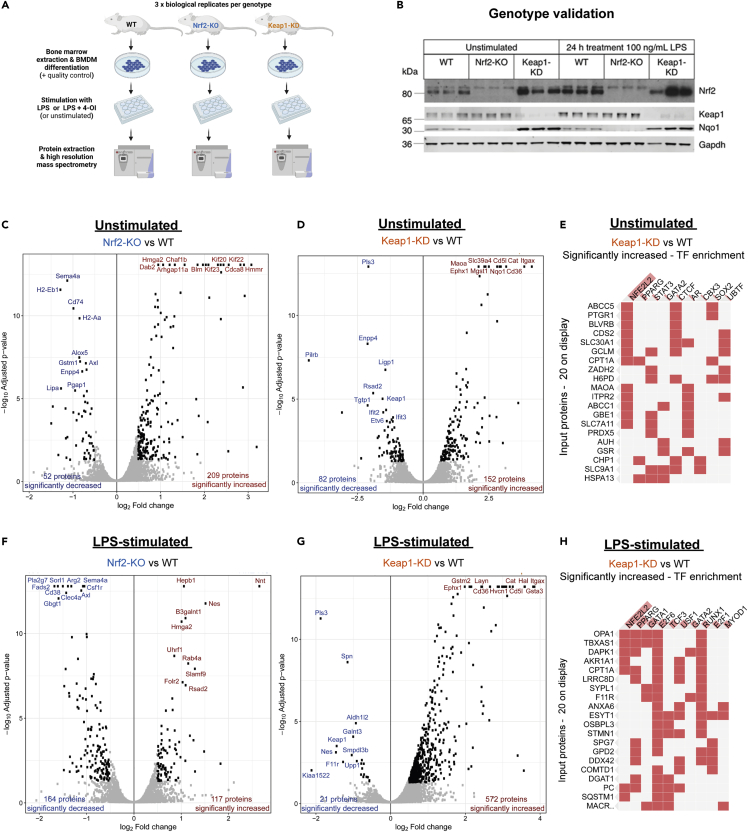
Figure 1.
Nrf2 remodels the proteome in resting and activated macrophages
(A) Experimental design and workflow.
(B) Validation of WT, Nrf2 and Keap1-KO genotypes (n = 3 biological replicates).
(C) Volcano plot of unstimulated Nrf2-KO compared to WT.
(D) Volcano plot of unstimulated Keap1-KD compared to WT.
(E) Transcription factor (TF) enrichment of increased targets in unstimulated Keap1-KD compared to WT.
(F) Volcano plot of LPS-stimulated Nrf2-KO compared to WT.
(G) Volcano plot of LPS-stimulated Keap1-KD compared to WT.
(H) TF enrichment of increased targets in LPS-stimulated Keap1-KD compared to WT.
(E, H) ORA by Enrichr. Top transcription factors ranked according to combined enrichment score (p value and Z score) using ENCODE and ChEA databases.
(C-D, F-G) (n = 3 biological replicates). Cut-offs - log2FC = 0.5; FDR <0.05, determined using t statistics.
(E, F) (combined score – p value and Z score)