprogramming and differentiation difficult. Reprogramming cell identity and altering cell fate has tremendous therapeutic potential. For example, patient-derived iPSCs were differentiated into RPE cells and transplanted for treatment of neovascular age-related macular degeneration [227]. Conventionally, somatic cells can be reprogrammed into iPSCs by expression of reprogramming transcription factors Oct4, Sox2, Klf4, and c-Myc [228]. Common strategies for cellular reprogramming of fibroblasts into iPSCs utilize non-CRISPR approaches, including viral, episomal, and RNA transfection methods [229]. Recently, two independent studies demonstrated fibroblast reprogramming into iPSCs using viral- and plasmid-based CRISPR activation approaches [230, 231]. Successful reprogramming to iPSCs requires extended activation (days to weeks) of reprogramming transcription factors. As such, RNPs, which are rapidly cleared through degradation pathways and lead to brief gene activation in the setting of CRISPR activation, are ineffective, and other delivery approaches should be favored. Similarly, the effective differentiation of iPSCs into various cell types often requires prolonged gene activation. Therefore, to this date, investigations have focused on inducible viral- and plasmid-based CRISPR activation approaches for the differentiation of iPSCs into specific lineages [232–234]. However, reintroduction of RNPs into target cells at regular intervals could overcome the challenges of transient activation while retaining the benefits of RNPs over DNA-based systems, such as eliminating the possibility of vector DNA integration into the host genome. Additionally, further exploration of stable Cas9 variants that allow for longer Cas9 half-life would be beneficial at extending the duration of RNP-mediated transcriptional control for potential use in cellular reprogramming and differentiation [235].
6.2. Clinical Translation of RNPs
The ability of RNPs to dramatically curtail off-target genome and epigenome editing make RNP-based approaches well-suited for clinical applications. However, off-target effects can still be present with RNPs, and they remain a barrier to the clinical translation of CRISPR-based therapeutics. In order to further reduce the off-target activity of Cas9 RNPs, one approach could be to promote Cas9 degradation. For example, fusion of ubiquitin-proteasomal degradation signals to Cas9 (Ubi-Cas9) led to reduced Cas9 half-life [236]. Notably, cynomolgus monkey zygotes injected with Ubi-Cas9 and WT-Cas9 mRNAs led to similar on-target editing efficiencies (74% and 77%, respectively), demonstrating that Ubi-Cas9 did not compromised on-target activity. While this report did not analyze off-target activity, the shorter half-life of Cas9 resulted in decreased mosaicism in monkey embryos, a signal of rapid Cas9 degradation. Exploration as to whether Ubi-Cas9 RNPs or other shorter half-life Cas9 RNPs can lower off-target Cas9 activity should be initiated.
In vivo delivery of RNPs is another major challenge in the clinical translation of RNP-based genome and epigenome editing applications. In order to mediate sufficient editing in vivo, RNPs must maintain stability during delivery and lead to sufficient internalization into target cells [237]. Importantly, in vivo editing may require selective targeting of cells, tissues, or organs. In these cases, the addition of ligands or antibodies unique to the target can be attached to delivery vehicles. One promising approach involves the packaging of RNPs into lipid nanoparticles (LNPs) followed by systemic administration [238]. As described in section 3.2.1, LNPs have the ability to maintain cargo stability and, upon endocytosis, release cargo into the cellular cytoplasm [97]. Moreover, RNP-packaged LNPs can be formulated to selectively target various organs [60]. Another strategy involves the local injection of lipid-based transfection agents, which was used to achieve local gene and base editing in the retina and cochlea [60, 100–104]. Further development of stable, tissue-specific LNPs will advance the ability to translate RNP-based editing approaches to the clinic.
Immunogenicity to Cas9 proteins remains of considerable interest and has implications on the application of CRISPR/Cas9 gene therapies [239–241]. Notably, a fraction of the human population is known to carry pre-existing immunity to Cas9 proteins, likely from past exposure to microbes [242–245]. Innate and adaptive immune responses may diminish the effects of CRISPR-based gene therapies and pose safety risks to patients. Given the transient nature of RNPs compared to other Cas9 delivery systems, immune responses and downstream effects may be limited. Further research into the comparison of Cas9 immune responses from DNA, mRNA, and RNP approaches should be initiated. Furthermore, therapeutic use of Cas9 variants from uncommon microbes may decrease the chances of pre-existing immunity in the general population and lead to favorable outcomes. Immune responses may be compounded in vivo with the use of potentially immunogenic virus-like particles for RNP delivery, emphasizing the benefit of nanoparticle vehicles for in vivo delivery applications.
6.3. Production of RNPs
As expanded upon in sections 2.1 and 2.2, the production of RNP components is a critical step in its use for genome and epigenome editing. Depending on the editing application, production of both Cas9 and sgRNA may be simple or difficult. For example, if unmodified WT-Cas9 is the desired Cas9 variant, it can be purchased from commercial vendors. Similarly, commonly used chemically modified sgRNAs (such as 2’-O-methyl 3’phosphorothioate at the three terminal nucleotides at both the 5’ and 3’ ends) are commercially available. In these instances, RNP production consists of sgRNA design, purchasing of Cas9 and sgRNA components, and a brief (~20 minutes) RNP assembly step (detailed in section 2.3).
However, some editing applications rely on Cas9 variants or sgRNA modifications that are not commercially available. Here, onsite production is required, which is accompanied by various challenges. S. pyogenes Cas9 is a large protein with a molecular weight of ~160 kDa. Cas9 fusion proteins weigh more, such as the BE3 base editor, which is ~200 kDa. The large size of Cas9 variants make production via E. coli-based expression systems and downstream purification challenging. For example, proteins may become insolubilized and lost during purification steps or a high purity product may not be obtained. Moreover, Cas9 must maintain its stability and biological activity throughout the purification process. Modified sgRNAs may also require onsite production via in vitro transcription (IVT). IVT is accompanied by various challenges, including incomplete transcription and the need to optimize reaction conditions. Onsite production of Cas9 and sgRNA requires significant time and effort, taking valuable resources away from RNP experimentation. The commercial availability of Cas9 variants as proteins would greatly enhance the field of RNP editing.
Another form of Cas9-mediated editing includes codelivery of Cas9 mRNA and sgRNA. Here, the production of Cas9 and sgRNA mirrors its production when used in RNPs, with both commercially available products and the need for onsite mRNA or sgRNA production when some Cas9 variants or sgRNA chemical modifications are desired. Cas9 mRNA is produced using IVT and faces similar challenges to sgRNA IVT.
Finally, Cas9 editing can be achieved by the delivery of DNA as plasmids or viral vectors. Various plasmids containing both Cas9 and sgRNA expression cassettes are available on Addgene, including base and prime editing Cas9 variants. As such, plamid-based approaches offer the most straightforward production steps. Therefore, in certain circumstances, such as when a novel Cas9 variant is being tested and is commercially unavailable, screening editing outcomes using a plasmid-based approach may be beneficial prior to production and experimentation with RNPs. In contrast to plasmids, production of viral vectors is laborious, time consuming, and hard to scale up.
In summary, RNP production can be both simple and challenging, depending on the editing application. However, excluding plasmid-based approaches, other delivery techniques also require time-intensive production of CRISPR components.
7. Concluding Remarks
The CRISPR/Cas9 system has revolutionized biological research since its introduction in the early 2010s. Recently, increasing attention is being paid to Cas9 RNP-based genome and epigenome editing approaches. This is partly due to its advantages over other delivery systems and improved clinical applicability. As reviewed, RNPs can be introduced to mammalian cells through various delivery platforms, including physical approaches, synthetic carriers, and virus-like particles. These approaches mediate editing in both in vitro and in vivo systems, expanding their use in various clinical applications. Compared to DNA- and RNA-based methods, RNPs lead to increased editing efficiencies, reduced off-target effects, and decreased cellular toxicity. These advantages have been demonstrated in a range of mammalian cell types with various editing mechanisms, including novel base and prime editing. However, challenges remain for the use of RNP-based approaches. Further research on Cas9 stability and immunogenicity, as well as LNP delivery systems, will empower RNPs to reach greater research and clinical applications.
Acknowledgements
We would like to thank Richard H. Smith, PhD, Andre Larochelle, MD, PhD, and Christi T. Salisbury-Ruf, PhD for their inputs and comments on the content and organization of this manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) Grants R01 EB027170-01 and UG3 TR002636-01. H.B. was supported by the NIH under Award Number T32GM008448. J.K. was supported by the National Science Foundation (NSF) Graduate Research Fellowships Program (GRFP) Grant DGE-184274.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare no conflict of interest.
References
- [1].Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, Genome editing with engineered zinc finger nucleases, Nat Rev Genet, 11 (2010) 636–646. [DOI] [PubMed] [Google Scholar]
- [2].Bogdanove AJ, Voytas DF, TAL effectors: customizable proteins for DNA targeting, Science, 333 (2011) 1843–1846. [DOI] [PubMed] [Google Scholar]
- [3].Khan SH, Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application, Mol Ther Nucleic Acids, 16 (2019) 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bhaya D, Davison M, Barrangou R, CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation, Annu Rev Genet, 45 (2011) 273–297. [DOI] [PubMed] [Google Scholar]
- [5].Wiedenheft B, Sternberg SH, Doudna JA, RNA-guided genetic silencing systems in bacteria and archaea, Nature, 482 (2012) 331–338. [DOI] [PubMed] [Google Scholar]
- [6].Terns MP, Terns RM, CRISPR-based adaptive immune systems, Curr Opin Microbiol, 14 (2011) 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gasiunas G, Barrangou R, Horvath P, Siksnys V, Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria, Proc Natl Acad Sci U S A, 109 (2012) E2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J, RNA-programmed genome editing in human cells, Elife, 2 (2013) e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, RNA-guided human genome engineering via Cas9, Science, 339 (2013) 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, Multiplex genome engineering using CRISPR/Cas systems, Science, 339 (2013) 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cho SW, Kim S, Kim JM, Kim JS, Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease, Nat Biotechnol, 31 (2013) 230–232. [DOI] [PubMed] [Google Scholar]
- [12].Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X, Generation of gene-modified mice via Cas9/RNA-mediated gene targeting, Cell Res, 23 (2013) 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering, Cell, 153 (2013) 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering, Cell, 154 (2013) 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 337 (2012) 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat Protoc, 8 (2013) 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chang HHY, Pannunzio NR, Adachi N, Lieber MR, Non-homologous DNA end joining and alternative pathways to double-strand break repair, Nat Rev Mol Cell Biol, 18 (2017) 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature, 533 (2016) 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR, Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage, Nature, 551 (2017) 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Porto EM, Komor AC, Slaymaker IM, Yeo GW, Base editing: advances and therapeutic opportunities, Nat Rev Drug Discov, 19 (2020) 839–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anzalone AV, Koblan LW, Liu DR, Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors, Nat Biotechnol, 38 (2020) 824–844. [DOI] [PubMed] [Google Scholar]
- [22].Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR, Search-and-replace genome editing without double-strand breaks or donor DNA, Nature, 576 (2019) 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Scholefield J, Harrison PT, Prime editing - an update on the field, Gene Ther, 28 (2021) 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes, Cell, 154 (2013) 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM, CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering, Nat Biotechnol, 31 (2013) 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tadic V, Josipovic G, Zoldos V, Vojta A, CRISPR/Cas9-based epigenome editing: An overview of dCas9-based tools with special emphasis on off-target activity, Methods, 164–165 (2019) 109–119. [DOI] [PubMed] [Google Scholar]
- [27].Goell JH, Hilton IB, CRISPR/Cas-Based Epigenome Editing: Advances, Applications, and Clinical Utility, Trends Biotechnol, 39 (2021) 678–691. [DOI] [PubMed] [Google Scholar]
- [28].Lino CA, Harper JC, Carney JP, Timlin JA, Delivering CRISPR: a review of the challenges and approaches, Drug Deliv, 25 (2018) 1234–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ates I, Rathbone T, Stuart C, Bridges PH, Cottle RN, Delivery Approaches for Therapeutic Genome Editing and Challenges, Genes (Basel), 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xu CL, Ruan MZC, Mahajan VB, Tsang SH, Viral Delivery Systems for CRISPR, Viruses, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, Loov C, Ingelsson M, Chamberlain JS, Corey DP, Aryee MJ, Joung JK, Breakefield XO, Maguire CA, Gyorgy B, High levels of AAV vector integration into CRISPR-induced DNA breaks, Nat Commun, 10 (2019) 4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Uren AG, Kool J, Berns A, van Lohuizen M, Retroviral insertional mutagenesis: past, present and future, Oncogene, 24 (2005) 7656–7672. [DOI] [PubMed] [Google Scholar]
- [33].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, DNA targeting specificity of RNA-guided Cas9 nucleases, Nat Biotechnol, 31 (2013) 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, Weng Z, Gao G, Xue W, Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses, Hum Gene Ther, 26 (2015) 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM, A multifunctional AAV-CRISPR-Cas9 and its host response, Nat Methods, 13 (2016) 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim S, Kim D, Cho SW, Kim J, Kim JS, Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins, Genome Res, 24 (2014) 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD, Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection, J Biotechnol, 208 (2015) 44–53. [DOI] [PubMed] [Google Scholar]
- [38].Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR, Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo, Nat Biotechnol, 33 (2015) 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang Y, Wang B, Xie H, Ren Q, Liu X, Li F, Lv X, He X, Cheng C, Deng R, Li J, Zhao J, Song Z, Gu F, Efficient Human Genome Editing Using SaCas9 Ribonucleoprotein Complexes, Biotechnol J, 14 (2019) e1800689. [DOI] [PubMed] [Google Scholar]
- [40].Rousseau BA, Hou Z, Gramelspacher MJ, Zhang Y, Programmable RNA Cleavage and Recognition by a Natural CRISPR-Cas9 System from Neisseria meningitidis, Mol Cell, 69 (2018) 906–914 e904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, McNeill MS, Yan S, Camarena J, Lee CM, Park SH, Wiebking V, Bak RO, Gomez-Ospina N, Pavel-Dinu M, Sun W, Bao G, Porteus MH, Behlke MA, A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells, Nat Med, 24 (2018) 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim D, Kim S, Kim JS, Directed evolution of CRISPR-Cas9 to increase its specificity, Nat Commun, 9 (2018) 3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O, Engineered CRISPR-Cas9 nuclease with expanded targeting space, Science, 361 (2018) 1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K, Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing, Nat Commun, 8 (2017) 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F, Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity, Cell, 154 (2013) 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC, Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects, Nat Methods, 11 (2014) 399–402. [DOI] [PubMed] [Google Scholar]
- [47].Trevino AE, Zhang F, Genome editing using Cas9 nickases, Methods Enzymol, 546 (2014) 161–174. [DOI] [PubMed] [Google Scholar]
- [48].Gopalappa R, Suresh B, Ramakrishna S, Kim HH, Paired D10A Cas9 nickases are sometimes more efficient than individual nucleases for gene disruption, Nucleic Acids Res, 46 (2018) e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression, Cell, 152 (2013) 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rees HA, Yeh WH, Liu DR, Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks, Nat Commun, 10 (2019) 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, Malik P, CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites, Nat Commun, 10 (2019) 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Brocken DJW, Tark-Dame M, Dame RT, dCas9: A Versatile Tool for Epigenome Editing, Curr Issues Mol Biol, 26 (2018) 15–32. [DOI] [PubMed] [Google Scholar]
- [53].DeWitt MA, Corn JE, Carroll D, Genome editing via delivery of Cas9 ribonucleoprotein, Methods, 121–122 (2017) 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH, Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells, Nat Biotechnol, 33 (2015) 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, Mou H, Oberholzer A, Ding J, Kwan SY, Bogorad RL, Zatsepin T, Koteliansky V, Wolfe SA, Xue W, Langer R, Anderson DG, Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing, Nat Biotechnol, 35 (2017) 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ryan DE, Taussig D, Steinfeld I, Phadnis SM, Lunstad BD, Singh M, Vuong X, Okochi KD, McCaffrey R, Olesiak M, Roy S, Yung CW, Curry B, Sampson JR, Bruhn L, Dellinger DJ, Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs, Nucleic Acids Res, 46 (2018) 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen Q, Zhang Y, Yin H, Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing, Adv Drug Deliv Rev, 168 (2021) 246–258. [DOI] [PubMed] [Google Scholar]
- [58].Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z, Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing, Angew Chem Int Ed Engl, 54 (2015) 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shapiro J, Tovin A, Iancu O, Allen D, Hendel A, Chemical Modification of Guide RNAs for Improved CRISPR Activity in CD34+ Human Hematopoietic Stem and Progenitor Cells, Methods Mol Biol, 2162 (2021) 37–48. [DOI] [PubMed] [Google Scholar]
- [60].Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ, Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing, Nat Commun, 11 (2020) 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, McAnally JR, Amoasii L, Mammen PPA, Bassel-Duby R, Olson EN, CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells, Sci Adv, 5 (2019) eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Qiu M, Glass Z, Chen J, Haas M, Jin X, Zhao X, Rui X, Ye Z, Li Y, Zhang F, Xu Q, Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3, Proc Natl Acad Sci U S A, 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang S, Shen J, Li D, Cheng Y, Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing, Theranostics, 11 (2021) 614–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Seki A, Rutz S, Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells, J Exp Med, 215 (2018) 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Oh SA, Seki A, Rutz S, Ribonucleoprotein Transfection for CRISPR/Cas9-Mediated Gene Knockout in Primary T Cells, Curr Protoc Immunol, 124 (2019) e69. [DOI] [PubMed] [Google Scholar]
- [66].Wiebking V, Lee CM, Mostrel N, Lahiri P, Bak R, Bao G, Roncarolo MG, Bertaina A, Porteus MH, Genome editing of donor-derived T-cells to generate allogenic chimeric antigen receptor-modified T cells: Optimizing alphabeta T cell-depleted haploidentical hematopoietic stem cell transplantation, Haematologica, 106 (2021) 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Webber BR, Lonetree CL, Kluesner MG, Johnson MJ, Pomeroy EJ, Diers MD, Lahr WS, Draper GM, Slipek NJ, Smeester BA, Lovendahl KN, McElroy AN, Gordon WR, Osborn MJ, Moriarity BS, Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors, Nat Commun, 10 (2019) 5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Johnson MJ, Laoharawee K, Lahr WS, Webber BR, Moriarity BS, Engineering of Primary Human B cells with CRISPR/Cas9 Targeted Nuclease, Sci Rep, 8 (2018) 12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hung KL, Meitlis I, Hale M, Chen CY, Singh S, Jackson SW, Miao CH, Khan IF, Rawlings DJ, James RG, Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells, Mol Ther, 26 (2018) 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu CM, Roth TL, Baglaenko Y, Ferri DM, Brauer P, Zuniga-Pflucker JC, Rosbe KW, Wither JE, Marson A, Allen CDC, Genetic engineering in primary human B cells with CRISPR-Cas9 ribonucleoproteins, J Immunol Methods, 457 (2018) 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Freund EC, Lock JY, Oh J, Maculins T, Delamarre L, Bohlen CJ, Haley B, Murthy A, Efficient gene knockout in primary human and murine myeloid cells by non-viral delivery of CRISPR-Cas9, J Exp Med, 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hiatt J, Cavero DA, McGregor MJ, Zheng W, Budzik JM, Roth TL, Haas KM, Wu D, Rathore U, Meyer-Franke A, Bouzidi MS, Shifrut E, Lee Y, Kumar VE, Dang EV, Gordon DE, Wojcechowskyj JA, Hultquist JF, Fontaine KA, Pillai SK, Cox JS, Ernst JD, Krogan NJ, Marson A, Efficient generation of isogenic primary human myeloid cells using CRISPR-Cas9 ribonucleoproteins, Cell Rep, 35 (2021) 109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, Uchida N, Hendel A, Narla A, Majeti R, Weinberg KI, Porteus MH, CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells, Nature, 539 (2016) 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lattanzi A, Meneghini V, Pavani G, Amor F, Ramadier S, Felix T, Antoniani C, Masson C, Alibeu O, Lee C, Porteus MH, Bao G, Amendola M, Mavilio F, Miccio A, Optimization of CRISPR/Cas9 Delivery to Human Hematopoietic Stem and Progenitor Cells for Therapeutic Genomic Rearrangements, Mol Ther, 27 (2019) 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bloomer H, Smith RH, Hakami W, Larochelle A, Genome editing in human hematopoietic stem and progenitor cells via CRISPR-Cas9-mediated homology-independent targeted integration, Mol Ther, 29 (2021) 1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zeng J, Wu Y, Ren C, Bonanno J, Shen AH, Shea D, Gehrke JM, Clement K, Luk K, Yao Q, Kim R, Wolfe SA, Manis JP, Pinello L, Joung JK, Bauer DE, Therapeutic base editing of human hematopoietic stem cells, Nat Med, 26 (2020) 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jackow J, Guo Z, Hansen C, Abaci HE, Doucet YS, Shin JU, Hayashi R, DeLorenzo D, Kabata Y, Shinkuma S, Salas-Alanis JC, Christiano AM, CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells, Proc Natl Acad Sci U S A, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Martin RM, Ikeda K, Cromer MK, Uchida N, Nishimura T, Romano R, Tong AJ, Lemgart VT, Camarena J, Pavel-Dinu M, Sindhu C, Wiebking V, Vaidyanathan S, Dever DP, Bak RO, Laustsen A, Lesch BJ, Jakobsen MR, Sebastiano V, Nakauchi H, Porteus MH, Highly Efficient and Marker-free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination, Cell Stem Cell, 24 (2019) 821–828 e825. [DOI] [PubMed] [Google Scholar]
- [79].Ruan J, Hirai H, Yang D, Ma L, Hou X, Jiang H, Wei H, Rajagopalan C, Mou H, Wang G, Zhang J, Li K, Chen YE, Sun F, Xu J, Efficient Gene Editing at Major CFTR Mutation Loci, Mol Ther Nucleic Acids, 16 (2019) 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang W, Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, Zhang Y, Taft RA, Buaas FW, Wang H, Delivery of Cas9 Protein into Mouse Zygotes through a Series of Electroporation Dramatically Increases the Efficiency of Model Creation, J Genet Genomics, 43 (2016) 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chen S, Lee B, Lee AY, Modzelewski AJ, He L, Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes, J Biol Chem, 291 (2016) 14457–14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hashimoto M, Yamashita Y, Takemoto T, Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse, Dev Biol, 418 (2016) 1–9. [DOI] [PubMed] [Google Scholar]
- [83].Ohtsuka M, Sato M, Miura H, Takabayashi S, Matsuyama M, Koyano T, Arifin N, Nakamura S, Wada K, Gurumurthy CB, i-GONAD: a robust method for in situ germline genome engineering using CRISPR nucleases, Genome Biol, 19 (2018) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS, Highly efficient RNA-guided base editing in mouse embryos, Nat Biotechnol, 35 (2017) 435–437. [DOI] [PubMed] [Google Scholar]
- [85].Camargo LSA, Owen JR, Van Eenennaam AL, Ross PJ, Efficient One-Step Knockout by Electroporation of Ribonucleoproteins Into Zona-Intact Bovine Embryos, Front Genet, 11 (2020) 570069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Alghadban S, Bouchareb A, Hinch R, Hernandez-Pliego P, Biggs D, Preece C, Davies B, Electroporation and genetic supply of Cas9 increase the generation efficiency of CRISPR/Cas9 knock-in alleles in C57BL/6J mouse zygotes, Sci Rep, 10 (2020) 17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hui SW, Overview of drug delivery and alternative methods to electroporation, Methods Mol Biol, 423 (2008) 91–107. [DOI] [PubMed] [Google Scholar]
- [88].Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, Lee HW, Kim JS, Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases, Genome Res, 24 (2014) 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Tang L, Zeng Y, Du H, Gong M, Peng J, Zhang B, Lei M, Zhao F, Wang W, Li X, Liu J, CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein, Mol Genet Genomics, 292 (2017) 525–533. [DOI] [PubMed] [Google Scholar]
- [90].Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, Van Dyken C, Li Y, Kang E, Park AR, Kim D, Kim ST, Gong J, Gu Y, Xu X, Battaglia D, Krieg SA, Lee DM, Wu DH, Wolf DP, Heitner SB, Belmonte JCI, Amato P, Kim JS, Kaul S, Mitalipov S, Correction of a pathogenic gene mutation in human embryos, Nature, 548 (2017) 413–419. [DOI] [PubMed] [Google Scholar]
- [91].Kumita W, Sato K, Suzuki Y, Kurotaki Y, Harada T, Zhou Y, Kishi N, Sato K, Aiba A, Sakakibara Y, Feng G, Okano H, Sasaki E, Efficient generation of Knock-in/Knock-out marmoset embryo via CRISPR/Cas9 gene editing, Sci Rep, 9 (2019) 12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Liu C, Zhang L, Liu H, Cheng K, Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications, J Control Release, 266 (2017) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N, Efficient intracellular delivery of native proteins, Cell, 161 (2015) 674–690. [DOI] [PubMed] [Google Scholar]
- [94].Kholosy WM, Visscher M, Ogink K, Buttstedt H, Griffin K, Beier A, Gerlach JP, Molenaar JJ, Geijsen N, de Boer M, Chatsisvili A, Simple, fast and efficient iTOP-mediated delivery of CRISPR/Cas9 RNP in difficult-to-transduce human cells including primary T cells, J Biotechnol, 338 (2021) 71–80. [DOI] [PubMed] [Google Scholar]
- [95].Ghasemiyeh P, Mohammadi-Samani S, Solid lipid nanoparticles and nanostructured lipid carriers as novel drug delivery systems: applications, advantages and disadvantages, Res Pharm Sci, 13 (2018) 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nat Rev Genet, 15 (2014) 541–555. [DOI] [PubMed] [Google Scholar]
- [97].Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC, Mahmoudi M, Cellular uptake of nanoparticles: journey inside the cell, Chem Soc Rev, 46 (2017) 4218–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Donahue ND, Acar H, Wilhelm S, Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine, Adv Drug Deliv Rev, 143 (2019) 68–96. [DOI] [PubMed] [Google Scholar]
- [99].Yu X, Liang X, Xie H, Kumar S, Ravinder N, Potter J, de X Mollerat du Jeu, J.D. Chesnut, Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX, Biotechnol Lett, 38 (2016) 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, Kim KE, Kim JH, Kim JS, Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration, Genome Res, 27 (2017) 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, Hu YJ, Hu JH, Thompson DB, Shu Y, Li Y, Wang H, Yang S, Xu Q, Polley DB, Liberman MC, Kong WJ, Holt JR, Chen ZY, Liu DR, Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents, Nature, 553 (2018) 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Rees HA, Komor AC, Yeh WH, Caetano-Lopes J, Warman M, Edge ASB, Liu DR, Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery, Nat Commun, 8 (2017) 15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR, In vivo base editing of post-mitotic sensory cells, Nat Commun, 9 (2018) 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Holmgaard AB, Askou AL, Jensen EG, Alsing S, Bak RO, Mikkelsen JG, Corydon TJ, Targeted Knockout of the Vegfa Gene in the Retina by Subretinal Injection of RNP Complexes Containing Cas9 Protein and Modified sgRNAs, Mol Ther, 29 (2021) 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sun S, Wang M, Knupp SA, Soto-Feliciano Y, Hu X, Kaplan DL, Langer R, Anderson DG, Xu Q, Combinatorial library of lipidoids for in vitro DNA delivery, Bioconjug Chem, 23 (2012) 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Takeda YS, Xu Q, Synthetic and nature-derived lipid nanoparticles for neural regeneration, Neural Regen Res, 10 (2015) 689–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fenton OS, Kauffman KJ, McClellan RL, Kaczmarek JC, Zeng MD, Andresen JL, Rhym LH, Heartlein MW, DeRosa F, Anderson DG, Customizable Lipid Nanoparticle Materials for the Delivery of siRNAs and mRNAs, Angew Chem Int Ed Engl, 57 (2018) 13582–13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lee SM, Cheng Q, Yu X, Liu S, Johnson LT, Siegwart DJ, A Systematic Study of Unsaturation in Lipid Nanoparticles Leads to Improved mRNA Transfection In Vivo, Angew Chem Int Ed Engl, 60 (2021) 5848–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hou X, Zaks T, Langer R, Dong Y, Lipid nanoparticles for mRNA delivery, Nat Rev Mater, (2021) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Battaglia L, Gallarate M, Lipid nanoparticles: state of the art, new preparation methods and challenges in drug delivery, Expert Opin Drug Deliv, 9 (2012) 497–508. [DOI] [PubMed] [Google Scholar]
- [111].Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair, Nat Biomed Eng, 1 (2017) 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM, Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing, ACS Nano, 11 (2017) 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY, Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours, Nat Biomed Eng, 2 (2018) 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Arvizo R, Bhattacharya R, Mukherjee P, Gold nanoparticles: opportunities and challenges in nanomedicine, Expert Opin Drug Deliv, 7 (2010) 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Patel SG, Sayers EJ, He L, Narayan R, Williams TL, Mills EM, Allemann RK, Luk LYP, Jones AT, Tsai YH, Cell-penetrating peptide sequence and modification dependent uptake and subcellular distribution of green florescent protein in different cell lines, Sci Rep, 9 (2019) 6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ju A, Lee SW, Lee YE, Han KC, Kim JC, Shin SC, Park HJ, EunKyeong Kim E, Hong S, Jang M, A carrier-free multiplexed gene editing system applicable for suspension cells, Biomaterials, 217 (2019) 119298. [DOI] [PubMed] [Google Scholar]
- [117].Yin J, Wang Q, Hou S, Bao L, Yao W, Gao X, Potent Protein Delivery into Mammalian Cells via a Supercharged Polypeptide, J Am Chem Soc, 140 (2018) 17234–17240. [DOI] [PubMed] [Google Scholar]
- [118].Yue H, Zhou X, Cheng M, Xing D, Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing, Nanoscale, 10 (2018) 1063–1071. [DOI] [PubMed] [Google Scholar]
- [119].Zhou W, Cui H, Ying L, Yu XF, Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing, Angew Chem Int Ed Engl, 57 (2018) 10268–10272. [DOI] [PubMed] [Google Scholar]
- [120].Tang H, Zhao X, Jiang X, Synthetic multi-layer nanoparticles for CRISPR-Cas9 genome editing, Adv Drug Deliv Rev, 168 (2021) 55–78. [DOI] [PubMed] [Google Scholar]
- [121].Lyu P, Wang L, Lu B, Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing, Life (Basel), 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lu B, Javidi-Parsijani P, Makani V, Mehraein-Ghomi F, Sarhan WM, Sun D, Yoo KW, Atala ZP, Lyu P, Atala A, Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing, Nucleic Acids Res, 47 (2019) e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lyu P, Javidi-Parsijani P, Atala A, Lu B, Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing, Nucleic Acids Res, 47 (2019) e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Choi JG, Dang Y, Abraham S, Ma H, Zhang J, Guo H, Cai Y, Mikkelsen JG, Wu H, Shankar P, Manjunath N, Lentivirus pre-packed with Cas9 protein for safer gene editing, Gene Ther, 23 (2016) 627–633. [DOI] [PubMed] [Google Scholar]
- [125].Lyu P, Lu Z, Cho SI, Yadav M, Yoo KW, Atala A, Kim JS, Lu B, Adenine Base Editor Ribonucleoproteins Delivered by Lentivirus-Like Particles Show High On-Target Base Editing and Undetectable RNA Off-Target Activities, CRISPR J, 4 (2021) 69–81. [DOI] [PubMed] [Google Scholar]
- [126].Mangeot PE, Risson V, Fusil F, Marnef A, Laurent E, Blin J, Mournetas V, Massourides E, Sohier TJM, Corbin A, Aube F, Teixeira M, Pinset C, Schaeffer L, Legube G, Cosset FL, Verhoeyen E, Ohlmann T, Ricci EP, Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins, Nat Commun, 10 (2019) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Shirley JL, de Jong YP, Terhorst C, Herzog RW, Immune Responses to Viral Gene Therapy Vectors, Mol Ther, 28 (2020) 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kosicki M, Tomberg K, Bradley A, Repair of double-strand breaks induced by CRISPRCas9 leads to large deletions and complex rearrangements, Nat Biotechnol, 36 (2018) 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nunez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JC, In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration, Nature, 540 (2016) 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Suzuki K, Izpisua Belmonte JC, In vivo genome editing via the HITI method as a tool for gene therapy, J Hum Genet, 63 (2018) 157–164. [DOI] [PubMed] [Google Scholar]
- [131].Adli M, The CRISPR tool kit for genome editing and beyond, Nat Commun, 9 (2018) 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, Leong KW, CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery, Chem Rev, 117 (2017) 9874–9906. [DOI] [PubMed] [Google Scholar]
- [133].Burnight ER, Gupta M, Wiley LA, Anfinson KR, Tran A, Triboulet R, Hoffmann JM, Klaahsen DL, Andorf JL, Jiao C, Sohn EH, Adur MK, Ross JW, Mullins RF, Daley GQ, Schlaeger TM, Stone EM, Tucker BA, Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration, Mol Ther, 25 (2017) 1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zaboikin M, Zaboikina T, Freter C, Srinivasakumar N, Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents, PLoS One, 12 (2017) e0169931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Heyer WD, Ehmsen KT, Liu J, Regulation of homologous recombination in eukaryotes, Annu Rev Genet, 44 (2010) 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lieber MR, The mechanism of human nonhomologous DNA end joining, J Biol Chem, 283 (2008) 1–5. [DOI] [PubMed] [Google Scholar]
- [137].Chapman JR, Taylor MR, Boulton SJ, Playing the end game: DNA double-strand break repair pathway choice, Mol Cell, 47 (2012) 497–510. [DOI] [PubMed] [Google Scholar]
- [138].Lin S, Staahl BT, Alla RK, Doudna JA, Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery, Elife, 3 (2014) e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Jang HK, Song B, Hwang GH, Bae S, Current trends in gene recovery mediated by the CRISPR-Cas system, Exp Mol Med, 52 (2020) 1016–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Prakash V, Moore M, Yanez-Munoz RJ, Current Progress in Therapeutic Gene Editing for Monogenic Diseases, Mol Ther, 24 (2016) 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A, p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells, Nat Med, 24 (2018) 939–946. [DOI] [PubMed] [Google Scholar]
- [142].Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J, CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response, Nat Med, 24 (2018) 927–930. [DOI] [PubMed] [Google Scholar]
- [143].Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A, Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems, Science, 353 (2016). [DOI] [PubMed] [Google Scholar]
- [144].Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR, Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity, Sci Adv, 3 (2017) eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kim D, Lim K, Kim ST, Yoon SH, Kim K, Ryu SM, Kim JS, Genome-wide target specificities of CRISPR RNA-guided programmable deaminases, Nat Biotechnol, 35 (2017) 475–480. [DOI] [PubMed] [Google Scholar]
- [146].Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A, Liu DR, Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction, Nat Biotechnol, 36 (2018) 843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, Simon A, Han T, Goswami S, Montgomery E, Thibado J, Kastenhuber ER, Sanchez-Rivera FJ, Shi J, Vakoc CR, Lowe SW, Tschaharganeh DF, Dow LE, Optimized base editors enable efficient editing in cells, organoids and mice, Nat Biotechnol, 36 (2018) 888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Rees HA, Wilson C, Doman JL, Liu DR, Analysis and minimization of cellular RNA editing by DNA adenine base editors, Sci Adv, 5 (2019) eaax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Grunewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK, Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors, Nature, 569 (2019) 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Grunewald J, Zhou R, Iyer S, Lareau CA, Garcia SP, Aryee MJ, Joung JK, CRISPR DNA base editors with reduced RNA off-target and self-editing activities, Nat Biotechnol, 37 (2019) 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, Wei Y, Hu X, Zeng R, Li Y, Zhou H, Guo F, Yang H, Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis, Nature, 571 (2019) 275–278. [DOI] [PubMed] [Google Scholar]
- [152].Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR, Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity, Nat Biotechnol, 38 (2020) 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Doman JL, Raguram A, Newby GA, Liu DR, Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors, Nat Biotechnol, 38 (2020) 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gaudelli NM, Lam DK, Rees HA, Sola-Esteves NM, Barrera LA, Born DA, Edwards A, Gehrke JM, Lee SJ, Liquori AJ, Murray R, Packer MS, Rinaldi C, Slaymaker IM, Yen J, Young LE, Ciaramella G, Directed evolution of adenine base editors with increased activity and therapeutic application, Nat Biotechnol, 38 (2020) 892–900. [DOI] [PubMed] [Google Scholar]
- [155].Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X, Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells, Nat Methods, 13 (2016) 1029–1035. [DOI] [PubMed] [Google Scholar]
- [156].Kluesner MG, Lahr WS, Lonetree CL, Smeester BA, Qiu X, Slipek NJ, Claudio Vazquez PN, Pitzen SP, Pomeroy EJ, Vignes MJ, Lee SC, Bingea SP, Andrew AA, Webber BR, Moriarity BS, CRISPR-Cas9 cytidine and adenosine base editing of splice-sites mediates highly-efficient disruption of proteins in primary and immortalized cells, Nat Commun, 12 (2021) 2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, Mammadov R, Adli M, CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations, Nat Methods, 14 (2017) 710–712. [DOI] [PubMed] [Google Scholar]
- [158].Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR, Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions, Nat Biotechnol, 35 (2017) 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, Savage DF, Liu DR, Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors, Nat Biotechnol, 37 (2019) 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, Joung JK, An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities, Nat Biotechnol, 36 (2018) 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Shin HR, See JE, Kweon J, Kim HS, Sung GJ, Park S, Jang AH, Jang G, Choi KC, Kim I, Kim JS, Kim Y, Small-molecule inhibitors of histone deacetylase improve CRISPR-based adenine base editing, Nucleic Acids Res, 49 (2021) 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H, Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos, Science, 364 (2019) 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].McGrath E, Shin H, Zhang L, Phue JN, Wu WW, Shen RF, Jang YY, Revollo J, Ye Z, Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing, Nat Commun, 10 (2019) 5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Kim D, Kim DE, Lee G, Cho SI, Kim JS, Genome-wide target specificity of CRISPR RNA-guided adenine base editors, Nat Biotechnol, 37 (2019) 430–435. [DOI] [PubMed] [Google Scholar]
- [165].Surun D, Schneider A, Mircetic J, Neumann K, Lansing F, Paszkowski-Rogacz M, Hanchen V, Lee-Kirsch MA, Buchholz F, Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors, Genes (Basel), 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Chemello F, Chai AC, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Atmanli A, Mireault AA, Liu N, Bassel-Duby R, Olson EN, Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing, Sci Adv, 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Schene IF, Joore IP, Oka R, Mokry M, van Vugt AHM, van Boxtel R, van der Doef HPJ, van der Laan LJW, Verstegen MMA, van Hasselt PM, Nieuwenhuis EES, Fuchs SA, Prime editing for functional repair in patient-derived disease models, Nat Commun, 11 (2020) 5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Liu Y, Li X, He S, Huang S, Li C, Chen Y, Liu Z, Huang X, Wang X, Efficient generation of mouse models with the prime editing system, Cell Discov, 6 (2020) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Kim Y, Hong SA, Yu J, Eom J, Jang K, Yoon S, Hong DH, Seo D, Lee SN, Woo JS, Jeong J, Bae S, Choi D, Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease, Cell Stem Cell, 28 (2021) 1614–1624 e1615. [DOI] [PubMed] [Google Scholar]
- [170].Liu P, Liang SQ, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K, Mir A, Sontheimer EJ, Gao G, Flotte TR, Wolfe SA, Xue W, Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice, Nat Commun, 12 (2021) 2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Petri K, Zhang W, Ma J, Schmidts A, Lee H, Horng JE, Kim DY, Kurt IC, Clement K, Hsu JY, Pinello L, Maus MV, Joung JK, Yeh JJ, CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells, Nat Biotechnol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK, CRISPR RNA-guided activation of endogenous human genes, Nat Methods, 10 (2013) 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Villamizar O, Waters SA, Scott T, Saayman S, Grepo N, Urak R, Davis A, Jaffe A, Morris KV, Targeted Activation of Cystic Fibrosis Transmembrane Conductance Regulator, Mol Ther, 27 (2019) 1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Liu J, Sun M, Cho KB, Gao X, Guo B, A CRISPR-Cas9 repressor for epigenetic silencing of KRAS, Pharmacol Res, 164 (2021) 105304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA, Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds, Cell, 160 (2015) 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Galonska C, Charlton J, Mattei AL, Donaghey J, Clement K, Gu H, Mohammad AW, Stamenova EK, Cacchiarelli D, Klages S, Timmermann B, Cantz T, Scholer HR, Gnirke A, Ziller MJ, Meissner A, Genome-wide tracking of dCas9-methyltransferase footprints, Nat Commun, 9 (2018) 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, Marson A, A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells, Cell Rep, 17 (2016) 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Rupp LJ, Schumann K, Roybal KT, Gate RE, Ye CJ, Lim WA, Marson A, CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells, Sci Rep, 7 (2017) 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Gomes-Silva D, Srinivasan M, Sharma S, Lee CM, Wagner DL, Davis TH, Rouce RH, Bao G, Brenner MK, Mamonkin M, CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies, Blood, 130 (2017) 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Wu Y, Zeng J, Roscoe BP, Liu P, Yao Q, Lazzarotto CR, Clement K, Cole MA, Luk K, Baricordi C, Shen AH, Ren C, Esrick EB, Manis JP, Dorfman DM, Williams DA, Biffi A, Brugnara C, Biasco L, Brendel C, Pinello L, Tsai SQ, Wolfe SA, Bauer DE, Highly efficient therapeutic gene editing of human hematopoietic stem cells, Nat Med, 25 (2019) 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Fananas-Baquero S, Quintana-Bustamante O, Dever DP, Alberquilla O, Sanchez-Dominguez R, Camarena J, Ojeda-Perez I, Dessy-Rodriguez M, Turk R, Schubert MS, Lattanzi A, Xu L, Lopez-Lorenzo JL, Bianchi P, Bueren JA, Behlke MA, Porteus M, Segovia JC, Clinically relevant gene editing in hematopoietic stem cells for the treatment of pyruvate kinase deficiency, Mol Ther Methods Clin Dev, 22 (2021) 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Newby GA, Yen JS, Woodard KJ, Mayuranathan T, Lazzarotto CR, Li Y, Sheppard-Tillman H, Porter SN, Yao Y, Mayberry K, Everette KA, Jang Y, Podracky CJ, Thaman E, Lechauve C, Sharma A, Henderson JM, Richter MF, Zhao KT, Miller SM, Wang T, Koblan LW, McCaffrey AP, Tisdale JF, Kalfa TA, Pruett-Miller SM, Tsai SQ, Weiss MJ, Liu DR, Base editing of haematopoietic stem cells rescues sickle cell disease in mice, Nature, 595 (2021) 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Cromer MK, Camarena J, Martin RM, Lesch BJ, Vakulskas CA, Bode NM, Kurgan G, Collingwood MA, Rettig GR, Behlke MA, Lemgart VT, Zhang Y, Goyal A, Zhao F, Ponce E, Srifa W, Bak RO, Uchida N, Majeti R, Sheehan VA, Tisdale JF, Dever DP, Porteus MH, Gene replacement of alpha-globin with beta-globin restores hemoglobin balance in beta-thalassemia-derived hematopoietic stem and progenitor cells, Nat Med, 27 (2021) 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Pavel-Dinu M, Wiebking V, Dejene BT, Srifa W, Mantri S, Nicolas CE, Lee C, Bao G, Kildebeck EJ, Punjya N, Sindhu C, Inlay MA, Saxena N, DeRavin SS, Malech H, Roncarolo MG, Weinberg KI, Porteus MH, Gene correction for SCID-X1 in long-term hematopoietic stem cells, Nat Commun, 10 (2019) 1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Kuo CY, Long JD, Campo-Fernandez B, de Oliveira S, Cooper AR, Romero Z, Hoban MD, Joglekar AV, Lill GR, Kaufman ML, Fitz-Gibbon S, Wang X, Hollis RP, Kohn DB, Site-Specific Gene Editing of Human Hematopoietic Stem Cells for X-Linked Hyper-IgM Syndrome, Cell Rep, 23 (2018) 2606–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].De Ravin SS, Reik A, Liu PQ, Li L, Wu X, Su L, Raley C, Theobald N, Choi U, Song AH, Chan A, Pearl JR, Paschon DE, Lee J, Newcombe H, Koontz S, Sweeney C, Shivak DA, Zarember KA, Peshwa MV, Gregory PD, Urnov FD, Malech HL, Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease, Nat Biotechnol, 34 (2016) 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Vavassori V, Mercuri E, Marcovecchio GE, Castiello MC, Schiroli G, Albano L, Margulies C, Buquicchio F, Fontana E, Beretta S, Merelli I, Cappelleri A, Rancoita PM, Lougaris V, Plebani A, Kanariou M, Lankester A, Ferrua F, Scanziani E, Cotta-Ramusino C, Villa A, Naldini L, Genovese P, Modeling, optimization, and comparable efficacy of T cell and hematopoietic stem cell gene editing for treating hyper-IgM syndrome, EMBO Mol Med, 13 (2021) e13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Gomez-Ospina N, Scharenberg SG, Mostrel N, Bak RO, Mantri S, Quadros RM, Gurumurthy CB, Lee C, Bao G, Suarez CJ, Khan S, Sawamoto K, Tomatsu S, Raj N, Attardi LD, Aurelian L, Porteus MH, Human genome-edited hematopoietic stem cells phenotypically correct Mucopolysaccharidosis type I, Nat Commun, 10 (2019) 4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Li D, Li X, Zhou WL, Huang Y, Liang X, Jiang L, Yang X, Sun J, Li Z, Han WD, Wang W, Genetically engineered T cells for cancer immunotherapy, Signal Transduct Target Ther, 4 (2019) 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Seif M, Einsele H, Loffler J, CAR T Cells Beyond Cancer: Hope for Immunomodulatory Therapy of Infectious Diseases, Front Immunol, 10 (2019) 2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Scott DW, Gene therapy for immunological tolerance: using ‘transgenic’ B cells to treat inhibitor formation, Haemophilia, 16 (2010) 89–94. [DOI] [PubMed] [Google Scholar]
- [192].Morgan RA, Gray D, Lomova A, Kohn DB, Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned, Cell Stem Cell, 21 (2017) 574–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: from T cell basic science to clinical practice, Nat Rev Immunol, 20 (2020) 651–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Cornu TI, Mussolino C, Muller MC, Wehr C, Kern WV, Cathomen T, HIV Gene Therapy: An Update, Hum Gene Ther, 32 (2021) 52–65. [DOI] [PubMed] [Google Scholar]
- [195].Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P, CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection, PLoS One, 9 (2014) e115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Li C, Guan X, Du T, Jin W, Wu B, Liu Y, Wang P, Hu B, Griffin GE, Shattock RJ, Hu Q, Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9, J Gen Virol, 96 (2015) 2381–2393. [DOI] [PubMed] [Google Scholar]
- [197].Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A, Generation of knock-in primary human T cells using Cas9 ribonucleoproteins, Proc Natl Acad Sci U S A, 112 (2015) 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, Sadelain M, Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection, Nature, 543 (2017) 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Freen-van Heeren JJ, Popovic B, Guislain A, Wolkers MC, Human T cells employ conserved AU-rich elements to fine-tune IFN-gamma production, Eur J Immunol, 50 (2020) 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Akidil E, Albanese M, Buschle A, Ruhle A, Pich D, Keppler OT, Hammerschmidt W, Highly efficient CRISPR-Cas9-mediated gene knockout in primary human B cells for functional genetic studies of Epstein-Barr virus infection, PLoS Pathog, 17 (2021) e1009117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Lee CZW, Kozaki T, Ginhoux F, Studying tissue macrophages in vitro: are iPSC-derived cells the answer?, Nat Rev Immunol, 18 (2018) 716–725. [DOI] [PubMed] [Google Scholar]
- [202].Baker PJ, Masters SL, Generation of Genetic Knockouts in Myeloid Cell Lines Using a Lentiviral CRISPR/Cas9 System, Methods Mol Biol, 1714 (2018) 41–55. [DOI] [PubMed] [Google Scholar]
- [203].von Levetzow G, Spanholtz J, Beckmann J, Fischer J, Kogler G, Wernet P, Punzel M, Giebel B, Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells, Stem Cells Dev, 15 (2006) 278–285. [DOI] [PubMed] [Google Scholar]
- [204].Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, Fischer A, A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency, N Engl J Med, 348 (2003) 255–256. [DOI] [PubMed] [Google Scholar]
- [205].Larochelle A, Dunbar CE, Hematopoietic stem cell gene therapy:assessing the relevance of preclinical models, Semin Hematol, 50 (2013) 101–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Bak RO, Dever DP, Porteus MH, CRISPR/Cas9 genome editing in human hematopoietic stem cells, Nat Protoc, 13 (2018) 358–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Salisbury-Ruf CT, Larochelle A, Advances and Obstacles in Homology-Mediated Gene Editing of Hematopoietic Stem Cells, J Clin Med, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Dever DP, Porteus MH, The changing landscape of gene editing in hematopoietic stem cells: a step towards Cas9 clinical translation, Curr Opin Hematol, 24 (2017) 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Ferrari S, Jacob A, Beretta S, Unali G, Albano L, Vavassori V, Cittaro D, Lazarevic D, Brombin C, Cugnata F, Kajaste-Rudnitski A, Merelli I, Genovese P, Naldini L, Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking, Nat Biotechnol, 38 (2020) 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Ferrari S, Vavassori V, Canarutto D, Jacob A, Castiello MC, Javed AO, Genovese P, Gene Editing of Hematopoietic Stem Cells: Hopes and Hurdles Toward Clinical Translation, Front Genome Ed, 3 (2021) 618378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Cromer MK, Vaidyanathan S, Ryan DE, Curry B, Lucas AB, Camarena J, Kaushik M, Hay SR, Martin RM, Steinfeld I, Bak RO, Dever DP, Hendel A, Bruhn L, Porteus MH, Global Transcriptional Response to CRISPR/Cas9-AAV6-Based Genome Editing in CD34(+) Hematopoietic Stem and Progenitor Cells, Mol Ther, 26 (2018) 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Hovnanian A, Rochat A, Bodemer C, Petit E, Rivers CA, Prost C, Fraitag S, Christiano AM, Uitto J, Lathrop M, Barrandon Y, de Prost Y, Characterization of 18 new mutations in COL7A1 in recessive dystrophic epidermolysis bullosa provides evidence for distinct molecular mechanisms underlying defective anchoring fibril formation, Am J Hum Genet, 61 (1997) 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Doudna J, Perspective: Embryo editing needs scrutiny, Nature, 528 (2015) S6. [DOI] [PubMed] [Google Scholar]
- [214].Sharma A, Scott CT, The ethics of publishing human germline research, Nat Biotechnol, 33 (2015) 590–592. [DOI] [PubMed] [Google Scholar]
- [215].Lea RA, Niakan KK, Author Correction: Human germline genome editing, Nat Cell Biol, 22 (2020) 135. [DOI] [PubMed] [Google Scholar]
- [216].Yasue A, Mitsui SN, Watanabe T, Sakuma T, Oyadomari S, Yamamoto T, Noji S, Mito T, Tanaka E, Highly efficient targeted mutagenesis in one-cell mouse embryos mediated by the TALEN and CRISPR/Cas systems, Sci Rep, 4 (2014) 5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Hashimoto M, Takemoto T, Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing, Sci Rep, 5 (2015) 11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Benati D, Patrizi C, Recchia A, Gene editing prospects for treating inherited retinal diseases, J Med Genet, 57 (2020) 437–444. [DOI] [PubMed] [Google Scholar]
- [219].Yu W, Wu Z, In Vivo Applications of CRISPR-Based Genome Editing in the Retina, Front Cell Dev Biol, 6 (2018) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, Dass A, Dhanapal V, Fennell TJ, Friedland AE, Giannoukos G, Gloskowski SW, Glucksmann A, Gotta GM, Jayaram H, Haskett SJ, Hopkins B, Horng JE, Joshi S, Marco E, Mepani R, Reyon D, Ta T, Tabbaa DG, Samuelsson SJ, Shen S, Skor MN, Stetkiewicz P, Wang T, Yudkoff C, Myer VE, Albright CF, Jiang H, Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10, Nat Med, 25 (2019) 229–233. [DOI] [PubMed] [Google Scholar]
- [221].Pan B, Askew C, Galvin A, Heman-Ackah S, Asai Y, Indzhykulian AA, Jodelka FM, Hastings ML, Lentz JJ, Vandenberghe LH, Holt JR, Geleoc GS, Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c, Nat Biotechnol, 35 (2017) 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Zetsche B, Volz SE, Zhang F, A split-Cas9 architecture for inducible genome editing and transcription modulation, Nat Biotechnol, 33 (2015) 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Liu KI, Ramli MN, Woo CW, Wang Y, Zhao T, Zhang X, Yim GR, Chong BY, Gowher A, Chua MZ, Jung J, Lee JH, Tan MH, A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing, Nat Chem Biol, 12 (2016) 980–987. [DOI] [PubMed] [Google Scholar]
- [224].Polstein LR, Gersbach CA, A light-inducible CRISPR-Cas9 system for control of endogenous gene activation, Nat Chem Biol, 11 (2015) 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Nihongaki Y, Kawano F, Nakajima T, Sato M, Photoactivatable CRISPR-Cas9 for optogenetic genome editing, Nat Biotechnol, 33 (2015) 755–760. [DOI] [PubMed] [Google Scholar]
- [226].Gamboa L, Phung EV, Li H, Meyers JP, Hart AC, Miller IC, Kwong GA, Heat-Triggered Remote Control of CRISPR-dCas9 for Tunable Transcriptional Modulation, ACS Chem Biol, 15 (2020) 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Mandai M, Kurimoto Y, Takahashi M, Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration, N Engl J Med, 377 (2017) 792–793. [DOI] [PubMed] [Google Scholar]
- [228].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 126 (2006) 663–676. [DOI] [PubMed] [Google Scholar]
- [229].Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, Cherry AB, Demirbas D, Tsankov AM, Zon LI, Rubin LL, Feinberg AP, Meissner A, Cowan CA, Daley GQ, A comparison of non-integrating reprogramming methods, Nat Biotechnol, 33 (2015) 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Liu P, Chen M, Liu Y, Qi LS, Ding S, CRISPR-Based Chromatin Remodeling of the Endogenous Oct4 or Sox2 Locus Enables Reprogramming to Pluripotency, Cell Stem Cell, 22 (2018) 252–261 e254. [DOI] [PubMed] [Google Scholar]
- [231].Weltner J, Balboa D, Katayama S, Bespalov M, Krjutskov K, Jouhilahti EM, Trokovic R, Kere J, Otonkoski T, Human pluripotent reprogramming with CRISPR activators, Nat Commun, 9 (2018) 2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM, Highly efficient Cas9-mediated transcriptional programming, Nat Methods, 12 (2015) 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Kwon JB, Vankara A, Ettyreddy AR, Bohning JD, Gersbach CA, Myogenic Progenitor Cell Lineage Specification by CRISPR/Cas9-Based Transcriptional Activators, Stem Cell Reports, 14 (2020) 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Wei S, Zou Q, Lai S, Zhang Q, Li L, Yan Q, Zhou X, Zhong H, Lai L, Conversion of embryonic stem cells into extraembryonic lineages by CRISPR-mediated activators, Sci Rep, 6 (2016) 19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Harrington LB, Paez-Espino D, Staahl BT, Chen JS, Ma E, Kyrpides NC, Doudna JA, A thermostable Cas9 with increased lifetime in human plasma, Nat Commun, 8 (2017) 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Tu Z, Yang W, Yan S, Yin A, Gao J, Liu X, Zheng Y, Zheng J, Li Z, Yang S, Li S, Guo X, Li XJ, Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos, Sci Rep, 7 (2017) 42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Wilson RC, Gilbert LA, The Promise and Challenge of In Vivo Delivery for Genome Therapeutics, ACS Chem Biol, 13 (2018) 376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Duan L, Ouyang K, Xu X, Xu L, Wen C, Zhou X, Qin Z, Xu Z, Sun W, Liang Y, Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing, Front Genet, 12 (2021) 673286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Mehta A, Merkel OM, Immunogenicity of Cas9 Protein, J Pharm Sci, 109 (2020) 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Crudele JM, Chamberlain JS, Cas9 immunity creates challenges for CRISPR gene editing therapies, Nat Commun, 9 (2018) 3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Gough V, Gersbach CA, Immunity to Cas9 as an Obstacle to Persistent Genome Editing, Mol Ther, 28 (2020) 1389–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA, Dejene B, Cieniewicz B, Romano R, Lesch BJ, Gomez-Ospina N, Mantri S, Pavel-Dinu M, Weinberg KI, Porteus MH, Identification of preexisting adaptive immunity to Cas9 proteins in humans, Nat Med, 25 (2019) 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyuz L, Reinke P, Volk HD, Schmueck-Henneresse M, High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population, Nat Med, 25 (2019) 242–248. [DOI] [PubMed] [Google Scholar]
- [244].Ferdosi SR, Ewaisha R, Moghadam F, Krishna S, Park JG, Ebrahimkhani MR, Kiani S, Anderson KS, Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes, Nat Commun, 10 (2019) 1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE, Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population, Mol Ther Methods Clin Dev, 10 (2018) 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]


