Abstract
The transcription factor hypoxia-inducible factor 1 (HIF1) is an important driver of cancer and is therefore an attractive drug target. Acriflavine (ACF) has been suggested to inhibit HIF1, but its mechanism of action is unknown. Here we investigated the interaction of ACF with DNA and long non-coding RNAs (lncRNAs) and its function in human endothelial cells. ACF promoted apoptosis and reduced proliferation, network formation, and angiogenic capacity. It also induced changes in gene expression, as determined by RNA sequencing (RNA-seq), which could not be attributed to specific inhibition of HIF1. A similar response was observed in murine lung endothelial cells. Although ACF increased and decreased a similar number of protein-coding genes, lncRNAs were preferentially upregulated under normoxic and hypoxic conditions. An assay for transposase accessibility with subsequent DNA sequencing (ATAC-seq) demonstrated that ACF induced strong changes in chromatin accessibility at lncRNA promoters. Immunofluorescence showed displacement of DNA:RNA hybrids. Such effects might be due to ACF-mediated topoisomerase inhibition, which was indeed the case, as reflected by DNA unwinding assays. Comparison with other acridine derivatives and topoisomerase inhibitors suggested that the specific function of ACF is an effect of acridinium-class compounds. This study demonstrates that ACF inhibits topoisomerases rather than HIF specifically and that it elicits a unique expression response of lncRNAs.
Keywords: long non-coding RNA, endothelial cells, angiogenesis, chromatin, topoisomerase inhibitor, hypoxia, acriflavine
Graphical abstract
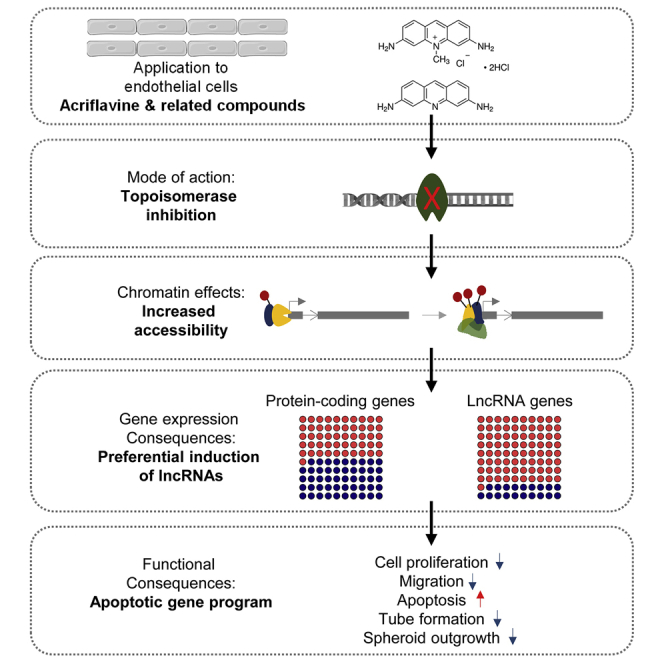
Seredinski et al. demonstrate that acriflavine inhibits topoisomerases rather than blocking hypoxia signaling by specifically inhibiting hypoxia-inducible factor. Acriflavine-induced changes in chromatin accessibility induced pronounced upregulation of long non-coding RNAs. Functionally, this resulted in inhibition of proliferation, migration, spheroid outgrowth, and network formation of human endothelial cells.
Introduction
The DNA double helix exists predominantly in the B form and possesses alternating minor and major grooves. Major grooves are flexible and accessible to DNA-interacting proteins and small molecules. For the latter, this is referred to as intercalation.1,2 Intercalation is commonly exploited for labeling of nucleic acids;3 it is associated with DNA damage and inhibition of transcription, which provide the basis for many drugs in cancer treatment.4 Intercalation sites become accessible when there are structural fluctuations of the DNA. The shape of the intercalation cavity is determined physicochemically and by DNA-associated proteins like transcriptions factors, nuclear receptors, or topoisomerases (TOPs).5,6 Intercalation is not random but induces local unwinding of DNA dependent on the intercalation site.7 Many intercalators can form ternary complexes of DNA, compound, and DNA-associated proteins, such as TOPs. These can be divided into two classes: type I TOPs relax double-stranded DNA (dsDNA) by cleaving a single strand of the DNA, whereas type II TOPs cleave double strands.8, 9, 10 Both are attractive targets in cancer treatment because they are crucial for transcription initiation, DNA replication, and chromosome segregation.11, 12, 13, 14
Acriflavine (ACF), an acridine dye, is a flat, aromatic, small molecule with strong anticancer properties whose application against bacteria was recommended in 1921.15 ACF intercalates into DNA and binds to TOPI and II.1,16 Formation of such a DNA-drug-TOP complex hinders re-ligation of DNA breaks, which causes a greater extent of cleaved DNA. Ultimately, this leads to apoptosis. Small molecules that deploy this mode of action are referred to as TOP poisons.6 Other proteins are also direct targets of ACF, exemplified by hypoxia-inducible factor (HIF) 1α and HIF2α. These subunits of the HIF1 dimer are important for adaptation to hypoxia. By direct binding to either subunit, ACF prevents dimerization with HIF1β and abolishes gene expression of, e.g., vascular endothelial growth factor (VEGF) and enzymes for glucose metabolism, leading to tumor growth arrest and inhibition of vascularization.17,18
Because of the ability of ACF to intercalate into DNA and poison TOPs, it can be inferred that ACF ultimately alters RNA expression. In addition to mRNA, cells express numerous other classes of RNA, some of which have been implicated in stress responses, architectural functions, or gene expression control.19, 20, 21 These functions are frequently executed by long non-coding RNAs (lncRNAs), which represent a large class of RNAs that are defined as being transcribed but not translated and that are longer than 200 nt.22,23
Considering the broad range of functions of lncRNAs and the fact that ACF is a potential supporting chemotherapeutic agent, it is essential to elucidate the effects of ACF on cells other than tumor cells to prevent potential side effects.
Here we investigated the physiological and molecular consequences of ACF treatment on human endothelial cells. In vitro angiogenesis, proliferation, and caspase assays paired with next-generation sequencing, DNA TOP assays, and conservation analysis revealed ACF as a potential tool to study lncRNA regulation. The resulting chromatin changes and drug specificity data could be highly informative for development of future RNA-targeting therapeutic agents.
Results
ACF alters the expression of specific human endothelial genes independent of HIF
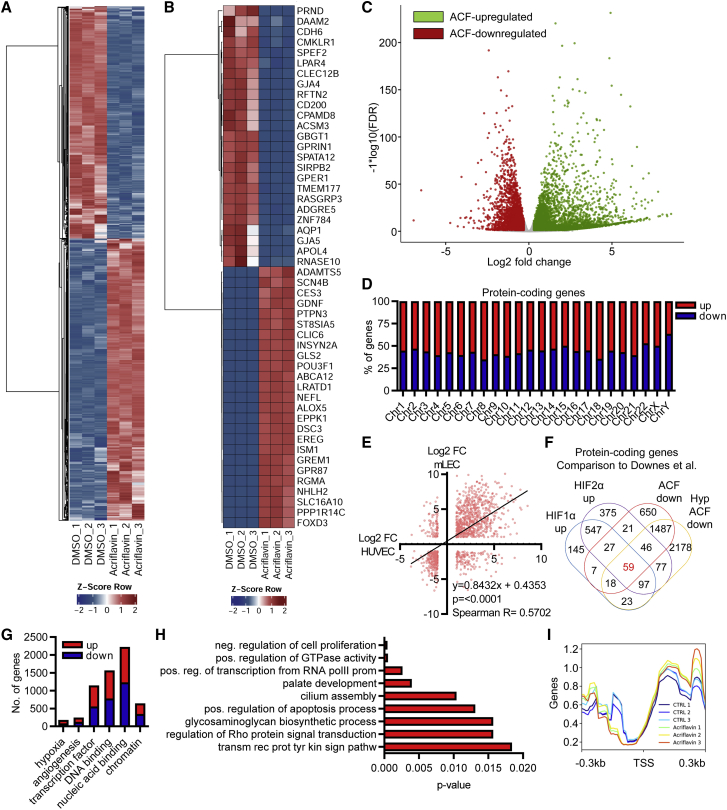
To investigate the molecular consequences of ACF treatment on human endothelial cells, gene expression studies were performed. RNA sequencing (RNA-seq) of three different batches of human endothelial cells (umbilical vein, pools of males and females, passage 4) with and without ACF stimulation revealed that ACF alters the expression of thousands of protein-coding genes across all chromosomes. These effects were similar among the batches, suggesting that they are a consequence of a specific function of ACF and not mediated by random interactions between ACF and the DNA (Figures 1A–1D; Table S1). To substantiate this concept, we hypothesized that the effect of ACF should be species independent and determined its effects on RNA expression of murine lung endothelial cells (mLECs) (Table S2). Indeed, a strong correlation of the human and murine RNA expression sets was observed (Figure 1E).
Figure 1.
ACF strongly changes gene expression
(A) Heatmap of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h) showing significantly (p-value adjusted (padj) < 0.05, log2 fold change (log2FC) >±0.3) altered protein-coding genes. DMSO served as a negative control. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (B) Heatmap as in (A). The top 25 up- and downregulated protein-coding genes are shown. (C) Volcano plot of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h), showing significantly altered protein-coding genes. Colored dots indicate significantly differentially expressed genes; padj (false discovery rate [FDR]) < 0.05, log2FC>±0.3). Red, ACF downregulated genes; green, ACF upregulated genes; gray, non-regulated genes (log2FC <±0.3). (D) Chromosomal distribution and percentage of protein-coding genes up- or downregulated with ACF. (E) Correlation analysis (linear regression) of protein-coding genes up- or downregulated with ACF (logFC > ±1.0) of HUVECs and murine lung endothelial cells (mLECs). (F) Venn diagram showing overlap of protein-coding genes significantly downregulated with ACF under normoxic or hypoxic (Hyp) conditions (this study) with genes significantly upregulated after HIF1α or HIF2α overexpression from Downes et al.24 in HUVECs. (G) Number of protein-coding genes up- or downregulated with ACF assigned to selected GO terms. (H) GO enrichment analysis with KOBAS2.0, indicating highly significant GO terms for protein-coding genes up- and downregulated with ACF. (I) Overlay analysis with deeptools2 of the reads with the transcription start sites (TSSs) of all genes, including 0.3 kb up- or downstream.
ACF has been suggested to be a potent inhibitor of HIF1.17 Thus, it could be expected that HIF1-dependent genes are among those regulated by ACF. To test this assumption, we additionally performed RNA-seq under hypoxic conditions (1% O2) with and without stimulation (Table S3; Figures S1A and S1B) with ACF and overlapped the ACF-responsive genes with previously published HIF-responsive protein-coding genes.24 This analysis showed that less than 10% of the ACF-responsive genes are also HIF dependent (Figure 1F). In line with this, classifying and counting up- and downregulated genes for certain Gene Ontology (GO) terms revealed that ACF did not primarily affect hypoxia-associated genes but, according to KOBAS2.0,25 acted on genes belonging to the GO terms “positive regulation of RNA polymerase II transcriptional activity,” “transcription factors,” or “DNA or nucleic acid binding” (Figures 1G, 1H, and S2). Indeed, overlaying the RNA-seq reads with the transcription start sites of all genes using deeptools226 revealed that ACF increased gene transcription in the sense direction (Figure 1I).
These data demonstrate that the response of endothelial cells to ACF is not restricted to HIF1 but still elicits a specific but cross-species gene expression response.
TopI and TopIIA inhibition by ACF
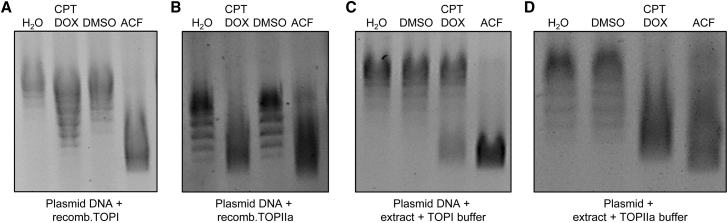
In addition to inhibiting HIF1, TOP inhibition and DNA intercalation have been suggested for ACF.1,16 To test this and to determine the strength of the possible TOP inhibition, we compared the inhibitory effect of ACF on DNA unwinding against the well-known TOPI and TOPIIA inhibitors camptothecin (CPT) and doxorubicin (DOX). TOP assays were performed using plasmid DNA together with recombinantly purified TOPI or TOPIIA or native human endothelial cell nuclear extracts. Application of ACF led to a strong shift of unwound DNA, and this effect was even stronger than that observed in response to the positive controls CPT and DOX (Figures 2A–2D). Thus, ACF is a potent TOPI and TOPIIA inhibitor.
Figure 2.
ACF is a TOPI and TOPIIA inhibitor
(A–D) Agarose gels of the TOP assays with 500 ng pUC19 vector with human recombinant TOPI (A), TOPIIA (B), or 2.5 μg nuclear extract containing native TOPs (C and D). Assays were performed in the presence of ACF or doxorubicin (DOX) and camptothecin (CPT) at a final concentration of 10 μM, and DNA was stained with ethidium bromide. H2O and DMSO served as positive controls showing TOP activity.
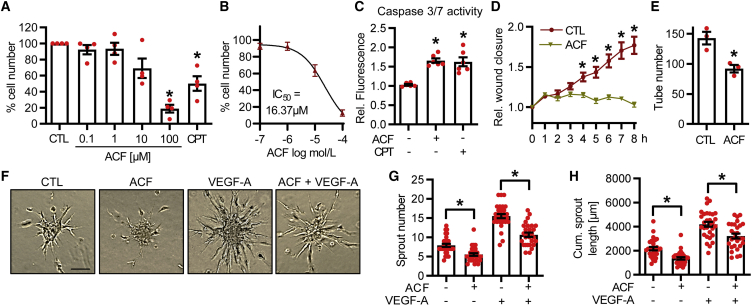
ACF inhibits cell proliferation and spheroid outgrowth
To identify the physiological consequences of ACF treatment on human endothelial cells, we first determined its effect on endothelial cell survival when applied at different concentrations. When applied for 16 h, ACF dose-dependently decreased cell survival, with an IC50 of approximately 16 μM (Figures 3A and 3B). The decrease in cell numbers was probably mediated by apoptosis because ACF also increased caspase-3/7 activity (Figure 3C). ACF also inhibited functional endothelial readouts. 10 μM ACF blocked endothelial migration in the scratch wound assay (Figure 3D) as well as angiogenic tube formation and spheroid outgrowth (Figures 3E–3H). The latter effects were observed under basal conditions as well as in VEGF-A-stimulated spheroids (Figures 3F–3H). Thus, ACF decreases endothelial angiogenic capacity and induces apoptosis.
Figure 3.
ACF has anti-proliferative, pro-apoptotic, and anti-angiogenic effects on human endothelial cells
(A) Percentage of HUVECs surviving different concentrations of ACF (0.1–100 μM, 16 h) or CPT (10 μM, 16 h). DMSO served as a negative control (CTL). n = 4, one-way ANOVA with Dunnett post hoc test. (B) Calculation of IC50, indicated by the percentage of HUVECs surviving different concentrations of ACF (−7 to −4 log mol/L, 16 h). n = 7, nonlinear regression model. (C) Caspase-3/7 activity assay of HUVECs treated with or without ACF (10 μM, 16 h) or CPT (10 μM, 16 h). n = 6, one-way ANOVA with Bonferroni post hoc test. (D) Relative wound closure of HUVECs treated with or without ACF (16 h, 10 μM). DMSO served as a negative CTL. n = 24, two-way ANOVA with Bonferroni post hoc test. (E) Matrigel assay of HUVECs treated with or without ACF (10 μM, 16 h). DMSO served as a negative CTL. n = 3, paired t test. (F–H) Spheroid outgrowth assay (F) and quantification of sprout numbers (G) and cumulative sprout lengths (H) of HUVECs treated with or without ACF (10 μM, 16 h). Cells were studied under basal conditions or after treatment with VEGF-A (30 ng/mL). The scale bar indicates 100 μm. n = 30. One-way ANOVA with Bonferroni post hoc test. Data are mean ± SEM. ∗p < 0.05.
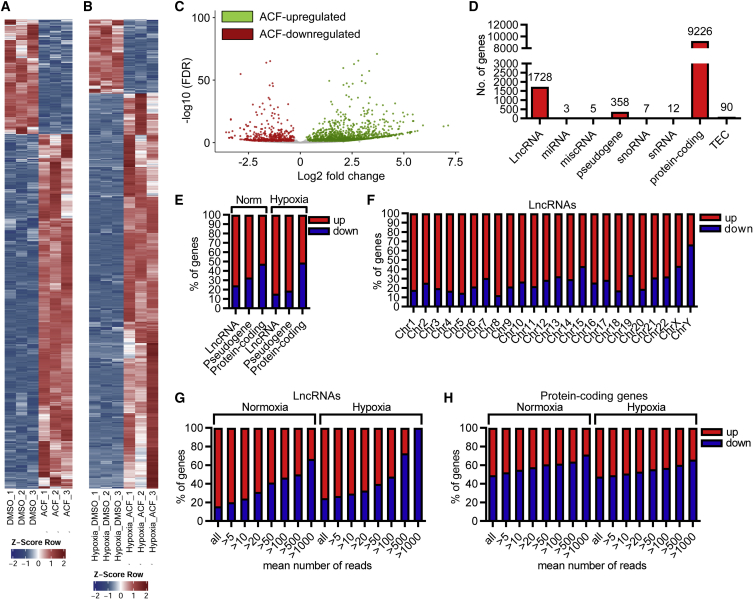
ACF increases expression of low-abundance lncRNAs
Because of the strong effects of ACF on mRNA expression, we wondered whether other groups of RNAs besides protein-coding mRNAs are affected similarly by ACF treatment. Analysis of the normoxic and hypoxic RNA-seq data for expression of lncRNAs revealed that ACF strongly altered the expression of hundreds of lncRNA genes (Figures 4A–4D and S3A). Surprisingly, lncRNA expression was predominantly increased compared with pseudogenes or protein-coding genes (Figure 4E), even after removing the lowly expressed genes (Figure S3B). Except for the sex chromosomes (and chromosome 15 under normoxic conditions), more than 70% of all ACF-affected lncRNAs were upregulated (Figures 4F and S3C). Normalization to the total abundance of lncRNAs demonstrated that the lowly expressed lncRNAs in particular were induced the most, whereas ACF decreased the expression of the highly abundant lncRNAs (Figures 4G and 4H). Thus, ACF has an effect on the precision of gene expression control regardless of normoxia or hypoxia. Because ACF increased gene expression of low-abundance lncRNAs, inhibition of highly abundant lncRNAs might be a consequence of displacement of permissive epigenetic signatures.
Figure 4.
ACF increases expression of numerous lncRNAs
(A and B) Heatmap of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h) under normoxic (A) or hypoxic conditions (B), showing significantly (padj <0.05, log2FC >±0.3) altered lncRNAs. DMSO served as a negative CTL. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (C) Volcano plot of RNA-seq, showing significantly altered lncRNAs under normoxia. Colored dots indicate genes significantly differentially expressed; padj (FDR) < 0.05, log2FC >±0.3. Red, ACF downregulated genes; green, ACF upregulated genes; gray, non-regulated genes (log2FC <±0.3). (D) Numbers of genes from different gene classes significantly altered with ACF under normoxic conditions. (E) Percentage of genes belonging to lncRNAs, pseudogenes, or protein-coding genes up- or downregulated with ACF under normoxic (Norm) or hypoxic conditions. (F) Chromosomal distribution and percentage of lncRNAs up- or downregulated with ACF under normoxic conditions. (G and H) Percentages of lncRNAs (G) and protein-coding genes (H) up- or downregulated by ACF under normoxic or hypoxic conditions, depending on filtering the data for the mean number of reads. All indicates no filter; >5 to >1000 reads indicates that only lncRNAs with read numbers higher than 5–1,000 were used for the analysis.
Application of ACF alters chromatin accessibility and displaces DNA:RNA hybrids
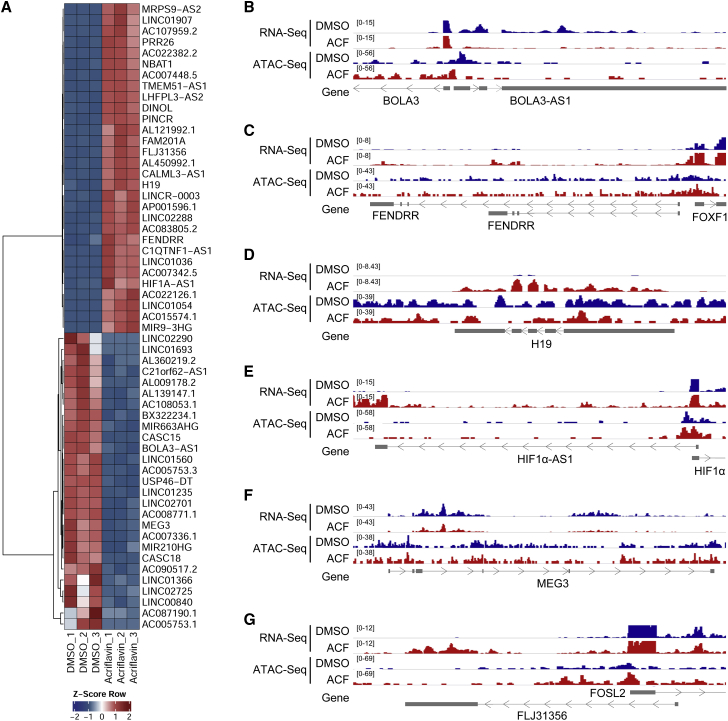
Analysis of the batch reproducibility of lncRNA expression revealed that the three endothelial cell batches used in this study responded uniformly to ACF, suggesting a specific mechanism on gene expression (Figures 5A and S3D). To study this aspect, the effect of ACF on chromatin accessibility was analyzed using an assay for transposase accessibility with subsequent DNA sequencing (ATAC-seq) with or without ACF treatment. As shown for a few selected lncRNA candidates, ACF induced drastic changes in chromatin accessibility, which resulted in corresponding changes in RNA expression (Figures 5B–5G). For example, the decrease in abundance of the lncRNAs BOLA3-AS1 and MEG3 was paralleled by shifts and decreased ATAC signals around the transcriptional start sites (Figures 5B and 5F). Accordingly, the increase in expression of the lncRNAs FENDRR, HIF1α-AS1, and FLJ31356 were associated with increased chromatin accessibility. In the case of FENDRR and HIF1α-AS1, this applied to the promoter and the transcriptional start site, whereas for FLJ31356, the ATAC signals in the first intron increased (Figures 5C, 5E, and 5G). Given the broad changes in cellular function, there were also exceptions. The expression of the lncRNA H19 was increased by ACF despite a decrease in chromatin accessibility in the gene promoter and gene body region (Figure 5D). This could potentially be mediated by changes in H19 stability or effects of ACF on important H19 regulatory elements.
Figure 5.
ACF alters chromatin accessibility on lncRNA loci
(A) Heatmap of RNA-seq of selected significantly altered lncRNAs after treatment of HUVECs with ACF (10 μM, 16 h) under normoxic conditions. DMSO served as a negative CTL. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (B–G) IGV original traces loaded of RNA-seq and ATAC-seq in HUVECs treated with ACF. Loci for BOLA3-AS1 (B; chr2(hg38):74,146,447–74,152,132), FENDRR (C; chr16(hg38):86,472,458–86,514,791), H19 (D; chr11(hg38):1,993,921–1,998,595), HIF1α-AS1 (E; chr14(hg38):61,679,952–61,697,350), MEG3 (F; chr14(hg38):100,822,031–100,862,947), and FLJ31356 (G; chr2(hg38):28,382,410–28,396,672) are shown. DMSO served as a negative CTL. Numbers in square brackets indicate data range values.
A study by Melé et al.27 showed that binding sites of the transcription factors (TF) GATA, JUN, FOS, FOXA and IRF4 are enriched at lincRNA promoters. Comparing the expression differences of these TFs in our RNA-seq data, we identified that GATA4, FOXA1, FOXA3, and IRF4 are indeed strongly upregulated under normoxia and hypoxia upon ACF stimulation (Table 1). These TFs could potentially play a role in ACF-mediated upregulation of many lncRNAs.
Table 1.
LogFC values of TFs previously published to be enriched in lincRNA promoters
| Gene | logFC ACF/DMSO normoxia | logFC ACF/DMSO hypoxia |
|---|---|---|
| GATA2 | +0.39 | +1.93 |
| GATA3 | +0.52 | +1.16 |
| GATA4 | +4.83 | +3.89 |
| GATA6 | −0.01a | −0.52 |
| FOXA1 | +2.38 | +4.41 |
| FOXA2 | not found | +5.87 |
| FOXA3 | +1.89 | +3.73 |
| JUN | −0.16 | −0.38 |
| JUNB | −0.12a | +1.40 |
| JUND | −0.44 | −0.36 |
| IRF4 | +6.46 | +9.17 |
| FOS | 0.00a | +4.88 |
| FOSB | +0.10a | +6.35 |
| FOSL1 | +0.56 | +0.93 |
| FOSL2 | −0.38 | −0.42 |
Not significant.
Many lncRNAs are tethered to chromatin and influence chromatin accessibility through hybrid structures with DNA, such as triple helices (triplexes) or R-loops.28 Given that ACF intercalates with DNA, it is tempting to speculate that such interactions are disturbed by ACF. Indeed, as observed by immunofluorescence with antibodies against DNA:RNA hybrids, ACF decreased DNA:RNA hybrids in the nucleus of human endothelial cells and resulted in displacement of the nuclear signal (Figure S4). This observation could be a possible explanation for the altered chromatin accessibility in response to ACF because DNA:RNA hybrids can recruit silencing complexes to the DNA.29
ACF effects are specific and distinct from other TOP inhibitors
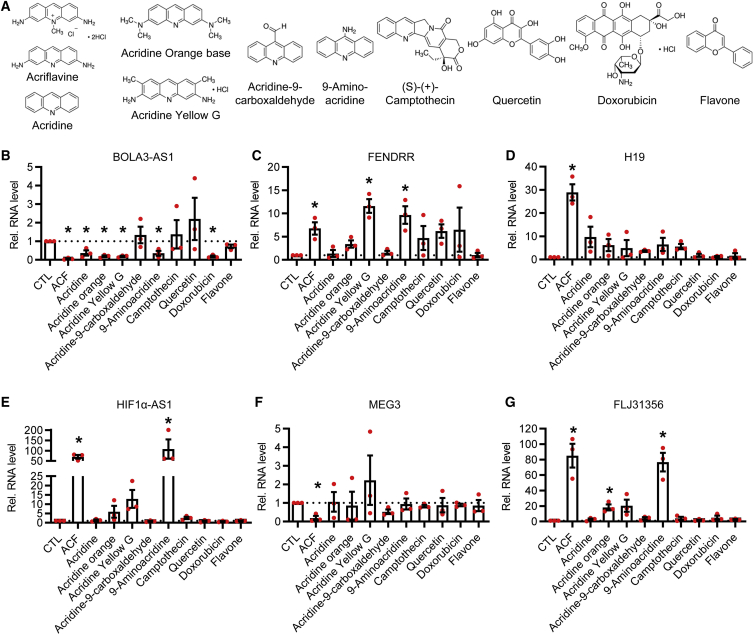
The data of the present study suggest that ACF, by binding DNA, affects processes requiring direct DNA interaction, like hybrid formation and TOP inhibition. To further dissect the mechanism, the effect of ACF on expression of 6 lncRNAs was compared with that of established TOP inhibitors as well as derivatives of ACF (Figure 6A). None of the compounds studied fully mimicked the effect of ACF (Figures 6B–6G). ACF belongs to the class of acridines, and the acridine derivatives acridine orange, acridine yellow G, and 9-aminoacridine elicited a similar but weaker effect on the lncRNAs BOLA3-AS1, FENDRR, and H19 than ACF (Figures 6B–6D). The lncRNAs FLJ31356 and HIF1α-AS1, in contrast, were selectively increased by ACF and 9-aminoacridine (Figures 6E and 6G). Interestingly, in all cases, acridine-9-carboxaldehyde caused the opposite effect or no effect whatsoever. The established TOP inhibitors CPT and DOX induced a differential effect on lncRNA expression and also differed in their effects on ACF. These data demonstrate that TOP inhibitors do not elicit a uniform effect on lncRNA expression. Moreover, compounds structurally similar to ACF act in a manner similar to ACF.
Figure 6.
Amino group substituents are a determining factor for the function of ACF
(A) Structural formulas of ACF and different derivatives of acridine and TOP inhibitors used. Images were taken from https://www.sigmaaldrich.com. (B–G) Relative lncRNA expression levels of BOLA3-AS1 (B), FENDRR (C), H19 (D), HIF1α-AS1 (E), MEG3 (F), and FLJ31356 (G) after treatment with the indicated reagents (16 h, 10 μM) in HUVECs. The CTL was set to 1. n = 3, paired t test. Error bars are defined as mean ± SEM. ∗p < 0.05.
Discussion
The present study demonstrates that treatment of human umbilical vein endothelial cells (HUVECs) with the TOP inhibitor ACF led to a strong gene upregulation, particularly of lncRNAs, which was often accompanied by changes in chromatin accessibility. Physiologically, ACF inhibited proliferation and angiogenesis and increased caspase activity. Compared with acridine, acridine derivatives, and known TOP poisons and DNA intercalators, ACF elicits a unique gene expression profile. In particular, ACF-responsive genes showed little overlap with HIF-responsive genes, indicating that ACF-dependent gene regulation is mostly independent to its postulated action on HIF. ACF resulted in similar expression changes for human and mouse genes, which is indicative of a cross-species sequence dependency of DNA intercalation.5,6
Besides protein-coding genes, a great number of lncRNAs were differentially expressed in response to ACF. Differentially expressed protein-coding genes were almost equally up- or downregulated, whereas lncRNAs were more than 70% upregulated on most of the chromosomes in response to ACF and lowly expressed lncRNAs were upregulated more than highly expressed lncRNAs. In contrast, there was no such trend for protein-coding genes. As with mRNAs, most lncRNAs are transcribed by RNA polymerase II (RNA Pol II), spliced, capped, and polyadenylated. In contrast to mRNAs, lncRNAs are less conserved, more frequently nuclearly localized, and relatively lowly expressed.28 For this reason, we have chosen a low cutoff of the base count in RNA-seq. Promoters of lncRNAs, especially their TF binding sites, seem to be well conserved.27 Interestingly, Melé et al.27 revealed that binding sites of some TFs were enriched at lincRNA promoters. These included TFs of the GATA family, JUN, FOS, FOXA, and IRF4. In our data, ACF increased RNA levels of GATA4, FOXA1, FOXA3, and IRF4 under normoxia and hypoxia, arguing that these TFs could potentially play a role in ACF-mediated upregulation of many lncRNAs.
ACF acts as a TOP poison, which results in an increased number of DNA strand breaks.6,16 TOP II alpha and beta are recruited to promoter sites, where they transiently induce DNA strand breaks that promote transcription.13 In contrast, an increased number of DNA breaks provokes a DNA damage response (DDR). DNA strand breaks are recognized by the meiotic recombination 11 homologue (MRE11)-RAD50-Nijmegen breakage syndrome protein (NBS1) complex.30 Via recruitment of a full pre-initiation complex, damage-induced lncRNAs (dilncRNAs) are transcribed. They function as guides but are also processed by DROSHA and DICER to produce DNA damage RNAs to recruit other DNA damage factors.31, 32, 33 Our findings suggest that ACF-induced DNA strand breaks potentially invoke induction of DDR processes.
ATAC-seq revealed that the regulatory effects of ACF in many cases paralleled changes in DNA accessibility. Intercalation leads to stretching and opening of DNA,34,35 which might be the reason for chromatin opening in response to ACF. DOX, a TOP II poison as well as DNA intercalator, is further known to evict histones, which leads to an impaired DDR,36 an aspect that has not yet been studied for ACF. The lncRNA BOLA3-AS1 was downregulated in response to ACF, and this was accompanied by a shift in open chromatin. Thus, ACF may induce changes in DNA topology, leading to downregulation of this lncRNA. The lncRNA BOLA3-AS1 is upregulated in endometrial cancer,37 which may justify an evaluation of ACF-like compounds for cancer therapy. In the case of the lncRNA FENDRR, ACF not only increased its expression but also chromatin accessibility upstream of its transcription start site. This is an important function to restore drug sensitivity and to inhibit proliferation of cancer cell lines.38 ACF also induced the well-studied lncRNA H19. The underlying mechanism, however, differs from that of the aforementioned lncRNAs because of ACF-induced chromatin closure on the H19 promoter. In HUVECs, H19 promoted angiogenesis when released by CD90+ cells,39 whereas its expression in some tumor types suppressed angiogenesis.40 Knockdown of MEG3 is associated with induction of DNA damage, elevated p53 levels, and expression of p53 target genes.41 This is in line with the cytotoxic effects observed for ACF in the present study; ACF decreased MEG3 expression as well as accessibility of chromatin at the MEG3 transcriptional start site. ACF also strongly upregulated HIF1α-AS1, a lncRNA upstream of HIF1A. ATAC-seq revealed a shift and an upcoming second peak of chromatin accessibility in the promoter region, which is shared with HIF1A. This is particularly interesting because HIF1α-AS1 is proapoptotic in HUVECs.42
We observed that the IC50 of ACF in HUVECs is 16.37 μM, whereas at 10 μM, cell numbers did not decrease significantly. This effect is different from cancer cells, which have a lower IC50 and appear to be more sensitive to ACF.16,43, 44, 45 This may suggest that HUVEC are more resistant to ACF or that cancer cells exhibit a particularly high sensitivity as a consequence of their decreased DNA stability, accumulation of mutations, state of dedifferentiation, or high proliferation rate. Additionally, we observed elevated levels of caspase-3/7 activity. It is interesting that caspase-3 in endothelial cells is not only associated with apoptosis46 but also with enhanced endothelial barrier function.47 Therefore, ACF might reduce the propensity of some cancers to metastasize. In line with previous studies,16,17 ACF impaired the angiogenic capacity of endothelial cells, and it is therefore attractive to speculate that dysregulation of the abovementioned lncRNAs may contribute to this effect. ACF might therefore be an interesting tool to link lncRNA expression and anticancer therapy.
To better understand the molecular function of ACF, we compared its effects with those of acridine and selected derivatives like acridine orange base, 9-aminoacridine, and acridine-9-carboxaldehyde as well as with known TOP inhibitors like quercetin, DOX, flavone, and CPT. In many cases, acridine and its derivatives caused similar but weaker effects than ACF. An exception was acridine-9-carboxaldehyde, which was largely inactive. Because acridine-9-carboxaldehyde is the only acridine-based compound with an aldehyde substituent, this may highlight the importance of amino-group substituents for the intercalator function of ACF with DNA.48 Interestingly, the gene expression changes in response to ACF were different from those elicited by other TOP inhibitors. Indeed, intercalation sites are compound specific because they are dependent on the surrounding nucleotide sequence and the local biophysical parameters.6,49
ACF is not only a useful tool to study lncRNA expression, it also has a unique effect on lncRNAs, which have been specifically linked to malignant disease. Thus, ACF might act as a lead compound for future anticancer drug development.
Materials and methods
Cell culture
Pooled HUVECs were purchased from PromoCell (C-12203, lot numbers 405Z013, 408Z014, and 416Z042; Heidelberg, Germany) and cultured at 37°C with 5% CO2 in a humidified incubator. The different batches were commercial pools of cells obtained from umbilical cords/umbilical veins (405Z013: 2 males, 1 female; 408Z014: 2 males, 1 female; 416Z042: 2 males, 2 females). Cell culture dishes were coated with gelatin (Sigma-Aldrich, G1890-500G) prior to cell seeding. Endothelial growth medium (EGM) was prepared by supplementing endothelial basal medium (EBM) with human recombinant epidermal growth factor (EGF), EndoCGS-heparin (PeloBiotech, Germany), 8% fetal calf serum (FCS; S0113, Biochrom, Germany), penicillin (50 U/mL) and streptomycin (50 μg/mL) (15140-122, Gibco/Life Technologies, USA). Biological replicates comprised at least three different batches from passage 4. mLECs were isolated as described previously.50 Briefly, the tissue was minced and digested with dispase at 37°C for 60 min. After washing and filtering, mLEC separation was done with CD144-coated Dynabeads. mLECs were cultured in DMEM/F12 supplemented with 20% FCS, penicillin (50 U/mL), streptomycin (50 μg/mL), and ECGF bovine + heparin (PeloBiotech) at 37°C with 5% CO2. For experiments with hypoxia, cells were incubated for 16 h in a SciTive workstation (Baker Ruskinn, Leeds, UK) at 1% O2 and 5% CO2
Materials
The following chemicals and concentrations were used for stimulation: human recombinant VEGF-A 165 (R&D Systems, 293-VE, 30 ng/mL), ACF (A8126, Sigma-Aldrich), acridine (A23609, Sigma-Aldrich), acridine orange base (235474, Sigma-Aldrich), acridine-9-carboxaldehyde (775525, Sigma-Aldrich), 9-aminoacridine (A7295, Sigma-Aldrich), (S)-(+)-CPT (C9911, Sigma-Aldrich), DOX (15,007, Cayman), flavone (F2003, Sigma-Aldrich), quercetin (PHR1488, Sigma-Aldrich), and acridine yellow G (199,508, Sigma-Aldrich).
RNA isolation, reverse transcription, and qRT-PCR
Total RNA isolation was performed using the RNA Mini Kit (Bio&Sell), followed by reverse transcription with SuperScript III reverse transcriptase (Thermo Fisher Scientific) and oligo(dT)23 together with random hexamer primers (Sigma). Copy DNA amplification was quantified by qRT-PCR using ITaq Universal SYBR Green Supermix with ROX as reference dye (Bio-Rad, 1725125) in an AriaMX cycler (Agilent Technologies). Human target genes were normalized to β-actin. Relative expression was calculated using the ΔΔCt method with AriaMX qPCR software (Agilent). Oligonucleotides used for amplification are listed in Table 2.
Table 2.
List of primers for qRT-PCR
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| β-actin | AAA GAC CTG TAC GCC AAC AC | GTC ATA CTC CTG CTT GCT GAT |
| FENDRR | GGC AAG AAG GAG AAC ATC AC | GGC TGC CTG ATT ACA AAG AC |
| MEG3 | GAG TGT TTC CCT CCC CAA GG | GCG TGC CTT TGG TGA TTC AG |
| BOLA3-AS1 | GGC CTG TTT CAT GGA TAG GG | CCG AGA CCT TGG AGT GTT TC |
| H19 | TCA AGC CTG GGC CTT TGA AT | GGC TGA TGA GGT CTG GTT CC |
| HIF1α-AS1 | CGG AGA AGA GAA GGA AAG AC | TAG GCA GAG ACG AGA TGA AC |
| FLJ31356 | GGA AGA AAG CCA ACC TAG AG | GGA AAG TCC AGG GAG ATA AC |
RNA-seq
For RNA-seq, total RNA isolation was performed using the RNA Mini Kit (Bio&Sell). RNA and library preparation integrity were verified with LabChip Gx Touch 24 (PerkinElmer). 1 μg of total RNA was used as input for the SMARTer Stranded Total RNA Sample Prep Kit – HI Mammalian (Clontech Laboratories). Sequencing was performed on the NextSeq500 instrument (Illumina) using v2 chemistry, resulting in an average of 90 Mio reads (for HUVECs) or 47 Mio reads (for mLECs) per library with a 1 × 75-bp single-end setup. The resulting raw reads were assessed for quality, adapter content, and duplication rates with FastQC.51 Trimmomatic was employed to trim reads after a quality drop below a mean of Q20 in a window of 10 nt.52 Only reads between 30 and 150 nt were cleared for further analyses. Trimmed and filtered reads were aligned against the Ensembl human genome version hg38 (ensemble release 101) using STAR 2.7.7a with the parameter “--outFilterMismatchNoverLmax 0.1” to increase the maximum ratio of mismatches to mapped length to 10%.53 For mLECs, Ensembl mouse genome version mm10 (GRCm38.p5) and STAR 2.6.0c were used. The number of reads aligning to genes was counted with the featureCounts tool from the Subread package.54 Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. Differentially expressed genes in HUVECs were identified using DESeq2 version 1.30.0 (for mLECs, version 1.18.1 was used).55 For HUVECs, only genes with a minimum fold change of ±1.23-fold (±0.3 on the log2 scale), a maximum Benjamini-Hochberg-corrected p value of 0.05, were deemed to be significantly differentially expressed. For Figure S3B, gene filtering for the base mean was changed from 2 to at least 20. The Ensembl annotation was enriched with Universal Protein Resource (UniProt) data (release 06.06.2014) based on Ensembl gene identifiers (activities at UniProt).56
For RNA-seq of HUVECs treated with hypoxia, 1 μg total RNA was used with the SMARTer Stranded Total RNA Sample Prep Kit – HI Mammalian (Takara). Sequencing was done on Nextseq2000 with v3 chemistry, resulting in average of 53 Mio reads. Analysis was performed as described elsewhere.57
ATAC-seq
50,000 HUVECs were used for ATAC library preparation using Tn5 Transposase from the Nextera DNA Sample Preparation Kit (Illumina). The cell pellet was resuspended in 50 μL PBS; mixed with 25 μL TD buffer, 2.5 μL Tn5, 0.5 μL 10% NP-40, and 22 μL H2O. It was incubated at 37°C for 30 min, followed by 30 min at 50°C together with 500 mM EDTA (pH 8.0). 100 μL of 50 mM MgCl2 was added for neutralization. Purification of DNA fragments was done with the MinElute PCR Purification Kit (QIAGEN). Amplification of the library together with indexing was performed as described elsewhere.58 Libraries were mixed at equimolar ratios and sequenced on the NextSeq500 platform using v2 chemistry. Reaper version 13-100 was employed to trim reads after a quality drop below a mean of Q20 in a window of 5 nt.59 Only reads above 15 nt were cleared for further analyses. These were mapped versus the hg19 version of the human genome with STAR 2.5.2 b using only unique alignments to exclude reads with uncertain arrangement. Reads were further deduplicated using Picard 2.6.060 to avoid PCR artifacts leading to multiple copies of the same original fragment. The MUSIC peak caller (version from December 2015)61 was employed in punctate mode to accommodate the range of peak widths typically expected for ATAC-seq. The maximum q value was set to 0.2, and the p value normalization window length was changed to 1,500. To determine thresholds for significant peaks, the data were inspected manually in IGV 2.3.52.62 Peaks overlapping ENCODE blacklisted regions (known misassemblies, satellite repeats) were excluded. Peaks were annotated with the promoter (transcription start site [TSS] ± 5,000 nt) of the gene most closely located to the center of the peak based on reference data from GENCODE v19. To be able to compare peaks in different samples, the resulting lists of significant peaks were overlapped and unified to represent identical regions. The counts per unified peak per sample were computed with BigWigAverageOverBed (UCSC Genome Browser Utilities; http://hgdownload.cse.ucsc.edu/downloads.html). Raw counts for unified peaks were submitted to DESeq2 (version 1.20.0) for normalization. Spearman correlations were produced to identify the degree of reproducibility between samples using R. To permit a normalized display of samples in IGV, the raw BAM files were normalized for sequencing depth (number of mapped deduplicated reads per sample) and noise level (number of reads inside peaks versus number of reads not inside peaks). Two factors were computed and applied to the original BAM files using bedtools genomecov, resulting in normalized BigWig files for IGV. To convert the data to the updated genome version hg38, bwtool was used.63
Spheroid outgrowth assay
Spheroid outgrowth assays in HUVECs were performed as described previously.64 Spheroids were stimulated in EGM containing 50 ng/mL VEGF-A 165 for 16 h. Images were generated with an Axiovert135 microscope (Zeiss, Germany), and quantitative analysis of sprout number and cumulative length was performed with AxioVision software (Zeiss).
Matrigel assay (tube formation)
1 × 105 HUVECs were incubated in EBM and 1% FCS with Matrigel Growth Factor Reduced (BD Biosciences) for 4 h as described previously.65 After stopping the reaction with 4% paraformaldehyde (PFA), images were taken on an Axiovert135 microscope (Zeiss). Image quantification was performed with AxioVision software.
Caspase-3/7 activity assay
The caspase-3/7 activity assays were carried out using 1 × 106 HUVECs stimulated with EGM supplemented with 10 μM ACF, DOX, or CPT for 16 h. DMSO at a ratio of 1:1,000 was used as negative control. The assay was performed using the SR-FLICA Caspase-3/7 Assay Kit (ImmunoChemistry Technologies, 931) following the manufacturer’s instructions. Briefly, cells were washed, and a 1:5 dilution of FLICA was added at a dilution of 1:30 to the cell suspension. After an incubation of 1 h, cells were washed three times with buffer provided by the kit, counted, and diluted to 3,000 cells/μL before measuring emission at 595 nm in a Infinite M200 Pro plate reader (Tecan, Männedorf, Switzerland). The percentage of apoptotic cells relative to the total number of cells was calculated.
TOP-assay
Nuclear lysates of 2 × 106 HUVECs were prepared as described previously.66 Protein concentrations were determined with the Bradford assay. 2.5 μg protein extract or 2 U TOP (human TOP IIα [TG2000-H1] or human TOP I [TG2005-RC1], both from TopoGEN) was pre-incubated with 500 ng pUC19 vector for 20 min at 37°C. H2O or DMSO served as a negative control. ACF, DOX, or CPT at a final concentration of 10 μM was added, and incubation was continued for 30 min at 37°C. Samples were loaded on a 1% agarose gel, stained with ethidium bromide, and visualized in a UV transilluminator.
Scratch wound assay
30,000 HUVECs were cultured in EBM containing FCS (8%) in ImageLock 96-well plates (Essen Bioscience). Cells were stimulated for 16 h with ACF (10 μM) or DMSO prior to generating homogeneous scratch wounds in the Incucyte WoundMaker (Essen Bioscience). Cell migration monitoring and the following analysis were carried out with the Scratch Wound Cell Migration Module of the Incucyte S3 Live Cell Analysis System (Essen Bioscience).
Immunofluorescence
HUVECs were seeded on 8-well immunofluorescence plates (Ibidi) and treated with ACF (10 μM, 16 h) or DMSO. Then cells were washed with PBS, fixed with 4% PFA, quenched with glycine (2%), and washed again with PBS. Permeabilization was done with 0.05% Triton X-100, followed by blocking with 3% BSA for 30 min. The cells were incubated at 4°C overnight with a 1:200 dilution of the primary antibody anti-DNA-RNA hybrid (S9.6) (Kerafast, ENH001). Cells were washed with 0.3% Tween 20 in PBS and incubated with a 1:500 dilution of secondary antibody for 30 min. Then four washing steps were performed: once with PBS (0.3% Tween, 0.1 mg/mL DAPI, 5 min), twice with PBS (0.3% Tween, 3 min), and once with pure PBS for 3 min. Cells were then washed again with 0.3% Tween and counterstained with DAPI. Images were captured with a laser confocal microscope (LSM800, Zeiss) and analyzed with ZEN lite software (Zeiss).
Statistics
Unless otherwise indicated, data are given as means ± SEM. Prism 8 or BiAS.10.12 was used for calculations. The latter was also used to test for normal distribution and similarity of variance. Bonferroni correction was applied in case of multiple testing. For multiple-group comparisons, ANOVA followed by post hoc testing was performed. For testing individual statistics of dependent samples, paired t test was used; in the case of unpaired samples, unpaired t test. Samples with non-normal distributions were probed by Mann-Whitney test. A p value of less than 0.05 was considered significant. n indicates the number of individual experiments.
Acknowledgments
We thank Katalin Pálfi and Cindy F. Höper for excellent technical assistance and Dominik Fuhrmann for help with the hypoxia chamber. This work was supported by the Goethe University Frankfurt am Main, the German Centre for Cardiovascular Research (DZHK, PostDoc Startup Grant 81X3200107), the DFG Excellence Cluster Cardiopulmonary Institute (CPI) EXS2026, DFG Transregio TRR267 (TPA04 and TPA06), and SFB1039 (TP A01 to R.P.B.).
Author contributions
S.S. and M.S.L. designed the experiments. S.S., F.B., S.G., J.A.O., J.I.P., F.F.L., E.P., and B.P.M. performed the experiments. S.S., S.G., R.G., and M.S.L. analyzed the data. S.S., S.G., T.W., and R.G. performed bioinformatics. S.K. helped with the chemical compound. S.S., R.P.B., and M.S.L. supervised the studies and wrote the manuscript. All authors interpreted the data and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.01.016.
Supplemental information
Data availability
The datasets of normoxic experiments (ATAC-seq GSE176554, RNA-seq GSE176555) have been deposited as a superseries and are available at NCBI GEO with the accession number GSE176556.
The RNA-seq dataset under hypoxic conditions has been deposited and is available at NCBI GEO with the accession number GSE186297.
References
- 1.Lerman L.S. Structural considerations in the interaction of DNA and acridines. J. Mol. Biol. 1961;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- 2.Comba P. Springer Netherlands; 2010. Structure and Function. [Google Scholar]
- 3.Mazzini G., Danova M. Fluorochromes for DNA staining and quantitation. Methods Mol. Biol. 2017;1560:239–259. doi: 10.1007/978-1-4939-6788-9_18. [DOI] [PubMed] [Google Scholar]
- 4.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 5.Hendry L.B., Mahesh V.B., Bransome E.D., Ewing D.E. Small molecule intercalation with double stranded DNA: implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat. Res. 2007;623:53–71. doi: 10.1016/j.mrfmmm.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Capranico G., Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 7.Egli M. In: Structure Correlation. Bürgi H.-B., Dunitz J.D., editors. Wiley; 1994. Structural patterns in nucleic acids; pp. 705–749. [Google Scholar]
- 8.Kirkegaard K., Wang J.C. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 9.Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 10.Goulaouic H., Roulon T., Flamand O., Grondard L., Lavelle F., Riou J.F. Purification and characterization of human DNA topoisomerase IIIalpha. Nucleic Acids Res. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Wang J.C., Liu L.F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl. Acad. Sci. U S A. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino A., Madden K.R., Lane W.S., Champoux J.J., Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 13.Ju B.-G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 14.Ramamoorthy M., Tadokoro T., Rybanska I., Ghosh A.K., Wersto R., May A., Kulikowicz T., Sykora P., Croteau D.L., Bohr V.A. RECQL5 cooperates with topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res. 2012;40:1621–1635. doi: 10.1093/nar/gkr844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning C.H., Cohen J.B., Gaunt R., Gulbransen R. Relationships between antiseptic action and chemical constitution with special reference to compounds of the pyridine, quinoline, acridine and phenazine series. Proc. R. Soc. B Biol. Sci. 1922;93:329–366. [Google Scholar]
- 16.Hassan S., Laryea D., Mahteme H., Felth J., Fryknäs M., Fayad W., Linder S., Rickardson L., Gullbo J., Graf W., et al. Novel activity of acriflavine against colorectal cancer tumor cells. Cancer Sci. 2011;102:2206–2213. doi: 10.1111/j.1349-7006.2011.02097.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee K., Zhang H., Qian D.Z., Rey S., Liu J.O., Semenza G.L. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U S A. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Broekgaarden M., Weijer R., Krekorian M., van den IJssel B., Kos M., Alles L.K., van Wijk A.C., Bikadi Z., Hazai E., van Gulik T.M., et al. Inhibition of hypoxia-inducible factor 1 with acriflavine sensitizes hypoxic tumor cells to photodynamic therapy with zinc phthalocyanine-encapsulating cationic liposomes. Nano Res. 2016;9:1639–1662. [Google Scholar]
- 19.Guil S., Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Valadkhan S., Valencia-Hipólito A. lncRNAs in stress response. Curr. Top. Microbiol. Immunol. 2016;394:203–236. doi: 10.1007/82_2015_489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G., Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70:1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 23.Uchida S., Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 24.Downes N.L., Laham-Karam N., Kaikkonen M.U., Ylä-Herttuala S. Differential but complementary HIF1α and HIF2α transcriptional regulation. Mol. Ther. 2018;26:1735–1745. doi: 10.1016/j.ymthe.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.-Y., Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melé M., Mattioli K., Mallard W., Shechner D.M., Gerhardinger C., Rinn J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Statello L., Guo C.-J., Chen L.-L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syed A., Tainer J.A. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018;87:263–294. doi: 10.1146/annurev-biochem-062917-012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelini F., Pitchiaya S., Vitelli V., Sharma S., Gioia U., Pessina F., Cabrini M., Wang Y., Capozzo I., Iannelli F., et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pessina F., Giavazzi F., Yin Y., Gioia U., Vitelli V., Galbiati A., Barozzi S., Garre M., Oldani A., Flaus A., et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francia S., Cabrini M., Matti V., Oldani A., Di d'Adda Fagagna F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016;129:1468–1476. doi: 10.1242/jcs.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bürgi H.-B., Dunitz J.D., editors. Structure Correlation. Wiley; 1994. [Google Scholar]
- 35.Biebricher A.S., Heller I., Roijmans R.F.H., Hoekstra T.P., Peterman E.J.G., Wuite G.J.L. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015;6:7304. doi: 10.1038/ncomms8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang B., Qiao X., Janssen L., Velds A., Groothuis T., Kerkhoven R., Nieuwland M., Ovaa H., Rottenberg S., van Tellingen O., et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y., Chen J., Ling J., Zhu X., Jiang P., Tang X., Zhou H., Li R. Construction of a glycolysis-related long noncoding RNA signature for predicting survival in endometrial cancer. J. Cancer. 2021;12:1431–1444. doi: 10.7150/jca.50413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szafranski P., Stankiewicz P. Long non-coding RNA FENDRR: gene structure, expression, and biological relevance. Genes. 2021;12:177. doi: 10.3390/genes12020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conigliaro A., Costa V., Lo Dico A., Saieva L., Buccheri S., Dieli F., Manno M., Raccosta S., Mancone C., Tripodi M., et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C., Li X., Zhao H., Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol. Cancer. 2016;15:62. doi: 10.1186/s12943-016-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shihabudeen Haider Ali M.S., Cheng X., Moran M., Haemmig S., Naldrett M.J., Alvarez S., Feinberg M.W., Sun X. LncRNA Meg3 protects endothelial function by regulating the DNA damage response. Nucleic Acids Res. 2019;47:1505–1522. doi: 10.1093/nar/gky1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Chen L., Li H., Yang J., Gong Z., Wang B., Zhao X. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Mol. Cell. Biochem. 2015;404:203–210. doi: 10.1007/s11010-015-2379-1. [DOI] [PubMed] [Google Scholar]
- 43.Weijer R., Broekgaarden M., Krekorian M., Alles L.K., van Wijk A.C., Mackaaij C., Verheij J., van der Wal A.C., van Gulik T.M., Storm G., et al. Inhibition of hypoxia inducible factor 1 and topoisomerase with acriflavine sensitizes perihilar cholangiocarcinomas to photodynamic therapy. Oncotarget. 2016;7:3341–3356. doi: 10.18632/oncotarget.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strese S., Fryknäs M., Larsson R., Gullbo J. Effects of hypoxia on human cancer cell line chemosensitivity. BMC Cancer. 2013;13:331. doi: 10.1186/1471-2407-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangraviti A., Raghavan T., Volpin F., Skuli N., Gullotti D., Zhou J., Asnaghi L., Sankey E., Liu A., Wang Y., et al. HIF-1α- targeting acriflavine provides long term survival and radiological tumor response in brain cancer therapy. Sci. Rep. 2017;7:14978. doi: 10.1038/s41598-017-14990-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boatright K.M., Salvesen G.S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Suresh K., Carino K., Johnston L., Servinsky L., Machamer C.E., Kolb T.M., Lam H., Dudek S.M., An S.S., Rane M.J., et al. A nonapoptotic endothelial barrier-protective role for caspase-3. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019;316:L1118–L1126. doi: 10.1152/ajplung.00487.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman H.M., Stallings W., Carrell H.L., Glusker J.P., Neidle S., Taylor G., Achari A. Molecular and crystal structure of an intercalation complex: proflavine-cytidylyl-(3',5')-guanosine. Biopolymers. 1979;18:2405–2429. [Google Scholar]
- 49.Webb M.R., Ebeler S.E. Comparative analysis of topoisomerase IB inhibition and DNA intercalation by flavonoids and similar compounds: structural determinates of activity. Biochem. J. 2004;384:527–541. doi: 10.1042/BJ20040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fleming I., Fisslthaler B., Dixit M., Busse R. Role of PECAM-1 in the shear-stress-induced activation of Akt and the endothelial nitric oxide synthase (eNOS) in endothelial cells. J. Cell Sci. 2005;118:4103–4111. doi: 10.1242/jcs.02541. [DOI] [PubMed] [Google Scholar]
- 51.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 52.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 55.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisegang M.S., Bibli S.-I., Günther S., Pflüger-Müller B., Oo J.A., Höper C., Seredinski S., Yekelchyk M., Schmitz-Rixen T., Schürmann C., et al. Pleiotropic effects of laminar flow and statins depend on the Krüppel-like factor-induced lncRNA MANTIS. Eur. Heart J. 2019;40:2523–2533. doi: 10.1093/eurheartj/ehz393. [DOI] [PubMed] [Google Scholar]
- 58.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis M.P.A., van Dongen S., Abreu-Goodger C., Bartonicek N., Enright A.J. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broad Institute . Broad Institute; 2019. Picard Toolkit. [Google Scholar]
- 61.Harmanci A., Rozowsky J., Gerstein M. MUSIC: identification of enriched regions in ChIP-Seq experiments using a mappability-corrected multiscale signal processing framework. Genome Biol. 2014;15:474. doi: 10.1186/s13059-014-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pohl A., Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korff T., Augustin H.G. Integration of endothelial cells in multicellular spheroids prevents apoptosis and induces differentiation. J. Cell Biol. 1998;143:1341–1352. doi: 10.1083/jcb.143.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Josipovic I., Fork C., Preussner J., Prior K.-K., Iloska D., Vasconez A.E., Labocha S., Angioni C., Thomas D., Ferreirós N., et al. PAFAH1B1 and the lncRNA NONHSAT073641 maintain an angiogenic phenotype in human endothelial cells. Acta Physiol. (Oxf.) 2016;218:13–27. doi: 10.1111/apha.12700. [DOI] [PubMed] [Google Scholar]
- 66.Nitiss J.L., Soans E., Rogojina A., Seth A., Mishina M. Topoisomerase assays. Curr. Protoc. Pharmacol. 2012 doi: 10.1002/0471141755.ph0303s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets of normoxic experiments (ATAC-seq GSE176554, RNA-seq GSE176555) have been deposited as a superseries and are available at NCBI GEO with the accession number GSE176556.
The RNA-seq dataset under hypoxic conditions has been deposited and is available at NCBI GEO with the accession number GSE186297.