Abstract
Invasive candidiasis poses a major healthcare threat. The human opportunistic fungal pathogen Candida glabrata, which causes mucosal and deep-seated infections, is armed with distinct virulence attributes, including a family of 11 glycosylphosphatidylinositol-linked aspartyl proteases, CgYapsins. Here, we have profiled total membrane proteomes of the C. glabrata wildtype and 11 proteases-deficient strain, Cgyps1-11Δ, by mass spectrometry analysis and uncovered a novel role for fungal yapsins in glucose sensing and homeostasis. Furthermore, through label-free quantitative membrane proteome analysis, we showed differential abundance of 42% of identified membrane proteins, with electron transport chain and glycolysis proteins displaying lower and higher abundance in Cgyps1-11Δ cells, compared with wildtype cells, respectively. We also demonstrated elevated glucose uptake and upregulation of genes that code for the low-glucose sensor CgSnf3, transcriptional regulators CgMig1 and CgRgt1, and hexose transporter CgHxt2/10 in the Cgyps1-11Δ mutant. We further elucidated a potential underlying mechanism through genetic and transcript measurement analysis under low- and high-glucose conditions and found CgSNF3 deletion to rescue high glucose uptake and attenuated growth of the Cgyps1-11Δ mutant in YPD medium, thereby linking CgYapsins with regulation of the CgSnf3-dependent low-glucose sensing pathway. Last, high ethanol production, diminished mitochondrial membrane potential, and elevated susceptibility to oxidative phosphorylation inhibitors point toward increased fermentative and decreased respiratory metabolism in the Cgyps1-11Δ mutant. Altogether, our findings revealed new possible glucose metabolism-regulatory roles for putative cell surface-associated CgYapsins and advanced our understanding of fungal carbohydrate homeostasis mechanisms.
Keywords: aspartic protease, glucose, respiration, proteomics, human pathogenic fungi, GPI-linked aspartyl proteases, glucose sensing and uptake, hexose transporters, membrane proteome, high-affinity glucose sensor Snf3
Abbreviations: COX, cytochrome C oxidase; ETP, electron transport; GO, Gene Ontology; GPI, glycosylphosphatidylinositol; TMF, total membrane fraction
Extracellular aspartyl proteases are key virulence factors in pathogenic fungi, as these play a critical role in host invasion and infection establishment (1, 2, 3). Based on their localization, fungal extracellular aspartyl proteases have been divided into two classes: secretory and glycosylphosphatidylinositol (GPI) linked, with the latter class possessing the GPI-anchor sequence motif in their C termini, which facilitates attachment to the cell wall and/or the cell membrane (1, 2, 3, 4). The secretory aspartyl proteases in human fungal pathogens have been associated with degradation of a wide spectrum of host proteins including E-cadherin, cathepsin D, mucin, clotting and complement factors, and extracellular matrix proteins, whereas the GPI-anchored aspartyl proteases have been implicated in fungal cell wall organization, processing of the fungal cell wall proteins, and modulating the adherence and damage to host cells as well as suppressing the host immune response (1, 2, 3, 5, 6).
Candida species are the most common fungal pathogens that cause invasive infections in immunocompromised human hosts, with these infections associated with high economic burden, morbidity, and mortality (7, 8). Five Candida species, Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, and Candida krusei, have been reported to contribute to more than 90% of Candida bloodstream infections (7, 9). C. glabrata is a haploid nondimorphic yeast that grows by budding (6). Depending upon the geographical region, C. glabrata is the second to fourth most frequently isolated Candida species from the blood (9, 10, 11, 12). It also causes superficial mucosal infections and has been associated with high resistance toward azole and echinocandin antifungal drugs (6, 12).
An increased prevalence of C. glabrata has been reported in diabetic patients, with elevated glucose levels associated with Candida overgrowth (13, 14, 15). The capability of C. glabrata to proliferate in host macrophages in vitro is dependent upon reconfiguration of the carbon metabolism, reorganization of the chromatin structure and induction of pexophagy and autophagy in yeast cells, and blocking the acidification of the phagolysosome and activation of the NLRP3 inflammasome-dependent secretion of the proinflammatory cytokine IL-1β in macrophages (5, 16, 17, 18, 19). Of note, glucose homeostasis has recently been reported to play an important role in inflammasome activation and macrophage survival in response to C. albicans infections (20, 21). However, extracellular glucose sensing and transport mechanisms in C. glabrata and their contribution to pathogenesis (22) are yet to be fully elucidated.
A family of 11 putative GPI-linked aspartyl proteases, which are commonly referred to as CgYapsins, is a major contributor to the pathogenesis of C. glabrata (5, 6). A set of 11 genes, CgYPS1–11, which reside on three different chromosomes, Chromosome A (CgYPS7), E (CgYPS2, YPS3–6, and YPS8–11), and M (CgYPS1), code for CgYapsins, with CgYPS3–6 and CgYPS8–11 genes forming a cluster of eight genes on chromosome E (5). A central role for CgYapsins in virulence has been attributed to their functions in maintenance of the cell wall structure and composition, vacuole, pH and energy homeostasis, stress survival, regulation of the secretome, suppression of the macrophage proinflammatory immune response, and colonization of the mice and fly hosts (19, 23, 24, 25, 26, 27). Using mutants lacking single or multiple CgYapsin-encoding genes, it has been shown that CgYps1–11 aspartyl proteases are essential for biofilm formation and survival of C. glabrata in human and murine macrophages and a mouse model of systemic candidiasis (5, 19).
Furthermore, although CgYapsins are thought to be largely functionally redundant, CgYps1 has been demonstrated to be the sole CgYapsin that is required to survive low pH and menadione-induced oxidative stress (23, 27). Furthermore, a unique role of CgYps1 in acid and menadione stress survival has been associated with its regulation of the ATPase activity of the plasma membrane proton pump, CgPma1, and NADH:quinone oxidoreductase activity of the flavodoxin-like protein, CgPst2, respectively (23, 27). CgYps1 has also recently been reported to be enriched in the plasma membrane fraction of cell lysates, along with its substrate CgPst2 (27). Furthermore, we have recently reported an unexpected role of CgYapsins in secretome modulation as the secretome of Cgyps1-11Δ mutant (lacks all 11 CgYapsins) was found to be 4.6-fold larger, compared with the wildtype secretome (26). We also identified eight CgYapsins including CgYps1 in the wildtype secretome (26), underscoring the versatile and probably environmental cue-dependent location of CgYapsins.
In view of assorted physiological roles, and essentiality of CgYapsins for pathogenesis of C. glabrata, we are interested in deciphering the underlying molecular basis. Toward this end, we here have studied the effect of CgYPS-11 gene loss on the composition of the total membrane proteome of logarithmic-phase C. glabrata cells and identified 262 and 189 proteins, via label-free quantitative membrane proteome analysis, which showed decreased and increased abundance in Cgyps1-11Δ mutant, respectively, as compared with wildtype cells. Furthermore, we correlated the high abundance of glycolysis proteins with impaired signaling of CgSnf3 (high-affinity glucose sensor)-dependent pathway, increased glucose uptake, and elevated fermentative metabolism in the Cgyps1-11Δ mutant. Altogether, our data unveil an unanticipated role of fungal GPI-anchored aspartyl proteases in regulating the extracellular glucose sensing mechanisms.
Results
Identification of global membrane proteomes of wildtype and Cgyps1-11Δ strains
We have recently shown that CgYapsins modulate the C. glabrata secretome, as the number of proteins secreted by the Cgyps1-11Δ mutant (lacks 11 putative GPI-linked aspartyl proteases) was 4.6-fold higher, compared with the wildtype (wt) strain (26). Since the conventional trafficking route, which proteins destined to be secreted undertake in membrane-bound vesicles, is from the endoplasmic reticulum to Golgi apparatus to cell membrane to the extracellular environment (28), we sought to extend our work further and profiled the total membrane proteome of the Cgyps1-11Δ mutant. For this, we performed global proteome analyses on total membrane fractions (TMFs) of log-phase grown cells of wt and Cgyps1-11Δ strains in duplicates. For global membrane proteomes, proteins that were identified in both biological replicate samples and represented by a minimum of two total peptides in each sample were selected for further analysis.
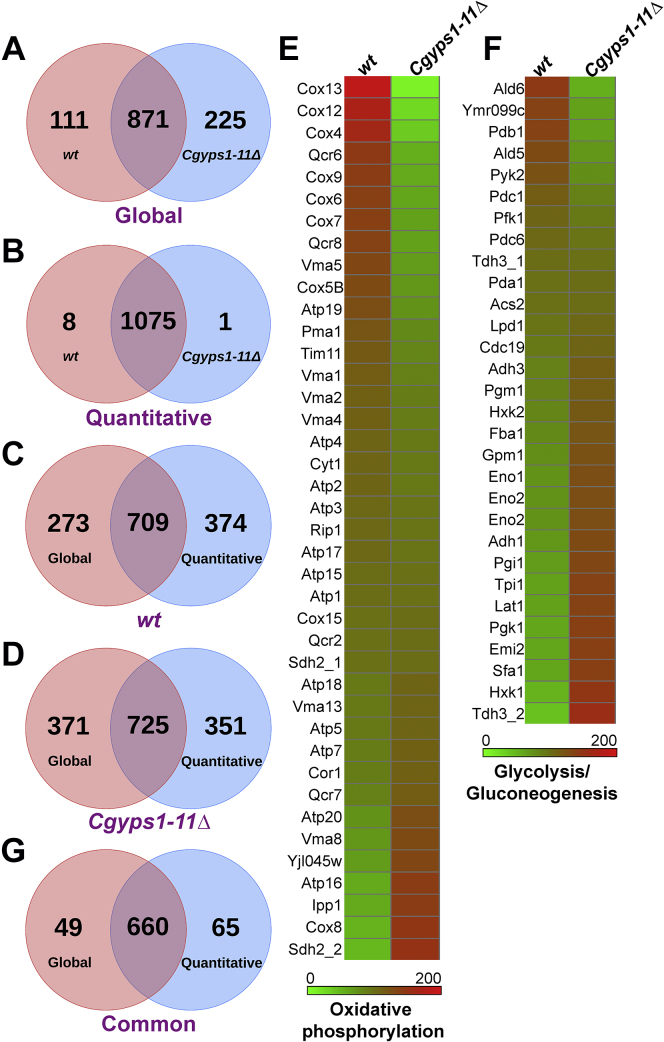
A total of 982 and 1096 proteins were found to constitute the membrane proteomes of wt and Cgyps1-11Δ strains, respectively, with 871 proteins being common to both proteomes (Fig. 1A and Tables S1 and S2). Although Gene Ontology (GO) Slim analysis revealed that, of 5294 C. glabrata ORFs, the products of 1172 genes (20.9% of total genes) belong to the Cellular Component term “Membrane (GO:0016020)” (http://www.candidagenome.org/download/go/go_slim/C_glabrata_CBS138_go_distribution.tab), 1202 proteins, using computational, high-throughput, and manually curated methods, were found to be annotated as membrane proteins (http://www.candidagenome.org/cgi-bin/search/featureSearch). This difference in the protein number could be due to the kind of prediction/annotation tools used in the two analyses. Of 1202 annotated membrane proteins, 300 (30.55%) and 385 (35.13%) proteins were identified in global membrane proteomes of wt and Cgyps1-11Δ strains, respectively (Table S3). GO enrichment analysis of identified proteins in wt and Cgyps1-11Δ membrane proteomes using the FungiFun tool (https://elbe.hki-jena.de/fungifun/) revealed the identified proteins to belong to varied biological processes, including Translation, Intracellular protein transport, Oxidation-reduction process, Ergosterol biosynthetic process, Protein folding, Protein N-linked glycosylation, and Glycolytic process (Tables S4 and S5). The subcellular localization of all identified proteins was predicted using the DeepLoc 1.0 Eukaryotic protein subcellular localization predictor tool (http://www.cbs.dtu.dk/services/DeepLoc-1.0/index.php), and the primary localization of 982 and 1096 proteins identified in wt and Cgyps1-11Δ membrane proteomes, respectively, is summarized in Table S6.
Figure 1.
Global membrane proteome analysis of C. glabrata wildtype and Cgyps1-11Δ strains.A, Venn diagram illustrating overlap between proteins identified in global membrane proteomes of wildtype (wt) and Cgyps1-11Δ strains. B, Venn diagram illustrating overlap between proteins identified in quantitative membrane proteomes of wt and Cgyps1-11Δ strains. C, Venn diagram illustrating overlap between proteins identified in global and quantitative membrane proteomes of the wt strain. D, Venn diagram illustrating overlap between proteins identified in global and quantitative membrane proteomes of the Cgyps1-11Δ mutant. E and F, heat map depicting abundance of proteins identified by quantitative membrane proteome profiling that belong to oxidative phosphorylation (E) and glycolysis/gluconeogenesis (F). Please note that CAGL0C03223g and CAGL0E03850g are labeled as Sdh2_1 and Sdh2_2, respectively, in E, and CAGL0G09383g and CAGL0J00451g are labeled as Tdh3_1 and Tdh3_2, respectively, in F. G, Venn diagram illustrating overlap between proteins identified in wt and Cgyps1-11Δ strains by both global and quantitative membrane proteome profiling.
The membrane proteome of a C. glabrata strain, ATCC 66032, has previously been identified, using the iTRAQ labeling approach, wherein a total of 624 proteins were identified in minimal medium–grown cultures (29). Of these 624 proteins, 361 (57.85%) and 407 (65.22%) proteins were present in membrane proteomes of wt and Cgyps1-11Δ strains, respectively, profiled in our study (Table S7), indicating a good overlap between the published and our dataset. The differences in proteins recovered in both analyses could be due to the usage of two different strains and rich/minimal growth medium, and sample preparation and analysis methods.
Comparative analysis of wt and Cgyps1-11Δ membrane proteomes
The strain-wise comparison of proteins identified in total membrane fractions of the wt and Cgyps1-11Δ strains revealed 871 common proteins, indicating 80% overlap between the two strains, with 111 and 225 proteins being exclusively present in the membrane proteomes of wt and Cgyps1-11Δ mutant, respectively (Fig. 1A and Tables S1 and S2). The enrichment analysis of GO terms in strain-specific proteins, using the DAVID Functional Annotation Tool (https://david.ncifcrf.gov/tools.jsp), showed 42-fold enrichment of GO:0033215 (iron assimilation by reduction and transport) and 12-fold enrichment of GO:0006491 (N-glycan processing) terms, in wt and Cgyps1-11Δ membrane proteomes, respectively (Table S8). This exclusive presence of 111 and 225 proteins in wt and Cgyps1-11Δ membrane proteomes, respectively, could arise from drastic changes in the protein abundance in the absence of 11 proteases. Therefore, we next conducted the label-free quantitative membrane proteomics to identify proteins whose levels were altered in the Cgyps1-11Δ mutant.
Label-free quantitative proteome analysis of membrane fractions of wt and Cgyps1-11Δ strains
Using label-free quantitative proteomics, we identified proteins in total membrane fractions, prepared from lysates of the log-phase grown wt and Cgyps1-11Δ cells, that showed differential abundance (≥1.5-fold change in levels) in the Cgyps1-11Δ mutant. This label-free membrane protein quantification analysis yielded 1083 and 1076 proteins that were identified in wt and Cgyps1-11Δ strains, respectively. A set of 1075 proteins was common between wt and Cgyps1-11Δ membrane fractions, whereas 8 and 1 proteins were present uniquely in wt and Cgyps1-11Δ, respectively (Fig. 1B and Table S9). Among 1075 proteins identified in TMFs of both wt and Cgyps1-11Δ strains, 1004 and 71 proteins were identified with high (<0.01 FDR) and medium (<0.05 FDR) confidence, respectively. The set of nine proteins, identified uniquely in the wt or Cgyps1-11Δ strain, was identified with high confidence. Of note, 22.2% (241) and 5% (54) of all identified proteins (1084) belonged to the Candida GO-SLIM Cellular Component term “Membrane” and “Plasma membrane,” respectively (http://www.candidagenome.org/cgi-bin/GO/goTermMapper). Similar to the global membrane proteome analysis, GO enrichment analysis of 1084 proteins identified in the label-free quantitative membrane proteomics, using the FungiFun tool, revealed the proteins to be involved in various processes including Translation, Glycolytic process, Oxidation-reduction process, rRNA processing, Tricarboxylic acid cycle, Intracellular protein transport, and Protein folding (Table S10).
Furthermore, of 1075 proteins, common to both wt and Cgyps1-11Δ strains, we identified proteins whose levels were altered by ≥1.5-fold in the Cgyps1-11Δ mutant. We found 262 and 189 proteins to display decreased and increased abundance, respectively, in TMF of the Cgyps1-11Δ mutant as compared with the wt TMF (Tables S11 and S12), whereas 624 proteins showed no change in abundance (Table S13). Therefore, overall, CgYPS-11 gene loss altered the abundance of 42% of identified total membrane proteins. GO analysis of differentially abundant membrane proteins, using the FungiFun and DAVID tools, revealed the enrichment of Intracellular protein transport, Ergosterol biosynthetic process and Mitochondrial electron transport, and Glycolytic process and Gluconeogenesis processes in proteins with lower and higher abundance, respectively, in the Cgyps1-11Δ mutant (Tables S14 and S15). To probe deeper into glucose metabolism proteins, we next performed the KEGG pathway analysis on all 1084 identified proteins and found 59 (5.4%), 40 (3.7%), and 30 (2.8%) proteins to belong to Carbon metabolism, Oxidative phosphorylation, and Glycolysis/Gluconeogenesis processes, respectively, and the abundance of these proteins in wt and Cgyps1-11Δ cells is shown as heat maps in Fig. S1 and Figure 1, E and F, respectively.
Next, we compared the global and quantitative membrane proteomes and found a set of 709 (∼72%) and 725 (∼66%) proteins to be common between the global and quantitative membrane proteomes of wt and Cgyps1-11Δ strains, respectively (Fig. 1, C and D). Among these 709 and 725 proteins, 660 proteins were common between both strains (Fig. 1G and Tables S16–S18). Of note, this set of 660 proteins detected in both global and quantitative membrane proteomes of wt and Cgyps1-11Δ strains are likely to represent bona fide membrane proteins that are expressed in YPD medium–grown log-phase C. glabrata cells. Moreover, the CGD GO Slim Mapper tool predicted 24% (161) of these 660 proteins to be membrane proteins, with 18.5%, 15.9%, and 15.5% proteins being associated with hydrolase activity, RNA binding, and transferase activity, respectively. Of importance, 25.8% (170) of these 660 proteins were mapped to the GO Biological Process Slim term “Transport,” indicating that a quarter of the identified membrane proteins are likely to be involved in the movement of molecules across membranes.
Mitochondrial morphology is not perturbed in the Cgyps1-11Δ mutant
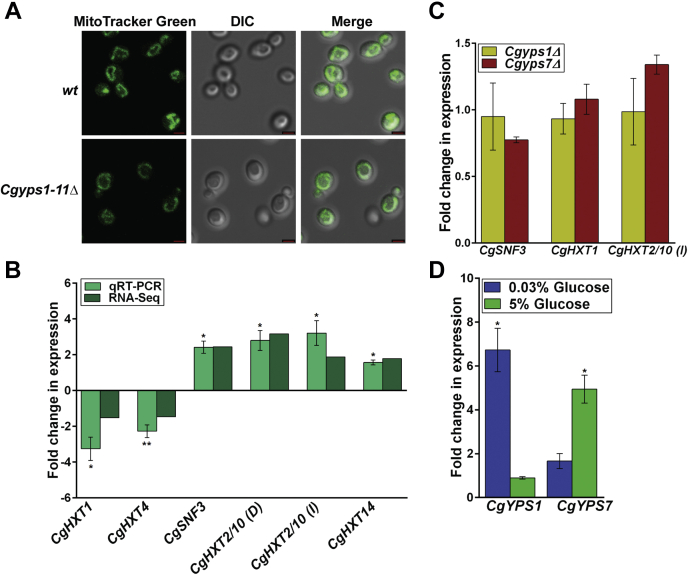
From the GO biological process enrichment analysis, we first focused on the mitochondrial electron transport (ETP) and pulled out proteins that are implicated in this process and were underrepresented in the membrane proteome of the Cgyps1-11Δ mutant (Fig. 1E and Table S11). These ETP proteins also included seven CgCox proteins, CgCox4, CgCox5B, CgCox6, CgCox7, CgCox9, CgCox12, and CgCox13 (Table S11), which represented different subunits of the cytochrome C oxidase (COX). Of note, COX (complex IV) is the terminal oxidase of the ETP chain in the inner membrane of the mitochondria, which reduces oxygen to produce water and facilitates ATP production (30). In this context, it is noteworthy that the Saccharomyces cerevisiae mutant lacking COX4 gene is known to have diminished viability in stationary phase and altered mitochondrial reticular network (31). Since the Cgyps1-11Δ mutant has previously been shown to lose viability in stationary phase growth conditions (5), we decided to examine the mitochondrial morphology, with the rationale that the low abundance of Cox proteins in Cgyps1-11Δ mutant may lead to a perturbation in the mitochondrial architecture. For this, we stained Cgyps1-11Δ and wt cells with the MitoTracker Green dye, as its localization to the mitochondria is independent of the mitochondrial membrane potential. We found robust mitochondrial network in both strains (Fig. 2A), which ruled out the possibility of any gross morphology defect in the mitochondrial structure upon loss of CgYps1–11 aspartyl proteases. Of note, since the Cgyps1-11Δ mutant is able to utilize nonfermentable carbon sources, glycerol and ethanol (23), the significance of diminished levels of Cox proteins on mitochondrial physiology is yet to be determined.
Figure 2.
Glucose sensing and transport genes are deregulated in the Cgyps1-11Δ mutant.A, MitoTracker Green–based mitochondrial morphology analysis. Representative maximum-intensity projection of Z-stack fluorescence confocal images showing mitochondrial network in YNB medium–grown, MitoTracker Green (100 nM)–stained log-phase cells of wt and Cgyps1-11Δ strains. The scale bar represents 2 μm. B, qRT-PCR-based expression analysis of indicated genes (two downregulated and four upregulated genes in the RNA-Seq experiment) in wt and Cgyps1-11Δ strains. Strains were grown to log-phase in YPD medium for 4 h, followed by RNA extraction using acid phenol. The cDNA synthesis reaction was set up with 500 ng total RNA, followed by qRT-PCR using appropriate primer sets, and gene transcript levels were measured using the 2−ΔΔCt method. Please note that CgHXT2/10 (D) and CgHXT2/10 (I) refer to CAGL0D02640g/CAGL0D02662g and CAGL0I00286g genes, respectively. Data (mean ± SEM, n = 3–4) were normalized against CgACT1 mRNA control and represent fold change in expression in Cgyps1-11Δ cells, compared with wt cultures (considered as 1.0). ∗p ≤ 0.05; ∗∗p ≤ 0.01, paired two-tailed Student's t test. C, qRT-PCR-based expression analysis of indicated genes in YPD medium–grown log-phase Cgyps1Δ and Cgyps7Δ cells. Data (mean ± SD, n = 2–3) were normalized against CgACT1 mRNA control and represent fold change in expression in Cgyps1Δ and Cgyps7Δ cells, compared with wt cultures (considered as 1.0). D, qRT-PCR-based expression analysis of CgYPS1 and CgYPS7 genes in log-phase wt cells that were grown in YNB medium containing low (0.03%), regular (2%), and high (5%) glucose for 2 h. Data (mean ± SEM, n = 3–4) were normalized against CgACT1 mRNA control and represent fold change in expression in low-glucose and high-glucose medium–grown wt cells, compared with regular-glucose medium–grown wt cells (considered as 1.0). ∗p ≤ 0.05, paired two-tailed Student's t test. DIC, differential interference contrast.
Genes involved in glucose sensing and transport are deregulated in the Cgyps1-11Δ mutant
Next, we focused on glycolytic process, identified in the GO biological process enrichment analysis, and pulled out proteins involved in this process that were overrepresented in the membrane proteome of the Cgyps1-11Δ mutant, as compared with the wt membrane proteome (Fig. 1F and Table S12). These proteins included enzymes, CgGpm1, CgPgi1, CgTpi1, CgFba1, CgPgk1, CgEno1, and CgEno2, that carry out different enzymatic reactions during breakdown of glucose by glycolysis, with some also performing the reverse reactions during glucose synthesis (22, 32). The enrichment of glycolysis and gluconeogenesis proteins in the mutant membrane proteome was particularly intriguing, in light of the decreased abundance of three hexose transporters, CgHxt1, CgHxt4, and CgHxt5, in the membrane proteome (Tables S11 and S19), and reported transcriptome profiles of the Cgyps1-11Δ mutant (19). We have previously analyzed the transcriptomes of log-phase cultures of wt and Cgyps1-11Δ strains by RNA-Seq approach and found genes involved in ion transport and sterol import to be downregulated and genes involved in carbohydrate metabolism, cell wall organization, and tricarboxylic acid cycle to be upregulated in the mutant (19).
Therefore, we next compared the quantitative membrane proteome results with RNA-Seq transcriptome data, wherein we found differential expression of three genes, CgSNF3 (CAGL0J09020g), CgHXT1 (CAGL0A01804g), and CgHXT14 (CAGL0M04103g), that code for the high-affinity glucose sensor, hexose transporter, and hexose transporter, respectively, in the Cgyps1-11Δ mutant (19). These genes have previously been implicated in glucose sensing in C. glabrata (33, 34). Of the three genes, CgSNF3 and CgHXT14 were upregulated, whereas CgHXT1 was downregulated (19). Since CgHxt1 also displayed decreased abundance in the total membrane proteome of the Cgyps1-11Δ mutant (Table S11), these data suggest that both gene and protein levels of CgHXT1 are downregulated in the mutant.
Ng et al. (33) have previously identified the sugar-receptor repressor pathway genes in C. glabrata, which are likely to be involved in glucose sensing. These include 11 hexose transporters, two glucose sensors CgSnf3 and CgRgt2, and two transcriptional regulators CgRgt1 and CgMig1 ((33, 34, 35); Table S19). For validation of our published RNA-Seq data by quantitative RT-PCR (qRT-PCR), we selected the aforementioned three genes, whose expression was differentially regulated in the RNA-Seq-based transcriptome profiling of the Cgyps1-11Δ mutant cells (19), as well as three other hexose transporter genes, CgHXT4 (CAGL0A01782g), CgHXT2/10 (D) (CAGL0D02640g/CAGL0D02662g), and CgHXT2/10 (I) (CAGL0I00286g), with CAGL0D02640g and CAGL0D02662g genes sharing 100% nucleotide identity ((33, 35); http://www.candidagenome.org/). The rationale behind selecting the latter set of three genes was their differential transcript levels in the Cgyps1-11Δ mutant; however, these were not classified as differentially expressed genes in our RNA-Seq analysis, as these did not meet one of the applied cutoff criteria, viz., ≥1.5-fold change in expression and a false discovery rate–adjusted p value of ≤0.05 to the RNA-Seq data (19). We found 2.8- and 2.3-fold lower transcript levels of CgHXT1 and CgHXT4 genes, respectively, whereas the expression of CgSNF3, CgHXT2/10 (D) (CAGL0D02640g/CAGL0D02662g), CgHXT2/10 (I) (CAGL0I00286g), and CgHXT14 genes was increased by 1.6- to 3.2-fold in the Cgyps1-11Δ mutant, compared with the wt strain (Fig. 2B). These data suggest that the loss of CgYapsins results in deregulated expression of genes involved in glucose sensing and uptake.
Since, of the 11 CgYapsins, CgYps1 and CgYps7 predominantly regulate many processes in vitro (19, 23, 24), we next determined whether the single deletion of CgYPS1 or CgYPS7 gene could lead to the perturbed expression of CgSNF3 and hexose transporter genes, CgHXT1 (CAGL0A01804g) and CgHXT2/10 (I) (CAGL0I00286g). We found transcript levels of CgSNF3, CgHXT1, and CgHXT2/10 (I) genes to be similar in wt and Cgyps1Δ and Cgyps7Δ mutants (Fig. 2C), indicating that CgYPS1 or CgYPS7 gene loss singly has no effect on the transcriptional regulation of these glucose homeostasis genes. This result could in part be due to functional redundancy among CgYapsins, as reported previously (19, 24).
Furthermore, to investigate the role of CgYapsins in cellular response to extracellular glucose, we checked if CgYPS1 and CgYPS7 gene expression is regulated by glucose concentration in the medium. Of note, CgYPS1 transcription is known to be activated in response to low pH and high temperature (23, 36). Since the rich YPD and synthetically defined minimal YNB media, used for routine culturing of C. glabrata strains, contain 2% glucose as carbon source, we used 0.03% and 5% glucose as glucose-limited and glucose-excess growth conditions, respectively. In this regard, it is noteworthy that 0.01% and 2% glucose have previously been reported as low-glucose and high-glucose environment, respectively, for C. glabrata (33). Transcript measurement by qRT-PCR revealed 7-fold activation of CgYPS1 gene upon exposure to low-glucose medium, and 5-fold activation of CgYPS7 gene in high-glucose medium (Fig. 2D). These data suggest that the cellular transcriptional response to environmental glucose levels involves activation of CgYPS1 and CgYPS7 genes, with CgYPS1 and CgYPS7 being activated during glucose-limited and glucose-excess conditions, respectively. In this context, it is noteworthy that C. glabrata is a crab-tree positive yeast, which prefers the fermentation mode of growth even in the presence of oxygen (22), and is assumed to encounter a glucose-limited environment in host phagocytic cells, including macrophages and neutrophils, with CgYps1 playing a key role in survival of the macrophage internal milieu (5, 17, 19, 37). Altogether, our data suggest that C. glabrata cells respond to glucose-limited and surplus conditions by activating the expression of CgYPS1 and CgYPS7 genes, respectively, and the Cgyps1-11Δ mutant, lacking 11 proteases, displays deregulated expression of genes involved in glucose sensing and transport.
CgSNF3 is required for virulence of C. glabrata
CgSnf3 is a high-affinity glucose sensor that is involved in sensing and transducing the low extracellular glucose concentration, and its loss is known to result in the transcriptional repression of four hexose transporter genes (34). CgSNF3 itself is transcriptionally activated in response to low extracellular glucose concentration (33). Therefore, an increased expression of the CgSNF3 gene in the YPD medium (contains 2% glucose)–grown Cgyps1-11Δ cells prompted us to examine whether CgSnf3-mediated signaling of glucose-limited environment is impaired in the mutant. We reasoned that, if elevated expression of genes coding for glucose transporters in the Cgyps1-11Δ mutant is due to higher CgSNF3 transcript levels, deletion of the CgSNF3 gene in the mutant strain could lead to decreased expression of HXT genes. For this, we created two deletion strains, Cgsnf3Δ (CgSNF3-deleted in wt background) and Cgsnf3Δyps1-11Δ (deleted for 12 genes, CgSNF3 and CgYPS1–11), and first characterized the Cgsnf3Δ mutant.
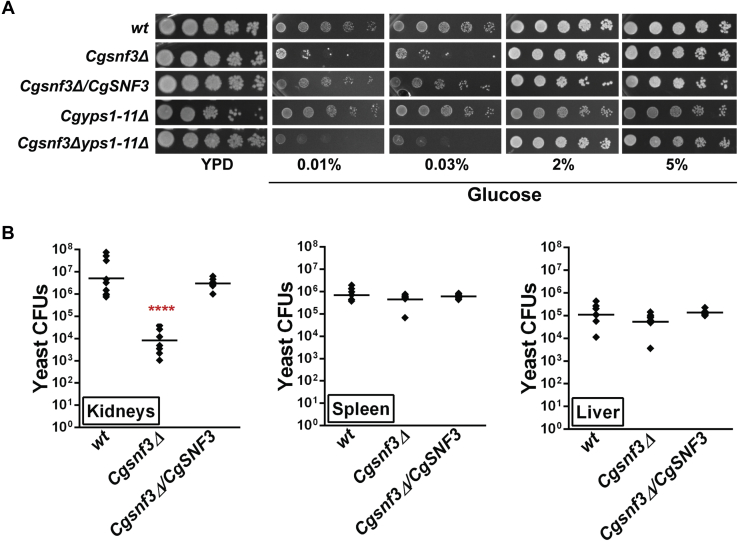
CgSnf3 has previously been shown to be required for growth in low-glucose medium (34); we, therefore, first verified phenotypes of the generated Cgsnf3Δ mutant strain. Similar to the earlier study (34), we found growth of the Cgsnf3Δ mutant to be highly attenuated in glucose-limited medium (0.01% and 0.03% glucose), which was restored upon ectopic expression of the CgSNF3 gene (Fig. 3A).
Figure 3.
CgSNF3 is required for virulence.A, serial dilution spotting analysis of wt, Cgsnf3Δ, Cgsnf3Δ/CgSNF3, Cgyps1-11Δ, and Cgsnf3Δyps1-11Δ strains in YPD (2% dextrose) or YNB medium containing indicated glucose concentrations. C. glabrata cultures were grown overnight in casamino acid medium and normalized to A600 of 1.0. After diluting cultures 10-fold serially in PBS, 3 μl was spotted on YPD medium or YNB medium containing 0.01%, 0.03%, 2%, and 5% glucose. Plates were incubated at 30 °C, and images were captured after 2 days. B, colony-forming unit–based survival analysis of the Cgsnf3Δ mutant. C. glabrata strains (100 μl cell suspension; 4 × 107 cells) were infected into the tail vein of 6- to 8-week-old female BALB/c mice. After 7 days, mice were sacrificed and three organs (kidneys, liver, spleen) were collected and homogenized in PBS. The homogenates were diluted in PBS, and appropriate dilutions were plated on penicillin- and streptomycin-containing YPD medium. The colony-forming units (CFUs) recovered from each organ of the individual mouse are represented by diamonds in graphs. The horizontal line bars represent the CFU geometric mean (n = 7–9) for each organ. Statistically significant differences in CFUs between kidneys of wt- and Cgsnf3Δ-infected mice are marked. ∗∗∗∗p < 0.0001; Mann-Whitney U test.
The Cgsnf3Δ mutant has previously been shown to form robust biofilms but diminished survival in macrophages (34). Therefore, to delineate the role of CgSnf3 in vivo, we checked the survival of the Cgsnf3Δ mutant in a murine model of systemic candidiasis. We recovered about 1000-fold lower fungal colony-forming units from kidneys of Cgsnf3Δ-infected BALB/c mice, compared with wt-infected BALB/c mice (Fig. 3B). This reduced renal fungal load was alleviated in mice infected with the Cgsnf3Δ mutant expressing CgSNF3 ectopically (Fig. 3B), indicating that the diminished fungal survival in murine kidneys is due to the lack of the CgSNF3 gene. Of note, since the glucose abundance is known to modulate the expression of various fungal pathogenesis-associated factors, including morphogenesis, biofilm formation, and stress resistance genes (22, 38, 39, 40), it is possible that both deregulated expression of such factors and impaired glucose homeostasis contribute to the diminished survival of the Cgsnf3Δ mutant in murine kidneys. Important, the fungal burden in liver and spleen was the same in both wt and Cgsnf3Δ infected mice (Fig. 3B). Therefore, besides verifying the role of CgSnf3 in growth in glucose-limited conditions, our data implicate for the first time CgSnf3 in virulence in mice in an organ-dependent manner.
CgSNF3 deletion rescues the growth defect of the Cgyps1-11Δ mutant in 2% glucose–containing medium
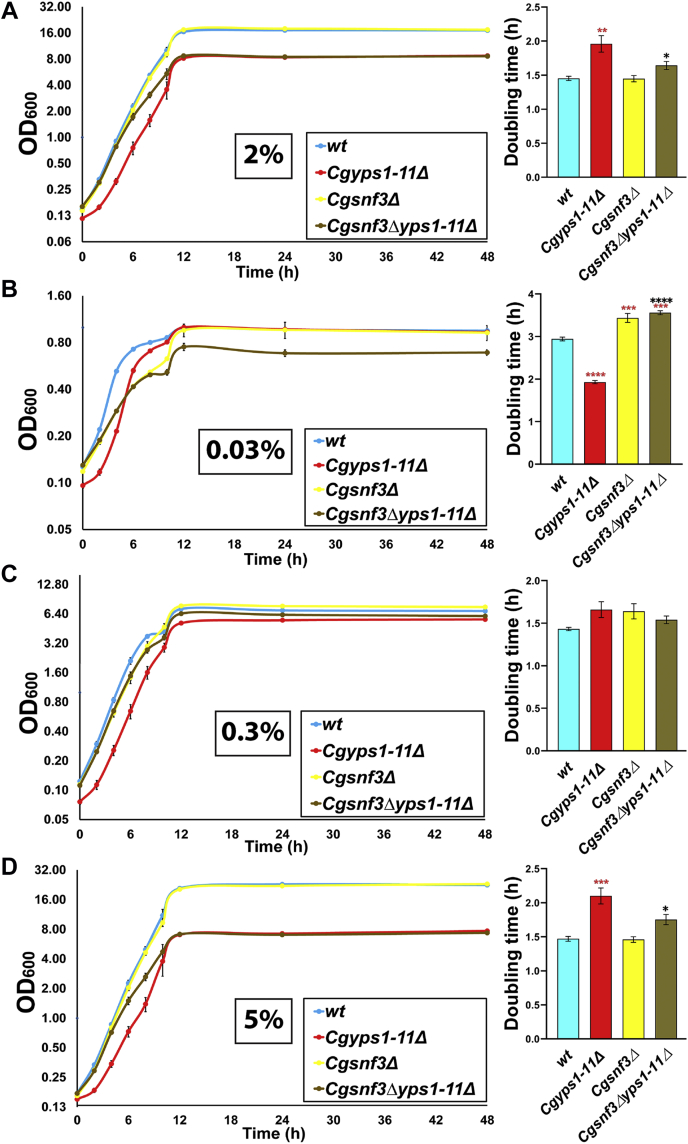
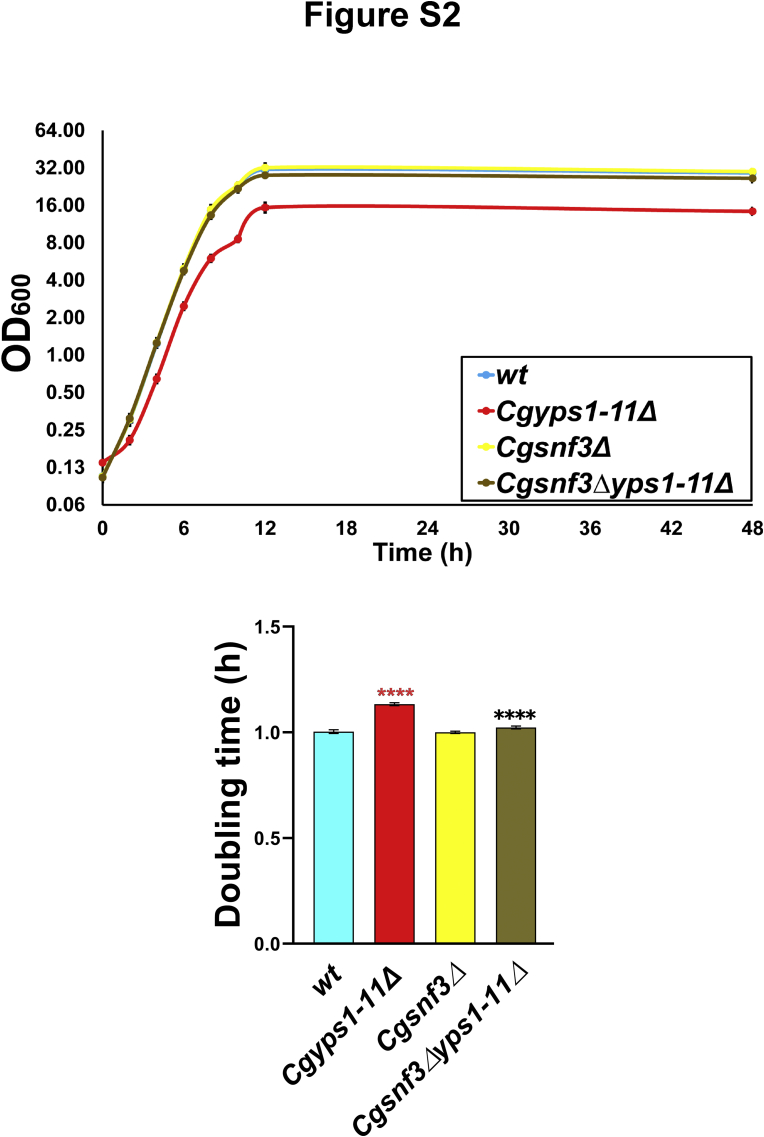
Next, we characterized the Cgsnf3Δyps1-11Δ mutant lacking 11 CgYapsins and CgSnf3 glucose sensor. We found this mutant to be growth defective in low-glucose medium, similar to the Cgsnf3Δ mutant (Fig. 3A). During phenotypic characterization of the generated mutants, we noticed that the Cgsnf3Δyps1-11Δ mutant grew better in 2% dextrose-containing YPD and YNB media, than the Cgyps1-11Δ mutant (Figs. 3A and S2), which is known to be a slow grower (5), indicating that CgSNF3 deletion confers a growth advantage to the Cgyps1-11Δ mutant. One possible reason for this better growth could be high basal transcript levels of CgSNF3, and the consequent activation of the high-affinity glucose uptake pathway even in the presence of surplus glucose in the environment. Consistent with this notion, growth attenuation of the Cgyps1-11Δ mutant, compared with the wt strain, was much less in medium containing low glucose (Fig. 3A). These data suggested that the slow growth phenotype of the Cgyps1-11Δ mutant is specific to the high-glucose medium and could be due to deregulated CgSnf3-mediated glucose signaling.
Therefore, to test this hypothesis, we next verified mutants’ growth profiles by conducting liquid medium–based growth time-course analyses in YNB medium containing 0.03%, 0.3%, 2%, and 5% glucose over the 48-h period. Expectedly, we found that, compared with wt cells, the Cgyps1-11Δ mutant exhibited 1.3-fold higher doubling time in YNB medium containing 2% glucose (Fig. 4A), whereas the Cgsnf3Δ mutant displayed 1.2-fold higher doubling time in YNB medium containing 0.03% glucose (Fig. 4B). In contrast, the growth rate of the Cgyps1-11Δ and Cgsnf3Δ mutants was same as that of the wt strain in glucose-limited (0.3%) and glucose-rich (5%) medium, respectively (Fig. 4, C and D). Intriguing, the doubling time of the Cgsnf3Δyps1-11Δ mutant population was substantially lower and higher than that of the Cgyps1-11Δ mutant population in medium containing 2% and 0.03% glucose, respectively (Fig. 4, A and B). Moreover, a pivotal requirement for CgSnf3 for growth in glucose-deficient medium was underscored by similar doubling times of the Cgsnf3Δ and Cgsnf3Δyps1-11Δ mutants in 0.03% glucose medium (Fig. 4B). Collectively, these data suggest that elevated environmental glucose levels lead to growth retardation in the Cgyps1-11Δ mutant and implicate CgYapsins in maintenance of glucose homeostasis probably via regulation of CgSnf3-dependent glucose sensing and signaling pathway.
Figure 4.
CgSNF3 gene loss confers growth advantage to Cgyps1-11Δ cells in medium containing 2% glucose. Time course analysis of wt, Cgyps1-11Δ, Cgsnf3Δ, and Cgsnf3Δyps1-11Δ strains. C. glabrata strains were grown overnight in YPD medium and inoculated at an initial A600 of 0.1 in YNB medium containing 2% (A), 0.03% (B), 0.3% (C), and 5% (D) glucose. Cultures were incubated at 30 °C with continuous shaking (200 rpm), and absorbance was monitored at regular intervals till 48 h. The absorbance (A600) values are plotted against time, and the growth period, corresponding to the log-phase (between 2 and 6 h), was used to determine the doubling time. Data represent mean ± SEM (n = 3–4). The one-way ANOVA with Tukey’s test was employed to determine the statistical significance of doubling time differences between strains. Red and black asterisks denote differences in doubling time between wt and mutants, and Cgyps1-11Δ and Cgsnf3Δyps1-11Δ mutants, respectively. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.
Transcriptional response to extracellular glucose is impaired in the Cgyps1-11Δ mutant
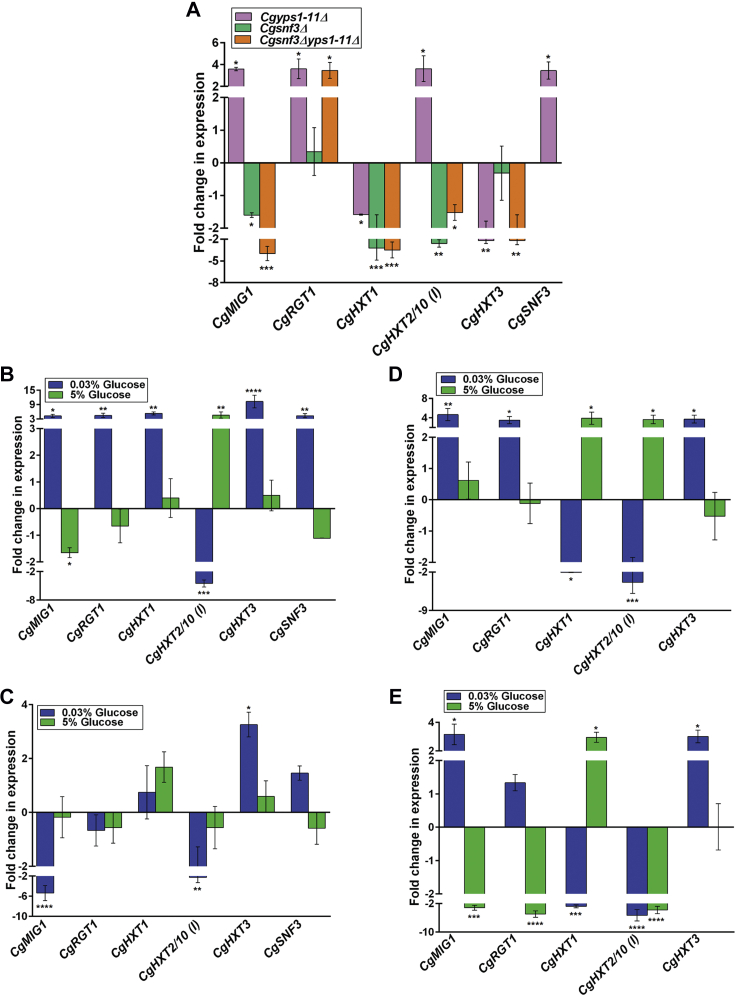
To probe deeper into CgSnf3-mediated glucose signaling in C. glabrata, we next examined the transcript levels of CgSNF3; three hexose transporter genes, CgHXT1, CgHXT2/10 (I), and CgHXT3 (CAGL0A02321g); CgRGT1 (codes for a glucose-responsive transcription factor that is regulated by the low-affinity glucose sensor CgRgt2); and CgMIG1 (encodes transcription factor involved in glucose repression) genes (Table S19) in all four mutant strains under low- (0.03%), regular- (2%), and high- (5%) glucose conditions. The rationale underlying the selection of these genes was their transcriptional regulation either in response to external glucose concentration or CgSNF3/CgYPS1–11 deletion. Of note, CgSNF3, CgRGT1, and CgHXT3 are known to be transcriptionally activated under low-glucose conditions while CgHXT1 and CgHXT3 are downregulated and upregulated, respectively, in the Cgsnf3Δ mutant (33, 34). Similarly, the CgMIG1 gene has been reported to be expressed at higher levels in 2% dextrose medium, as compared with no-glucose medium (33). Last, as presented earlier (Fig. 2B), CgSNF3 and CgHXT2/10, and CgHXT1 genes are induced and repressed, respectively, in the Cgyps1-11Δ mutant.
As shown in Figure 5A, we found CgMIG1, CgRGT1, CgSNF3, and CgHXT2/10 (I) gene expression to be upregulated, whereas the transcription of CgHXT1 and CgHXT3 genes was downregulated in YNB medium–grown Cgyps1-11Δ cells, compared with YNB medium–grown wt cells (Fig. 5A). Contrarily, transcript levels of CgMIG1 and CgHXT2/10 genes were lower in the Cgsnf3Δ mutant, compared with wt cells (Fig. 5A). Furthermore, although CgSNF3 gene deletion had no effect on CgRGT1 and CgHXT3 gene expression, it led to diminished expression of the CgHXT1 gene (Fig. 5A). In this context, it is noteworthy that Ng et al. (34) have previously found lower, higher, and similar transcript levels of CgMIG1 and CgHXT1, CgHXT3, and CgHXT2/10 and CgRGT1 genes, respectively, in the Cgsnf3Δ mutant, compared with wt cells. The reason for this discrepancy in CgHXT3 and CgHXT2/10 gene expression in theirs and our data is not understood and warrants further investigation.
Figure 5.
The response to extracellular glucose is impaired in the Cgyps1-11Δ mutant.A, qRT-PCR-based expression analysis of indicated genes in YNB medium–grown log-phase wt, Cgyps1-11Δ, Cgsnf3Δ, and Cgsnf3Δyps1-11Δ cells. Data (mean ± SEM, n = 3–4) were normalized against CgACT1 mRNA control and represent fold change in expression in mutant cells, compared with wt cultures (considered as 1.0). ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001, one-way ANOVA with uncorrected Fisher’s LSD test. B–E, qRT-PCR-based expression analysis of indicated genes in log-phase wt (B), Cgyps1-11Δ (C), Cgsnf3Δ (D), and Cgsnf3Δyps1-11Δ (E) cells that were grown in YNB medium containing low (0.03%), regular (2%), and high (5%) glucose for 2 h. Data (mean ± SEM, n = 3–4) were normalized against CgACT1 mRNA control and represent fold change in expression in low-glucose and high-glucose medium–grown cells, compared with regular-glucose medium–grown cells of each strain (considered as 1.0). ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001, one-way ANOVA with uncorrected Fisher’s LSD test.
Intriguing, the Cgsnf3Δyps1-11Δ mutant displayed transcriptional profiles that were in-between the two mutants Cgsnf3Δ and Cgyps1-11Δ, with higher and lower transcript levels of CgRGT1, and CgMIG1, CgHXT1, CgHXT2/10 (I) ,and CgHXT3 genes, respectively, in the Cgsnf3Δyps1-11Δ mutant (Fig. 5A). Collectively, these data implicate CgSnf3 in transcriptional activation of CgMIG1 and CgHXT2/10 (I) genes, whereas CgRGT1 and CgHXT3 gene expression is probably not regulated by CgSnf3. Therefore, CgRGT1 induction and CgHXT3 repression in Cgyps1-11Δ and Cgsnf3Δyps1-11Δ mutants is likely to be dependent upon the signaling pathway regulated by other glucose sensor/s.
Since the human host niches, blood, vagina, and urine, have varied glucose levels, and sugar sensing mechanisms are pivotal to microbial growth (22, 41, 42), we next examined the response of C. glabrata to extracellular glucose by measuring the expression of CgMIG1, CgRGT1, CgSNF3, CgHXT1, CgHXT2/10 (I), and CgHXT3 genes in regular- (2%), low- (0.03%), and high- (5%) glucose medium–grown cells. Moreover, to determine the role of CgYapsins and CgSnf3 in glucose sensing and transport, we examined transcript levels in Cgyps1-11Δ, Cgsnf3Δ, and Cgsnf3Δyps1-11Δ strains as well. We observed that wt cells respond to glucose starvation by inducing the expression of CgMIG1, CgRGT1, CgSNF3, CgHXT1, and CgHXT3 genes and repressing the transcription of CgHXT2/10 (I) gene (Fig. 5B). Contrarily, growth in the surplus-glucose medium led to transcriptional downregulation and upregulation of CgMIG1 and CgHXT2/10 (I) genes, respectively, while having no effect on expression of the remainder genes (Fig. 5B). These data suggest a reciprocal regulation of CgMIG1 and CgHXT2/10 (I) genes based on the extracellular glucose abundance and preclude CgRGT1, CgSNF3, CgHXT1, and CgHXT3 genes from cellular transcriptional response to the high-glucose environment. Of note, Ng et al. (33) also have previously shown upregulation of CgMIG1, CgRGT1, CgSNF3, and CgHXT3 in response to low glucose. However, we noticed one difference in the low-glucose-responsive gene expression data, which was for the CgHXT1 gene, with CgHXT1 being upregulated and downregulated in our and their study, respectively, upon exposure to low-glucose conditions (33), which could in part be due to different growth conditions (30 °C versus 37 °C).
Furthermore, the Cgyps1-11Δ mutant could not respond to low-glucose (0.03% glucose) medium via transcriptional activation of CgMIG1, CgRGT1, and CgSNF3 genes (Fig. 5C), although these genes had higher basal transcript levels in the mutant (Fig. 5A). In fact, CgMIG1 transcript levels were lower in low-glucose–grown Cgyps1-11Δ cells, compared with 2% glucose–grown Cgyps1-11Δ cells (Fig. 5C). Despite having deregulated basal expression of CgHXT2/10 (I) and CgHXT3 genes, the Cgyps1-11Δ mutant, similar to wt cells, showed low-glucose–responsive upregulation and downregulation of CgHXT3 and CgHXT2/10 (I) genes, respectively (Fig. 5C). Of note, CgHXT1 gene expression remained unchanged in low-glucose–grown Cgyps1-11Δ cells, compared with 2% glucose–grown Cgyps1-11Δ cells (Fig. 5C). Altogether, these data suggest that the loss of CgYapsins results in an impaired transcriptional response to glucose limitation (Fig. 5C). Furthermore, the transcriptional response of the Cgyps1-11Δ mutant to high glucose was dissimilar to that of the wt strain for CgMIG1 and CgHXT2/10 (I) genes, as CgMIG1 and CgHXT2/10 (I) genes were not downregulated and upregulated, respectively, in high-glucose–grown Cgyps1-11Δ, compared with 2% glucose–grown Cgyps1-11Δ cells (Fig. 5C), underscoring that the loss of CgYapsins also affects the cellular transcriptional response to surplus glucose.
The transcriptional response of the Cgsnf3Δ mutant to low glucose was similar to that of the wt strain except for the CgHXT1 gene, which was downregulated in the mutant (Fig. 5D), implicating CgSnf3 in both basal and glucose-limitation-responsive activation of CgHXT1 gene expression. However, this wildtype-like transcriptional response to the low-glucose environment was unexpected for the Cgsnf3Δ mutant, as Cgsnf3Δ cells could not grow well in glucose-limited conditions (Figs. 3A and 4B). This anomaly could be due to altered expression of other hexose transporters in the mutant. Alternatively, it may stem from differences in the growth conditions (continuous glucose starvation [serial dilution spotting-based growth analysis] versus 2-h growth in low-glucose medium [growth condition for qRT-PCR experiment]), and, thus, a systematic kinetic study of gene expression in glucose-limited conditions may shed light on these two possibilities.
The Cgsnf3Δ mutant also mounted wt-like transcriptional response to high glucose for the CgHXT2/10 (I) gene, whose levels were elevated in 5% glucose–grown Cgsnf3Δ cells, compared with 2% glucose–grown Cgsnf3Δ cells (Fig. 5D). However, contrary to wt cells, CgMIG1 gene expression remained unchanged between 2% and 5% glucose–grown Cgsnf3Δ cells (Fig. 5D). Unlike wt cells, the Cgsnf3Δ mutant displayed increased expression of the CgHXT1 gene in response to high glucose (Fig. 5D). These data suggest that CgSnf3 is required for transcriptional downregulation of CgMIG1 in the surplus-glucose environment; however, the high-glucose–induced expression of CgHXT1 in the Cgsnf3Δ mutant is likely to be mediated by another transcriptional regulator.
Although the Cgsnf3Δyps1-11Δ mutant responded to glucose limitation by activating CgMIG1 gene expression, it was, like the Cgyps1-11Δ mutant, deficient in activating CgRGT1 (Fig. 5E), thereby pointing toward a CgSnf3-indepdendent role of CgYapsins in regulating CgRGT1 gene expression. Of note, the Cgsnf3Δyps1-11Δ mutant exhibited low-glucose–responsive transcriptional downregulation of the CgHXT1 gene, similar to the Cgsnf3Δ mutant (Fig. 5E). Furthermore, like in wt, Cgyps1-11Δ, and Cgsnf3Δ strains, CgHXT2/10 (I) and CgHXT3 transcription was downregulated and upregulated, respectively, in response to low glucose (Fig. 5E). The transcriptional response of the Cgsnf3Δyps1-11Δ mutant to high glucose was complex, and identical to neither Cgsnf3Δ nor Cgyps1-11Δ mutant for genes analyzed, and involved downregulation of all genes except for CgHXT1 and CgHXT3 genes, whose expression was higher and similar, respectively, between 5% and 2% glucose–grown Cgsnf3Δyps1-11Δ cells (Fig. 5E), suggesting a complex interplay between CgSnf3 and CgYapsin-dependent regulation of glucose sensing and uptake genes.
Since yeast cells are known to respond to low- and high-glucose environmental conditions by upregulation and downregulation, respectively, of the high-affinity glucose transporter genes (43, 44), we draw seven major conclusions from our qRT-PCR analysis. First, both the high-affinity, low-glucose sensor CgSnf3 and the transcriptional regulators CgMig1 and CgRgt1, are transcriptionally activated in response to glucose starvation (Fig. 5B). Second, CgHxt1 and CgHxt3, and CgHxt2/10 (I) are likely to represent high-affinity and low-affinity glucose transporters, respectively, with the CgHXT2/10 (I) gene also showing glucose-responsive reciprocal transcriptional regulation (induction in surplus-glucose and repression in low-glucose environment) (Fig. 5B). Third, the transcriptional response of C. glabrata toward surplus environmental glucose involves downregulation of CgMIG1 and upregulation of CgHXT2/10 (I) (Fig. 5B). Fourth, despite CgSnf3’s requirement for growth in low-glucose medium (Figs. 3A and 4B), the Cgsnf3Δ mutant is not drastically impaired in mounting an appropriate transcriptional response to external glucose concentrations at early stages (2 h) of glucose starvation (Fig. 5D). Fifth, although CgMIG1 expression is controlled by CgSnf3-dependent glucose sensing pathway under regular-glucose (2%) condition, the transcriptional regulation of CgRGT1 and CgHXT3 is independent of the CgSnf3 pathway (Fig. 5A). Sixth, the reduced expression of CgMIG1 and CgHXT2/10 (I) genes in the Cgsnf3Δyps1-11Δ mutant (Fig. 5A) may contribute to better growth of the Cgsnf3Δyps1-11Δ mutant in the 2% glucose-containing medium, as compared with the Cgyps1-11Δ mutant (Figs. 3A, 4A and S2). Finally, the high basal transcript levels of low-glucose–responsive genes, CgMIG1, CgRGT1, and CgSNF3, in the Cgyps1-11Δ mutant (Fig. 5, A and B) suggest that the loss of CgYapsins impairs the cellular ability to sense environmental glucose, with the high-glucose environment probably being perceived as a low-glucose environment, and that, this perturbed glucose sensing and uptake pathway adversely affects the Cgyps1-11Δ cell physiology.
The Cgyps1-11Δ mutant displays elevated glucose uptake
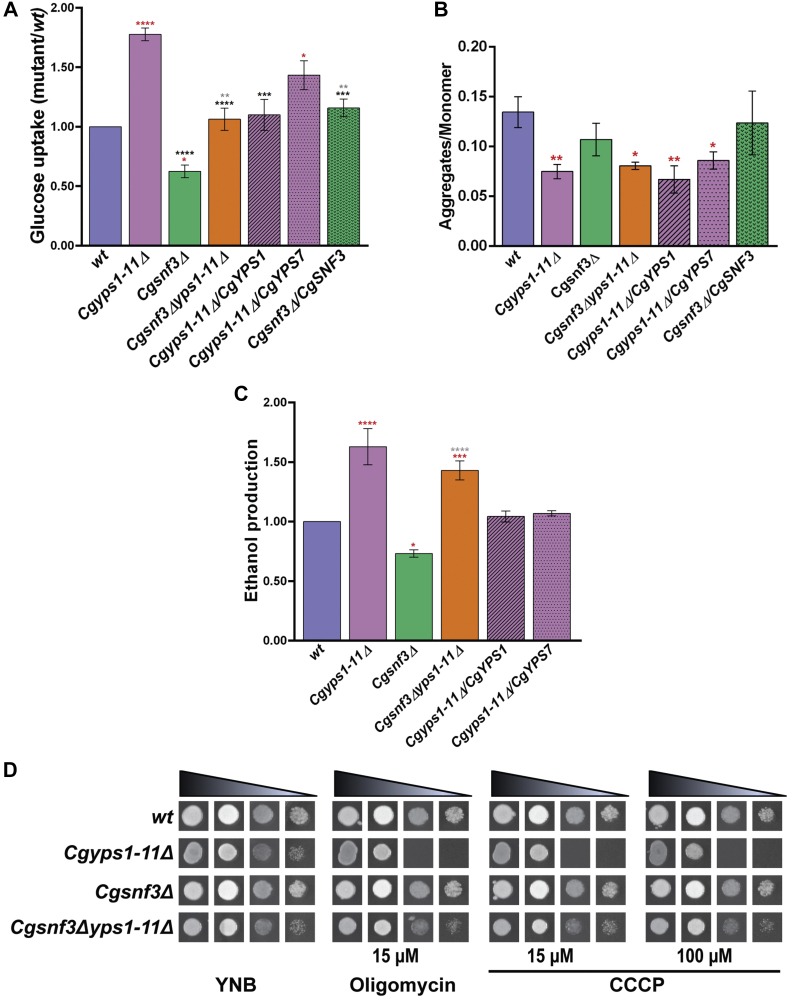
Next, to better understand the link among CgYapsins, CgSnf3, and glucose homeostasis, we performed four experiments. First, we measured glucose uptake in mutant strains using the fluorescent glucose analog 2-NBDG and found 1.8-fold higher glucose uptake in the Cgyps1-11Δ mutant, compared with wt cells (Fig. 6A), which is consistent with an increased expression of the high-affinity glucose sensing system in the mutant. Of note, the ectopic expression of CgYPS1 and CgYPS7 could fully and partially complement the elevated glucose uptake, respectively, in the Cgyps1-11Δ mutant (Fig. 6A). As a control, we also checked glucose uptake in the Cgsnf3Δ mutant and found it to be 1.6-fold lower than that in wt cells, which was restored back to wt levels upon ectopic expression of the CgSNF3 gene (Fig. 6A). Deletion of the CgSNF3 gene in the Cgyps1-11Δ mutant led to diminished glucose uptake, which was similar to that of the wt strain but more than that of the Cgsnf3Δ mutant (Fig. 6A), thereby implicating both CgSnf3-dependent and CgSnf3-independent pathways in controlling glucose uptake in the Cgsnf3Δyps1-11Δ mutant. These results are consistent with our qRT-PCR-based gene expression data (Fig. 5) and suggest that glucose uptake in C. glabrata is regulated by the CgSnf3 sensor and the elevated glucose uptake in Cgyps1-11Δ mutant is largely dependent upon CgSnf3.
Figure 6.
The Cgyps1-11Δ mutant displays higher glucose uptake.A, uptake of 2-NBDG ([2-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose) in indicated C. glabrata strains, as determined by spectrofluorimetry. Glucose-starved cells were incubated with 2-NBDG (100 μM) for 1 h at 30 °C, and the fluorescence emission was recorded at 540 nm, under excitation at 465 nm. Data (mean ± SEM, n = 3–5) were normalized against the wt fluorescence values (considered as 1.0) and represent fold change in NBDG uptake in mutant strains, compared with the wt strain. Red asterisks denote differences in the glucose uptake between wt and indicated strains, black asterisks denote differences between Cgyps1-11Δ and indicated strains, and gray asterisks denote differences between Cgsnf3Δ and indicated strains. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001, one-way ANOVA with Tukey’s test. B, JC-1 dye–based assessment of mitochondrial membrane potential. Log-phase cells were stained with JC-1 (20 μM) dye and washed with PBS, and fluorescence of J-aggregates (red) and monomers (green) was recorded at 550 nm excitation/emission 600 nm and excitation/emission 485 nm/535 nm, respectively. The ratio of red fluorescence (J aggregates) to green fluorescence (monomer) was calculated for each strain and plotted. Data represent mean ± SEM (n = 3–4). Red asterisks denote differences between wt and indicated strains. ∗p ≤ 0.05; ∗∗p ≤ 0.01, one-way ANOVA with Tukey’s test. C, ethanol measurement in the culture broth. Ethanol in the culture medium of indicated strains was extracted using dibutyl phthalate, followed by potassium dichromate oxidation of ethanol. The amount of ethanol in the culture medium was calculated from the standard curve, and data (mean ± SEM, n = 3–5) were normalized against ethanol produced by the wt strain (considered as 1.0). Data represent fold change in ethanol production in mutant strains, compared with the wt strain. Red and gray asterisks denote uptake differences between wt and indicated strains, and Cgsnf3Δ and Cgsnf3Δyps1-11Δ mutants, respectively. ∗p ≤ 0.05; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001, one-way ANOVA with Tukey’s test. D, liquid medium–based growth analysis of wt, Cgyps1-11Δ, Cgsnf3Δ, and Cgsnf3Δyps1-11Δ strains in the presence of indicated inhibitors. Cultures were inoculated at an initial A600 of 0.25 and grown in medium lacking (YNB) or containing oligomycin (15 μM) and carbonyl cyanide m-chlorophenylhydrazone (CCCP; 15 and 100 μM). After 12 h, cultures were diluted in PBS, and 3 μl of undiluted and 10-, 100-, and 1000-fold-diluted cultures were spotted on YNB medium. Plates were incubated at 30 °C, and images were captured after 1 day.
Second, we explored the possibility if elevated glucose uptake in the Cgyps1-11Δ mutant has any effect on mitochondrial functions. For this, we checked the mitochondrial membrane potential using the lipophilic JC-1 dye, which, upon entry into the mitochondria, displays a shift from green to red fluorescence, with green and red fluorescence emitted by native/free dye molecules and J-dye aggregates, respectively, with J-aggregates forming in the energized and negatively charged mitochondria (45). JC-1 dye accumulation in the mitochondria is dependent upon the mitochondrial membrane potential, with the depolarized mitochondria with reduced membrane potential exhibiting a lower fluorescence shift from green to red (45). Therefore, the reduced red/green fluorescence ratio of JC-1 is used as an indicator of the depolarization of the mitochondrial membrane. To measure the mitochondrial membrane potential in wt and Cgyps1-11Δ, we stained log-phase cells with JC-1 dye and measured the red and green fluorescence in excitation/emission, 550 nm/600 nm and 485 nm/535 nm, respectively. We found the ratio of red to green fluorescence to be 1.8-fold lower in the Cgyps1-11Δ mutant, compared with the wt strain (Fig. 6B), indicating diminished JC-1 aggregate formation, probably owing to a decline in the mitochondrial membrane potential. Of note, ectopic expression of neither CgYPS1 nor CgYPS7 could restore the reduced red to green fluorescence ratio in the Cgyps1-11Δ mutant (Fig. 6B), underscoring the contribution of multiple CgYapsins to maintenance of the mitochondrial membrane potential. Furthermore, the Cgsnf3Δ mutant displayed wt-like JC-1 accumulation pattern, whereas the Cgsnf3Δyps1-11Δ mutant was defective in fluorescence shift from red to green and exhibited a 1.7-fold diminished ratio of red to green fluorescence (Fig. 6B). Altogether, these data suggest that, despite normal mitochondrial morphology (Fig. 2A), the mitochondrial membrane potential is significantly impaired in the Cgyps1-11Δ mutant and CgYapsins may play a role in mitochondrial homeostasis.
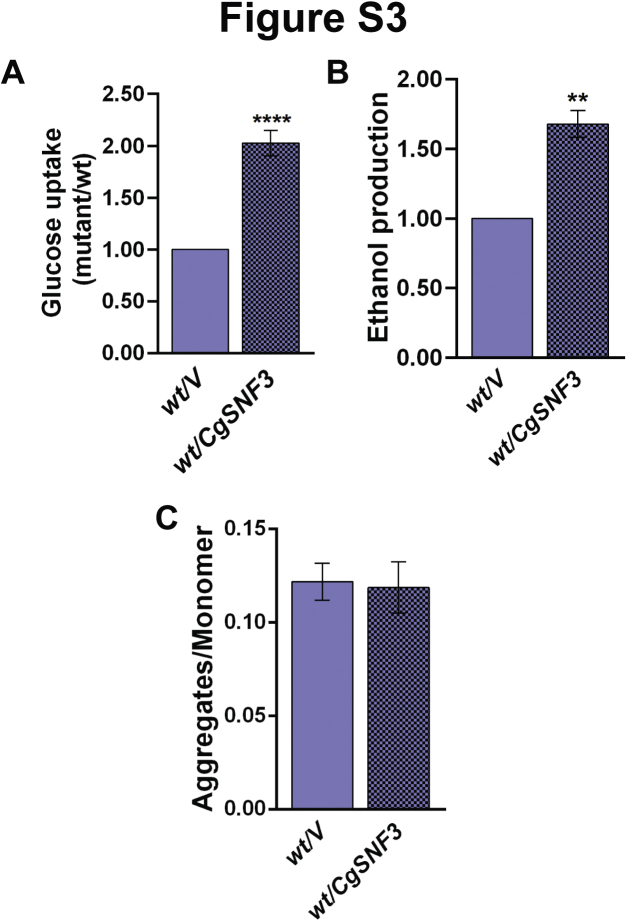
Based on the above data, we next reasoned that the defective glucose sensing pathway and the depolarized mitochondria in the Cgyps1-11Δ mutant may impact the cellular energy metabolic pathways, viz., fermentation and respiration. To test this hypothesis, we first measured the amount of ethanol produced in the culture media of wt and Cgyps1-11Δ cells. For this, ethanol was extracted using the nonalcoholic solvent dibutyl phthalate, followed by ethanol oxidation with potassium dichromate. We found 1.6-fold higher levels of ethanol in the culture supernatant of Cgyps1-11Δ cells, compared with the wt culture medium (Fig. 6C), suggestive of an increased flux through glycolysis in Cgyps1-11Δ cells, which may lead to higher production of ethanol from the glycolysis-derived pyruvate. Intriguing, CgYPS1 and CgYPS7 expression could rescue increased ethanol production in Cgyps1-11Δ cells, whereas the Cgsnf3Δyps1-11Δ mutant displayed higher, higher, and similar ethanol production, compared with wt, Cgsnf3Δ, and Cgyps1-11Δ cells, respectively (Fig. 6C). Cgsnf3Δ mutant cells exhibited 25% less ethanol production compared with wt cells (Fig. 6C). Although this result implicates CgSnf3 in fermentative metabolism in C. glabrata, the similar ethanol production in Cgyps1-11Δ and Cgsnf3Δyps1-11Δ mutants indicate that CgSNF3 is unlikely to be the sole contributor to the increased fermentative metabolism in the Cgyps1-11Δ mutant. To corroborate these results further, we investigated the effect of CgSNF3 overexpression on glucose uptake, mitochondrial potential, and ethanol production in C. glabrata. We found that the overexpression of CgSNF3 in wt cells mimicked the behavior of the Cgyps1-11Δ mutant for glucose uptake and ethanol production, as the wt/CgSNF3 strain exhibited a 2-fold elevated glucose uptake (Fig. S3A) and 1.5-fold higher ethanol production (Fig. S3B), as compared with wt cells expressing the empty vector. Of note, CgSNF3 overexpression had no effect on the mitochondrial membrane potential, as the ratio of red to green fluorescence of the JC-1 dye was similar between wt cells carrying empty vector and overexpressing CgSNF3 (Fig. S3C). Together, these data suggest that CgSnf3 is a critical component of glucose uptake and homeostasis system, and higher CgSNF3 transcript levels in the Cgyps1-11Δ mutant are likely to account for perturbed glucose homeostasis while mitochondrial perturbation may not be solely dependent upon CgSnf3.
Finally, since the cellular capacity to activate respiration has previously been reported to undergo a steady deterioration in the continued presence of glucose (46), and we found Cgyps1-11Δ cells to exhibit elevated glucose uptake (Fig. 6A), diminished mitochondrial membrane potential (Fig. 6B), and reduced ATP levels (24), we next hypothesized that the respiratory metabolism may be impaired in Cgyps1-11Δ cells. To test this, we examined the effect of oxidative phosphorylation inhibition on the viability of Cgyps1-11Δ cells. Liquid media-based growth analysis revealed an increased sensitivity of the Cgyps1-11Δ mutant to inhibitors oligomycin (mitochondrial ATP synthase inhibitor) and carbonyl cyanide m-chlorophenylhydrazone (CCCP; oxidative phosphorylation uncoupler) (Fig. 6D). The Cgsnf3Δyps1-11Δ mutant exhibited much better growth in the presence of oxidative phosphorylation inhibitors than the Cgyps1-11Δ mutant (Fig. 6D), suggesting that loss of the high-affinity glucose sensing system partially compensates for impaired oxidative phosphorylation in the Cgyps1-11Δ mutant. Altogether, our data raise the possibility that the respiratory mode of energy production may be crippled in the Cgyps1-11Δ mutant, which may lead to an overreliance on the fermentative metabolism for growth. Of note, a S. cerevisiae strain displaying reduced glucose uptake, due to the presence of a chimeric hexose transporter, has been reported to exhibit entirely respiratory metabolism even in the presence of high glucose (47). In this context, it is noteworthy that the increased and diminished abundance of glycolytic enzymes and ETP proteins, respectively, in the membrane proteome (Fig. 1, E and F) and the altered mitochondrial membrane potential (Fig. 6B) in Cgyps1-11Δ mutant also point toward the possibility of accelerated fermentative and inadequate respiratory metabolism in the mutant. However, it remains to be determined whether sensing of the glucose-rich environment as the glucose-poor environment by Cgyps1-11Δ cells is a cause or an effect of elevated glycolytic flux. Also, if and how glucose missensing affects the transcriptional regulation of fermentative and respiratory metabolic genes warrants further investigation.
Altogether, our data yielded four new findings. First, CgSnf3 is involved in glucose sensing, as the mutant lacking the CgSNF3 gene showed reduced glucose uptake (Fig. 6A) and altered expression of genes involved in glucose sensing and transport (Fig. 5A). Second, the diminished abundance of hexose transporters, CgHxt1 and CgHxt4, in the membrane proteome of the Cgyps1-11Δ mutant (Table S11) could be due to their transcriptional downregulation in the mutant (Fig. 5A). Third, the glucose and mitochondria homeostasis are perturbed in the Cgyps1-11Δ mutant, which could in part be attributed to the impaired CgSnf3-dependent glucose sensing. Fourth, the dysfunctional mitochondria in the Cgyps1-11Δ mutant either emerge from or lead to inadequate respiratory metabolism during cellular growth.
In conclusion, we demonstrate unequivocally for the first time Yapsin-dependent modulation of glucose homeostasis in any pathogenic fungus, and our findings have wide applications in design of strategies to better control fungal infections and understand glucose metabolism pathways.
Discussion
Glucose metabolism is essential for life, as it plays a central role in generation of energy and biosynthetic building blocks. Glucose homeostasis in fungi is maintained by a finely tuned system consisting of glucose sensors, transporters, and metabolic enzymes (43). Based on the environmental glucose abundance, expression of the hexose transporter–encoding genes is controlled by specific transcription factors of the extracellular glucose sensing pathways (43, 44). Since the human host niches that pathogenic fungi are exposed to vary widely in glucose levels, viz., blood (2–30 mM), vaginal fluids (5–149 mM), and urine (0–0.8 mM), glucose sensing mechanisms are pivotal to fungal virulence (41, 42). Therefore, the ability of Candida species to sense and appropriately respond to extracellular glucose is likely to contribute to their success as both bloodstream and urinary and vaginal tract pathogens (6, 7, 8, 22).
The pathogenic yeast C. glabrata is known to have two plasma membrane glucose sensors, CgSnf3 and CgRgt2, which are involved in sensing glucose-deficient and glucose-surplus environmental conditions, respectively, by regulating the expression of hexose transporters through transcription factors CgRgt1 and CgMig1 in response to glucose abundance (33, 34). Although the mechanistic basis for glucose sensing in C. glabrata is still elusive, the glucose transport and homeostasis mechanisms are well defined in its evolutionarily close relative S. cerevisiae (44, 48). Glucose repression, which involves deregulation of alternate carbon source utilization genes in the presence of the preferred carbon source glucose, plays a key role in glucose metabolic regulation in S. cerevisiae (43, 44). Furthermore, the hexose transporters, which have varied (high, moderate, and low) affinity for glucose, primarily import glucose via facilitated diffusion, with the Snf3/Rgt2-Rgt1 pathway regulating their gene expression (22, 43, 44). Rgt1 in S. cerevisiae is known to inhibit the expression of hexose transporter–encoding genes (HXTs), in the presence of glucose, via formation of a repressor complex with Mth1, Std1, Ssn6, and Tup1 corepressors (43, 44, 48). The binding of glucose to the plasma membrane glucose sensors Rgt2 and Snf3 results in casein kinase I (Yck1/Yck2)-mediated phosphorylation of Mth1 and Std1, which leads to their proteasomal degradation, thereby preventing the binding of Rgt1 to the HXT gene promoters (22, 43, 44, 48). Snf3 and Rgt2 serve as high- and low-affinity glucose receptors, respectively, with Snf3 being required for low-glucose–induced transcriptional activation of moderate-affinity glucose transporters, Hxt2 and Hxt4, and Rgt2 being involved in elevating expression of the low-affinity hexose transporter, Hxt1 (22, 43, 44, 48). Another transcriptional repressor Mig1, which contains Cys2His2 zinc finger DNA-binding motif, is involved in transcriptional downregulation of its target moderate- and high-affinity hexose transporter–encoding genes when glucose is present in high abundance, along with general corepressors, Ssn1 and Tup1 (22, 43, 44, 48). Under glucose starvation conditions, Mig1 is phosphorylated by Snf1 kinase, which inhibits its repressor activity and leads to its export from the nucleus (22, 43, 44, 48). With this multilayered, tightly controlled glucose sensing and uptake system, S. cerevisiae is able to grow over a wide range of environmental glucose concentrations (22, 44).
C. glabrata possesses orthologs of S. cerevisiae glucose sensors, transporters, and transcriptional regulators (22, 33, 34, 35). While CgSnf3 and CgRgt2, and CgRgt1 and CgMig1 are postulated to act as glucose sensors and transcription factors, respectively, in C. glabrata, its 11 hexose transporters (CgHxts) also share similarity with their S. cerevisiae counterparts (22, 33, 34, 35). Moreover, the upregulation of CgHXT3 and CgHXT5 genes under low-glucose growth conditions (33) suggests that these probably are high-affinity hexose transporters. Furthermore, consistent with fungal glucose sensors possessing long (about 200 aa) cytoplasmic C-terminal tails containing a 25-amino-acid sequence motif, which is involved in extracellular glucose signaling (22, 43), CgRgt2 and CgSnf3 also carry one copy of this motif in their C-terminal tails (33). Of note, the cytoplasmic C-terminal tail is absent in fungal hexose transporters (22, 35, 43). In the current study, we have shown that CgSnf3 is required for both uptake of glucose (Fig. 6A) and transcriptional regulation of glucose homeostasis genes (Fig. 5A), and our results place CgSnf3 at the forefront of glucose transport regulatory mechanisms. Furthermore, our finding of the nonresponsiveness of CgSNF3 gene expression to the high environmental glucose (Fig. 5D) is in accordance with its role primarily in sensing glucose-limited conditions (34).
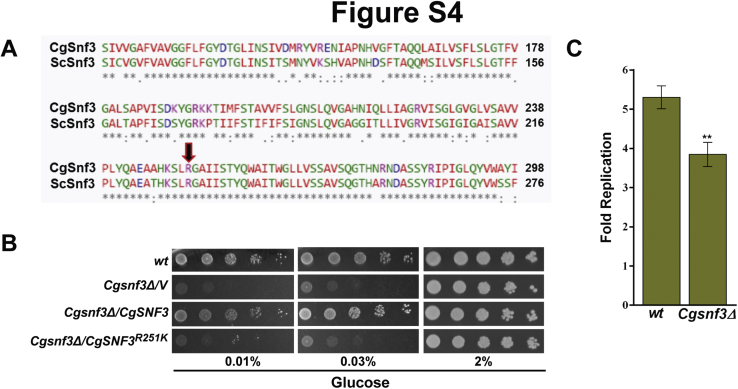
Despite the similar composition of glucose sensing pathways in S. cerevisiae and C. glabrata (22, 35), the individual pathway constituents may not function in an identical manner. For example, substitution of the conserved arginine 251 residue with lysine (R251K) in CgSnf3 led to impaired functions of CgSnf3 in survival under low environmental glucose conditions (Fig. S4, A and B), whereas the same mutation (R229K) rendered S. cerevisiae Snf3 to be constitutively active (49). Similarly, CgRGT1 gene expression is regulated in response to glucose abundance in C. glabrata (Fig. 5B; (33)), contrary to its S. cerevisiae counterpart (50). Furthermore, our transcript profiling of glucose homeostasis genes suggests a complex regulation of glucose sensing pathways, with CgMIG1 and CgRGT1 transcription being activated in response to both high and low glucose, and low-glucose environment, respectively (Fig. 5B). Our data also suggest that CgSnf3 is likely to be the major but not the sole sensor and/or transducer of glucose-limited environment, as the Cgsnf3Δ mutant, despite having low basal transcription of CgMIG1 (Fig. 5A), was able to activate CgMIG1 gene expression in response to glucose starvation (Fig. 5D). Moreover, consistent with the previous study (34), the Cgsnf3Δ mutant displayed diminished intracellular replication in human THP-1 macrophages, compared with wildtype cells (Fig. S4C), with the macrophage internal milieu being a glucose-deficient environment (6). Altogether, these data raise the possibility that the extracellular sensing and homeostasis of glucose in C. glabrata is probably maintained by partially functionally redundant proteins. Moreover, variations in glucose metabolic pathways between S. cerevisiae and C. glabrata may have evolved owing to different habitats of these two yeasts. In this context, it is noteworthy that, unlike S. cerevisiae, C. glabrata can utilize only two sugars, glucose and trehalose, as fermentable carbon sources (22).
Macrophages play an important role in control of Candida infections, with glucose metabolism being central to metabolic reconfiguration of both macrophages and Candida cells (6, 51). Recently, it has been shown that macrophages and C. albicans compete for the limited amount of glucose available by turning on their glycolysis pathway (20). This activation of the glucose utilization pathway is an important determinant for the outcome of C. albicans–macrophage interaction, as C. albicans utilizes the depleted-glucose environment to trigger macrophage cell death (20). Although the role of glucose metabolism in C. glabrata–macrophage interaction is yet to be investigated in depth, transcriptome profiling studies have shown that C. glabrata induces gluconeogenesis and the glyoxylate cycle, and represses the glycolysis pathway, upon phagocytosis by macrophages (5, 6). Furthermore, genes coding for CgHxt3 and CgHxt5 hexose transporters and CgMig1 transcriptional repressor were upregulated in macrophage-internalized C. glabrata cells (5), which is consistent with their gene activation in a low-glucose environment under laboratory growth conditions (Fig. 5B) (33). Genes coding for CgHxt2/10 (D) (CAGL0D02640p/CAGL0D02662p), CgHxt4, and CgHxt6/7 (CAGL0A00737p) hexose transporters, and CgStd1 (CAGL0L10043p) transcriptional repressor, were found to be downregulated in macrophage-ingested C. glabrata cells, which may arise from reprogramming of the glucose metabolism in the macrophage internal milieu (5). Of note, the mutant lacking 11 CgYapsins, Cgyps1-11Δ, which exhibited increased basal levels of CgHXT2/10 (D), CgSNF3, CgRGT1, and CgMIG1 and decreased basal levels of CgHXT1 and CgHXT4 gene transcripts (Figs. 2B and 5A), has been reported to be killed, whereas C. glabrata wildtype cells undergo 5- to 7-fold replication in macrophages (5, 18, 19). Although the intracellular survival defect of the Cgyps1-11Δ mutant has been attributed to NLRP3 inflammasome activation resulting in increased IL-1β production in THP-1 macrophages (19), our data collectively raise the possibility of glucose sensing and uptake mechanisms contributing to proliferation of C. glabrata in macrophages.
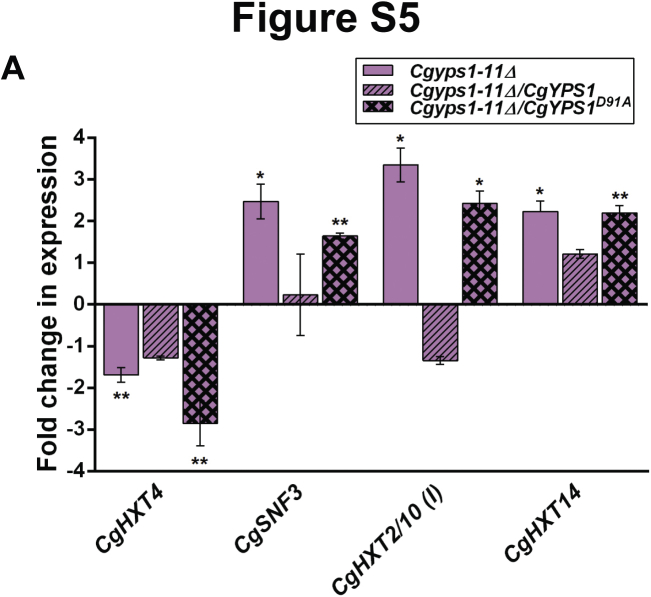
CgYapsins are known to work redundantly to ensure proper functioning of the cell wall and maintenance of vacuole and energy homeostasis (19, 24). Although CgYPS1 expression was sufficient to rescue the defect in both intracellular survival and proliferation of the Cgyps1-11Δ mutant in human THP-1 macrophages, CgYPS1 gene deletion only dampened the intracellular replication of C. glabrata (19), thereby pointing toward the role of other CgYapsins in interaction with macrophages. Consistently, seven CgYPS (CgYPS2, 4, 5, and 8–11) genes were upregulated in macrophage-ingested C. glabrata cells (5). The findings that Cgyps1Δ and Cgyps7Δ mutants do not exhibit deregulated expression of glucose sensing and uptake genes (Fig. 2C) and the CgypsCΔ (lacks nine proteases, CgYps2, 3–6, and 8–11) mutant grows like wildtype strain in YPD medium containing 2% glucose (5) suggest that CgYapsins are likely to perform functionally redundant roles in maintenance of glucose homeostasis. In this context, it is worth noting that GPI-anchored aspartyl proteases in S. cerevisiae (Yps 1–3, 6, 7) and C. albicans (Sap9 and 10) have not been implicated in glucose homeostasis (4, 52). Our genetic and transcriptional data suggest that glucose sensing is impaired in the Cgyps1-11Δ mutant, with a glucose-rich environment being perceived as a glucose-poor environment, and the dysregulated CgSnf3-dependent glucose sensing pathway partly accounts for this misperception (Figure 3, Figure 4, Figure 5, Figure 6). Although the mechanistic basis of CgYapsin-mediated regulation of glucose metabolism remains to be determined, it is possible that the glucose sensors, CgSnf3 and CgRgt2, in the plasma membrane are substrates of CgYapsins, with their processing potentially by CgYapsins generating a signal for activation of the glucose response pathway. In this context, it is worth noting that the plasma membrane flavodoxin-like protein CgPst2 has recently been shown to be a target of the CgYps1 protease (27). Furthermore, our preliminary results suggest that the proteolytic activity of CgYapsins is likely to be required for their role in glucose homeostasis, as ectopic expression of the catalytically active CgYPS1 but not of the catalytically inactive CgYPS1D91A (carries alanine in place of the catalytic residue aspartic acid and lacks proteolytic activity (27)) could restore the deregulated expression of glucose sensing and transport genes (CgSNF3, CgHXT2/10 (I), CgHXT4 and CgHXT14) in the Cgyps1-11Δ mutant to wt levels (Fig. S5). However, CgYapsin substrates involved in the erroneous glucose sensing and regulation of glucose metabolism are yet to be identified.
In summary, we have profiled the membrane proteomes of C. glabrata wildtype and virulence-attenuated Cgyps1-11Δ mutant, performed label-free quantitative proteome analysis to explore the differential abundance of membrane proteins in these two strains, and found diminished abundance of three hexose transporters, CgHxt1, CgHxt4, and CgHxt5 in the mutant strain (Tables S11 and S19). We further showed that hexose transporters in C. glabrata are transcriptionally regulated in response to the environmental glucose availability and CgYapsins regulate the CgSnf3-dependent low-glucose sensing pathway as well as the mitochondrial metabolism either directly or indirectly (Fig. 7). Furthermore, the depolarized mitochondria and the increased fermentative metabolism in the Cgyps1-11Δ mutant, which may adversely affect both cell growth and virulence, are likely to be the physiological manifestations of high and low abundance of glycolysis and mitochondrial transport chain membrane proteins, respectively, in the mutant (Fig. 7). Since the intracellular proliferation of C. glabrata in macrophages is probably dependent upon an ability to switch rapidly from fermentative to respiratory metabolism, and the converse, in response to the presence of varied available carbon sources, a defect in such reprogramming may be fatal for Cgyps1-11Δ cells. We speculate that, in addition to energy and building block generation, remodeling of the carbon metabolism in host phagocytic cells may aid C. glabrata survive other host-elicited antifungal responses including reactive oxygen species production, metal ion limitation, and inflammatory responses. Of note, NLRP3 inflammasome activation has recently been shown to be incited in response to C. albicans-mediated glucose starvation in macrophages (21).
Figure 7.
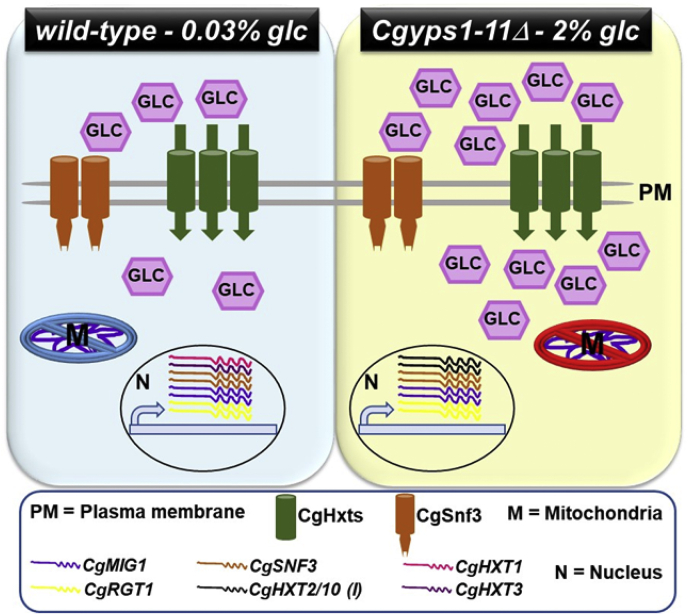
A schematic illustration of key findings of the study. The loss of CgYapsins impairs the ability of C. glabrata cells to sense the external glucose concentration. The Cgyps1-11Δ mutant perceives the 2% glucose environment (YPD/YNB medium) as glucose-poor environment, which results in transcriptional activation of genes coding for CgSnf3 glucose sensor, CgMig1 and CgRgt1 transcription factors, and CgHxt2/10 (I) (CAGL0I00286p) hexose transporter, which possibly leads to higher glucose uptake and perturbed glucose homeostasis. Contrarily, wildtype cells respond to a low-glucose environment (0.03% glucose) by elevating the expression of CgSNF3, CgMIG1, CgRGT1, and CgHXT1 and CgHXT3 (code for hexose transporters) genes, which probably facilitates glucose import, and glucose homeostasis is maintained. Of note, CgHXT2/10 (I) is transcriptionally repressed in glucose-starved wildtype cells, whereas CgHXT1 and CgHXT3 genes are transcriptionally repressed in 2% glucose–grown Cgyps1-11Δ cells. In addition, proteins belonging to glycolysis and oxidative phosphorylation are over- and underrepresented, respectively, in total membrane proteome of the Cgyps1-11Δ mutant, as compared with the wt cells, which may contribute partly to depolarized mitochondria and elevated ethanol production in the mutant. Altogether, these data underscore a critical requirement for CgYapsins in glucose metabolism in C. glabrata.
Altogether, our study provides an in-depth analysis of the membrane proteome of C. glabrata for the first time and uncovers a pivotal role for CgYapsins in glucose homeostasis. Our data suggest that CgYapsins could be a key determinant of energy (fermentative or respiratory) metabolism owing to their actions in glucose sensing mechanisms and regulating levels of mitochondrial membrane proteins.
Experimental procedures
Strains and growth analysis
C. glabrata strains used in the study (Table S20) were derived from the C. glabrata vaginal isolate BG2 and routinely cultured in YPD medium at 30 °C. The YPD medium contained 1% yeast extract, 2% peptone, and 2% dextrose, whereas the YNB medium contained 0.67% yeast nitrogen base and 2% dextrose. The medium containing varied glucose concentration (0.01%–5%) was prepared by adding the requisite amount of dextrose to yeast nitrogen base with ammonium sulfate for YNB medium, before autoclaving the medium. C. glabrata deletion strains, Cgsnf3Δ and Cgsnf3Δyps1-11Δ, were generated using the homologous recombination–based strategy with nourseothricin resistance–conferring gene nat1 or hygromcyin resistance–conferring gene hph1 as the selection marker (27). For generation of the Cgsnf3Δyps1-11Δ strain that lacks 12 genes, CgSNF3 and CgYPS1–11, the Cgyps1Δ mutant (CgYPS1 gene replaced hph1 gene) was used as the parent strain and CgSNF3 and CgYPS2–11 genes were sequentially deleted from its genome. First, the fragment containing CgSNF3-5′UTR-nat1-CgSNF3-3′UTR was amplified from the genomic DNA of Cgsnf3Δ mutant using the Phusion High-Fidelity DNA Polymerase and transformed into the Cgyps1Δ strain. Transformants were selected for nourseothricin resistance, and CgSNF3 gene deletion was confirmed by PCR. The Cgyps1Δsnf3Δ strain was made hygromycin sensitive by flipping out the hygromycin cassette using Flp recombinase, followed by replacement of the CgYPS7 gene with the hph1 gene in its genome. This step was repeated two more times, first to delete the CgYPS2 gene, followed by replacement of the CgYPS-Cluster (CgYPS3–6, 7–11) genes with the hph1 gene (5).
For CgSNF3 gene cloning, the full-length CgSNF3 ORF (2.57 kb) was amplified using the Phusion High-Fidelity DNA Polymerase, digested with restriction enzymes EcoRI and SalI, and cloned in the pGRB2.2 plasmid under the PGK1 promoter. For mutating arginine 251 residue to lysine in CgSnf3, mutagenic primers were used to amplify the pRK1593 plasmid containing CgSNF3, followed by DpnI digestion and self-ligation. The replacement of the arginine codon by lysine codon in the CgSNF3 gene was confirmed by sequencing. The oligos used in the current study are listed in Table S21.
For growth analysis in liquid medium, C. glabrata strains were grown in YPD medium for 12 to 16 h and inoculated in fresh YNB medium at an A600 of 0.1, followed by incubation at 30 °C with constant shaking. The culture aliquot was taken out at regular time intervals and absorbance at 600 nm was recorded. The A600 values were plotted against time, and the doubling time of each strain during logarithmic (log) phase was calculated from the slope of the curve.
Total membrane fraction isolation
For identification of the membrane proteome, the total membrane fraction was isolated from two biological replicate samples, as described (53). Briefly, wt and Cgyps1-11Δ strains were grown overnight in the YPD medium and inoculated at an initial cell density corresponding to an A600 of 0.1 into the fresh YPD medium. After 4 h growth at 30 °C, cells were harvested, washed twice using ice-cold water, normalized to 20 A600, and suspended in 0.1 M Tris (pH 10.7), 5 mM EDTA, 2 mM dithiothreitol (DTT), and 1× protease inhibitor cocktail solution. Cells were lysed using 0.5-mm acid-washed glass beads, and cell homogenates were diluted in buffer containing 0.1 M Tris-HCl (pH 8.0), 0.33 M sucrose, 5 mM EDTA, and 2 mM DTT, followed by centrifugation at 1000g for 3 min at 4 °C. The supernatant was centrifuged again at 3000g for 5 min at 4 °C to remove unbroken cells and cell debris, if any. Next, the supernatant was centrifuged at 19,000g for 45 min at 4 °C to obtain the total membrane fraction pellets. The pellets were washed with the wash buffer (0.1 M Tris-HCl [pH 8.0], 0.33 M sucrose, 5 mM EDTA, and 2 mM DTT) and suspended in membrane suspension buffer containing 20% glycerol (v/v), 0.1 mM EDTA, 0.1 mM DTT, and 10 mM Tris-HCl (pH 7.5), followed by protein quantification using the BCA protein assay kit.
Mass spectrometry analysis
For global proteome analysis of total membrane fractions, the membrane fractions (200 μg) were run on a 10% SDS-PAGE, till the bromophenol blue dye front reached 1 cm into the resolving gel. The gel was stained with Coomassie Brilliant Blue, and gel pieces containing proteins were cut and sent to Taplin Biological Mass Spectrometry Facility, Harvard Medical School, Boston, USA, for protein identification using LC-MS/MS (liquid chromatography-mass spectrometry) analysis. At the Taplin facility, proteins were digested with trypsin using in-gel digestion at 37 °C and analyzed using the Orbitrap mass spectrometer. The acquired peptide fragmentation data were analyzed using the Sequest software, run against the UniProt C. glabrata reference database, and identified peptides were filtered to 1% false discovery rate, as described (26).
The label-free quantitative mass spectrometry analysis of membrane proteomes was conducted at Valerian Chem Pvt Ltd, New Delhi, India. Total membrane fractions of wt and Cgyps1-11Δ strains were prepared as mentioned above, with one change in the membrane pellet suspension buffer, which contained 6 M Gn-HCL and 0.1 M Tris (pH 8.8). These samples, in duplicates, were sent on dry ice to Valerian Chem Pvt Ltd for label-free relative protein quantification, following LC-MS, using the Minora Feature Detector Node of the Proteome Discoverer 2.2 with default settings. At Valerian Chem, protein samples were reduced with TCEP solution, followed by trypsin digestion and resolution of the peptide mixture (1.0 μg) on a 25-cm-long PicoFrit column (360 μm outer diameter, 75 μm inner diameter, 10 μm tip) containing 1.8 μm of C18 resin, as described (26). The peptides were eluted with a 0% to 40% gradient of buffer containing 95% acetonitrile and 0.1% formic acid at a flow rate of 300 nl/min for 100 min. The sample analysis was carried out using the EASY-nLC 1000 system (Thermo Scientific) coupled to the QExactive mass spectrometer (Thermo Scientific) equipped with nanoelectrospray ion source, and the data-dependent top10 method was used to acquire MS data. The raw data were analyzed using the Uniprot C. glabrata reference database, with precursor and fragment mass tolerances set at 10 ppm and 0.5 Da, respectively, for Sequest search. The peptide spectrum match and the protein false discovery rate were set to 0.01, and the peptide spectrum matches with high and medium confidence were selected. The log2 ratio of ≥0.6 (≥1.5-fold change) for protein abundance in the Cgyps1-11Δ mutant to that in the wt strain was used to identify proteins that showed increased or decreased abundance in the mutant.
Of note, two biological replicates per strain were used for mass spectrometry experiments; the experimental details of the mass spectrometry runs and analysis are provided in Supporting Experimental Procedures, and the mass spectrometry search parameters for global and quantitative membrane proteome analysis are listed in Tables S22 and S23, respectively.
Microscopy analysis
The MitoTracker Green dye was used to study the mitochondrial architecture as its localization to the mitochondria is independent of the mitochondrial membrane potential. YNB medium–grown log-phase cells (1.0 A600) of wt and Cgyps1-11Δ strains were collected and suspended in Hepes buffer (10 mM Hepes [pH 7.4] and 5% glucose) containing 100 nM MitoTracker Green. After 30 min incubation at room temperature in the dark, cells were washed with PBS, suspended in the Hepes buffer without MitoTracker Green, and visualized using Leica TCS SP8 confocal microscope (63/1.52NA objective). For mitochondrial morphology analysis, Z-stack images were acquired throughout the cell at 0.5-μM intervals. A set of 11 and 10 images for wt and Cgyps1-11Δ, respectively, of the entire Z-stack were projected into a single image of maximum intensity using the Leica LAS-X software.
For JC-1 staining, YPD medium–grown log-phase cells (1.0 A600) were stained with JC-1 dye (20 μM) for 10 min at 30 °C. After three washes with PBS, cells were suspended in PBS and transferred to a 96-well black opaque bottom plate. The fluorescence values of J-aggregates and monomers were recorded by excitation/emission at 550 nm/600 nm and 485 nm/535 nm, respectively. The fluorescence intensity values of unstained cells were subtracted from those of stained cells, and the ratio of red to green fluorescence was determined for each strain.
Quantitative reverse transcription PCR
For glucose-responsive gene expression analysis, C. glabrata strains were grown in YNB medium for 4 h to log phase and cells were collected. After washes with PBS, cells were inoculated in the fresh YNB medium containing 0.03%, 2%, or 5% dextrose at an A600 of 1.0. After 2 h growth at 30 °C in an incubator shaker, cells were harvested and washed with chilled diethyl dicarbonate (DEPC)-treated water. Total RNA was isolated using the acid phenol extraction method and digested with DNase I. The DNase I treatment was given to eliminate residual DNA, if any. The reverse transcriptase (SuperScript III First-Strand Synthesis System for RT-PCR) was used to synthesize cDNA from DNase I-digested RNA (500 ng), followed by quantitative PCR using the SYBER Green qPCR Master Mix with appropriate set of primers. CgACT1 gene expression was used to normalize the expression of glucose sensing and transport genes. Differences in gene expression were calculated by the comparative CT (2−ΔΔCT) method, using two types of comparison, either 2% dextrose–grown mutant versus 2% dextrose–grown wt cells or 2% dextrose–grown versus 0.03%/5% dextrose–grown cells of a C. glabrata strain. A fold change of ≥1.5 was employed as a cutoff criterion to identify differentially expressed genes in different strains or varied growth conditions.
BALB/c mice infection assay
For virulence analysis, 6- to 8-week-old female BALB/c mice were infected with C. glabrata strains. Briefly, overnight YPD medium–grown C. glabrata cultures were harvested, washed twice with PBS, and suspended in PBS at an A600 of 20. A volume of 100 μl cell suspension (4 × 107 cells) was injected into tail veins of the BALB/c mice, and mice were monitored for 7 days. At seventh day post infection, mice were euthanized by carbon dioxide inhalation and three organs, kidneys, liver, and spleen, were collected. Organs were homogenized in sterile PBS, and appropriate dilutions were plated on the penicillin- and streptomycin-containing YPD medium. Mice infection procedures were approved by the Institutional Animal Ethics Committee (EAF/RK/CDFD/22) and were conducted at the Experimental Animal Facility of Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India.
Glucose uptake assay
C. glabrata strains were grown overnight in YPD medium and A600 was recorded. Cultures were inoculated in the glucose starvation medium (no-glucose YNB medium supplemented with 5% glycerol and 2% ethanol) (54) at an initial A600 of 0.1 to 0.15. After incubating for about 48 h at 30 °C, and once the A600 reached ∼1.5, cells of each strain were harvested, washed with milliQ water, and suspended in 100 μl YNB medium (without dextrose) in two sets. 2-NBDG ([2-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose; 100 μM) was added to one set, and the other set was left as such to be used as the autofluorescence control. The cells were incubated at 30 °C for 1 h, washed twice with milliQ water, and suspended in milliQ water. Cell suspensions were transferred to a black 96-well plate, and fluorescence emission was recorded at 540 nm, under excitation at 465 nm. The relative glucose uptake was calculated by dividing the fluorescence intensity values of mutants/complemented strains by fluorescence intensity values of the wt strain (taken as 1.0).
Ethanol measurement
C. glabrata strains were inoculated in YPD medium at an A600 of 0.3 and grown for about 4 h, till the A600 reached between 2.2 and 2.8 for each strain. Cultures were spun down, and the supernatant (3 ml) was transferred to a 15-ml tube containing dibutyl phthalate (3 ml). After vigorous vortex mixing (five times, 30 s each), tubes were centrifuged at 5000 rpm for 5 min, and 2.5 ml of the lower phase was collected. After adding 2.5 ml of potassium dichromate (0.37 M; prepared in 5 M H2SO4), tubes were vortexed vigorously (three times, 30 s each), followed by centrifugation at 5000 rpm for 5 min and collection of the lower phase (200 μl) in a 96-well plate. The plate was incubated at 37 °C for 20 min, and the absorbance was recorded at 595 nm. The amount of ethanol in the culture medium was calculated from the standard curve, prepared by using a range (0.1%–10%) of ethanol concentrations in YPD medium.
Gene Ontology analysis
Accession numbers of proteins, identified by mass spectrometry analysis, were used to retrieve the corresponding C. glabrata systematic ORFs (CAGL IDs) from the UniProt Database (https://www.uniprot.org/). The CAGL IDs were used to retrieve the S. cerevisiae orthologs and C. glabrata gene description from the Candida Genome Database (http://www.candidagenome.org/) using the “Batch Download” option. The S. cerevisiae systematic ORFs and gene description were downloaded from the Yeastract repository. (http://www.yeastract.com/). The DeepLoc1.0 (http://www.cbs.dtu.dk/services/DeepLoc-1.0/index.php) server with default settings was used to determine the intracellular localization of identified proteins. Venn diagrams were prepared using the Lucid Chart (www.lucidchart.com) server. The membrane protein prediction and functional enrichment analysis was performed using the GO Slim Mapper of the Candida Genome Database (CGD) (http://www.candidagenome.org/cgi-bin/GO/goTermMapper), and FungiFun (https://elbe.hki-jena.de/fungifun/) and DAVID tools (https://david.ncifcrf.gov/tools.jsp), respectively.
Other procedures
The serial dilution spotting assay and human THP-1 macrophage infection assay was performed as described (18, 27).
Statistical analysis
The Student’s t test, one-way analysis of variance (ANOVA) with uncorrected Fisher’s least significant difference (LSD) test, one-way ANOVA with Tukey’s test, and Mann-Whitney U test were used for data analysis, using the GraphPad Prism software, and p value ≤0.05 was considered statistically significant.
Data availability
The global and quantitative membrane mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD029614 and PXD029629, respectively.
Supporting information
This article contains supporting information (5, 55, 56, 57, 58, 59, 60).
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
We are indebted to Sandip Patra and S. Surya Vamshi for their help in generating CgSNF3-deleted strains and serial dilution spotting analysis, respectively. We also thank personnel of the Sophisticated Equipment Facility of CDFD for their assistance with microscopy experiments. We are grateful to Ross Tomaino, Taplin MS facility, and Gagan Deep Jhingan, Valerian Chem Pvt. Ltd for their assistance with mass spectrometry analysis.
Author contributions
M. R. conceptualization; M. R. and F. A. methodology; M. R. and F. A. validation; M. R., F. A., and R. K. formal analysis; M. R. and F. A. investigation; R. K. resources; M. R., F. A., and R. K. data curation; M. R., F. A., and R. K. writing – original draft; R. K. writing – review & editing; M. R., F. A., and R. K. visualization; R. K. supervision; R. K. funding acquisition.
Funding and additional information
This work was fully supported by the DBT/Wellcome Trust India Alliance Senior Fellowship to R. K. (IA/S/15/1/501831; www.indiaalliance.org/). F. A. is a recipient of Research Fellowship sponsored by the University Grants Commission, New Delhi, India.
Edited by Qi Qun Tang
Footnotes
Present address for Mubashshir Rasheed: MRC Centre for Medical Mycology, University of Exeter, Geoffrey Pope Building, Stocker Road, Exeter EX4 4QD, UK.
Supporting information
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Supplemental Figure S4.
Supplemental Figure S5.
References
- 1.Naglik J.R., Challacombe S.J., Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yike I. Fungal proteases and their pathophysiological effects. Mycopathologia. 2011;171:299–323. doi: 10.1007/s11046-010-9386-2. [DOI] [PubMed] [Google Scholar]
- 3.Rapala-Kozik M., Bochenska O., Zajac D., Karkowska-Kuleta J., Gogol M., Zawrotniak M., Kozik A. Extracellular proteinases of Candida species pathogenic yeasts. Mol. Oral Microbiol. 2018;33:113–124. doi: 10.1111/omi.12206. [DOI] [PubMed] [Google Scholar]
- 4.Gagnon-Arsenault I., Tremblay J., Bourbonnais Y. Fungal yapsins and cell wall: A unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 2006;6:966–978. doi: 10.1111/j.1567-1364.2006.00129.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaur R., Ma B., Cormack B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar K., Askari F., Sahu M.S., Kaur R. Candida glabrata: A lot more than meets the eye. Microorganisms. 2019;7:39. doi: 10.3390/microorganisms7020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarty T.P., Pappas P.G. Invasive candidiasis. Infect. Dis. Clin. North Am. 2016;30:103–124. doi: 10.1016/j.idc.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi (Basel) 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaller M.A., Diekema D.J., Turnidge J.D., Castanheira M., Jones R.N. Twenty years of the SENTRY antifungal surveillance program: Results for Candida species from 1997-2016. Open Forum Infect. Dis. 2019;6:S79–S94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti A., Sood P., Rudramurthy S.M., Chen S., Kaur H., Capoor M., Chhina D., Rao R., Eshwara V.K., Xess I., Kindo A.J., Umabala P., Savio J., Patel A., Ray U., et al. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015;41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 11.Lamoth F., Lockhart S.R., Berkow E.L., Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018;73:i4–i13. doi: 10.1093/jac/dkx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Astvad K.M.T., Johansen H.K., Røder B.L., Rosenvinge F.S., Knudsen J.D., Lemming L., Schønheyder H.C., Hare R.K., Kristensen L., Nielsen L., Gertsen J.B., Dzajic E., Pedersen M., Østergård C., Olesen B., et al. Update from a 12-year nationwide fungemia surveillance: Increasing intrinsic and acquired resistance causes concern. J. Clin. Microbiol. 2017;56 doi: 10.1128/JCM.01564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray D., Goswami R., Banerjee U., Dadhwal V., Goswami D., Mandal P., Sreenivas V., Kochupillai N. Prevalence of Candida glabrata and its response to boric acid vaginal suppositories in comparison with oral fluconazole in patients with diabetes and vulvovaginal candidiasis. Diabetes Care. 2007;30:312–317. doi: 10.2337/dc06-1469. [DOI] [PubMed] [Google Scholar]
- 14.Khatib R., Johnson L.B., Fakih M.G., Riederer K., Briski L. Current trends in candidemia and species distribution among adults: Candida glabrata surpasses C. albicans in diabetic patients and abdominal sources. Mycoses. 2016;59:781–786. doi: 10.1111/myc.12531. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues C.F., Rodrigues M.E., Henriques M. Candida sp. infections in patients with Diabetes mellitus. J. Clin. Med. 2019;8:76. doi: 10.3390/jcm8010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roetzer A., Gratz N., Kovarik P., Schüller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell. Microbiol. 2010;12:199–216. doi: 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seider K., Brunke S., Schild L., Jablonowski N., Wilson D., Majer O., Barz D., Haas A., Kuchler K., Schaller M., Hube B. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J. Immunol. 2011;187:3072–3086. doi: 10.4049/jimmunol.1003730. [DOI] [PubMed] [Google Scholar]
- 18.Rai M.N., Balusu S., Gorityala N., Dandu L., Kaur R. Functional genomic analysis of Candida glabrata-macrophage interaction: Role of chromatin remodeling in virulence. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasheed M., Battu A., Kaur R. Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response. J. Biol. Chem. 2018;293:6410–6433. doi: 10.1074/jbc.M117.813741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucey T.M., Verma J., Harrison P.F., Snelgrove S.L., Lo T.L., Scherer A.K., Barugahare A.A., Powell D.R., Wheeler R.T., Hickey M.J., Beilharz T.H., Naderer T., Traven A. Glucose homeostasis is important for immune cell viability during Candida challenge and host survival of systemic fungal infection. Cell Metab. 2018;27:988–1006.e7. doi: 10.1016/j.cmet.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucey T.M., Verma J., Olivier F.A.B., Lo T.L., Robertson A.A.B., Naderer T., Traven A. Metabolic competition between host and pathogen dictates inflammasome responses to fungal infection. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Ende M., Wijnants S., Van Dijck P. Sugar sensing and signaling in Candida albicans and Candida glabrata. Front. Microbiol. 2019;10:99. doi: 10.3389/fmicb.2019.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bairwa G., Kaur R. A novel role for a glycosylphosphatidylinositol-anchored aspartyl protease, CgYps1, in the regulation of pH homeostasis in Candida glabrata. Mol. Microbiol. 2011;79:900–913. doi: 10.1111/j.1365-2958.2010.07496.x. [DOI] [PubMed] [Google Scholar]
- 24.Bairwa G., Rasheed M., Taigwal R., Sahoo R., Kaur R. GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem. J. 2014;458:323–334. doi: 10.1042/BJ20130757. [DOI] [PubMed] [Google Scholar]
- 25.Quintin J., Asmar J., Matskevich A.A., Lafarge M.-C., Ferrandon D. The Drosophila Toll pathway controls but does not clear Candida glabrata infections. J. Immunol. 2013;190:2818–2827. doi: 10.4049/jimmunol.1201861. [DOI] [PubMed] [Google Scholar]
- 26.Rasheed M., Kumar N., Kaur R. Global secretome characterization of the pathogenic yeast Candida glabrata. J. Proteome Res. 2020;19:49–63. doi: 10.1021/acs.jproteome.9b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battu A., Purushotham R., Dey P., Vamshi S.S., Kaur R. An aspartyl protease-mediated cleavage regulates structure and function of a flavodoxin-like protein and aids oxidative stress survival. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delic M., Valli M., Graf A.B., Pfeffer M., Mattanovich D., Gasser B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 2013;37:872–914. doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- 29.Pais P., Costa C., Pires C., Shimizu K., Chibana H., Teixeira M.C. Membrane proteome-wide response to the antifungal drug clotrimazole in Candida glabrata: Role of the transcription factor CgPdr1 and the Drug:H+ antiporters CgTpo1_1 and CgTpo1_2. Mol. Cell. Proteomics. 2016;15:57–72. doi: 10.1074/mcp.M114.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maréchal A., Meunier B., Lee D., Orengo C., Rich P.R. Yeast cytochrome c oxidase: A model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim. Biophys. Acta. 2012;1817:620–628. doi: 10.1016/j.bbabio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubinski A.F., Camasta R., Soule T.G.B., Reed B.H., Glerum D.M. Consequences of cytochrome c oxidase assembly defects for the yeast stationary phase. Biochim. Biophys. Acta Bioenerg. 2018;1859:445–458. doi: 10.1016/j.bbabio.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Boonekamp F.J., Dashko S., van den Broek M., Gehrmann T., Daran J.-M., Daran-Lapujade P. The genetic makeup and expression of the glycolytic and fermentative pathways are highly conserved within the Saccharomyces genus. Front. Genet. 2018;9:504. doi: 10.3389/fgene.2018.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng T.S., Mohd Desa M.N., Sandai D., Chong P.P., Than L.T.L. Phylogenetic and transcripts profiling of glucose sensing related genes in Candida glabrata. Jundishapur J. Microbiol. 2015;8 doi: 10.5812/jjm.25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng T.S., Chew S.Y., Rangasamy P., Mohd Desa M.N., Sandai D., Chong P.P., Than L.T.L. SNF3 as high affinity glucose sensor and its function in supporting the viability of Candida glabrata under glucose-limited environment. Front. Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palma M., Seret M.-L., Baret P.V. Combined phylogenetic and neighbourhood analysis of the hexose transporters and glucose sensors in yeasts. FEMS Yeast Res. 2009;9:526–534. doi: 10.1111/j.1567-1364.2009.00511.x. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki T., Izumikawa K., Yamauchi S., Inamine T., Nagayoshi Y., Saijo T., Seki M., Kakeya H., Yamamoto Y., Yanagihara K., Miyazaki Y., Yasuoka A., Kohno S. The glycosylphosphatidylinositol-linked aspartyl protease Yps1 is transcriptionally regulated by the calcineurin-Crz1 and Slt2 MAPK pathways in Candida glabrata. FEMS Yeast Res. 2011;11:449–456. doi: 10.1111/j.1567-1364.2011.00734.x. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda Y., Tsai H.-F., Myers T.G., Bennett J.E. Transcriptional profiling of Candida glabrata during phagocytosis by neutrophils and in the infected mouse spleen. Infect. Immun. 2013;81:1325–1333. doi: 10.1128/IAI.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ene I.V., Brunke S., Brown A.J.P., Hube B. Metabolism in fungal pathogenesis. Cold Spring Harb. Perspect. Med. 2014;4:1–22. doi: 10.1101/cshperspect.a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.d’Enfert C., Janbon G. Biofilm formation in Candida glabrata: What have we learnt from functional genomics approaches? FEMS Yeast Res. 2016;16 doi: 10.1093/femsyr/fov111. [DOI] [PubMed] [Google Scholar]
- 40.Ng T.S., Desa M.N.M., Sandai D., Chong P.P., Than L.T.L. Growth, biofilm formation, antifungal susceptibility and oxidative stress resistance of Candida glabrata are affected by different glucose concentrations. Infect. Genet. Evol. 2016;40:331–338. doi: 10.1016/j.meegid.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Ehrström S., Yu A., Rylander E. Glucose in vaginal secretions before and after oral glucose tolerance testing in women with and without recurrent vulvovaginal candidiasis. Obstet. Gynecol. 2006;108:1432–1437. doi: 10.1097/01.AOG.0000246800.38892.fc. [DOI] [PubMed] [Google Scholar]
- 42.Johnston L., Wang G., Hu K., Qian C., Liu G. Advances in biosensors for continuous glucose monitoring towards wearables. Front. Bioeng. Biotechnol. 2021;9:733810. doi: 10.3389/fbioe.2021.733810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolland F., Winderickx J., Thevelein J.M. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 2001;26:310–317. doi: 10.1016/s0968-0004(01)01805-9. [DOI] [PubMed] [Google Scholar]
- 44.Kim J.-H., Roy A., Jouandot D., Cho K.H. The glucose signaling network in yeast. Biochim. Biophys. Acta. 2013;1830:5204–5210. doi: 10.1016/j.bbagen.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivandzade F., Bhalerao A., Cucullo L. Analysis of the mitochondrial membrane potential using the cationic JC-1 dye as a sensitive fluorescent probe. Bio Protoc. 2019;9 doi: 10.21769/BioProtoc.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerulus B., Jariani A., Perez-Samper G., Vermeersch L., Pietsch J.M., Crane M.M., New A.M., Gallone B., Roncoroni M., Dzialo M.C., Govers S.K., Hendrickx J.O., Galle E., Coomans M., Berden P., et al. Transition between fermentation and respiration determines history-dependent behavior in fluctuating carbon sources. Elife. 2018;7 doi: 10.7554/eLife.39234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otterstedt K., Larsson C., Bill R.M., Ståhlberg A., Boles E., Hohmann S., Gustafsson L. Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 2004;5:532–537. doi: 10.1038/sj.embor.7400132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Busti S., Coccetti P., Alberghina L., Vanoni M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors (Basel) 2010;10:6195–6240. doi: 10.3390/s100606195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozcan S., Dover J., Rosenwald A.G., Wölfl S., Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozcan S., Leong T., Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traven A., Naderer T. Central metabolic interactions of immune cells and microbes: Prospects for defeating infections. EMBO Rep. 2019;20 doi: 10.15252/embr.201947995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albrecht A., Felk A., Pichova I., Naglik J.R., Schaller M., De Groot P., Maccallum D., Odds F.C., Schäfer W., Klis F., Monod M., Hube B. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 2006;281:688–694. doi: 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- 53.Fernandes A.R., Peixoto F.P., Sá-Correia I. Activation of the H+-ATPase in the plasma membrane of cells of Saccharomyces cerevisiae grown under mild copper stress. Arch. Microbiol. 1998;171:6–12. doi: 10.1007/s002030050671. [DOI] [PubMed] [Google Scholar]
- 54.Roy A., Dement A.D., Cho K.H., Kim J.-H. Assessing glucose uptake through the yeast hexose transporter 1 (Hxt1) PLoS One. 2015;10 doi: 10.1371/journal.pone.0121985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 56.Peng J., Gygi S.P. Proteomics: The move to mixtures. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 57.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 58.Cormack B.P., Falkow S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics. 1999;151:979–987. doi: 10.1093/genetics/151.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Las Peñas A., Pan S.J., Castaño I., Alder J., Cregg R., Cormack B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frieman M.B., McCaffery J.M., Cormack B.P. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 2002;46:479–492. doi: 10.1046/j.1365-2958.2002.03166.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The global and quantitative membrane mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD029614 and PXD029629, respectively.