Summary
Background
Colon cancer (CC) is the leading cause of tumour-related death worldwide. SnoRNA plays a critical role in the tumour microenvironment. The tumour microenvironment can be shaped by tumour-infiltrating immune cells, which control the destiny of immunotherapy efficacy. This study uniquely focused on snoRNAs derived from immune cells to identify new biomarkers for immune landscape.
Methods
A novel computational framework was initiated for identifying tumour immune infiltration-associated snoRNAs (TIIsno) signatures and developed a TIIsno score model from integrative snoRNA profiling analysis of 21 purified immune cell lines, 43 colon cancer cell lines, and three datasets (training, test, real-world validation set).
Findings
Our study found that a high TIIsno score was associated with poor CC prognosis. TIIsno scores were seen to be negatively correlated with (I) the infiltration level of most immune cells, (II) the inhibitory immune checkpoints expression level, and (III) the immune score. These findings, taken together with the observation that TIIsno score is lower in MSI-H patients, suggests that patients with a low TIIsno score may have a better response to immunotherapy.
Interpretation
In conclusion, we successfully identified TIIsno and constructed a TIIsno score model, a new potential biomarker of immunotherapy response, which can effectively predict the prognosis of CC patients as well.
Funding
National Key R & D Program of China, National Natural Science Foundation of China, key projects from the Nature Science Foundation of Hunan Province, projects from Beijing CSCO Clinical Oncology Research Foundation, Fundamental Research Funds for the Central Universities of Central South University.
Keywords: SnoRNA, Immunotherapy, Colon cancer, Immune infiltration, Immune checkpoint
Research in context.
Evidence before this study
Colon cancer (CC) is the leading cause of tumour-related death among people. MSI-H/dMMR are the major predictors of immune checkpoint inhibitor therapy efficacy. However, in the 10-15% of colorectal cancer (CRC) patients who carry MSI-H/dMMR, only 30-50% of can benefit from immunotherapy. Apart from MSI-H/dMMR, no other coding genes have been identified as biomarkers for use in CRC immunotherapy. SnoRNA plays a critical role in the tumour microenvironment. At the same time, the tumour microenvironment can be shaped by tumour-infiltrating immune cells, which control the destiny of immunotherapy efficacy.
Added value of this study
We developed a novel computational framework for (I) the identification of tumour immune infiltration-associated snoRNAs (TIIsno) signature, and (II) the development of a TIIsno score model. Our study found that a high TIIsno score was associated with poor CC prognosis. TIIsno scores were seen to be negatively correlated with (I) the infiltration level of most immune cells, (II) the inhibitory immune checkpoints expression level, and (III) the immune score. In addition, the TIIsno score of MSI-H patients is lower than that in MSS/MSI-L, suggesting that patients with a low TIIsno score may have a better response to immunotherapy. Multiple immune-related pathways were discovered to be down-regulated in patients with a high TIIsno score.
Implications of all available evidence
The TIIsno score is an independent prognostic factor for colon cancer patients. Furthermore, it can be a potential biomarker that assists with screening the dominant population of immunotherapy patients, but further verification by the immunotherapy cohort is required.
Alt-text: Unlabelled box
Introduction
Colon cancer (CC) is the leading cause of tumour-related death worldwide.1 For this reason, clinicians are calling for more effective treatments for colon cancer. Immune checkpoint inhibitors (ICIs) have been approved as the first line treatment for metastatic colorectal cancer (CRC) with the molecular type of high microsatellite instability (MSI-H) or mismatch repair deficiency (dMMR).2 MSI-H/dMMR are the major predictors of immune checkpoint inhibitor therapy efficacy. However, in the 10-15% of CRC patients who carry MSI-H/dMMR, only 30-50% of can benefit from immunotherapy.3, 4, 5, 6 Due to the extensive therapy limitations, there is an urgent need to identify new immunotherapy biomarkers in colon cancer patients, to reveal resistance mechanisms and to seek potential targets for enhancing immunotherapy efficacy.
Apart from MSI-H/dMMR, no coding genes have been identified that could be used for CRC immunotherapy, suggesting that it may be worthwhile to shift the focus to other genetic information, such as non-coding RNAs.7 The efficacy of immunotherapy for cancer is closely related to the tumour microenvironment.8 Small nucleolar RNA (snoRNA) is a kind of non-coding RNAs conserved in eukaryotes. Its main function is to modify small nuclear RNA (snRNA) and ribosomal RNA (rRNA), a small number of snoRNAs are also involved in the processing and maturation of rRNA.9 There are two main types of snoRNAs: box C/D snoRNAs and box H/ACA snoRNAs. The box C/D snoRNAs catalyze the 2′-O-ribose methylation of rRNA,10 and box H/ACA snoRNAs are involved in the pseudouridylation of rRNAs.11 In addition to the above two main snoRNAs, there is also a type of small Cajal body-specific RNAs (scaRNAs), which is located in the Cajal body and whose main function is to participate in the post-transcriptional modification of small nuclear RNA (snRNA).12 Although we have a clearer understanding of the core functions of snoRNA, recent studies have discovered diverse new functions of snoRNA, such as guiding rRNA acetylation, tRNA methylation, regulating mRNA abundance, regulating variable splicing, etc.13 SnoRNA is widely involved in regulating the biological processes of lung, prostate, liver, colorectal cancer, and many other tumours, by affecting tumour proliferation, invasion, and metastasis.14, 15, 16 In addition, sno-derived RNAs (sdRNAs) are prevalent molecular markers of cancer immunity,17 snoRNA‐derived nuclear RNA 3 (sdnRNA‐3) has been shown to regulate the function of tumour-associated macrophages,18 sdRNA derived from SNORD63 can regulate the mRNA stability of interleukin 4 (IL-4), thereby affecting the development of Th2 lymphocytes.19 Robert J. Motzer et al. divided 823 cases of renal cell carcinoma into 7 molecular subtypes, the subtype with high snoRNA expression showed the longest PFS when treated with atezolizumab + bevacizumab.20 Together, this information shows that, snoRNA plays a critical role in the tumour microenvironment. However, the roles of snoRNA in tumour-immune interactions of colon cancer remain largely unknown. Given the tumour microenvironment can be shaped by tumour-infiltrating immune cells, which control the destiny of immunotherapy efficacy, this study focused on the snoRNAs derived from immune cells to identify new biomarkers for immunotherapy.21
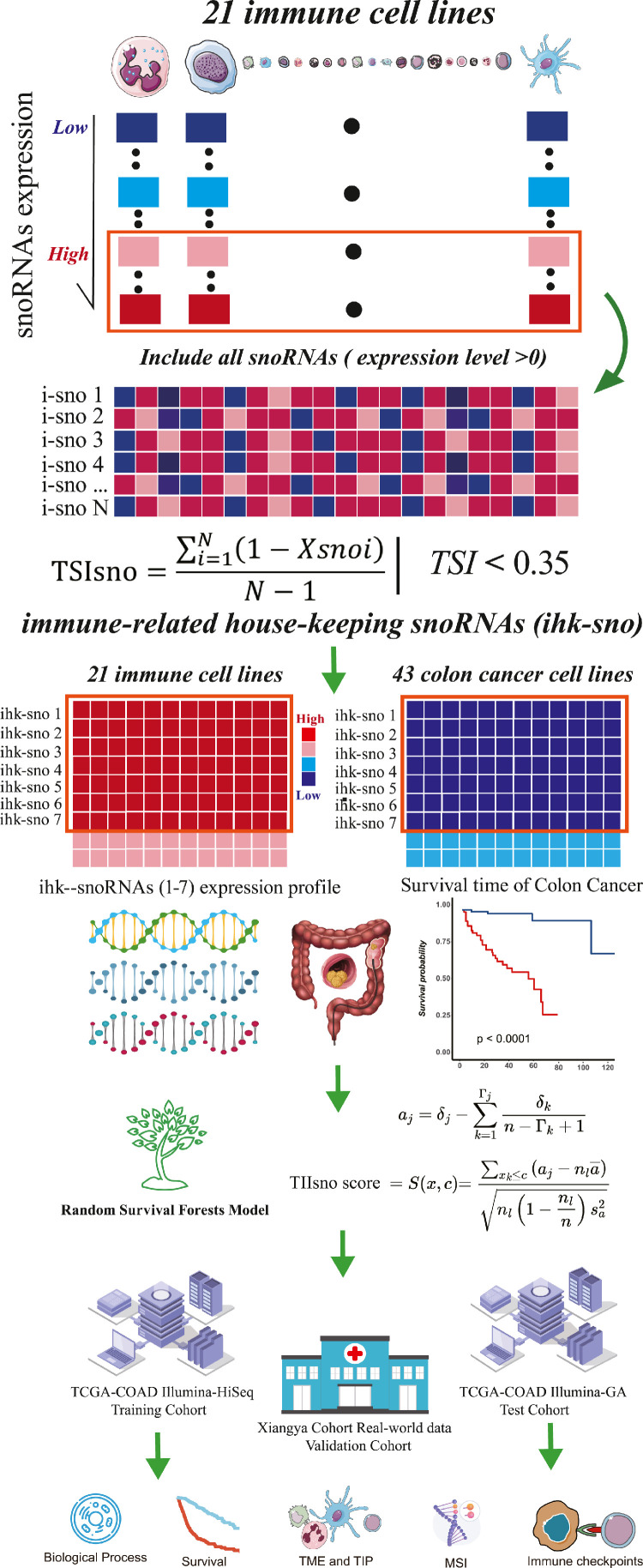
In this study, we initiated a novel computational framework for (I) the identification of tumour immune infiltration-associated snoRNAs (TIIsno) signature, and (II) the development of a TIIsno score model. The model was generated from integrative snoRNA profiling analysis of purified immune cell lines, colon cancer cell lines, and three datasets (training set, test set, real-world validation set) derived from bulk colon cancer tissues and survival data (Figure 1). First, for each immune cell line, we obtained the candidate immune-related snoRNAs base on the expression level. The immune-related house-keeping snoRNAs (ihk-sno) were then identified. Furthermore, The ihk-sno were selected as TIIsno according to their given expression levels in immune cells and colon cancer cells. A prognostic signature analysis was then performed and a TIIsno score model was developed. Lastly, the influence of TIIsno on CC prognosis and immunotherapy was comprehensively investigated. Our work explored the potential importance of TIIsno score as a new predictive biomarker for prognosis and immunotherapy response in colon cancer.
Figure 1.
The computational framework for identifying tumour immune infiltration-associated snoRNAs (TIIsno) signature and developing a TIIsno score model. (1) For each immune cell line, all the snoRNAs (expression level >0) were obtained as candidate immune-related snoRNAs. (2) Immune-related house-keeping snoRNAs (ihk-sno) were identified based on tissue specific index (TSI). (3) Those ihk-sno which are upregulated in immune cell lines and downregulated in colon cancer cell lines were selected as TIIsno. (4) A prognostic analysis by focusing on TIIsno and the overall survival time was performed via random survival forest. A TIIsno score model was developed according to the results of random survival forest. (5) The influence of the TIIsno on the prognosis and immunotherapy in colon cancer was comprehensively investigated.
Methods
Data Source
Gene-Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo): The RNA-Sequencing (RNA-Seq) data of human immune cell lines were downloaded from GEO database using GEOquery or manually. (GSE114765,22 GSE133145,23 GSE13563524 and GSE107011).25 The Cancer Cell Line Encyclopedia project (CCLE, https://portals.broadinstitute.org/ccle): The RNA-Seq data of colon cancer cell lines were downloaded from CCLE database based on histology subtype. The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/): The RNA-Seq data and the clinical information of colon cancer of TCGA were downloaded from the UCSC Xena data portal.26 (TCGA-COAD-IIIumina-HiSeq and TCGA-COAD-IIIumina-GA)
Processing of RNA-Seq Data
All counts or fragments per kilobase of transcript per million (FPKM) values were transformed to transcripts per kilobase million (TPM) values under GENCODE annotation (https://www.gencodegenes.org/) version 34 for further analysis. Samples with TPM data were directly used for further analysis.
Purified immune cell lines RNA-Seq data
All immune cell line samples, within any intervention, have been removed. xCell algorithm27 was performed to further confirm immune cell subtypes according to the maximum enrichment score. A total of 188 samples, from 21 subtypes and 9 main categories of immune cells, were enrolled in this study. Combat from sva package were used for removing batch effects cross immune cell samples.
Identification of tumour immune infiltration-associated snoRNAs (TIIsno) and development of TIIsno score model
We developed a novel computational framework for identifying TIIsno signature and developing TIIsno score model by integrative snoRNA profiling analysis of purified immune cell lines, colon cancer cell lines, bulk colon cancer tissues and survival data as follows (Figure 1):
-
(1)
For each immune cell line, all the 533 snoRNAs (expression level >0) were obtained as candidate immune-related snoRNAs.
- (2)
Where N was the total number of immune cell samples and is the expression profile component normalized by the maximal component value. The TSI ranges from 0 to 1. The snoRNA is classified as “general immune snoRNA” when the value is 0, while it is classified as “one immune cell-specific snoRNA” when the value is 1. 23 snoRNAs universally highly expressed in most of immune cell types were defined as “ihk-sno”.
-
(3)
7 ihk-sno that are upregulated in immune cell lines and downregulated in colon cancer cell lines were selected as TIIsno.
-
(4)
A prognostic analysis by focusing TIIsno and the overall survival time was performed via random survival forest.30 TIIsno score model was developed according to the results of random survival forest.
The log-rank score test for splitting survival trees is described in Hothorn and Lausen (2003).31 In this rule, assume the -variable has been ordered so that. Now, compute the “ranks” for each survival time where. Thus,
where . Note that is the index of the order for . The log-rank score test is defined as
where and are the sample mean and sample variance of, respectively. Log-rank score splitting defines the measure of node separation by. Maximizing this value over and yields the best split.
Evaluation of the immunological characteristics of the tumour immunology microenvironment in colon cancer
The marker genes of tumour-infiltrating immune cell types were obtained from a study by Charoentong et al,32 which analyzed 366 microarrays of immune cells collected from 37 studies. The 19 inhibitory immune checkpoints were obtained from studies by Auslander et al33 and Hu et al.34 The hallmarks of cancer analysis was performed by gene set variation analysis (GSVA) R package base on the “c2.cp.hallmark.v7.1.symbols” gene sets. Gene ontology (GO) enrichment was performed via single-sample gene-set enrichment analysis (ssGSEA) base on the “c5.all.v7.1.symbols”gene sets, which were download from MSigDB. The tumour gene mutation burden (TMB), neoantigens, MSI status, immune score, and immune subtype were obtained from the TCGA dataset or the studies base on TCGA dataset.
Xiangya real-world cohort patients and follow-up
Tissue samples of 72 patients diagnosed with colon cancer from October 2011 to November 2019 in Xiangya Hospital of Central South University were collected to establish a tissue bank. The demographic characteristics, cancer stages, and pathological reports were obtained from the electronic medical records system. A retrospective cohort was preformed such that as of October 1, 2020, a total of 72 patients were included. Survival analysis and multivariate Cox regression were conducted after follow-up.
Real-time quantitative PCR (qPCR) of TIIsno in Xiangya real-world cohort
The expression levels of TIIsno in Xiangya real-world cohort were measured by qPCR. Frozen tissue samples of 72 patients diagnosed with colon cancer from our tissue bank were preservation in liquid nitrogen. Total RNA was extracted from frozen tissues using TRIzol Reagent (Invitrogen,Carlsbad, CA). The reverse transcription and real-time quantitative PCR was performed using miDETECT A Track miRNA qRT-PCR Starter Kit (Ribobio, Guangzhou, China) according to the manufacturer's instructions. Real-time quantitative PCR was performed in triplicate on a QuantStudio™ 7 Real-Time PCR system (Applied Biosystems, Carlsbad, CA). All snoRNAs specific forward primers were designed and synthesized by Ribobio (Guangzhou, China). U6 was used as an internal control and reversed primer was applied in miDETECT A Track miRNA qRT-PCR Starter Kit. Relative expression levels of snoRNAs were calculated according the 2−△△CT method.
Immunohistochemistry staining
We collected the pathological tissues of 69 patients in Xiangya real-world cohort. Formalin-fixed, paraffin-embedded tissue array slides were used to detect Programmed death-ligand 1 (PD-L1; also called B7-H1 or CD274) and cytotoxic T-lymphocyte antigen-4 (CTLA4) protein expression. Briefly, after deparaffinization and rehydration, EDTA buffer (G1203; Servicebio, Wuhan, China) was used for heat-induced epitope retrieval. Endogenous peroxidase activity was inhibited for 25 minutes with 3% hydrogen peroxide (Annjet, Shandong, China). Nonspecific binding was blocked with 3% BSA (G5001; Servicebio) for 30 minutes at room temperature. The slides were then incubated overnight at 4°C with CD274 (PDL1) and CTLA4 rabbit polyclonal antibody at a dilution of 1:200 and 1:300, respectively (CD274: catalog no. ab213524, Abcam, USA; CTLA4: catalog no. ab237712, Abcam, USA). Next, the slides were incubated with Goat Anti-rabbit IgG/HRP antibody (G1215; Servicebio) for 50 minutes at 37°C. 3-3’-diaminobenzidine was used for coloration, and hematoxylin was used for counterstaining.
CD274 and CTLA4 staining were defined as positive when colon cells or immune cells showed cytomembrane staining. A staining score was defined by adding the staining intensity score and the positive staining percentage score. The staining intensity was categorized into 3 grades: score 1, yellow; score 2, light brown; score 3, brown. Positive staining percentage patterns were categorized into 4 groups: score 1, 0% to 25% staining of colon cells or immune cells; score 2, 25% to 50% staining of colon cells or immune cells; score 3, 50% to 75% staining of colon cells or immune cells; score 4, 75% to 100% staining of colon cells or immune cells. The percentage and intensity scores were added as the final results.
Statistical analysis
Correlations between variables were explored using Spearman coefficients. Continuous variables fitting a normal distribution between binary groups were compared using a t-test. Otherwise, the Kruskal-Wallis test were applied. “limma” package and empirical Bayesian approach were used for performing differential analysis on enrichment results or expression level. Survival analysis was based on the Kaplan-Meier method and log-rank test; univariate and multivariable Cox regression were used for calculating hazard ratios (HRs) and identifying independent prognostic factors. Receiver operating characteristic (ROC) curves were used to assess the specificity and sensitivity of the TIIsno score based on the pROC package. The cut off points of the TIIsno score was determined using the surv_cutpoint algorithm. P values or adjusted p values less than 0.05 were considered statistically significant. Data processing, analysis, and visualization were performed in R (3.6.3).
Ethics statement
This study was reviewed and approved by the Xiangya Hospital Medical Ethics Committee of Central South University (No.201905131), and got the consent from all participates.
Role of funding source
Funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Identification of tumour immune infiltration-associated snoRNAs
To maximize the obtained expression levels of immune cell-related snoRNA, we screened the immune cell data set in the GEO database for the past 10 years, and identified 4 data sets (GSE14765, GSE133145, GSE135635, GSE107011). These data sets contained a total of 188 RNA-Seq cell samples, including 9 major categories and 21 sub-classification immune cell types (Figure S1a, Figure 2a-b). Then, we sorted the expression level of all snoRNAs and snoRNAs with expression level greater than 0 as the candidate snoRNAs (Figure 1, Figure S1a-b). In order to identify snoRNAs most expressed by immune cells, known as housekeeping snoRNAs, we calculated the TSI score of the candidate snoRNAs. SnoRNAs with a lower TSI score are expressed in more immune cells, suggesting that they play a basic biological role in most immune cells. We used different TSI cut-off values to reduce dimensionality and screen out housekeeping snoRNAs. In addition, we collected RNA-Seq data from 43 types of colon cancer cell lines using the CCLE and ranked snoRNAs expression level in all tumours (Figure S1c). We performed the analysis in 0, 0.1, 0.15, 0.20, 0.25, 0.30 and 0.35. When it came to 0.35, the TIIsno score model has the most satisfactory AUC in OS and predicting capacity of immune microenvironment. After comparing the expression levels of housekeeping snoRNAs in immune cells and tumour cell lines, we identified 7 snoRNAs (SNORD59A, SNORD63B, SNORD100, SNORD99, SNORD63, SNORD12C, SNORD19) whose expression levels were significantly reduced in tumours and increased in immune cells. We defined these snoRNAs as tumour immune infiltration-associated snoRNAs (TIIsno) (Figure 1).
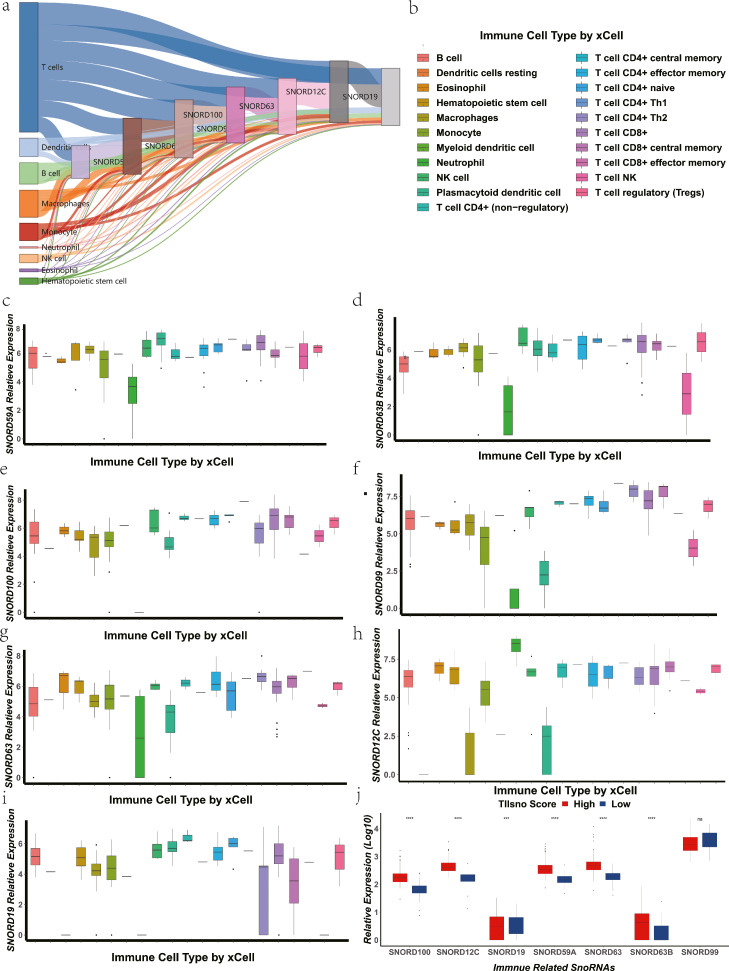
Figure 2.
The expression level of TIIsno in immune cells. (a) The origin and expression levels of TIIsno in 9 main categories of immune cells. (b) The 21 subtypes of immune cells in xCell algorithm. (c-i) The expression level of TIIsno in 21 subtypes of immune cells. (c) SNORD59A, (d) SNORD63B, (e) SNORD100, (f) SNORD99, (g) SNORD63, (h) SNORD12C, (i) SNORD19. (j) The expression level of TIIsno in different TIIsno score groups. (* p<0.05, ** p<0.01, *** p<0.001, N.S: no significant difference, Kruskal-Wallis test).
We have performed the analysis in the 43 types of colon cancer cell lines, and the results showed the expression levels of the 7 snoRNAs were pretty low, which is consistent with our previous analyses as expected. (Figure S4). To further confirm the expression level of TIIsno in immune cells, we analyzed the expression and origin of TIIsno. The results showed that each TIIsno was expressed in all 9 immune cell categories (Figure 2a). Most tumour immune infiltration-associated snoRNAs were highly expressed within the 21 immune cell subtypes simultaneously, suggesting that we have successfully screened out the housekeeping snoRNAs of immune cells (Figure 2b-i).
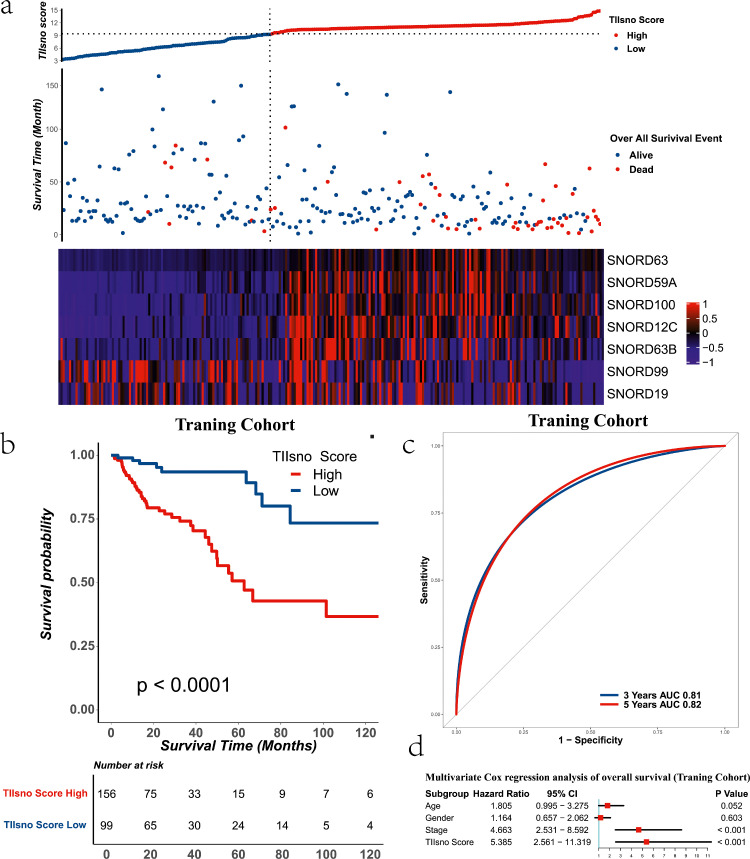
Development of TIIsno score model
To explore the role of tumour immune infiltration-associated snoRNAs in colon cancer, we obtained snoRNAs expression and survival data from colon cancer patients using TCGA, TCGA-COAD-IIIumina-HiSeq was selected as the training set and a TIIsno score model was constructed using a random survival forests (RSF) model (Figure 1). Meanwhile, the surv_cutpoint algorithm was used to obtain the best TIIsno score cut-off value, which was confirmed to be 9.392. Patients with a TIIsno score above the cut-off value were defined as the high group, while patients with a TIIsno score below the cut-off value were defined as the low group. At the same time, the expression levels of SNORD99 and SNORD19 are inversely proportional to the TIIsno score, while the expression levels of SNORD59A, SNORD63B, SNORD100, SNORD63 and SNORD12C are directly proportional to the TIIsno score (Figure 3a). We found that with the exception of SNORD99, expression levels of other TIIsno were significantly different between high TIIsno score group and low TIIsno score group (Figure 2j).
Figure 3.
The TIIsno score model and survival time of colon cancer in the training cohort. (a) The cut-off value of the TIIsno score according to the survival time (n=255). (b) Overall survival time in different TIIsno score groups (n=255, log-rank test). (c) AUC of 3-year and 5-year survival rate. (d) Multivariable cox regression analysis of overall survival (n=255).
In addition, the survival analysis of the TIIsno score model showed that the prognosis of the high TIIsno score group was poorer than that of the low TIIsno score group (Figure 3b, Table 1), with HR: 5.385 (2.561-11.319), P<0.001 of multivariate COX regression analysis (Figure 3d, Table 1). Furthermore, the predicted Area under the ROC curve (AUC) of the 3-years survival rate and 5-years survival rate were 0.81 and 0.82, respectively, indicating that our model has considerable predictive power (Figure 3c).
Table 1.
Cox regression analysis of overall survival in the different datasets.
| Variables | subgroup | Patients | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P Value | HR | 95% CI | P Value | |||
| TCGA-COAD Illumina-HiSeq | ||||||||
| Age | >65 | 160 | 1.262 | 0.727 - 2.19 | 0.409 | 1.805 | 0.995 - 3.275 | 0.052 |
| ≤65 | 95 | 1 | 1 | |||||
| Gender | Male | 136 | 1.574 | 0.905 - 2.738 | 0.108 | 1.164 | 0.657 - 2.062 | 0.603 |
| Female | 119 | 1 | 1 | |||||
| Stage | III-IV | 86 | 3.455 | 1.991 - 5.997 | < 0.001 | 4.663 | 2.531 - 8.592 | < 0.001 |
| I-II | 169 | 1 | 1 | |||||
| TIIsno Score | High | 156 | 4.416 | 2.142 - 9.103 | < 0.001 | 5.385 | 2.561 - 11.319 | < 0.001 |
| Low | 99 | 1 | 1 | |||||
| TCGA-COAD Illumina-GA | ||||||||
| Age | >65 | 80 | 1.919 | 0.655 – 5.620 | 0.235 | 1.926 | 0.637 – 5.822 | 0.246 |
| ≤65 | 38 | 1 | 1 | |||||
| Gender | Male | 61 | 0.589 | 0.257 - 1.349 | 0.211 | 0.729 | 0.308 - 1.724 | 0.472 |
| Female | 57 | 1 | 1 | |||||
| Stage | III-IV | 33 | 1.914 | 0.850 - 4.313 | 0.117 | 3.324 | 1.41 - 7.839 | 0.006 |
| I-II | 85 | 1 | 1 | |||||
| TIIsno Score | High | 40 | 11.597 | 3.932 - 34.202 | < 0.001 | 14.273 | 4.57 - 44.571 | < 0.001 |
| Low | 78 | 1 | 1 | |||||
| Xiangya cohort | ||||||||
| Age | >65 | 24 | 1.213 | 0.487-3.021 | 0.678 | 0.957 | 0.380-2.406 | 0.925 |
| ≤65 | 48 | 1 | 1 | |||||
| Gender | Male | 35 | 0.941 | 0.382-2.317 | 0.895 | 0.961 | 0.389-2.375 | 0.931 |
| Female | 37 | 1 | 1 | |||||
| Stage | III-IV | 31 | 4.843 | 1.833-12.800 | 0.001 | 3.496 | 1.301-9.391 | 0.013 |
| I-II | 41 | 1 | 1 | |||||
| TIIsno Score | High | 45 | 6.374 | 1.471-27.62 | 0.0133 | 5.223 | 1.176-23.202 | 0.03 |
| Low | 27 | 1 | 1 | |||||
Low TIIsno score shapes an inflamed microenvironment in colon cancer
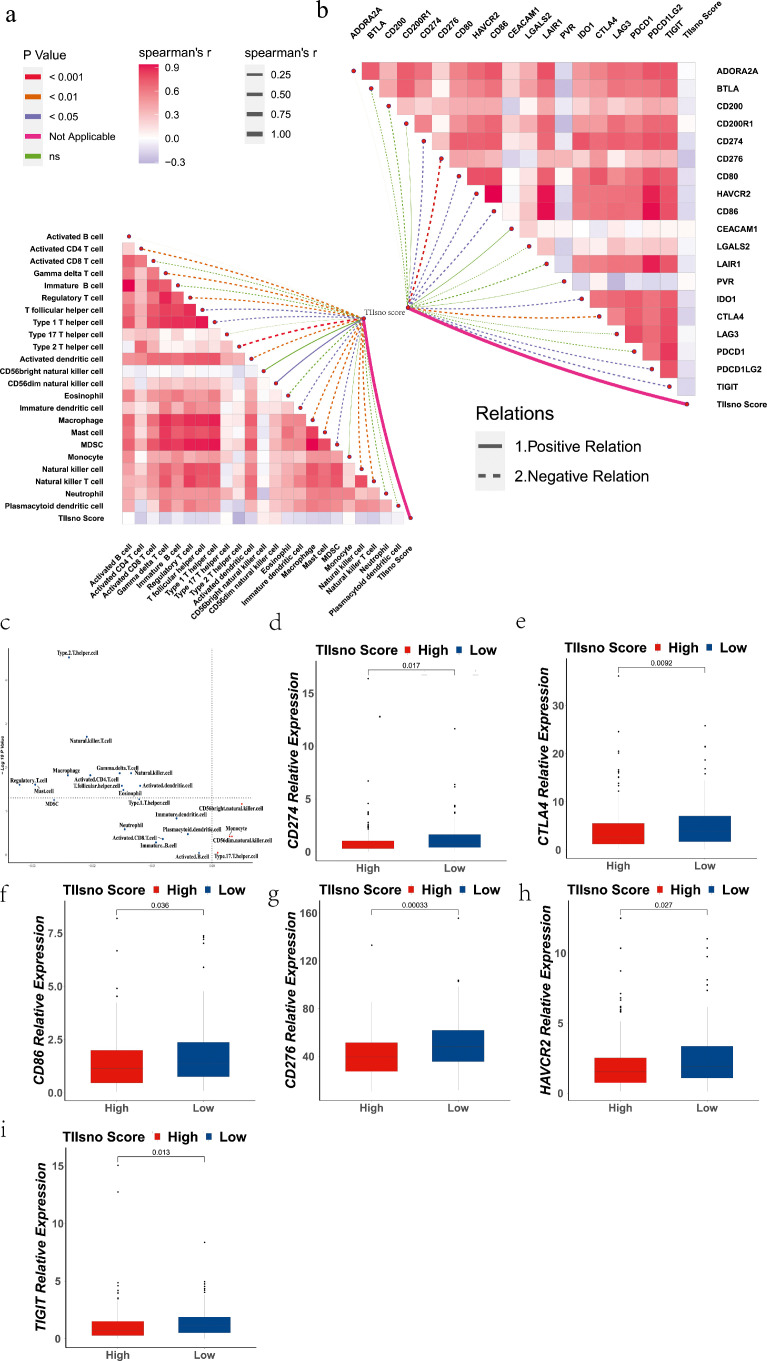
To evaluate the role of TIIsno in the immune microenvironment of colon cancer, we explored the relationship between TIIsno and (1) immune cell infiltration, and (2)immune checkpoints. We used ssGSEA to calculate the activation level of 23 types of tumour immune infiltrating cell-related gene sets according to Charoentong et al32 (Table S1). We then analyzed the correlation between the TIIsno score and the activation level of tumour immune infiltrating cells. This analysis showed that there were significant correlations between 14 types of tumour immune infiltrating cells and TIIsno score. All types of tumour immune infiltrating cells were negatively correlated, with the exception of the CD56dim natural killer cell, which was positively correlated. (Rank by the spearman's r: Type 2 T helper cell (Th2), Activated CD4+ T cell, Natural killer T cell (NKT), Gamma delta T cell, Mast cell, Macrophage, Activated dendritic cell (DC), Natural killer cell (NK), Regulatory T cell (Treg), T follicular helper cell (Tfh), Immature dendritic cell, Myeloid-derived suppressor cells (MDSC), Type 1 T helper cell (Th1)) (Figure 4a, Table S2). In addition, we also analyzed the correlation between the expression levels of 19 inhibitory immune checkpoint genes and the TIIsno score. The 19 inhibitory immune checkpoints were obtained from studies by Auslander et al24 and Hu et al.25 The results showed that there were correlations between the expression level of 9 immune checkpoints and TIIsno score. The 9 checkpoints were all negatively correlated. (Rank by the spearman's r: CD276, CTLA4, TIGIT, CD86, HAVCR2, CD274, IDO1, PDCD1LG2, CD80) (Figure 4b, Table S2).
Figure 4.
The correlation of immune infiltration level, inhibitory immune checkpoint, and TIIsno score. (a) The correlation of immune infiltration level and TIIsno score. (b) The correlation of inhibitory immune checkpoint and TIIsno score. (c) The differences of tumour immune infiltrating cells between the two TIIsno score groups. (d-i) The differences of inhibitory immune checkpoints between the two TIIsno score groups. (d) CD274, (e) CTLA4, (f) CD86, (g) CD276, (h) HAVCR2, (i) TIGIT (n=255, Kruskal-Wallis test).
To further compare the differences of tumour immune infiltrating cells between high TIIsno score group and low TIIsno score group, we used the normalized enrichment scores algorithm. The results showed that the activation level of 11 immune cells in the high TIIsno score group significantly decreased (Rank by the LogFC: Regulatory T cell, Mast cell, Macrophage, Type.2 T helper cell, Natural killer T cell, Activated CD4+ T cell, Gamma delta T cell, T follicular helper cell, Eosinophil, Natural killer cell, Activated dendritic cell) (Figure 4c). These immune cell infiltrating levels are all negatively correlated with the TIIsno score in the above analysis, suggesting that the results of our two analyses are consistent. Based on the correlation analysis between the TIIsno score and immune checkpoints, we further analyzed the expression of immune checkpoints in the high TIIsno score group and the low TIIsno score group. However, the results showed different findings from the previously explained analysis, as there are only 6 immune checkpoints whose expression levels are significantly different between the high and low TIIsno score groups (CD274, CTLA4, CD86, CD276, HAVCR2, TIGIT) (Figure 4d-k, Figure S2).
TIIsno score is a potential biomarker for immunotherapy response
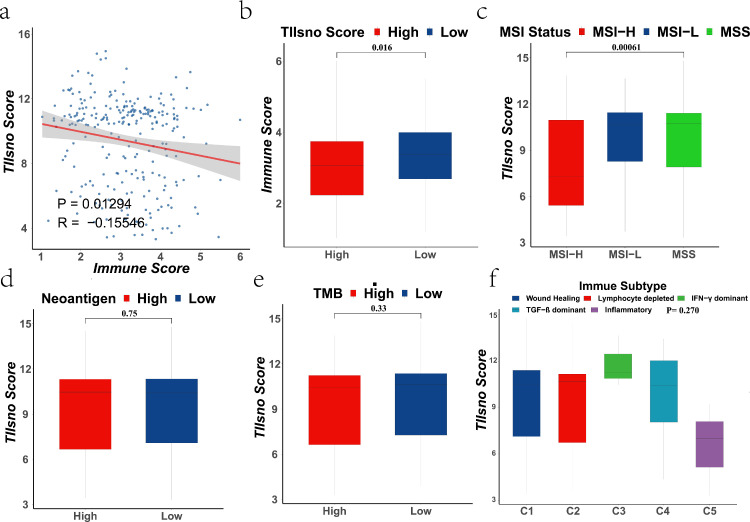
To further explore the relationship between TIIsno and immunotherapy, we analyzed the relationship between TIIsno score and (I) immune score, (II) MSI status, (III) neoantigen, (IV) TMB, and (V) immune subtype. We found that the TIIsno score was correlated with immune score and MSI status, while the neoantigen, TMB, and immune subtype were not significantly different between the high and low TIIsno score groups (Figure 5). TIIsno score is negatively correlated with immune score (R=-0.155, P=0.0129), and immune score is lower in the high TIIsno score group compared to low TIIsno score group (Figure 5a-b). In addition, the TIIsno score of MSI-H patients was lower than that of MSI-L and MSS patients (Figure 5c). Our analysis indicated that patients with a low TIIsno score may be more likely to benefit from immunotherapy. This is consistent previous results, where were we demonstrated, that patients with low TIIsno scores have an inflamed microenvironment in colon cancer (Figure 4). These findings suggest that, TIIsno score may be a potential biomarker for immunotherapy response.
Figure 5.
The correlation between the indicators of immunotherapy response and TIIsno score. (a-b) The correlation of immune score and TIIsno score (n=255, Kruskal-Wallis test). (c) The correlation of MSI status and TIIsno score (n=255, Kruskal-Wallis test). (d) The correlation of neoantigen and TIIsno score (n=255, Kruskal-Wallis test). (e) The correlation of TMB and TIIsno score (n=255, Kruskal-Wallis test). (f) The correlation of immune subtype and TIIsno score (n=255, Kruskal-Wallis test).
Possible mechanisms of TIIsno in colon cancer
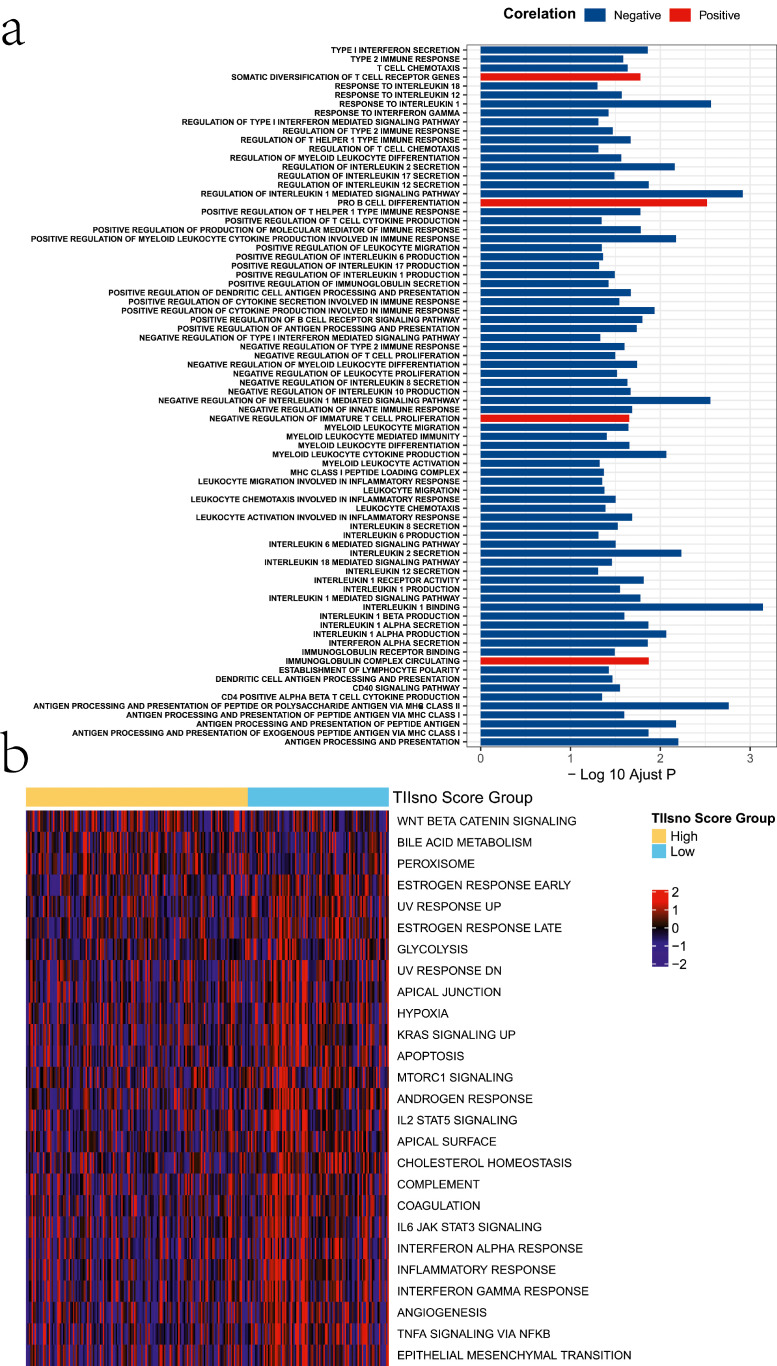
To explore the possible biological functions and mechanisms of TIIsno, we conducted a GO analysis. In the GO analysis, we first calculated the activation level of the GO function for each sample using ssGSEA, followed by a comparison between the high and low TIIsno score groups. We found differences among a total of 78 biological functions, most of which were immune-related. 75 biological functions were down-regulated in the high TIIsno score group while 3 biological functions were up-regulated. In the high TIIsno score group, negative_regulation_of_interleukin_1_mediated_signaling_pathway was downregulated to the greatest extent, while the somatic_diversification_of_T_cell_receptor_genes were upregulated to the greatest extent (Figure 6a, Table S3).
Figure 6.
Possible mechanisms of TIIsno in colon cancer. (a) The activation level of GO terms in the high TIIsno score group compared to the low TIIsno score group via ssGSEA. (b) The difference of hallmark pathways of cancer in the two TIIsno score groups.
To explore other roles of TIIsno in colon cancer, we selected 50 hallmark pathways of cancer to calculate the degree of pathway activation and enrich them using GSVA. The results showed that 26 hallmark pathways differed between the two TIIsno score groups. Only 3 hallmark pathways had greater activation in the high TIIsno score group, while the other 23 had a higher degree of activation degree in the low TIIsno score group (Figure 6b, Table S4).
The validation cohorts for TIIsno score model
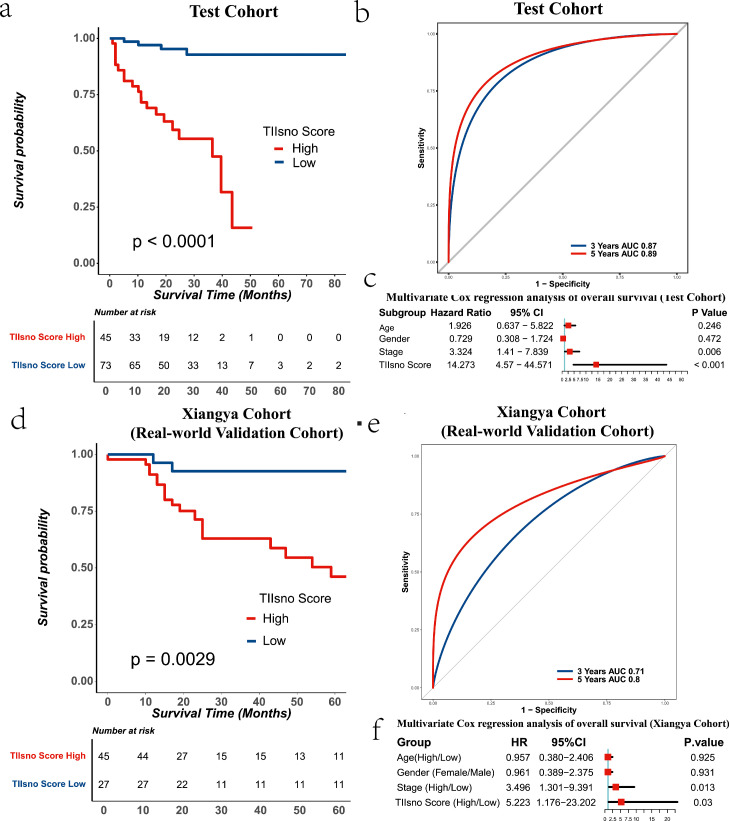
To verify the role of the TIIsno score model in colon cancer, we selected two other independent data sets for analysis (TCGA-COAD-IIIumina-GA cohort, Xiangya real-world cohort). The survival analysis based on the two data sets showed that the high TIIsno score group had a poorer prognosis than the low TIIsno score group, which was consistent with the results from the training set. At the same time, the TIIsno score model has demonstrated excellent predictive performance whether the data had been analyzed with log-rank or multivariate COX regression. Surprisingly, in the TCGA-COAD-IIIumina-GA cohort, the AUC of the 3-year survival rate and the 5-year survival rate are 0.87 and 0.89, respectively, values which are higher than that seen in our training set. In the Xiangya real-world cohort, the AUC of 3-year survival rate and 5-year survival rate are 0.71 and 0.80, respectively, findings that are approximately the same as those seen in the training set. Survival analysis verification testified that the TIIsno score model was a great independent prognostic factor in colon cancer (Figure 7, Table 1, the expression levels of TIIsno in Xiangya real-world cohort were showed in Figure S3.).
Figure 7.
The TIIsno score model and survival time of colon cancer in a test cohort and xiangya real-world validation cohort. (a) Overall survival time in different TIIsno score groups in a test cohort (n=118, log-rank test). (b) AUC of 3-year and 5-year survival rate in a test cohort. (c) Multivariable cox regression analysis of overall survival in test cohort (n=118). (d) Overall survival time in different TIIsno score groups in the xiangya real-world validation cohort (n=72, log-rank test). (e) AUC of 3-year and 5-year survival rate in the xiangya real-world validation cohort. (f) Multivariable cox regression analysis of overall survival in the xiangya real-world validation cohort (n=72).
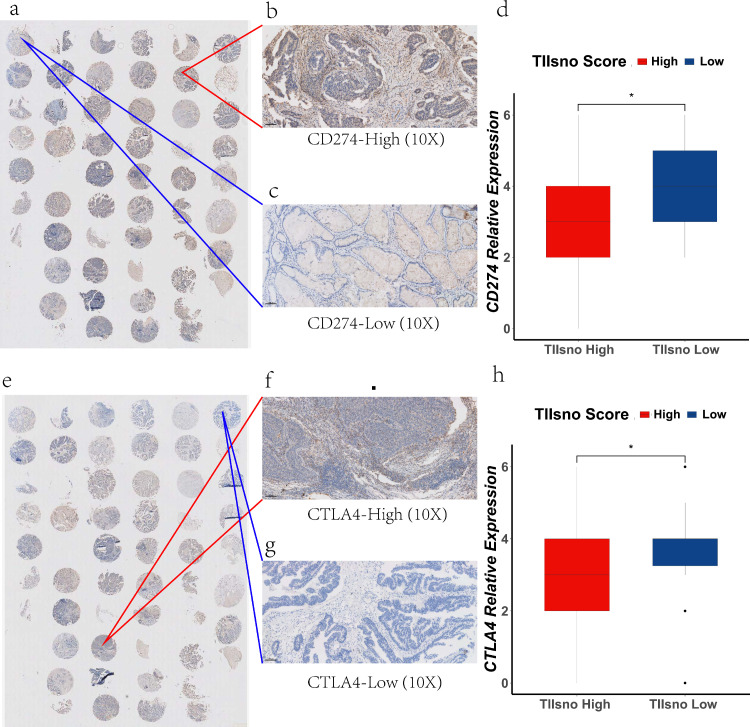
To evaluate the predictive value of the TIIsno score model on immunotherapy response, we selected CD274 and CTLA4, two immune checkpoints approved for clinical treatment, for further verification of the model. We collected pathological tissues from the 69 patients in Xiangya real-world cohort. Tissue was used to make tissue arrays and for immunohistochemical staining. The results show that both CD274 and CTLA4 were highly expressed in the low TIIsno score group compared with the high TIIsno score group, a finding consistent with the results of our bio-informatics analysis. Our research further verified that patients with a low TIIsno score could benefit from immunotherapy to a greater degree than those with a high TIIsno score. (Figure 8). As such, TIIsno score may be a potential biomarker for immunotherapy response.
Figure 8.
The expression level of CD274 and CTLA4 in the xiangya real-world validation cohort. (a-d) The expression level of CD274 in two TIIsno score groups. (e-h) The expression level of CTLA4 in two TIIsno score groups. Representative data of 3 independent experiments. Scale bar:100 μm. (n=69, * p<0.05, student's t test).
The host genes of TIIsno signature
To investigate if the host genes can recapitulate its snoRNAs, we have checked the host gene of our TIIsno signature. There are 6 host genes: ATP5F1B, RPS12, SNHG12, HSPA9, ZFAS1, GNL3 (Table S6). (SNORD 63 and SNORD 63B have the same host gene) We have analyzed the expression level of 6 host genes in the 43 types colon cancer cell lines, the results showed most of the 6 host genes have a high expression level, especially RPS12 and ATP5F1B (host gene of SNORD100 and SNORD59A), while all the TIIsno signature have a low expression level (Figure S5). At the same time, we conducted the analysis of host genes signature base on the RSF model. The survival analysis of the host genes score model showed that the prognosis of the high host gene score group was poorer than that of the low TIIsno score group in training cohort, however, there is no significant difference in test cohort. What's more, the AUC is lower than that in TIIsno signature (Figure S6a-d). In the analysis of the immune microenvironment, the host gene score has little positive connection with immune cells and immune checkpoints, while TIIsno score has negative connection with immune cells and immune checkpoints, which is distinct (Figure S6e-f). Our analyses indicated that the role of TIIsno signature and its host genes may be different in colon cancer.
Discussion
We successfully identified 7 TIIsno through this study: SNORD59A, SNORD63B, SNORD100, SNORD99, SNORD63, SNORD12C, SNORD19. We tried to use COX regression to develop the score model, but the AUC showed low predictive power, and we therefore used random survival forest model instead. (Table S5). Based on TIIsno expression level and survival data, the TIIsno score model established by RSF showed that patients with high TIIsno score had shorter survival time when compared with the low TIIsno patients. In addition, the model was demonstrated to have good predictive power. Meanwhile, TIIsno score is negatively correlated with the (I) infiltration level of most immune cells, (II) the inhibitory immune checkpoints expression level, and (III) the immune score. In addition, the TIIsno score of MSI-H patients is lower than that in MSS/MSI-L, suggesting that patients with a low TIIsno score may have a better response to immunotherapy. Multiple immune-related pathways were discovered to be down-regulated in patients with a high TIIsno score. Our research suggests that the TIIsno score is an independent prognostic factor for colon cancer patients. Furthermore, it can be a potential biomarker that assists with screening the dominant population of immunotherapy patients.
There are 3 snoRNAs that have been reported among our identified 7 TIIsnos. SNORD59A was a positive prognostic factor in bladder cancer.35 Shang et al36 found that SNORD63 was elevated in the plasma and tumour tissues of patients with clear cell renal cell carcinoma, such that SNORD63 can be used as a diagnostic marker. The expression of SNORD12C was significantly increased in colorectal cancer,37 and it played an important role in tumourigenesis, through the ZFAS1-NOP58-SNORD12C/78-EIF4A3/LAMC2 signaling axis.38 At this time, there are no detailed reports or related studies regarding the other 4 TIIsno.
The immune microenvironment is closely related to the prognosis of tumours and the efficacy of immunotherapy.21 A high level of immune cell infiltration is a positive prognostic factor for colorectal cancer, and patients with high levels of immune cell infiltration are also the dominant populations of immunotherapy.39 In our research, TIIsno score is negatively correlated with the infiltration level of most immune cells, which may be the reason for the poor prognosis of patients with a high TIIsno score. Activated CD4+ T cells can recruit cytotoxic T cells together with dendritic cells to enhance anti-tumour immunity in immunotherapy.40 NKT cells are also an anti-tumour immune cell, as higher NKT cell infiltration level is associated with better prognosis of cancer patients. At the same time, PD-1 inhibitors can decrease the emergence of type I NKT anergy, thereby enhancing the anti-tumour effect of NKT.41 PD-1 can inhibit the anti-tumour effect of gamma delta T cells, suggesting that gamma delta T cells have a positive effect on anti-PD1 therapy.42 NK cells are prone to failure under the regulation of immune checkpoints, however the use of immune checkpoint inhibitors can effectively restore activity of NK cells, enhancing the anti-tumour effect.43 In breast cancer, immune checkpoint blockade activates Tfh cells in the anti-tumour response.44 Modulation of tumour-infiltrating myeloid cells by IFN-γ-producing Th1 effector cells is partially responsible for the success of ICIs therapy.45 Given the level of infiltration of these immune cells in colon cancer is negatively correlated with TIIsno score, TIIsno score can be used as an immunotherapy biomarker. For this reason, patients with a low TIIsno score are more likely to benefit from immune checkpoint inhibitors therapy.
MSI-H is the only biomarker that can guide the immunotherapy of colorectal cancer. However, there are still a large number of patients with MSI-H that do not respond to immunotherapy.4 A possibility reason for this finding is that MSI-H cannot completely distinguish the anti-tumour immune microenvironment from the suppressed tumour immune microenvironment.46 Unfortunately, the TIIsno score cannot either. Infiltration levels of some immune cells involved in immunosuppression were also negatively correlated with TIIsno score, such as Th2, Treg, mast cell, and macrophage. However, survival analysis showed that the prognosis of the low TIIsno score group was much better than that of the high TIIsno score group, suggesting that anti-tumour immune cells play a major role in the low TIIsno score group. In addition, we found that colon cancer had a different immune environment from other tumours in our analysis, indicating that the immunotherapy strategy of colon cancer needs to be adjusted, such as the combination of multiple immune targets. In this way, it can not only enhance the effect of anti-tumour immune cells, but also regulate immunosuppressive cells to polarize towards the anti-tumour direction. MSI-H, an immunotherapy biomarker, does not integrate survival status. Although MSI-H can identify patients that have high levels of immune infiltration, which contains immunosuppressive cells, such that it may result in poor treatment effects in some patients.46 Unlike the MSI status, we combined survival data to construct the TIIsno score model, which can be used as an effective prognostic indicator. Furthermore, the selected patients with high levels of immune infiltration also have a good prognosis, suggesting that anti-tumour immunity plays a major role in these patients, and they are more likely to benefit from immunotherapy, further verification by the immunotherapy cohort is required.
In the low TIIsno score group, as the immune cell infiltration level increased, the expression level of immune checkpoints such as CD274 and CTLA4 also increased, a result verified by our real-world cohort. CD274 and CTLA4 are currently in clinical use.47 Increased expression of CD274 demonstrates a better therapeutic effect in the contemporary immune checkpoint inhibitor therapy,48 suggesting that TIIsno score can predict the response rate of immunotherapy. However, this finding is both an opportunity and a challenge. Given the expression of immune checkpoints is synergistic, the increase in the expression of other immune checkpoints also hinders immunotherapy. Our research once again suggests that immune checkpoint inhibitors should be applied to multiple targets in colon cancer.
However, our research also has limitations. It is better to prepare the small RNA library in the detection process of small RNAs. However, there are no enough specific snoRNA datasets to perform the analysis. Indeed, the biggest limitation of RNA-Seq is that it may not detect all snoRNAs, and the key problem is some small RNA may be washed out. However, RNA-Seq can identify some snoRNAs, which has been confirmed by many researchers. Meanwhile, we have done our best to reduce the bias caused by RNA-Seq by ruling out the datasets that washed the small RNA out. What's more, our analyses indicated that the role of TIIsno signature and its host genes may be different in colon cancer. To investigate the connections of host genes and the 7 snoRNAs, the molecular mechanism researches will be needed.
In conclusion, by integrating expression data from cell lines and tumour tissues, as well as clinical information from databases and a real-world cohort, we successfully identified TIIsno, and constructed a TIIsno score model that can effectively predict the prognosis of colon cancer patients. TIIsno score is a potential biomarker for immunotherapy response and may provide new clues for the diagnosis and treatment of colon cancer.
Contributors
C.C., Y.P., H.S. and S.Z. Y.H. designed the study. C.C., Y.P., E.S., R.W, Q.H, Y.C., P.L., C.G., Z.F., L.G., Y.L, Y.Z, and X.Z collected the data and performed the major analysis. S.Z. and H.S. supervised the study. C.C. and Y.P. analyzed and interpreted the data. E.S. and Z.F. did the statistical analysis. C.C., Y.P., C.G. and Y.L. drafted the manuscript. All authors read and approved the final manuscript.
Declaration of interests
None of the authors have any potential financial conflicts of interest related to this manuscript.
Acknowledgments
Acknowledgements
This study was supported by grants from the National Key R & D Program of China (No. 2018YFC1313300), National Natural Science Foundation of China (No.: 81070362, 81172470, 81372629, 81772627, 81874073 & 81974384), key projects from the Nature Science Foundation of Hunan Province (No. 2015JC3021 & 2016JC2037), the projects from Beijing CSCO Clinical Oncology Research Foundation (No. Y-HR2019-0182, Y-2019Genecast-043), and the Fundamental Research Funds for the Central Universities of Central South University (2020zzts273, 2019zzts797).
In addition, we want to show our appreciates to by Zirconicusso/Freepik for providing the materials for making Figure 1.
Data sharing statement
All the public datasets can be downloaded in the Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), the Cancer Cell Line Encyclopedia project (CCLE, https://portals.broadinstitute.org/ccle), and the Gene Expression Omnibus (GEO) (https://www.ncbi. nlm.nih.gov/geo/) database.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103866.
Contributor Information
Hong Shen, Email: hongshen2000@csu.edu.cn.
Shan Zeng, Email: zengshan2000@csu.edu.cn.
Ying Han, Email: yinghan@csu.edu.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1) doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Romero D. New first-line therapy for dMMR/MSI-H CRC. Nat Rev Clin Oncol. 2021;18(2):63. doi: 10.1038/s41571-020-00464-y. [DOI] [PubMed] [Google Scholar]
- 3.Asaoka Y., Ijichi H., Koike K. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;373(20):1979. doi: 10.1056/NEJMc1510353. [DOI] [PubMed] [Google Scholar]
- 4.Overman M.J., McDermott R., Leach J.L., et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overman M.J., Lonardi S., Wong K.Y.M., et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. official journal of the American Society of Clinical Oncology. [DOI] [PubMed] [Google Scholar]
- 6.Schrock A.B., Ouyang C., Sandhu J., et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30(7):1096–1103. doi: 10.1093/annonc/mdz134. [DOI] [PubMed] [Google Scholar]
- 7.Mannoor K., Shen J., Liao J., Liu Z., Jiang F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer. 2014;13:104. doi: 10.1186/1476-4598-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBerardinis R.J. Tumor microenvironment, metabolism, and immunotherapy. N Engl J Med. 2020;382(9):869–871. doi: 10.1056/NEJMcibr1914890. [DOI] [PubMed] [Google Scholar]
- 9.Dieci G., Preti M., Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94(2):83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henras A.K., Soudet J., Gérus M., et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65(15):2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henras A.K., Dez C., Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol. 2004;14(3):335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Bratkovič T., Božič J., Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020;48(4):1627–1651. doi: 10.1093/nar/gkz1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crea F., Quagliata L., Michael A., et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol Oncol. 2016;10(5):693–703. doi: 10.1016/j.molonc.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Zheng J., Chen P., Liu Q., Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;90:705–712. doi: 10.1016/j.biopha.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Fang X., Yang D., Luo H., et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9(3):243–255. doi: 10.1093/jmcb/mjw048. [DOI] [PubMed] [Google Scholar]
- 17.Chow R.D., Chen S. Sno-derived RNAs are prevalent molecular markers of cancer immunity. Oncogene. 2018;37(50):6442–6462. doi: 10.1038/s41388-018-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y., Shi Q., Shen Q., Zhang Q., Cao X. Dicer-independent snRNA/snoRNA-derived nuclear RNA 3 regulates tumor-associated macrophage function by epigenetically repressing inducible nitric oxide synthase transcription. Cancer Commun. 2021;41(2):140–153. doi: 10.1002/cac2.12131. (Lond) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong F., Zhou N., Wu K., et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43(21):10474–10491. doi: 10.1093/nar/gkv954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motzer R.J., Banchereau R., Hamidi H., et al. Molecular subsets in renal cancer determine outcome to checkpoint and angiogenesis blockade. Cancer Cell. 2020;38(6) doi: 10.1016/j.ccell.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X., Li J., Zou J., et al. Association of germline variants in natural killer cells with tumor immune microenvironment subtypes, tumor-infiltrating lymphocytes, immunotherapy response, clinical outcomes, and cancer risk. JAMA Netw Open. 2019;2(9) doi: 10.1001/jamanetworkopen.2019.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilipow K., Scamardella E., Puccio S., et al. Antioxidant metabolism regulates CD8+ T memory stem cell formation and antitumor immunity. JCI Insight. 2018;3(18) doi: 10.1172/jci.insight.122299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraud-Gatineau A., Coya J.M., Maure A., et al. The antibiotic bedaquiline activates host macrophage innate immune resistance to bacterial infection. Elife. 2020;9 doi: 10.7554/eLife.55692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillen M.R., Pandit A., Blokland S.L.M., et al. Plasmacytoid DCs from patients with sjögren's syndrome are transcriptionally primed for enhanced pro-inflammatory cytokine production. Front Immunol. 2019;10:2096. doi: 10.3389/fimmu.2019.02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaco G., Lee B., Xu W., et al. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26(6) doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman M.J., Craft B., Hastie M., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aran D., Hu Z., Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanai I., Benjamin H., Shmoish M., et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21(5):650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 29.Sun J., Zhang Z., Bao S., et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishwaran H., Kogalur U. Random forests for survival, regression and classification (RF-SRC). R package version 16. 2014. http://CRAN.R-project.org/package=randomForestSRC.
- 31.Hothorn T., Lausen B. On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal. 2003;43(2):121–137. [Google Scholar]
- 32.Charoentong P., Finotello F., Angelova M., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Auslander N., Zhang G., Lee J.S., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J., Yu A., Othmane B., et al. Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics. 2021;11(7):3089–3108. doi: 10.7150/thno.53649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He R.Q., Huang Z.G., Zhai G.Q., et al. Small nucleolar RNAs (snoRNAs)-based risk score classifier predicts overall survival in bladder carcinoma. Med Sci Monit. 2020;26 doi: 10.12659/MSM.926273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang X., Song X., Wang K., et al. SNORD63 and SNORD96A as the non-invasive diagnostic biomarkers for clear cell renal cell carcinoma. Cancer Cell Int. 2021;21(1):56. doi: 10.1186/s12935-020-01744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L., Liang X.Z., Deng Y., et al. Prognostic value of small nucleolar RNAs (snoRNAs) for colon adenocarcinoma based on RNA sequencing data. Pathol Res Pract. 2020;216(6) doi: 10.1016/j.prp.2020.152937. [DOI] [PubMed] [Google Scholar]
- 38.Wu H., Qin W., Lu S., et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19(1):95. doi: 10.1186/s12943-020-01201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L., Wang C., Qiu X., Pu X., Chang P. Colorectal cancer immune infiltrates: significance in patient prognosis and immunotherapeutic efficacy. Front Immunol. 2020;11:1052. doi: 10.3389/fimmu.2020.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borst J., Ahrends T., Bąbała N., Melief C.J.M., Kastenmüller W. CD4 T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18(10):635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 41.Nair S., Dhodapkar MV. Natural killer T cells in cancer immunotherapy. Front Immunol. 2017;8:1178. doi: 10.3389/fimmu.2017.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gustafsson K., Herrmann T., Dieli F. Editorial: understanding gamma delta T cell multifunctionality - towards immunotherapeutic applications. Front Immunol. 2020;11:921. doi: 10.3389/fimmu.2020.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C., Hu Y., Shi C. Targeting natural killer cells for tumor immunotherapy. Front Immunol. 2020;11:60. doi: 10.3389/fimmu.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollern D.P., Xu N., Thennavan A., et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191–1206. doi: 10.1016/j.cell.2019.10.028. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Lozano-Ruiz B., Yang F.M., Fan D.D., Shen L., González-Navajas JM. The multifaceted role of Th1, Th9, and Th17 cells in immune checkpoint inhibition therapy. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.625667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deschoolmeester V., Baay M., Lardon F., Pauwels P., Peeters M. Immune cells in colorectal cancer: prognostic relevance and role of MSI. Cancer Microenviron. 2011;4(3):377–392. doi: 10.1007/s12307-011-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Dai Z., Wu W., et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res CR. 2021;40(1):184. doi: 10.1186/s13046-021-01987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kloten V., Lampignano R., Krahn T., Schlange T. Circulating tumor cell PD-L1 expression as biomarker for therapeutic efficacy of immune checkpoint inhibition in NSCLC. Cells. 2019;8(8) doi: 10.3390/cells8080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.