Summary
In clinical trials, oral supplementation with nicotinamide riboside (NR) fails to increase muscle mitochondrial respiratory capacity and insulin sensitivity but also does not increase muscle NAD+ levels. This study tests the feasibility of chronically elevating skeletal muscle NAD+ in mice and investigates the putative effects on mitochondrial respiratory capacity, insulin sensitivity, and gene expression. Accordingly, to improve bioavailability to skeletal muscle, we developed an experimental model for administering NR repeatedly through a jugular vein catheter. Mice on a Western diet were treated with various combinations of NR, pterostilbene (PT), and voluntary wheel running, but the metabolic effects of NR and PT treatment were modest. We conclude that the chronic elevation of skeletal muscle NAD+ by the intravenous injection of NR is possible but does not affect muscle respiratory capacity or insulin sensitivity in either sedentary or physically active mice. Our data have implications for NAD+ precursor supplementation regimens.
Subject areas: Drugs, Molecular physiology, Transcriptomics
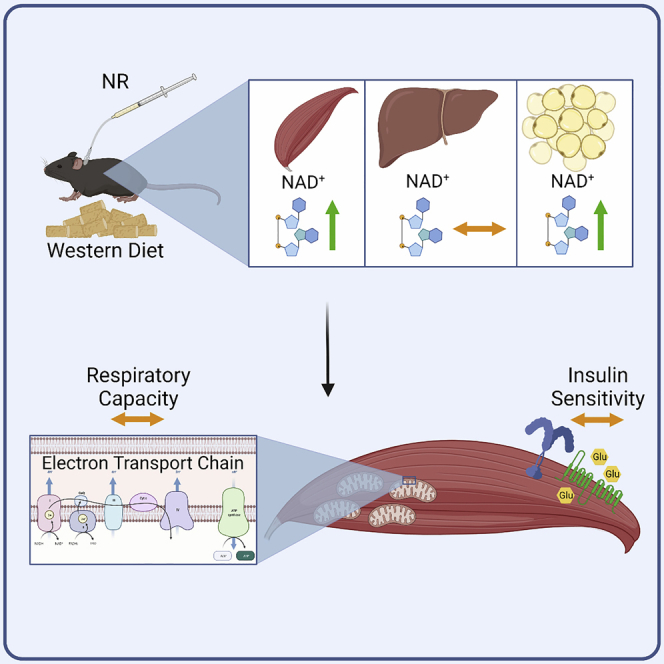
Graphical abstract
Highlights
-
•
A model was developed for daily intravenous NR injections
-
•
Intravenous NR stably elevates NAD+ of skeletal muscle and adipose, but not liver
-
•
Voluntary running and intravenous NR synergize to boost mouse skeletal muscle NAD+
-
•
NR did not impact skeletal muscle insulin sensitivity or respiratory capacity
Drugs; Molecular physiology; Transcriptomics
Introduction
NAD+ is an important co-factor for glycolysis, the tricarboxylic acid cycle as well as oxidative phosphorylation in the mitochondria. As such, it is no surprise that changes in NAD+ levels have been suggested to influence metabolism (Imai, 2010). In fact, lowered levels of NAD+ have been associated with aging (Massudi et al., 2012; Clement et al., 2019), obesity (Jukarainen et al., 2016), mitochondrial dysfunction (Basse et al., 2021), lowered insulin secretion (Revollo et al., 2007) as well as hepatic inflammation and impaired insulin sensitivity (Zhou et al., 2016). In contrast, when NAD+ was increased in mouse skeletal muscle by overexpressing the rate-limiting NAD+ salvage enzyme NAMPT, it resulted in increased expression of several mitochondrial genes, increased mitochondrial respiratory capacity, and a concomitant boost in exercise capacity (Costford et al., 2018; Brouwers et al., 2018). This suggests that NAD+ availability could improve muscle metabolism, but since this type of genetic modification cannot be performed in humans, other strategies must be utilized to elevate NAD+ levels. One such strategy is to supplement with precursors such as nicotinamide riboside (NR). Accordingly, there have been attempts to elucidate the effect of oral NR supplementation in humans, but none of them were able to demonstrate a beneficial effect of NR supplementation on mitochondrial respiratory capacity in muscle (Dollerup et al., 2020; Elhassan et al., 2019; Remie et al., 2020). Strikingly, while this approach presumably increases flux through the NAD metabolome, the studies universally failed to elevate muscle NAD+ levels, which could explain the lack of benefits to respiratory capacity. Similarly, in animal experiments, oral NR elevates NAD+ levels in the liver (Dall et al., 2019; Wang et al., 2018) and NR supplementation also increases NAD+ levels in various cell types in culture (Agerholm et al., 2018; Dall et al., 2019; Wilk et al., 2020), but the bioavailability of orally supplemented NR to skeletal muscle appears low (Frederick et al., 2016). The reason for this may be the degradation of NR in the intestine as well as first-pass metabolism in the liver, since NR given as an acute intravenous injection is taken up and incorporated into skeletal muscle as NAD+ within hours (Liu et al., 2018). In spite of this, the metabolic consequences of chronically elevated levels of NAD+ in skeletal muscle mediated by NR remain unexplored. As a result, we decided to investigate the feasibility of using intravenous NR supplementation to chronically elevate skeletal muscle NAD+ levels and to determine the putative effects of this on insulin sensitivity, mitochondrial respiratory capacity, and gene expression in skeletal muscle.
In addition to its role in bioenergetics, NAD+ is also a necessary co-substrate for the protein SIRT1, so in this exploratory study, we included pterostilbene (PT) as a secondary treatment to investigate the potential for synergy between PT and NR. PT is a naturally occurring analog of resveratrol, with approximately four times higher bioavailability (Kapetanovic et al., 2011). Interestingly, both resveratrol and PT have been predicted to bind in the same pocket of SIRT1 (Guo et al., 2016), and they share many of the same beneficial effects with regards to longevity and metabolic health (Li et al., 2018). To increase the potential for the therapeutic effect of NR and PT, all mice were fed a Western diet (WD) prior to and during the intervention period. Furthermore, we included voluntary wheel running (VWR) as a treatment to elucidate potential differences between active and inactive muscle as well as to increase blood flow to the skeletal muscle under the assumption that it would facilitate delivery of NR and PT.
Results
Oral pterostilbene enters circulation but bioavailability to muscle is low
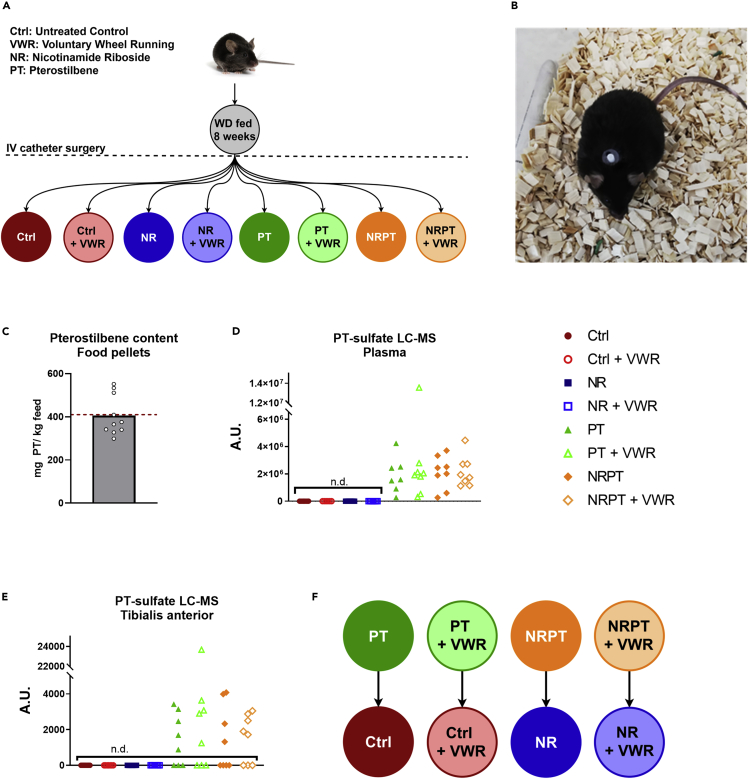
The experimental setup utilized here was a classical 23 full-factorial setup with the three factors NR, PT, and VWR, originally resulting in eight experimental groups (Figure 1A). All mice had a vascular access button implanted (Figure 1B, which was connected to a jugular vein catheter to facilitate intravenous delivery of saline or NR. To validate our method for supplementing PT, we first confirmed the presence of expected amounts of PT in the diet. In the 10 diet pellets we tested, we found an average of 405.28 mg PT per kg diet, corresponding to ∼99% of the expected amount (Figure 1C). Next, we assessed levels of PT in plasma taken at termination. We did not detect any PT in plasma (data not shown), but instead discovered a compound with a mass signature identical to PT-sulfate in the samples from every single mouse receiving PT in the diet, while none was detected in the other groups (Figure 1D). Similarly, we did not detect PT in skeletal muscle (data not shown), but here PT-sulfate was only detected in 19 of 32 muscle samples from PT groups (Figure 1E). PT-sulfate is not commercially available, so we were not able to quantify the amounts, but the relatively low intensities of the LC-MS peaks from muscle, as well as the many samples where PT-sulfate was not detected, suggest low bioavailability to skeletal muscle. This is consistent with a previous report on rats (Azzolini et al., 2014). In our initial statistical analyses, PT showed no effect in any tested parameter, with the exception of a seemingly detrimental interaction between PT and VWR in the analysis of submaximally activated mitochondrial respiration, which we return to in the discussion. The daily dosage of ∼30 mg PT per kg bodyweight is relatively high compared to previous reports on the PT treatment of mice, albeit with different endpoints (Liu et al., 2017; Elango et al., 2016). Owing to the low bioavailability to skeletal muscle, as well as the apparent lack of metabolic benefits, we decided to merge the PT groups into their respective control groups (Figure 1F) for all following figures, except the one detailing submaximal mitochondrial respiration (Figure 5C). This was done to improve the visual clarity and readability of the figures, and our original 23 full factorial experiment setup was chosen with exactly such a possibility in mind (Natoli and Oimoen, 2019; NIST/SEMATECH, 2003). In the interest of transparency, all original figures with the full set of eight groups have been included as supplemental figures (Figures S1, S2, S3, S4, and S5).
Figure 1.
Oral pterostilbene enters circulation but bioavailability to muscle is low
(A) Schematic overview of the group setup. C57BL/6NTac mice were fed a WD for 8 weeks, then underwent surgery with the insertion of a jugular catheter connected to a vascular access button. After recovery, the mice were split into eight groups, creating a full-factorial experiment setup with the three factors; nicotinamide riboside (NR), pterostilbene (PT), and voluntary wheel running (VWR). Blue and orange groups received NR (100 mg/kg/day), green and orange groups received PT (410 mg pr. Kg diet), and hollow groups had access to VWR.
(B) Top-down picture of C57BL/6NTac mouse with a Vascular Access Button for the facilitation of intravenous NR administration.
(C) PT content in food pellets. 10 food pellets were randomly selected and tested for PT content. The dotted red line shows the expected amount of PT. Data are presented as mean with overlain data points.
(D) Relative amounts of PT-sulfate in plasma as measured by LC-MS. Samples, where PT-sulfate could not be detected, have been marked with n.d.
(E) Relative amounts of PT-sulfate in TA muscle as measured by LC-MS and normalized to tissue mass. Samples, where PT-sulfate could not be detected, have been marked with n.d.
(F) Schematic representation of the merging of PT groups into their respective control groups.
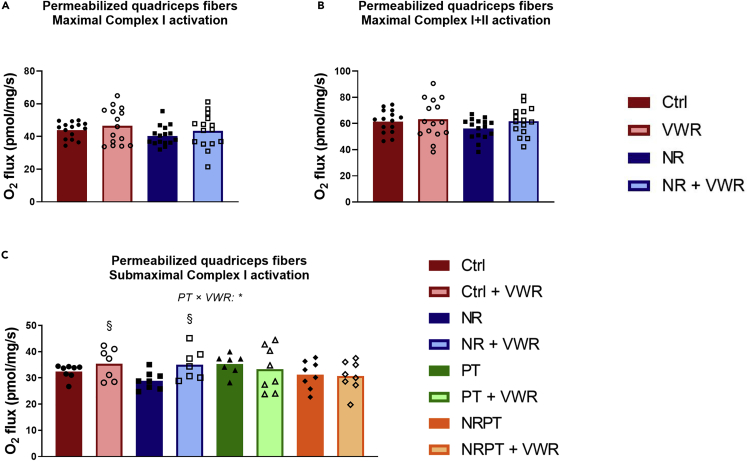
Figure 5.
Neither NR nor VWR increases maximal respiratory capacity in permeabilized muscle fibers.
Oxygen consumption of permeabilized muscle fibers from the superficial lateral part of quadriceps muscle
(A) Incubated in the presence of pyruvate, malate, glutamate, and saturating levels of ADP (5 mM), leading to the maximal activation of Complex I.
(B) Incubated in the presence of pyruvate, malate, glutamate, saturating levels of ADP and succinate, leading to the maximal activation of Complex I and Complex II.
(C) Incubated in the presence of pyruvate, malate, glutamate, and subsaturating levels of ADP (0.1 mM), leading to the submaximal activation of Complex I.
All data were corrected for residual oxygen consumption and are presented as mean with overlain data points. Two-way ANOVA was performed for (A) and (B). Three-way ANOVA was performed for (C), and Tukey’s test was employed to explore the interaction. Statistical significance was indicated in the following manner: Effects of VWR: p < 0.05 = §. Interactions are written out in the figure and noted with: p < 0.05 = ∗. The symbols indicate a simple effect of VWR within groups that did not receive PT. See also Figure S4.
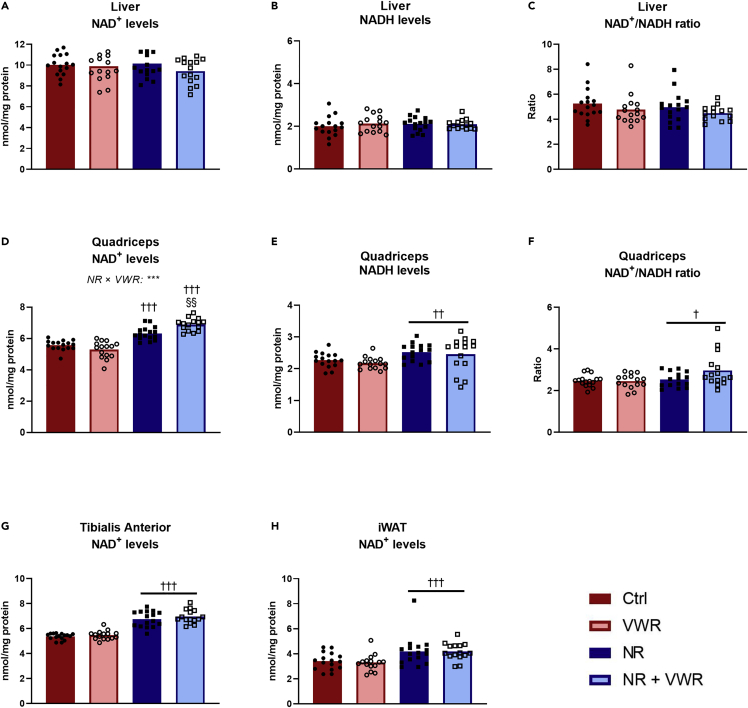
Daily intravenous nicotinamide riboside injections lead to a sustained NAD+ increase in peripheral tissues but not in the liver
To validate our method of administering NR, we performed two pilot experiments. First, we compared seven consecutive days of intravenous delivery versus gavage delivery of 200 mg/kg NR. This resulted in a ∼140% increase in TA NAD+ levels with intravenous delivery (p <0.001), whereas oral gavage failed to boost NAD compared to the control group (p = 0.544) (Figure S1A). This was seemingly explained by intravenous NR delivery giving rise to a ∼36-fold increase in TA NR content (p = 0.005), whereas gavage did not increase NR content (p = 0.996) (Figure S1B). Together, these findings reinforce the observation that orally supplemented NR is not directly bioavailable to skeletal muscle, but that this can be circumvented by intravenous delivery. We then tested a lower dose of intravenous NR (100 mg/kg), which lead to an NAD+ increase in EDL muscle of ∼50% after 1 h (p = 0.001), and used this model to demonstrate that NAD+ levels were increased in EDL muscle for at least 12 h after the last intravenous injection (p = 0.006) (Figure S1C). Thus, with this NR administration method having completed initial validation, we proceeded with our study. Oral NR supplementation usually increases liver NAD+ as we and others have previously demonstrated (Dall et al., 2019; Wang et al., 2018; Liu et al., 2018). However, in our model, there were no changes in either NAD+ (Figure 2A), NADH (Figure 2B), or the NAD+/NADH ratio (Figure 2C) in the liver, at the time of termination. In quadriceps muscle, there was a remarkable interaction between NR and VWR (p < 0.001) in NAD+ levels (Figure 2D). In sedentary mice, the daily NR injections increased NAD+ levels by ∼13% (p < 0.001), whereas NR increased NAD+ levels by ∼30% (p < 0.001) in mice with access to running wheels. This intriguing interaction suggests that physical activity either enhances the uptake of NR or helps maintain the elevated NAD+ in quadriceps muscle. NADH levels in quadriceps were slightly increased with NR (Figure 2E, p = 0.003), but not enough to prevent an increase in the ratio between NAD+ and NADH (Figure 2F, p = 0.029). This latter effect seemed to be largely caused by the group that both received NR and had access to VWR, but there was no statistical support for an interaction (p = 0.077). In the TA muscle (Figure 2G), NAD+ levels increased by an average of ∼27% (p < 0.001), but there was no effect of VWR. In iWAT, we observed an increase in NAD+ levels of ∼25% (p < 0.001, Figure 2H), illustrating that this method of NR administration increases NAD+ levels in multiple peripheral tissues. All of the above-mentioned changes were completely independent of whether the animals received PT or not (Figures S1D–S1K). Importantly, NR injections were given every 24 h during the intervention period and tissues were collected 16–20 h after the last injection of NR, indicating that animals receiving NR had sustained NAD+ increases in peripheral tissues throughout the intervention period. This also allows for the possibility that liver NAD+ levels did increase with NR but returned to normal prior to termination.
Figure 2.
Daily intravenous NR injections lead to sustained NAD+ increase in peripheral tissues but not in the liver
Various tissues were analyzed for NAD+ and NADH content after four weeks of daily injections of either saline or nicotinamide riboside at a dosage of 100 mg/kg bodyweight. Tissues were harvested between 16 and 20 h after the last injection.
(A) Liver NAD+ levels.
(B) Liver NADH levels.
(C) Liver NAD+/NADH-ratio.
(D) Quadriceps muscle NAD+ levels.
(E) Quadriceps muscle NADH levels.
(F) Quadriceps muscle NAD+/NADH-ratio.
(G) Tibialis anterior NAD+ levels.
(H) Inguinal white adipose tissue (iWAT) NAD+ levels.
All data were normalized to the protein content of the sample and are presented as mean with overlain data points. Two-way ANOVA was performed for all figures, and Tukey’s test was employed to explore the interaction in (D) Statistical significance was indicated in the following manner: Effects of intravenous NR: p < 0.05 = †, p < 0.01 = ††, p < 0.001 = †††. Effects of VWR: p < 0.01 = §§. Interactions are written out in the figures and noted with: p < 0.001 = ∗∗∗. In cases without statistical interactions, the above symbols indicate main effects, while in cases with statistical interactions, the symbols indicate simple effects compared to the appropriate control group. See also Figure S1.
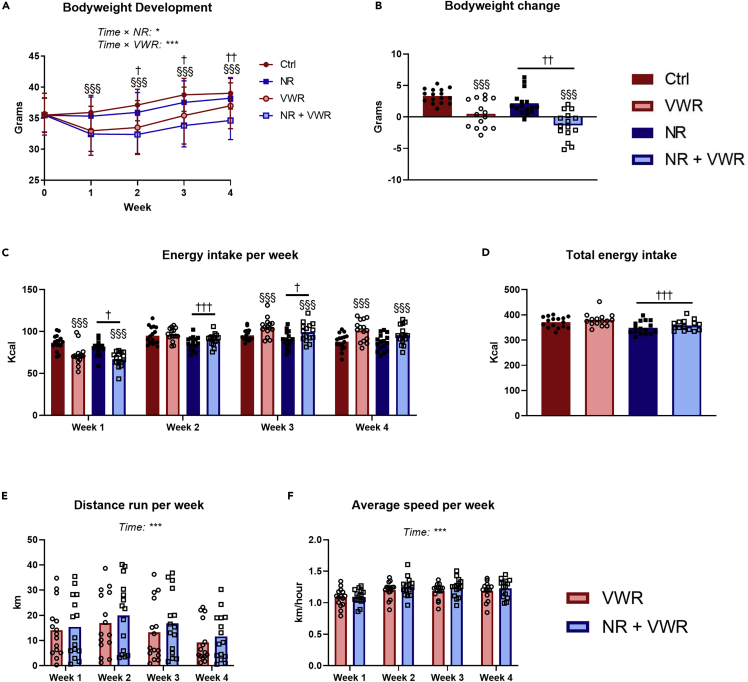
Nicotinamide riboside and voluntary wheel running protects additively against Western diet-induced weight gain
During the intervention, VWR protected against weight gain as indicated by an interaction between VWR and time (Figure 3A, p < 0.001). This appeared to be caused by a significant loss of weight during the first week of the intervention, after which the weight development of the VWR groups ran roughly parallel to that of the sedentary mice. Similarly, but to a lesser degree, NR injections protected against weight gain as indicated by the interaction between NR and time (Figure 3A, p = 0.046). This difference was significant after 2 weeks of the intervention and persisted for the remainder of the study. Looking at the total change in bodyweight over the course of the intervention (Figure 3B), it became apparent that the effects of VWR and NR (p <0.001 and p = 0.003, respectively) were additive such that the group receiving both treatments on average lost weight during the intervention. In support of this, we also observed differences in energy intake during the intervention. Administration of NR caused a reduced energy intake during weeks 1, 2 and 3 (Figure 3C), which can also be appreciated from the total energy intake during the intervention (Figure 3D, p < 0.001). This suggests a somewhat mild anorexigenic effect of NR when delivered intravenously. The VWR groups had lower energy intake than the sedentary groups during week 1, but then exhibited a gradual relative increase week by week, culminating with them having a higher energy intake during weeks 3 and 4 (Figure 3C). Intriguingly, judging by total energy intake (Figure 3D) there was no effect of VWR, so it seems that during the intervention period, the early reduction in energy intake was corrected by the end of week 4. Throughout the intervention, both groups changed their running distance (Figure 3E, p < 0.001) and average running velocity (Figure 3F, p < 0.001), but there was no effect of NR in these regards, despite the increased NAD+ levels in skeletal muscle. All the above observations were independent of PT supplementation (Figures S2A–S2F).
Figure 3.
NR and VWR protects additively against Western diet-induced weight gain
(A) Weekly measures of bodyweight during the intervention.
(B) Total change in bodyweight over the four weeks of treatments.
(C) Weekly energy intake.
(D) Total energy intake over the 4 weeks of treatment.
(E) Weekly running distance for groups with access to voluntary wheel running.
(F) Average running speed during each week for groups with access to voluntary wheel running.
All data are presented as mean with overlain data points, except A, which is presented as mean + SD for visual clarity. Three-way repeated-measures ANOVA was performed for (A), and interactions with time were explored by assessing contrasts between time points. Two-way ANOVA was performed for (B), (C), (D), (E) and (F). Statistical significance was indicated in the following manner: Effects of intravenous NR: p < 0.05 = †, p < 0.01 = ††, p < 0.001 = †††. Effects of VWR: p < 0.001 = §§§. Interactions as well as additional effects are written out in the figures and noted with: p < 0.05 = ∗, p < 0.001 = ∗∗∗. In cases without statistical interactions, the above symbols indicate main effects, while in cases with statistical interactions, the symbols indicate simple effects compared to the appropriate control group. See also Figure S2.
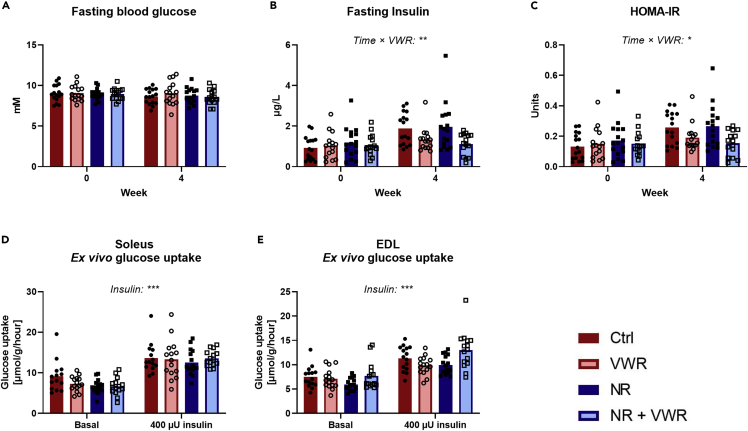
Voluntary wheel running, but not nicotinamide riboside, protects against a Western diet-induced increase in fasting insulin
Fasting blood glucose levels remained unchanged during 4 weeks of WD feeding, irrespective of treatment with either NR or VWR (Figure 4A). In contrast, we observed an interaction between time and VWR (p = 0.005) in determining fasting insulin levels (Figure 4B). This interaction showed that VWR partially protected against the increase in fasting insulin detectable in sedentary groups. In sedentary groups, fasting insulin increased by an average of ∼82%, whereas it only increased by an average of ∼17% in mice with access to VWR. However, there was no effect of NR to modulate insulin levels. To assess the extent of insulin resistance, we utilized the homeostatic model assessment for insulin resistance (HOMA-IR, Figure 4C). In accordance with fasting insulin, HOMA-IR showed that VWR, as expected, protected against the development of whole-body insulin resistance (p = 0.011), but this effect was independent of NR, and NR on its own did not affect HOMA-IR.
Figure 4.
VWR, but not NR, protects against a Western diet-induced increase in fasting insulin
(A) 2 h fasting blood glucose levels before and after four weeks of treatments.
(B) 2 h fasting insulin levels before and after four weeks of treatments.
(C). Homeostatic model-assessment of insulin resistance (HOMA-IR) before and after four weeks of treatment, as calculated based on fasting blood glucose and fasting insulin levels.
(D) Glucose uptake rate in Krebs-Ringer incubated soleus muscle with and without insulin.
(E) Glucose uptake rate in Krebs-Ringer incubated EDL muscle with and without insulin.
All data are presented as mean with overlain data points. Three-way repeated-measures ANOVA was performed for all figures. Statistical significance of interactions as well as additional effects are written out in the figures and noted with: p < 0.05 = ∗, p < 0.01 = ∗∗, p < 0.001 = ∗∗∗. See also Figure S3.
Soleus and EDL muscles were isolated and incubated to determine basal and insulin-stimulated glucose uptake ex vivo. In soleus, insulin increased glucose uptake as expected (p < 0.001, Figure 4D), but neither NR nor VWR gave rise to an interaction with insulin. The same pattern was observed in EDL (p < 0.001), so the insulin sensitivity in these muscles was unaffected by NR and VWR. All of the above observations were independent of PT supplementation (Figures S3A–S3E).
Neither nicotinamide riboside nor voluntary wheel running increases maximal respiratory capacity in permeabilized muscle fibers
Besides soleus and EDL, we also excised the superficial lateral part of the quadriceps muscle to permeabilize fibers for the assessment of mitochondrial respiratory capacity. We tested the maximal activation of Complex I in the electron transport chain by supplying pyruvate, malate, glutamate, and saturating levels of ADP (5 mM), but found that neither NR nor VWR treatment had any effect in this regard (Figure 5A). Addition of Cytochrome C at this point did not affect respiration, confirming that the outer mitochondrial membranes were intact (data not shown). Subsequently, we added succinate to test the maximal coupled activity of Complex I and Complex II (Figure 5B), but again found no effects of either NR or VWR. These readouts were also unaffected by PT supplementation (Figures S4A and S4B). Intriguingly, when a subsaturating concentration of ADP (0.1 mM) was used, we observed an interaction between PT and VWR (p = 0.043). In the groups without PT, VWR increased oxygen consumption modestly (p = 0.027), whereas PT supplementation appeared to abolish this effect. As a result, the data are presented here with all eight original groups (Figure 5C).
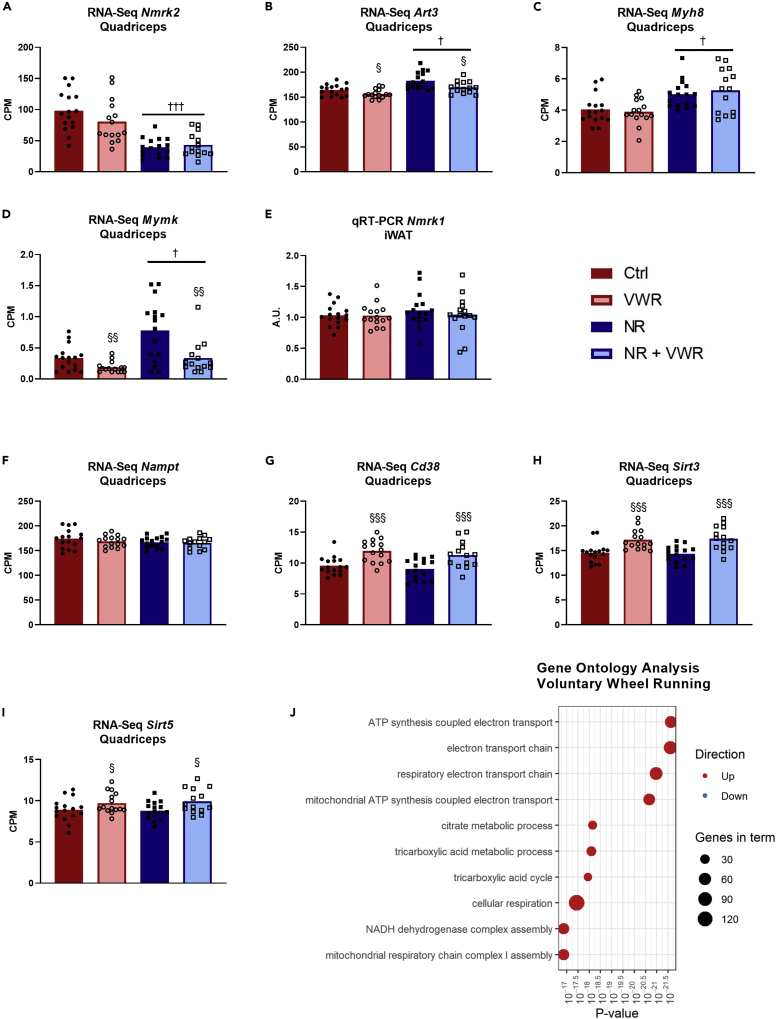
The gene expression profile of quadriceps muscle is largely unaffected by nicotinamide riboside
We decided to assess the quadriceps muscle gene expression profile by RNA sequencing. Based on q-values with a cutoff of 0.05, there were no interactions between NR, PT, and VWR at the transcriptional level, allowing us to focus on individual treatment effects. A total of four protein-coding genes changed with NR. These were Nmrk2, Art3, Myh8, and Mymk (Figures 6A–6D). Interestingly, the Nmrk2 transcript, which is translated to NRK2, the protein responsible for phosphorylating NR into nicotinamide mononucleotide (NMN) in muscle (Bieganowski and Brenner, 2004), was reduced by ∼65% with NR (q < 0.001, Figure 6A). Art3, which encodes an ADP-ribosyltransferase that utilizes NAD+ to ADP-ribosylate and inactivate target proteins (Glowacki et al., 2002) was upregulated with NR (Figure 6B, q = 0.018), but downregulated by VWR (q = 0.019). Together, these changes in Nmrk2 and Art3 expression may indicate that the quadriceps muscle fibers were experiencing an NAD+ excess.
Figure 6.
The gene expression profile of quadriceps muscle is largely unaffected by NR.
Expression of (A) Nmrk2, (B) Art3, (C) Myh8, and (D) Mymk transcripts in counts per million (CPM) from the full mRNA sequencing of quadriceps muscle.
(E) Expression of Nmrk1 transcript in iWAT from qRT-PCR analysis.
(F–I) Expression of Nampt, Cd38, Sirt3, and Sirt5 transcripts in counts per million (CPM) from the full mRNA sequencing of quadriceps muscle.
(J) Top 10 most significantly changed gene ontology terms associated with genes affected by voluntary wheel running. The size of the point indicates the number of genes in the term. The color indicates whether genes in the term tend to be up-regulated or down-regulated. Note that the x axis is on a logarithmic scale.
All data except J are presented as mean with overlain data points. The statistical analysis used for (A), (B), (C), (D), (F), (G), (H), and (I) can be found in the Method details under mRNA sequencing. Two-way ANOVA was performed for (E). Statistical significance was indicated in the following manner: Effects of intravenous NR: p < 0.05 = †, p < 0.001 = †††. Effects of VWR: p < 0.05 = § p < 0.01 = §§§. The symbols indicate the main effects. See also Figure S5.
The other two genes that were changed in NR groups, Mymk (Figure 6C) and Myh8 (Figure 6D), were both upregulated by NR (q = 0.032 and 0.037, respectively). Mymk is translated to the protein Myomaker, which is specifically expressed in myoblast and satellite cells and is important for the development as well as the regeneration of muscle cells (Millay et al., 2013, 2014). Interestingly, Mymk expression, similarly to Art3 was also downregulated by VWR (q = 0.006). Myh8 translates into MYH8 and is also referred to as MyHC-Perinatal. As the name suggests, this type of myosin heavy chain is mainly expressed in fetal skeletal muscle but is also expressed in regenerating muscle (Schiaffino et al., 2015). After discovering that the major change with NR in skeletal muscle was a reduction in Nmrk2 expression, we wanted to investigate whether there was a similar pattern in iWAT. As Nmrk2 is only expressed in heart and skeletal muscle, we used qRT-PCR to analyze the expression of the homolog Nmrk1 in iWAT, but it was unaffected by both NR and VWR (Figure 6E). Together, these data suggest only minor effects of NR on gene expression in quadriceps muscle despite the elevated NAD+ levels.
In contrast to NR, VWR leads to changes in muscle gene expression of 2,245 protein-coding genes with a q-value cutoff of 0.05. VWR did not affect any of the genes classically associated with NAD+ production: Nmrk1, Nmrk2, Nmnat1, Nmnat3, Nadsyn1, Naprt, Qprt, and Nampt (Figure 6F) but increased the expression of some genes associated with NAD+ consumption, namely: Cd38, Sirt3 and Sirt5 (Figures 6G–6I). We performed a gene ontology (GO) analysis of genes differentially expressed by VWR and found the top ten GO-terms to be upregulated by VWR, and to be fully in line with the expected effects of physical activity on muscle physiology (Figure 6J). PT did not affect the expression of a single gene in quadriceps muscle, nor did it affect any of the measurements above (Figures S5A–S5J).
Discussion
We designed this study to determine whether stably elevated NAD+ levels in skeletal muscle would affect insulin sensitivity or mitochondrial function in mice fed a WD and whether PT would interact with NR on these readouts. To accomplish this, mice received daily NR injections intravenously to bypass intestinal degradation and first-pass metabolism in the liver and make NR directly available to peripheral tissues such as skeletal muscle. PT was given through the diet, owing to its insolubility in water. We successfully increased NAD+ levels not only in skeletal muscle but also iWAT. This was not simply an acute effect around the time of the injection, but rather a sustained increase throughout the intervention period, as illustrated by the increased NAD+ levels 16–20 h after the final injection. In contrast, NAD+ levels in liver were unchanged by NR at this timepoint, which could be a result of the higher NAD+ turnover in this tissue (McReynolds et al., 2021). It has previously been suggested that oral NR supplementation increases NAD+ in various tissues (Canto et al., 2012), but a more recent report was unable to reproduce the increase in skeletal muscle (Frederick et al., 2020). Our pilot experiments support previous publications showing that NR, at least in the intact version, does not reach skeletal muscle when supplied orally (Liu et al., 2018; Frederick et al., 2016). In addition, our results suggest that the peripheral tissues were able to take up NR directly and that the usually observed accumulation in the liver (Wang et al., 2018; Dall et al., 2019; Canto et al., 2012) is mainly a result of the route of administration.
We observed a difference between muscle types in response to intravenous NR and VWR. In the TA muscle, NR increased NAD+ levels, but this effect was not augmented by wheel running, whereas in quadriceps, the magnitude of the increase in NAD+ levels by NR was larger in VWR than in sedentary groups. Interestingly, in quadriceps, the expression of genes encoding NAD+ producing enzymes was unaffected by VWR, whereas some genes encoding NAD+ consuming enzymes were upregulated by VWR. This seemingly opposes the notion that physical activity induces changes in the tissue to accommodate higher levels of NAD+. On the other hand, it supports the hypothesis that physical activity may increase the delivery of intravenous NR to skeletal muscle by increasing the blood flow. Thus, we speculate that the observed difference between muscle types could be a result of differences in recruitment and/or activity patterns between quadriceps and TA during wheel running. Nevertheless, despite elevated NAD+ levels, the overall physiological impact of intravenous NR was mild. In fact, it is remarkable that the increased NAD+ availability in skeletal muscle and white adipose tissue during the 4-week intervention of this study did not have a larger impact on overall physiology. NR did appear to be slightly anorexigenic, which lead to modest protection against weight gain, but it is noteworthy that the mice were injected immediately prior to the onset of the dark phase in which >80% of their energy intake usually occurs (Kohsaka et al., 2007). Thus, although the injection did not visibly affect the animals, any discomfort associated with the dose of NR could in principle account for the observed anorexigenic effect.
One of our primary endpoints was glucose homeostasis. Blood glucose remained unchanged during the intervention for all groups, and while fasting insulin and thus our index of insulin resistance generally increased during the 4-week intervention, only VWR had a protective effect in this regard. Investigating ex vivo insulin sensitivity in soleus and EDL muscles, as representatives of fast- and slow-twitch muscles, respectively (Barclay et al., 1993; Ciapaite et al., 2015), we found no insulin-sensitizing effects of either NR, VWR, or the combination. We have previously observed that a similar VWR protocol failed to increase insulin sensitivity in chow-fed mice (Basse et al., 2018). This lack of response could be related to mode, duration, timing, and frequency of VWR compared with other more structured exercise regimens. Taken together, our data suggest that VWR improves whole-body insulin sensitivity without affecting muscle insulin sensitivity per se.
When oxidative phosphorylation was analyzed in permeabilized muscle fibers, we found no effects of either NR or VWR to increase maximal respiratory capacity, although we did notice a tendency for a reduction with NR. Oral NR supplementation has been tested in humans with similar readouts, but did not lead to actual increases in muscle NAD+ and had no effect on mitochondrial respiration (Elhassan et al., 2019; Remie et al., 2020; Dollerup et al., 2020). In mice, overexpression of Nampt in skeletal muscle has been shown to improve mitochondrial respiratory capacity, which was associated with elevated NAD+ levels (Brouwers et al., 2018; Costford et al., 2018). In contrast, another report showed that Nampt overexpression in skeletal muscle elevated mouse muscle NAD+ levels, but did not improve mitochondrial function (Frederick et al., 2015). Interestingly, increases in NAM and NMN levels were reported in one of the studies (Costford et al., 2018), but not the other (Frederick et al., 2015). This indicates that the genetic manipulation of Nampt may affect fluxes in other NAD+-dependent pathways that potentially lead to specific physiological adaptations. Thus, changes in respiratory capacity may not result from increased NAD+ levels or even Nampt expression per se, but may be an effect of the specific type of genetic manipulation utilized to overexpress Nampt. Another example that the genetic manipulation of NAD+ biosynthetic or degrading pathways affects the whole NAD metabolome and has clear physiological consequences shows that knockdown of nicotinamide N-methyltransferase in adipose tissue increases NAMPT and NAD+ levels and profoundly affects energy expenditure in mice (Kraus et al., 2014). Thus, although the main physiological endpoints in the present study are negative, our data are not confounded by side effects from genetic manipulation, and they provide novel insights into the dynamics of NAD+ precursor supplementation.
During the analysis of mitochondrial respiration, we did observe an interesting interaction of PT, despite the low bioavailability to skeletal muscle and the lack of effect on any other tested parameter. Specifically, when subsaturating levels of ADP were used for the analysis, VWR increased oxygen consumption, but only in the groups that did not receive PT. There is the possibility that this is a type I error, but it is also possible that this is a legitimate effect of PT that we cannot fully appreciate with the current setup owing to the extremely small effect sizes associated with VWR. Though it does seem puzzling that PT would affect skeletal muscle respiratory capacity without affecting the expression of even a single gene, it could relate to PT’s antioxidative properties. Elucidating this would demand a new study with a comprehensive exercise regimen and preferably in a species more receptive to the effects of exercise training.
Transcriptomic analysis on quadriceps showed no interactions between the treatments, no effect of PT at all, and only a few genes changed with NR despite the increased NAD+ pool. However, the downregulation of the Nmrk2 transcript does suggest that muscle cells are taking steps to avoid the conversion of NR to nicotinamide mononucleotide. This could be a passive response to the increased availability of NAD+ precursor or it could be an active response to avoid NAD+ overload, which raises the question of whether daily intravenous NR injection overloads skeletal muscle with NAD+. The tendency toward a reduction in respiratory capacity on quadriceps fibers does hint at that possibility. This is supported by the profound reduction of Nmrk2 mRNA, the upregulation of Art3 mRNA, and perhaps the slightly anorexigenic effect of NR. In line with this, a recent study focusing on HeLa cells showed that continued exposure to extracellular NAD+ resulted in only a transient increase in intracellular NAD+ levels followed by a return to physiological levels (Buonvicino et al., 2021). This reset was accompanied by a reduced expression of genes important for the uptake and utilization of extracellular NAD+. Interestingly, when we assessed Nmrk1 expression in iWAT, it remained unaffected, and as iWAT does not express Nmrk2, this tissue did not appear to limit NR utilization in the same manner as muscle. Another interesting discovery of the mRNA sequencing analysis was the increased Mymk and Myh8 expression in quadriceps of NR groups. Both Mymk and Myh8 expressions are associated with the regeneration of skeletal muscle (Millay et al., 2014; Schiaffino et al., 2015). As regeneration is most often secondary to an insult, the overexpression of Mymk and Myh8 in response to NR supplementation is likely an additional indicator that the skeletal muscle does not respond well to the treatment. However, if the increased amount of NAD+ was highly toxic or a serious detriment to the muscle cells, we would have expected markers of inflammation or apoptosis to appear in the mRNA sequencing analysis as well as a more profound overall physiological impact. In contrast to our data, a human study reported a specific transcriptional signature with a total of ∼885 differentially expressed protein-coding genes in muscle after oral NR treatment (Elhassan et al., 2019). However, it should be noted that the human study reported uncorrected p values and that the FDR-corrected values from their supplemental material revealed only six differentially expressed protein-coding genes. For comparison, our intravenous NR supplementation resulted in 1071 protein-coding genes with p <0.05, but only four remained statistically significant after FDR correction. Of further note, there were no overlaps in FDR-corrected differentially expressed protein-coding genes between the two studies, but this is likely attributable to the differences in species or supplementation methods.
In conclusion, we successfully and stably increased the NAD+ pool in skeletal muscle and white adipose tissue by the daily intravenous injection of NR, but the physiological effects of this were modest. Chronically elevating NAD+ levels in white adipose tissue and either active or sedentary skeletal muscle was not associated with improvements in muscle respiratory capacity or insulin sensitivity.
Limitations of the study
We did not include a test of maximal exercise capacity owing to the potential interference with our primary endpoints. Similarly, we decided to forgo a strict exercise regimen in favor of focusing on VWR as a means of keeping skeletal muscle active rather than training it for increased load. Accordingly, we make no attempt to conclude on the potential effects of NR or PT on exercise capacity, or on the effect of strict exercise on our primary endpoints. Additionally, as we were interested in the chronical effects of NR supplementation, animals were sacrificed 16–20 h after the last injection of NR or saline. Therefore, we provide no data or conclusions on the acute effects of NR supplementation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Mouse Insulin ELISA kit | Mercodia | 10-1247-01 |
| Pierce BCA protein assay kit | Thermo Fisher Scientific | 23225 |
| RNeasy kit | Qiagen | 74104 |
| iScript cDNA synthesis kit | BIO-RAD | 1708890 |
| TruSeq Stranded mRNA | Illumina | 20020595 |
| Deposited data | ||
| RNA-seq data | This paper | GEO: GSE194193 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6NTac | Taconic | B6-M |
| Mouse: I-Pax7CreERT2-R26RtdTomato+ | Own breeding | N/A |
| Oligonucleotides | ||
| Primer 18s Forward | This paper | AGTCCCTGCCCTTTGTACACA |
| Primer 18s Reverse | This paper | GATCCGAGGGCCTCACTAAAC |
| Primer Gapdh Forward | This paper | GCACAGTCAAGGCCGAGAAT |
| Primer: Gapdh Reverse | This paper | GCCTTCTCCATGGTGGTGAA |
| Primer Tbp Forward | This paper | ACCCTTCACCAATGACTCCTATG |
| Primer Tbp Reverse | This paper | TGACTGCAGCAAATCGCTTGG |
| Primer Nmrk1 Forward | This paper | AGAGCTTGCAGAAGCACCTTCC |
| Primer Nmrk1 Reverse | This paper | CATCCAACAGGAAACTGCTGACA |
| Software and algorithms | ||
| OTOFControl | Bruker Daltonics | v6.0 |
| Compass HyStar | Bruker Daltonics | v5.0 |
| MzMine | Pluskal et al. (2010) | v2.53 |
| STAR aligner | Dobin et al. (2013) | v2.7.3a |
| FeatureCounts | Liao et al. (2014) | v2.0.0 |
| edgeR | Robinson et al. (2010) | v3.30.3 |
| JMP | SAS | Pro 15 |
| Prism | GraphPad | v8.4.3 |
| Other | ||
| Mouse jugular vein catheter | Instech | C20PU-MJV1458 |
| Vascular Access Button | Instech | VABM1B/25 |
| Handling tool for magnetic mouse Vascular Access Buttons | Instech | VABMG |
| Pinport Injectors | Instech | PNP3M-50 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact Jonas Thue Treebak (jttreebak@sund.ku.dk)
Materials availability
This study did not generate new unique reagents. Nicotinamide riboside and pterostilbene was provided us by Elysium Health and as per the MTA may not be distributed.
Experimental model and subject details
Ethical approval
All animal experiments were in accordance with the European directive 2010/63/EU of the European Parliament and of the Council for the protection of animals used for scientific purposes. Ethical approval was given by the Danish Animal Experiments Inspectorate (#2019-15-0201-01630).
Animal care and use
Sixty-four male C57BL/6NTac mice were acquired from Taconic Laboratories at 5 weeks of age. Mice were kept at a 12 h light/dark-cycle, 20–22°C, 50 % humidity, and were single-housed after one week of acclimation. Mice had access to ad libitum feed and water for the entirety of the experiment, and fresh feed was provided twice weekly. For 8 weeks, the animals received a WD (Research diet, D18053102 – a variant of D12079B utilizing lard and soy bean oil as fat sources). The mice then underwent surgery and were given one week to recover, after which the intervention period of 4 weeks started. During this intervention period, the WD fed mice were split into 8 groups (Figure 1A). NR groups (blue and orange) were given daily intravenous injections with pH-adjusted and sterile filtered NR (Elysium Health, USA) at a dose of 100 mg/kg, while the other groups received injections with equimolar saline. PT groups (green and orange) were switched to a near-identical WD designed to contain 410 mg PT (Elysium Health, USA) per kg diet, while the other groups were kept on the regular WD. VWR groups (hollow) were given access to running wheels. Each group consisted of eight mice and the 23 full-factorial setup ensured that every single group of animals had all necessary control groups present. Furthermore, to facilitate the daily intravenous injections, we inserted a Vascular Access Button (Figure 1B) connected to a jugular catheter in each mouse, which allowed quick and low-stress injections of NR or saline. The surgery is outlined below. Running wheels were unlocked at all hours and were checked daily for blockages. Voluntary running distance and speed was monitored weekly. Running wheels were locked prior to the last dark phase before termination to avoid any confounding effects of acute physical activity.
Validation of the experimental model
We performed two experiments to determine the validity of our NR supplementation model. All mice were fed a chow diet and, if destined for intravenous delivery of NR or saline, underwent the same jugular catheter surgery as for the main study, outlined below. Experiments were conducted after a week of recovery. To compare oral delivery of NR to intravenous delivery, we acquired 15 male CBL57/6NTac mice from Taconic at 8 weeks of age and split them into 3 groups. One group was left undisturbed and acted as control. One group received daily oral gavage with NR at a dose of 200 mg/kg, and one group received daily intravenous injection with NR at a dose of 200 mg/kg. After 7 consecutive treatment days, mice were terminated 1 h after the last gavage or injection and tissues were frozen in liquid nitrogen. We used liquid chromatography-mass spectrometry (LC-MS) to assess the level of NR and NAD+ in TA from these animals. To test the stability of the increase in NAD+ upon intravenous NR, we utilised 15 male I-Pax7CreERT2-R26RtdTomato+ mice on a CBL57/6NTac background that were leftovers from our own breeding. These mice were ∼30 weeks of age. These 15 mice were split in three groups of 5. One group received saline and 2 groups received intravenous NR at a dose of 100 mg/kg for 5 consecutive days. The saline control group and one of the NR groups were terminated 1 h after the final intravenous injection. The remaining NR group was terminated 12 h after the last injection. From these mice, extensor digitorum longus (EDL) muscles were harvested for analysis of NAD+ content.
Method details
Surgery
All surgeries were performed under aseptic conditions using pre-sterilised materials and instruments. Anesthesia was induced with 2.5% isoflurane in oxygen and maintained with 1.5% isoflurane in oxygen, and the depth of anesthesia was assessed before and during surgery by checking of reflexes. Fur was removed around the two incision points (upper back and neck) and the skin was disinfected with ethanol/chlorohexidine. Mice were subcutaneously injected with Carprofen (10 mg/kg, Norodyl Vet, Scanvet, Denmark) for analgesia and a mixture of Lidocaine (10 mg/kg, Xylocain, AstraZeneca, UK) and Bupivacaine (2.5 mg/kg, Marcaine, Orifarm, Denmark) at the incision sites for local anesthesia. A 2 Fr PU-catheter (C20PU-MJV1458; Instech, USA) was inserted into the right jugular vein and externalised using a Vascular Access Button (VABM1B/25; Instech, USA) placed on the upper back of the animal. The catheter was locked with heparinised saline (200 IU/mL). Mice were placed in clean cages after surgery and had voluntary thermal support in the form of a heat pad under approximately half the cage for the following 24 h. Recovery of the animals was monitored the following week, ensuring no large weight loss (>10% bodyweight) or signs of pain based on the mouse grimace scale (Langford et al., 2010). A single mouse was removed from the study at this point due to a suspected stroke during surgery. The rest had an average weight loss during the recovery week of ∼2.8% and none went past the threshold of 10%, or showed signs of pain. One additional mouse was removed from the study due to infection at the Vascular Access Button during the second intervention week.
Administration of nicotinamide riboside
Immediately prior to the onset of the dark phase every day of the intervention period, mice were given an intravenous injection of 100 mg/kg pH-adjusted and sterile filtered NR or an equal volume of equimolar saline through the vascular access button. The catheter was subsequently locked with heparinised saline (200 IU/mL).
Blood glucose homeostasis
One day prior to the onset of intervention as well as one day prior to termination, blood samples were taken for assessment of HOMA-IR. Mice were fasted for 2 h in the early light-phase, after which they were bled from the tail to assess blood glucose using a Contour XT glucometer (Bayer Health Care, Germany). 25 μL blood was collected using EDTA-coated minivettes (Sarstedt, Germany). Blood was centrifuged for 10 min at 1,000 g in a precooled centrifuge (4°C) to isolate plasma, which was snap-frozen in liquid nitrogen and stored at −80°C until further processing. Plasma was then used to analyze insulin levels with the Mouse Insulin ELISA kit (Mercodia, Sweden), according to manufacturer’s instructions. HOMA-IR was calculated using the formula: .
Termination of experiment
During termination the mice were anesthetized using Avertin (2,2,2-Tribromoethanol and 2-Methyl-2-butanol 99%) (Sigma-Aldrich, USA). The soleus and EDL muscles were first isolated and used for measurement of ex vivo glucose uptake (described below). Then the superficial lateral part of the quadriceps (mainly vastus lateralis) was isolated and stored in BIOPS buffer; [10 mM Ca-EGTA buffer, 0.1 μM free calcium, 20 mM imidazole, 20 mM taurine, 50 mM K-MES, 0.5 mM DTT, 6.56 mM MgCl2, 5.77 mM ATP, 15 mM phosphocreatine, pH 7.1 at 0°C] for later analysis of mitochondrial respiratory capacity. Blood was drawn for plasma isolation and subsequently the mice were cervically dislocated and the following tissues were harvested, snap-frozen in liquid nitrogen and stored at −80°C: Liver, Quadriceps muscle, tibialis anterior (TA) muscle and inguinal white adipose tissue (iWAT).
NAD+ and NADH measurement
To assess NAD+ levels, tissue was lysed in 400 μL 0.6 M perchloric acid (HClO4) using steel beads and a Tissuelyzer II (Qiagen, Germany). The samples were then centrifuged for 2 min at 13,000 g and then supernatant was moved to a new tube and was diluted 300–800x in 100 mM Na2HPO4. A reaction mix was made, consisting of 5,000 μL of 200 mM Na2HPO4, 10 μL of flavinmononucleotide (Sigma-Aldrich, USA), 200 μL of absolute ethanol, 1,200 μL of 750 U/mL Alcohol dehydrogenase (Sigma-Aldrich, USA), 26 μL of 50 U/mL Diaphorase (Sigma-Aldrich, USA), 5 μL of 5 mg/mL Resazurin (Sigma-Aldrich, USA), 100 μL 1M nicotinamide and 3500 μL H2O. 100 μL of this reaction mix was added to 100 μL diluted sample. Samples were measured on a Hidex Sense (Hidex, Finland) by repeated fluorescence measurement once per minute for 30 min with excitation 544 nm and emission 580 nm. Readout slopes from samples were compared to the slopes of a standard curve. The pellets from the centrifugation step were dissolved in 0.2 M NaOH by heating to 95°C and the concentration of protein was measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific, USA) to be used for normalization.
To assess NADH levels a similar protocol was used with the following exceptions. Tissue was lysed in 0.1 M NaOH, heated to 70°C for 10 min and centrifuged similarly to the NAD+ assay. A small sample was taken out for protein measurement. The rest of the supernatant was then diluted 40 times in 10 mM Tris-HCl (pH = 8), mixed with reaction mix, and measured similarly to what was described for the NAD+ assay.
Liquid chromatography-mass spectrometry analysis
Plasma was prepared for LC-MS in the following way. 20 μL of plasma was aliquoted in 1.5 mL Eppendorf tubes. Aliquots were mixed with 20 μL of 0.5 mg/L 3-hydroxy tridecanoic acid (Merck, Darmstadt, Germany) used as internal standard and 100 μL of methanol. The suspension was vortexed and left on ice for 30 min to precipitate proteins. Afterwards, samples were centrifuged at 11,292 g for 3 min, 4°C, and thereafter 100 μL of supernatant was collected in LC vials and evaporated to dryness. Dry extract was reconstituted with 40 μL of solvent composed of methanol and water (V:V = 1:1). Samples were vortexed and kept on −20°C until analysis. All samples were double randomised before injection. Method repeatability was determined by repeatedly analyzing a pooled sample made from equal volume of each plasma sample. Pterostilbene standard (Merck, Darmstadt, Germany) was injected in the sequence in four different concentration levels. Skeletal muscle tissue was prepared for LC-MS in the following way. Frozen tissue was put in a TT05M XT tissueTUBE (Covaris, Massachusetts, USA); which withstand cryogenic conditions and pressure. The bag was kept in liquid nitrogen while the tissue was crushed into a powder with CPO2 (Covaris, Massachusetts, USA). Powder was transferred into cryo-vials. Exact mass of tissue was weighed and used for peak area normalization. Subsequently, the tissue powder was mixed with 40 μL of internal standard and 320 μL of methanol. The suspension was vortexed and sonicated in an ice-cold ultrasound bath for 15 min in order to facilitate metabolite extraction. Afterwards, it was left for 15 min on ice for proteins to precipitate. The suspension was centrifuged at 11,292 g for 3 min, 4°C. 200 μL of extract was evaporated to dryness, the rest was used for the pooled sample. Dried extract was reconstituted in the same way as plasma samples. For the chromatographic separation, an Agilent 1290 Infinity II liquid chromatograph was used while the detection was made with a Bruker Tims TOF Pro mass spectrometer (Bruker Daltonics, Bremen, Germany). Separation was made with a Waters TSS T3 10 cm x 2.1 x 1.8 μm column. Mobile phase A consisted of water with 0.1% formic acid while mobile phase B was composed of acetonitrile and isopropanol (V:V = 3:1) with 0.1% formic acid. Mobile phase gradient started with 3% of mobile phase B, and thereafter increased to 100% B over the course of 9 min. Thereafter it was held for 5 min and for final re-equilibration to initial conditions. The flow-rate was 400 μL/min. Column temperature was kept at 40°C and injection volume was 4 μL. Ions were generated by negative electrospray ionization (ESI). Full-scan MS spectra acquisition rates were set to 2 Hz, and the resolution was approximately 60,000. For mass calibration 50 μL internal celebrant of 10 mM Na-format was injected at the beginning of each analysis. Data acquisition was performed with OTOFControl version 6.0 and Bruker Compass HyStar version 5.0 (Bruker Daltonics, Bremen, Germany). Data extraction was made with MzMine 2.53 (Pluskal et al., 2010) in a targeted way. A mass feature with the same mass as PT-sulfate was detected. To verify the identity of the peak, tandem mass spectrometry was performed. The tandem spectrum was compared with native corresponding spectrum for the PT standard. The spectra showed similarities that enabled us to tentatively identify the metabolite as PT-sulfate. Quantitative results were expressed as area under the chromatographic peak for plasma or mass normalised area under the peak for muscle tissue (AU).
For NAD+ and NR analysis, muscle tissue was prepared in a similar way as described above. Approximately 20 mg of tissue powder was aliquoted and mixed together with 380 μL of methanol and water (V:V = 4:1) and 20 μL of internal standard solution in methanol at a concentration of 10 mg/L. Internal standard solution contained: nicotinic-d4 acid, nicotinamide-d4, N-methylnicotinamide-d4, D-tryptophan-d5 (CDN isotopes, QC, Canada), β-NAD-d4, adenosine-13C5 (TRC, Toronto, Canada) and adenosine-13C10,15N5 5′-triphosphate (Merck, Darmstadt, Germany). Protein precipitation and centrifugation was made as described above. 200 μL of supernatant was evaporated under a stream of nitrogen to dryness and re-dissolved with 50 μL of methanol and water (V:V = 4:1). Samples were kept on −20°C until injection. The same LC-MS instrument as above was used for NAD+ and NR measurements. Metabolites were separated using a Waters ACQUITY premier BEH Amide 1.7 VanGuard FIT 2.1 × 150 mm column. Mobile phase B was water and acetonitrile (V:V = 1:9) with added 10 mM MH4Ac and 5 μM medronic acid. Mobile phase A was water with added 10 mM MH4Ac and 5 μM medronic acid. Flow was 0.2 mL/min. Gradient started with 90% of mobile phase B and it was held steady for 2 min. After that it was decreased to 55% in 2 min where it was again held steady for 2 min. Additional 6 min were used for re-equilibration. Column temperature was 40°C and injection volume was 2 μL. Acquisition was made with ESI positive ionization with scan speed of 1 Hz. Data extraction was made with MzMine 2.53 in a targeted way. Dilution series of NAD+ and NR standards was used for metabolite identification and quantification. Exact mass and characteristic retention time were used for feature integration. Extracted features were normalised with internal standard that showed highest R2 when applied to calibration curve. Results were normalised by the weight of the originally used tissue powder aliquot.
Glucose uptake measurements in skeletal muscle ex vivo
Soleus and EDL muscles were incubated as previously described (Basse et al., 2018). In short, the muscles were acclimated in Krebs-Ringer buffer with added 0.1% BSA, 8 mM mannitol and 2 mM pyruvate in a Muscle Strip Myograph system (820MS, DMT, Denmark) for 10 min. They were then incubated with or without 400 μU/mL insulin (Actrapid, Novo Nordisk, Denmark) for 20 min under 5 mN resting tension, with the muscle from one leg acting as control for the corresponding muscle from the other leg. This was followed by 10 min incubation with added 3H-2-deoxyglucose (PerkinElmer, USA) for assessment of glucose uptake and 14C-mannitol (PerkinElmer, USA) to account for extracellular fluid. Subsequently, the muscles were snap-frozen in liquid nitrogen. Frozen muscle tissues were lysed using steel beads and a Tissuelyzer II (Qiagen, Germany) in DML-buffer [pH 7.4, 10% glycerol, 1% IGEPAL, 150 mM NaCl, 50 mM HEPES, 20 mM β-glycerophosphate, 10 mM NaF, 1 mM EDTA, 1 mM EGTA, 1 mM sodium butyrate, 2 mM sodium orthovanadate, 20 mM sodium-pyrophosphate, 5 mM nicotinamide, 4 μM Thiamet G, and protease inhibitors (no. S8820; SigmaFast)]. The muscle lysates and media samples were then mixed into vials containing Ultima Gold scintillation liquid (PerkinElmer, USA) and DPM of both 3H-2-deoxyglucose and 14C-mannitol was counted on a Hidex 300SL (Hidex, Finland). Total protein content of each sample was measured prior to the addition of scintillation liquid and was used for normalization.
Mitochondrial respiration
Mitochondrial respiratory capacity was measured in permeabilised skeletal muscle fibers using high-resolution respirometry (Oxygraph, Oroboros, Innsbruck, Austria). Muscle fibers were manually dissected using needles in ice-cold BIOPS buffer kept on ice. The fibers were then permeabilised by gentle agitation on ice for 30 min in BIOPS with 50 μg/mL of saponin, this was followed by rinsing the fibers twice for 10 min in ice-cold respiration medium; MiR05 [0.5 mM EGTA, 3 mM MgCl2 ‧ 6 H2O, 60 mM lactobionic acid, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 110 D-sucrose, 1 g/L BSA, pH 7.1 at 37°C]. The muscle fibers were then weighed (Mettler Toledo, XS105) and added to the respirometry chamber containing MiR05. All measurements were done in duplicates, with hyperoxygenation (450–200 nmol/mL) to avoid oxygen limitations and at 37°C. The following protocol was applied. State 2 respiration was assessed by adding 2 mM malate, 10 mM glutamate, 5 mM pyruvate, state 3 respiration with complex I linked substrates was reached by addition of ADP (0.1 and 5 mM). The integrity of the outer mitochondrial membrane was ensured by adding 10 μM cytochrome c, and then 10 mM succinate was added to evaluate maximal coupled respiration with complex I and II linked substrates. Finally, 2.5 μM antimycin A was added to inhibit complex III.
RNA isolation
1 mL TRIzol reagent (Life Technologies, USA) was added to each tissue and each sample was homogenised through the use of steel beads and a Tissuelyzer II (Qiagen, Germany). Beads were collected, and 200 μL chloroform was added and mixed thoroughly. After a 5 min incubation, the tubes were spun at 12,000 g for 15 min in a precooled centrifuge. The aqueous supernatant was mixed in two volumes of 99% ethanol in new tubes and stored at −20°C overnight. These samples were then added to RNA columns from an RNeasy kit (Qiagen, Germany) and isolated according to manufacturer’s instructions. We included the optional DNase digestion steps. RNA concentration was measured by Nanodrop (Thermo Fisher Scientific, USA), and RNA integrity was tested on a Bioanalyzer instrument (Agilent Technologies, USA) according to manufacturer’s instructions. All RIN values were above our preselected minimum of 8.
mRNA sequencing
mRNA sequencing was performed by the Single-Cell Omics platform at the Novo Nordisk Foundation Center for Basic Metabolic Research, University of Copenhagen.
mRNA sequencing libraries were prepared using the Illumina TruSeq Stranded mRNA protocol (Illumina, USA). Poly-A containing mRNAs were purified by poly-T attached magnetic beads, fragmented, and cDNA was synthesised using Super-Script III Reverse Transcriptase (Thermo Fisher Scientific, USA). cDNA was adenylated to prime for adapter ligation and after a clean-up using AMPure beads (Beckman coulter, USA), DNA fragments were amplified using PCR followed by a final clean-up. Libraries were quality-controlled once more using a Bioanalyzer instrument (Agilent Technologies, USA) and subjected to 52-bp paired-end sequencing on a NovaSeq 6000 (Illumina, USA). A total of 1.6 billion reads was generated.
The STAR aligner (Dobin et al., 2013) v2.7.3a was used to align reads to the mm10 mouse genome with the GENCODE Comprehensive gene annotations (Frankish et al., 2019) vM22 gene model. Reads were summarised onto the same gene model using featureCounts (Liao et al., 2014) v2.0.0. Tests for differential expression was performed using edgeR (Robinson et al., 2010) v3.30.3 using the recommended quasi-likelihood negative binomial generalised log-linear model with model of the form “∼ group + RIN” where RIN is the measured RNA Integrity Number and group encodes the 4 (or 8) combinations of NR, VWR (and PT). Differential expression was found by comparing the 4 (or 8) levels encoded by group as recommended by the edgeR manual. Resulting p values were corrected by false discovery rate (FDR) and q-values were reported. Gene ontology enrichments were found using the camera (Wu and Smyth, 2012) function, which is part of the edgeR package. Only gene ontology categories with between 5 and 500 genes were investigated.
qRT-PCR
cDNA was synthesised from RNA using the iScript cDNA synthesis kit (BIO-RAD, USA) according to manufacturer’s instructions. cDNA was diluted 10x and mixed with Brilliant III Ultra-Fast SYBR Green qPCR Master Mix (Agilent Technologies, USA) as well as forward and reverse primers surrounding the regions of interest. The protocol for qRT-PCR was 5 min at 95°C followed by 40 cycles of 5 s 95°C, 10 s annealing at variable temperatures (58–64°C) and 10 s elongation at 72°C. For every analysis we included a melting curve and a standard curve to ensure a single amplification product as well as a linear relationship between log(concentration) of the cDNA and the resulting CT values. For each sample, the expression level of the target gene, Nmrk1, was normalised to the geometric mean of the expression of three housekeeping genes: 18s, Gapdh and Tbp after factoring in exact amplification levels calculated from the respective standard curves. The expression mean of the control group was defined as having a value of 1.
Quantification and statistical analysis
For statistical analysis, we used SAS JMP Pro 15. For analyses without repeated measurements, we used the Standard Least Squares personality, resulting in 2(/3)-way ANOVA. Post-hoc testing was done with the Tukey HSD function. For analyses with repeated measurements, the MANOVA personality was used with Repeated Measures (RM) as response specification, resulting in 3(/4)-way RM ANOVA. For the entire study, excepting the measures of running distance and velocity, sample sizes were: n (treatment main effects) = 30–32, n (simple effects) = 15–16. For the measures of running distance and velocity specifically, sample sizes were: n = 15. The specific analysis used for mRNA sequencing data can be found in the Method details under mRNA sequencing. Statistical significance was indicated in the following manner: Effects of intravenous NR: p < 0.05 = †, p < 0.01 = ††, p < 0.001 = †††. Effects of VWR: p < 0.05 = §, p < 0.01 = §§, p < 0.001 = §§§. Interactions as well as additional effects are written out in the figures and noted with: p < 0.05 = ∗, p < 0.01 = ∗∗, p < 0.001 = ∗∗∗. In cases without statistical interactions, the above symbols indicate main effects, while in cases with statistical interactions, the symbols indicate simple effects compared to the appropriate control group. For the pilot experiments sample sizes were n = 5. Effects of intravenous NR were indicated in the following manner: p < 0.05 = †, p < 0.01 = ††, p < 0.001 = †††, whereas differences between groups receiving oral vs intravenous delivery were denoted: p < 0.05 = #, p < 0.01 = ##, p < 0.001 = ###. Statistical testing for these experiments was done in Prism 8.4.3. Ordinary one-way ANOVA was performed with Tukey’s multiple comparisons test.
Acknowledgments
We acknowledge Lars Roed Ingerslev, Mette Carlsen Mohr, Mie Mechta, and the Single-Cell Omics platform at the CBMR for technical and computational expertise and support for the transcriptomic analysis by RNA sequencing. We further acknowledge Elysium Health for supplying nicotinamide riboside and pterostilbene. The graphical abstract was created with BioRender.com. Financial support for this study was provided by Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR). CBMR is an independent Research Center at the University of Copenhagen and is partially funded by an unrestricted donation (NNF18CC0034900) from the Novo Nordisk Foundation. This work was further supported by a PhD scholarship to M.V.D. from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation (NNF17SA0031406). Additional support was provided by the A.P. Møller Foundation to M.V.D. through their program, Fonden til Lægevidenskabens Fremme, specifically to cover the cost associated with mRNA sequencing. Lastly, the study was supported financially by Elysium Health.
Author contributions
Conceptualization, M.V.D., R.W.D., and J.T.T.; Methodology, M.V.D., T.S.N.; Investigation, M.V.D., T.S.N., A.L.B., S.C., K.T., and S.L.; Writing – Original Draft, M.V.D.; Visualization, M.V.D.; Writing – Review & Editing, M.V.D, T.S.N., A.L.B., S.C., K.T., T.M., R.W.D., S.L., and J.T.T.; Funding Acquisition, M.V.D., J.T.T.; Resources, T.M, R.W.D., S.L., J.T.T.; Supervision, T.M., S.L., and J.T.T.
Declaration of interests
This study was supported financially by Elysium Health. R.W.D. is Scientific Director with Elysium Health.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103863.
Supplemental information
Data and code availability
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Remaining data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agerholm M., Dall M., Jensen B.A.H., Prats C., Madsen S., Basse A.L., Graae A.S., Risis S., Goldenbaum J., Quistorff B., et al. Perturbations of NAD(+) salvage systems impact mitochondrial function and energy homeostasis in mouse myoblasts and intact skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2018;314:E377–E395. doi: 10.1152/ajpendo.00213.2017. [DOI] [PubMed] [Google Scholar]
- Azzolini M., La Spina M., Mattarei A., Paradisi C., Zoratti M., Biasutto L. Pharmacokinetics and tissue distribution of pterostilbene in the rat. Mol. Nutr. Food Res. 2014;58:2122–2132. doi: 10.1002/mnfr.201400244. [DOI] [PubMed] [Google Scholar]
- Barclay C.J., Constable J.K., Gibbs C.L. Energetics of fast- and slow-twitch muscles of the mouse. J. Physiol. 1993;472:61–80. doi: 10.1113/jphysiol.1993.sp019937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse A.L., Agerholm M., Farup J., Dalbram E., Nielsen J., Ortenblad N., Altintas A., Ehrlich A.M., Krag T., Bruzzone S., et al. Nampt controls skeletal muscle development by maintaining Ca(2+) homeostasis and mitochondrial integrity. Mol. Metab. 2021;53:101271. doi: 10.1016/j.molmet.2021.101271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse A.L., Dalbram E., Larsson L., Gerhart-Hines Z., Zierath J.R., Treebak J.T. Skeletal muscle insulin sensitivity show circadian rhythmicity which is independent of exercise training status. Front. Physiol. 2018;9:1198. doi: 10.3389/fphys.2018.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieganowski P., Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Brouwers B., Stephens N.A., Costford S.R., Hopf M.E., Ayala J.E., Yi F., Xie H., Li J.L., Gardell S.J., Sparks L.M., Smith S.R. Elevated nicotinamide phosphoribosyl transferase in skeletal muscle augments exercise performance and mitochondrial respiratory capacity following exercise training. Front. Physiol. 2018;9:704. doi: 10.3389/fphys.2018.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonvicino D., Ranieri G., Pittelli M., Lapucci A., Bragliola S., Chiarugi A. SIRT1-dependent restoration of NAD+ homeostasis after increased extracellular NAD+ exposure. J. Biol. Chem. 2021;297:100855. doi: 10.1016/j.jbc.2021.100855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y., Fernandez-Marcos P.J., Yamamoto H., Andreux P.A., Cettour-Rose P., et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapaite J., van den Berg S.A., Houten S.M., Nicolay K., Van Dijk K.W., Jeneson J.A. Fiber-type-specific sensitivities and phenotypic adaptations to dietary fat overload differentially impact fast- versus slow-twitch muscle contractile function in C57BL/6J mice. J. Nutr. Biochem. 2015;26:155–164. doi: 10.1016/j.jnutbio.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Clement J., Wong M., Poljak A., Sachdev P., Braidy N. The plasma NAD(+) metabolome is dysregulated in “normal” aging. Rejuvenation Res. 2019;22:121–130. doi: 10.1089/rej.2018.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costford S.R., Brouwers B., Hopf M.E., Sparks L.M., Dispagna M., Gomes A.P., Cornnell H.H., Petucci C., Phelan P., Xie H., et al. Skeletal muscle overexpression of nicotinamide phosphoribosyl transferase in mice coupled with voluntary exercise augments exercise endurance. Mol. Metab. 2018;7:1–11. doi: 10.1016/j.molmet.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall M., Trammell S.A.J., Asping M., Hassing A.S., Agerholm M., Vienberg S.G., Gillum M.P., Larsen S., Treebak J.T. Mitochondrial function in liver cells is resistant to perturbations in NAD(+) salvage capacity. J. Biol. Chem. 2019;294:13304–13326. doi: 10.1074/jbc.RA118.006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollerup O.L., Chubanava S., Agerholm M., Sondergard S.D., Altintas A., Moller A.B., Hoyer K.F., Ringgaard S., Stodkilde-Jorgensen H., Lavery G.G., et al. Nicotinamide riboside does not alter mitochondrial respiration, content or morphology in skeletal muscle from obese and insulin-resistant men. J. Physiol. 2020;598:731–754. doi: 10.1113/JP278752. [DOI] [PubMed] [Google Scholar]
- Elango B., Dornadula S., Paulmurugan R., Ramkumar K.M. Pterostilbene ameliorates streptozotocin-induced Diabetes through enhancing antioxidant signaling pathways mediated by Nrf2. Chem. Res. Toxicol. 2016;29:47–57. doi: 10.1021/acs.chemrestox.5b00378. [DOI] [PubMed] [Google Scholar]
- Elhassan Y.S., Kluckova K., Fletcher R.S., Schmidt M.S., Garten A., Doig C.L., Cartwright D.M., Oakey L., Burley C.V., Jenkinson N., et al. Nicotinamide riboside augments the aged human skeletal muscle NAD(+) metabolome and induces transcriptomic and anti-inflammatory signatures. Cell Rep. 2019;28:1717–1728 e6. doi: 10.1016/j.celrep.2019.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick D.W., Davis J.G., Davila A., JR., Agarwal B., Michan S., Puchowicz M.A., Nakamaru-Ogiso E., Baur J.A. Increasing NAD synthesis in muscle via nicotinamide phosphoribosyltransferase is not sufficient to promote oxidative metabolism. J. Biol. Chem. 2015;290:1546–1558. doi: 10.1074/jbc.M114.579565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick D.W., Loro E., Liu L., Davila A., JR., Chellappa K., Silverman I.M., Quinn W.J.,3R.D., Gosai S.J., Tichy E.D., Davis J.G., et al. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 2016;24:269–282. doi: 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick D.W., Mcdougal A.V., Semenas M., Vappiani J., Nuzzo A., Ulrich J.C., Becherer J.D., Preugschat F., Stewart E.L., Sevin D.C., Kramer H.F. Complementary NAD(+) replacement strategies fail to functionally protect dystrophin-deficient muscle. Skelet Muscle. 2020;10:30. doi: 10.1186/s13395-020-00249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacki G., Braren R., Firner K., Nissen M., Kuhl M., Reche P., Bazan F., Cetkovic-Cvrlje M., Leiter E., Haag F., Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11:1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhang L., Li F., Hu C.P., Zhang Z. Restoration of sirt1 function by pterostilbene attenuates hypoxia-reoxygenation injury in cardiomyocytes. Eur. J. Pharmacol. 2016;776:26–33. doi: 10.1016/j.ejphar.2016.02.052. [DOI] [PubMed] [Google Scholar]
- Imai S. “Clocks” in the NAD World: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim. Biophys. Acta. 2010;1804:1584–1590. doi: 10.1016/j.bbapap.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukarainen S., Heinonen S., Ramo J.T., Rinnankoski-Tuikka R., Rappou E., Tummers M., Muniandy M., Hakkarainen A., Lundbom J., Lundbom N., et al. Obesity is associated with low NAD(+)/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. J. Clin. Endocrinol. Metab. 2016;101:275–283. doi: 10.1210/jc.2015-3095. [DOI] [PubMed] [Google Scholar]
- Kapetanovic I.M., Muzzio M., Huang Z., Thompson T.N., Mccormick D.L. Pharmacokinetics, oral bioavailability, and metabolic profile of resveratrol and its dimethylether analog, pterostilbene, in rats. Cancer Chemother. Pharmacol. 2011;68:593–601. doi: 10.1007/s00280-010-1525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y., Turek F.W., Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kraus D., Yang Q., Kong D., Banks A.S., Zhang L., Rodgers J.T., Pirinen E., Pulinilkunnil T.C., Gong F., Wang Y.C., et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford D.J., Bailey A.L., Chanda M.L., Clarke S.E., Drummond T.E., Echols S., Glick S., Ingrao J., Klassen-Ross T., Lacroix-Fralish M.L., et al. Coding of facial expressions of pain in the laboratory mouse. Nat. Methods. 2010;7:447–449. doi: 10.1038/nmeth.1455. [DOI] [PubMed] [Google Scholar]
- Li Y.R., Li S., Lin C.C. Effect of resveratrol and pterostilbene on aging and longevity. Biofactors. 2018;44:69–82. doi: 10.1002/biof.1400. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu L., Su X., Quinn W.J., 3rd, Hui S., Krukenberg K., Frederick D.W., Redpath P., Zhan L., Chellappa K., White E., et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–1080.e5. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang X., Han L., Ye F., Liu M., Fan W., Zhang K., Kong Y., Zhang J., Shi L., et al. Pterostilbene alleviates polymicrobial sepsis-induced liver injury: possible role of SIRT1 signaling. Int. Immunopharmacol. 2017;49:50–59. doi: 10.1016/j.intimp.2017.05.022. [DOI] [PubMed] [Google Scholar]
- Massudi H., Grant R., Braidy N., Guest J., Farnsworth B., Guillemin G.J. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS ONE. 2012;7:e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcreynolds M.R., Chellappa K., Chiles E., Jankowski C., Shen Y., Chen L., Descamps H.C., Mukherjee S., Bhat Y.R., Lingala S.R., et al. NAD(+) flux is maintained in aged mice despite lower tissue concentrations. Cell Syst. 2021;12:1160–1172.e4. doi: 10.1016/j.cels.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay D.P., O'Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., Olson E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay D.P., Sutherland L.B., Bassel-Duby R., Olson E.N. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28:1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli C., Oimoen S. Classical designs: full factorial designs [online]. Scientific test & analysis techniques center of excellence. 2019. https://www.afit.edu/STAT/passthru.cfm?statfile=103
- NIST/SEMATECH Two-level full factorial designs [Online]. e-Handbook of Statistical Methods. 2003. https://www.itl.nist.gov/div898/handbook/pri/section3/pri3331.htm
- Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remie C.M.E., Roumans K.H.M., Moonen M.P.B., Connell N.J., Havekes B., Mevenkamp J., Lindeboom L., De Wit V.H.W., van de Weijer T., Aarts S., et al. Nicotinamide riboside supplementation alters body composition and skeletal muscle acetylcarnitine concentrations in healthy obese humans. Am. J. Clin. Nutr. 2020;112:413–426. doi: 10.1093/ajcn/nqaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo J.R., Korner A., Mills K.F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R.R., et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Mccarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Rossi A.C., Smerdu V., Leinwand L.A., Reggiani C. Developmental myosins: expression patterns and functional significance. Skelet Muscle. 2015;5:22. doi: 10.1186/s13395-015-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wan T., Ye M., Qiu Y., Pei L., Jiang R., Pang N., Huang Y., Liang B., Ling W., et al. Nicotinamide riboside attenuates alcohol induced liver injuries via activation of SirT1/PGC-1alpha/mitochondrial biosynthesis pathway. Redox Biol. 2018;17:89–98. doi: 10.1016/j.redox.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A., Hayat F., Cunningham R., Li J., Garavaglia S., Zamani L., Ferraris D.M., Sykora P., Andrews J., Clark J., et al. Extracellular NAD(+) enhances PARP-dependent DNA repair capacity independently of CD73 activity. Sci. Rep. 2020;10:651. doi: 10.1038/s41598-020-57506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.C., Yang X., Hua X., Liu J., Fan M.B., Li G.Q., Song J., Xu T.Y., Li Z.Y., Guan Y.F., et al. Hepatic NAD(+) deficiency as a therapeutic target for non-alcoholic fatty liver disease in ageing. Br. J. Pharmacol. 2016;173:2352–2368. doi: 10.1111/bph.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Remaining data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.