Summary
Background
Imaging of subclinical atherosclerosis improves cardiovascular risk prediction on top of traditional risk factors. However, cardiovascular imaging is not universally available. This work aims to identify circulating proteins that could predict subclinical atherosclerosis.
Methods
Hypothesis-free proteomics was used to analyze plasma from 444 subjects from PESA cohort study (222 with extensive atherosclerosis on imaging, and 222 matched controls) at two timepoints (three years apart) for discovery, and from 350 subjects from AWHS cohort study (175 subjects with extensive atherosclerosis on imaging and 175 matched controls) for external validation. A selected three-protein panel was further validated by immunoturbidimetry in the AWHS population and in 2999 subjects from ILERVAS cohort study.
Findings
PIGR, IGHA2, APOA, HPT and HEP2 were associated with subclinical atherosclerosis independently from traditional risk factors at both timepoints in the discovery and validation cohorts. Multivariate analysis rendered a potential three-protein biomarker panel, including IGHA2, APOA and HPT. Immunoturbidimetry confirmed the independent associations of these three proteins with subclinical atherosclerosis in AWHS and ILERVAS. A machine-learning model with these three proteins was able to predict subclinical atherosclerosis in ILERVAS (AUC [95%CI]:0.73 [0.70–0.74], p < 1 × 10−99), and also in the subpopulation of individuals with low cardiovascular risk according to FHS 10-year score (0.71 [0.69–0.73], p < 1 × 10−69).
Interpretation
Plasma levels of IGHA2, APOA and HPT are associated with subclinical atherosclerosis independently of traditional risk factors and offers potential to predict this disease. The panel could improve primary prevention strategies in areas where imaging is not available.
Keywords: Subclinical atherosclerosis, Proteomics, Biomarkers, IGHA2, APOA, HPT
Abbreviations: CV, cardiovascular; SA, subclinical atherosclerosis; PESA, Progression of Early Subclinical Atherosclerosis study; AWHS, Aragon Worker Health Study; FHS, Framingham Heart Study; CACS, coronary artery calcium score; MS, mass spectrometry; PIGR, polymeric immunoglobulin receptor; IGHA1 and IGHA2, immunoglobulin heavy constant alpha 1 and 2; IGHG1,IGHG2, IGHG3 and IGHG4, immunoglobulin heavy constant gamma 1, 2, 3 and 4; IGKC, immunoglobulin kappa constant; IGLC2, immunoglobulin lambda constant 2; IGHD, immunoglobulin heavy constant delta; HPT, haptoglobin; HEP2, heparin cofactor 2; APOA, apolipoprotein(a); GELS, gelsolin; CD5L, CD5 antigen-like; Lp(a), lipoprotein(a)
Research in context.
Evidence before this study
Atherosclerosis is the leading cause of death worldwide, and early prevention is the best approach to fight this pandemic disease. Cardiovascular risk assessment is based on equations that use traditional risk factors. Identification of atherosclerosis by non-invasive imaging has been shown to improve risk stratification over classical equations. Clinical practice guidelines recommend screening for atherosclerosis by imaging and using these measures as risk modifiers, especially in low-moderate risk individuals. However, cardiovascular imaging is not universally available. The identification of plasma biomarkers closely associated with subclinical atherosclerosis could overcome this limitation and make it possible to improve risk prediction in a wider scale. In this regard, existing biomarkers to date, such as C-reactive protein, provide limited additional value.
Added value of this study
This is the largest study to date exploring the association between plasma protein levels and subclinical atherosclerosis by using high-throughput, unbiased quantitative proteomics. The discovery phase was undertaken in a population of 444 individuals from the PESA cohort study undergoing simultaneous imaging and blood testing at two different timepoints three years apart. Validation of these findings was also performed by proteomics in an external cohort (350 individuals from the AWHS cohort study). These analyses allowed the identification of a panel of three biomarker proteins that are significantly associated with the presence of subclinical atherosclerosis even after adjusting for traditional risk factors. Quantification of these proteins by immunoturbidimetry, a fast and simple technology, was shown to predict subclinical atherosclerosis in a large sample composed of 2999 individuals from a third cohort (ILERVAS). Subclinical atherosclerosis could also be predicted by this panel in individuals with low cardiovascular risk.
Implications of all the available evidence
Measuring plasma levels of IGHA2, APOA and HPT by immunoturbidimetry provides valuable information on the presence of subclinical atherosclerosis. This approach may thus provide an affordable alternative to cardiovascular imaging for the identification of subclinical atherosclerosis. These measures can be used for cardiovascular risk reclassification in low to moderate-risk individuals in primary cardiovascular prevention.
Alt-text: Unlabelled box
Introduction
Atherosclerosis is the leading cause of death worldwide. The natural history of atherosclerosis starts in childhood, involves a protracted subclinical phase and diagnosis usually occurs in advanced stage or following a cardiovascular (CV) event. Early prevention is thus the best approach to fight this pandemic disease. To guide actions for prevention, clinical practice guidelines recommend individual risk assessment by using algorithms that are based on traditional CV risk factors.1 However, there is a substantial variation in the amount of atherosclerosis among individuals belonging to the same risk category,2,3 and interest has emerged in the use of non-invasive imaging techniques for screening atherosclerotic burden to improve CV risk assessment.
Numerous works have shown that the detection of coronary calcification or carotid plaques using non-invasive imaging tools improves risk prediction and reclassification compared with only conventional risk factors.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Indeed, current guidelines (2021 ESC guidelines) consider coronary artery calcium score and carotid plaque detection as risk modifiers in CV risk assessment, specially in individuals with calculated CV risks based on the major conventional risk factors around the decisional thresholds.14 However, there are important concerns regarding availability and cost-effectiveness,14 as well as expertise requirements and radiation exposure for the routine screening of atherosclerosis with imaging modalities. In this context, the concept of using circulating biomarkers to improve the assessment of CV risk is not new. The high-sensitivity C-reactive protein has shown consistency across large prospective studies as a risk factor integrating multiple metabolic and low-grade inflammatory factors, but its contribution to the existing methods of CV risk assessment has limited additional value.14 Today, mass spectrometry (MS)-based proteomics offers the opportunity of detecting novel protein biomarkers directly related to the presence, extension and/or progression of atherosclerosis. To the best of our knowledge, unbiased deep quantitative proteomics has not been used previously to study associations between circulating plasma protein levels and subclinical atherosclerosis (SA) in large enough populations (e.g., more than 100 individuals).
In this work, we performed a case-control study aimed to detect novel protein biomarkers of SA using unbiased deep MS-based proteomics on plasma from 444 asymptomatic, middle aged men with or without SA from the PESA study.4 The results were internally validated by repeating the proteomics in the same population after a 3-year follow up. The identified biomarkers were also externally validated by proteomics in an external cohort of 350 individuals from the AWHS study.15 Finally, three selected proteins were also validated using a commercially available turbidimetric test in the AWHS cohort and in a large cohort of 2999 individuals from the ILERVAS study.16
Methods
Study populations and assessment of subclinical atherosclerosis
For the discovery phase, a nested case-control study within the prospective PESA cohort17 was designed (PESA-V1). The PESA cohort was composed by healthy, middle-aged employees of the Banco de Santander Headquarters in Madrid (Spain).4 In PESA, the presence of atherosclerotic plaques was assessed by two-dimensional vascular ultrasound of carotids, infrarenal abdominal aorta, and iliofemoral arteries and by non-contrast cardiac computed tomography.17,18 Plaques were defined as a focal protrusion into the arterial lumen of thickness >0.5 mm or >50% of the surrounding intima-media thickness (IMT), or a diffuse thickness >1.5 mm measured between the media-adventitia and intima-lumen interfaces. Plaque thickness was calculated by summing plaque areas from the two thickest plaques measured in each vascular territory: carotids, abdominal aorta and iliofemoral arteries.4,19 Coronary artery calcification score (CACS) was measured using the Agatston scoring method; any Agatston score ≥1 is considered indicative of atherosclerosis.4 Cases (222) were selected among participants with 3 or more vascular territories affected, and controls (222) among participants with ≤1 vascular territory affected. Controls were individually matched with cases based on a hierarchical single summary score including, in order of importance, age (caliper: 3 years), diabetes, smoking, dyslipidemia and hypertension. Due to the low prevalence of subclinical atherosclerosis in women in PESA,17 the study was restricted to men. To optimize statistical power to detect significant protein alterations in plasma, the discovery phase of the study was restricted to men. For internal validation, plasma samples from the same individuals were collected three years later (PESA-V2), except two cases and their matched controls that did not renew their consent for ‘omics’ analysis.
The external validation set was designed within the AWHS cohort15,20 as a nested case-control study, also restricted to men. The AWHS cohort was composed by workers of a large car assembly plant in Figueruelas (Zaragoza, Spain).15 Similar to PESA, the presence of plaques in both carotid and femoral arteries was determined using linear high-frequency 2-dimensional ultrasound probes.20 Plaques were defined as a focal structure protruding ≥0.5 mm into the lumen or reaching a thickness ≥50% of the surrounding intima. CACS was also analyzed following the Agatston method. Cases (175) were selected among individuals with 3 or more vascular territories affected, and controls (175) among participants with no vascular territory affected. Controls were individually matched with cases based on a hierarchical single summary score including age (caliper: 3 years), diabetes, smoking, dyslipidemia and hypertension.
The final validation was performed in a subpopulation from the ongoing ILERVAS study.21,22 ILERVAS is based on a middle-aged population with low-to-moderate cardiovascular risk, randomized to undergo or not vascular ultrasound examination.21,22 This cohort was composed by subjects from 30 primary health care centers in Lleida (Spain).21 The presence of plaques was explored in carotid and femoral arteries by two-dimensional ultrasound.21 Plaque was defined as a focal intima-media thickness ≥ 1.5 mm protruding into the lumen.21,22 All subjects (men and women) with vascular ultrasound examination that completed biochemical analysis and had either 0 (1404 controls) or ≥ 3 vascular territories affected (1595 cases) were selected.
Protein digestion for proteomics
Plasma samples were collected in tubes with K-EDTA, subjected to centrifugation at 1000 g at 4 ˚C for 10 min, and stored at -80. 5 µl of plasma of each individual (corresponding to about 300 µg of protein measured by NanoDrop 1000, Thermo Fisher), were mixed with 5 µl of a buffer containing 50 mM Tris, 2 % SDS and 100 mM DTT, and boiled for 5 min for protein denaturation. Proteins were then subjected to filter-aided digestion (Nanosep Centrifugal Devices with Omega Membrane-10 K, PALL) according to manufacter's instructions. Briefly, 320 µl of urea were added to each sample to dilute SDS according to manufacter's instructions, and they were transferred to the filter and subjected to centrifugation at 14,000 g for 10 min. Cysteine residues were blocked with 50 mM iodoacetamide for 1 h at room temperature under darkness. After two washes with 100 µl urea followed by two washes with 100 µl of 100 mM ammonium bicarbonate pH 8.8, proteins were digested with trypsin (1:30-trypsin:protein, Promega) overnight at 37 ˚C. After protein digestion, peptides were eluted from the filter in two different steps using 40 µl of ammonium bicarbonate and 50 µl of 500 mM NaCl, respectively. Peptides were acidified with 25% trifluoroacetic acid (TFA) to a final concentration of 1%, desalted with Oasis cartridges (Waters) following manufacture's instructions, Speed-vac dried and stored at -20 ˚C.
TMT labeling
Peptides were subjected to multiplexed isobaric labeling with 10-plex TMT reagents (Thermo Fisher Scientific) following manufacter's instructions. Briefly, peptide concentration was measured using DirectDetect Infrared Spectrometer (Merck) and 50 µg of peptides from each sample were labeled.23,24 After labeling, peptides from all the samples in each TMT experiment were mixed in the same tube, acidified with 25%TFA to a final concentration of 1% and desalted with Oasis cartridges (Waters). Aliquots of 1/10 (in volume) were saved for direct MS analysis (without peptide fractionation). Labeled peptide aliquots were Speed-vac dried and stored at -20 ˚C. Samples were processed in batches containing a maximum of 15 TMT experiments (120 samples). Each TMT experiment contained the samples from 8 individuals and two channels were reserved for reference internal standards constructed by pooling all the samples from each batch. Labeling and subsequent LC-MS analysis was performed in a blind manner.
Peptide fractionation
Peptides were fractionated using the high pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) according to manufacter's instructions. Briefly, cartridges were washed with 50% and 100% acetonitrile (ACN), and equilibrated with 0.1% of TFA. 100 µg of peptides were resuspended in TFA 0.1 % and loaded into the cartridges. Peptides were then eluted into five fractions with increasing amounts of ACN: Fr1 (12.5% ACN), Fr2 (15% ACN), Fr3 (17.5% ACN), Fr4 (20% ACN) and Fr5 (50% ACN). Fractions were Speed-vac dried and stored -20 ˚C until MS analysis.
LC-MS analysis
Each fraction of labeled peptide samples was analyzed using an Easy-nLC 1200 system (Thermo Fisher Scientific) coupled via a nanoelectrospray ion source (Thermo Fisher Scientific, Bremen, Germany) to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) for PESA-V1 cohort, and to a Q-exactive HF Hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific) for PESA-V2 and AWHS cohorts. C18-based reverse phase separation was performed using a PepMap 100 C18 5 μm 0.3 × 5 mm as trapping column (Thermo Fisher Scientific) and a PepMap RSLC C18 EASY-Spray column 50 cm × 75 μm ID as analytical column (Thermo Fisher Scientific). Peptides were loaded in buffer A (0.1% of formic acid in water (v/v)) and eluted with a 338 min linear gradient of buffer B (100% ACN, 0.1% formic acid (v/v)) at 200 nl/min. Mass spectra were acquired in data-dependent manner, with an automatic switch between MS and MS/MS using a top-speed adquisition mode method. MS spectra were acquired in the Orbitrap analyser using full ion-scan mode with a mass range of 400–1500 mass-to-charge (m/z) and 70,000 FT resolution. The automatic gain control target was set at 2 × 105 with 50 ms maximun injection time. MS/MS was performed in the top-speed adquisition mode with 3 s cycle time. HCD fragmentation was performed at 30% of normalized collision energy and MS/MS spectra were analysed at a 60,000 resolution in the Orbitrap.
Protein identification
For peptide identification MS/MS spectra were searched with the SEQUEST HT algorithm implemented in Proteome Discoverer 2.1 (Thermo Scientific) against a Uniprot database comprised human protein sequences (July 2014), using trypsin digestion with a maximum of 2 missed cleavages, using as fixed modifications Cys carbamidomethylation (57.021464 Da) and TMT labeling at N-terminal end and Lys (229.162932 Da), and Met oxidation (15.994915) as dynamic modification. Precursor mass tolerance was set at 800 ppm and fragment mass tolerance at 0.03 Da; precursor charge range was set to 2–4. Results were analyzed using the probability ratio method25 and the false discovery rate (FDR) was calculated based on the search of results against the corresponding decoy database using the refined method,26 with an additional filter for precursor mass tolerance of 15 ppm.27 1% FDR was used as criterion for peptide identification. The three biomarkers identified in this work (HPT, IGHA2 and APOA) were unambiguously identified by unique peptides (Tables S1–S3).
Protein quantification
Protein quantification and statistical and systems biology analysis were performed using the models previously developed in our laboratory28,29 with the SanXoT software package.30 Quantitative information was extracted from the MS/MS spectra of TMT-labeled peptides. Peptide quantification was analyzed using the WSPP model, which uses raw quantifications as input data and computes the protein log2-fold changes for each individual with respect to the average of the values of the two reference internal standard samples. In this model protein log2-ratios are expressed as standardized variables in units of standard deviation according to their estimated variances (Zq values).
Immunoturbidimetry
Plasma levels of IGHA2, HPT and APOA were measured by immunoturbidimetric assays (LK088.OPT, NK058.OPT and LK098.OPT, respectively, from The Binding Site) using the Binding Site Optilite analyzer in a blinded manner.
Machine learning
For classification of individuals with subclinical atherosclerosis, we used a distributed random forest (RF) model, an ensemble method well established in the diagnostic prediction.32 The RandomForestClassifier method from the ensemble Scikit-Learn module was used to implement the RF model. Optimal values for RF hyperparameters were obtained using 10-fold cross-validation for AUC optimization on the test datasets using the Python library Scikit-Learn library.33 Hyperparameter tuning was performed sequentially using the RandomizedSearchCV module, to find an initial naïve range of values for the different RF hyperparameters,34 and the GridSearchCV module, to obtain the optimal combination of specific values to maximize performance. AUC calculation for the training and test sets was done applying the roc_auc_score function. The class imbalance problem was avoided using the StratifiedKFold method, which preserves the percentage of samples for each class in all the folds. The primary outcome of the RF model was a continuous variable between 0 and 1 describing the probability of having SA.
Statistics
Correlation of protein Zq values with plaque thickness or with CACS was analyzed by Pearson's method. Adjustment for multiple hypothesis testing was performed by controlling for the False Discovery Rate (FDR).35 Linear and logistic regression models were tested using SPSS software (IBM, Armonk, New York). Associations were expressed as standardized odds ratios (ORs) with 95% confidence intervals (CI). The C-statistic or area under the receiver operating characteristic (ROC) curve (AUC) was used as a measure of predictive power. Comparison of AUC for different models was performed according to the method of DeLong.36
Ethics
Ethical committee advice and patient informed consent were obtained (Instituto de Salud Carlos III Ethics Committee (PESA), the Central Institutional Review Board of Aragón (CEICA) (AWHS) and the Ethics Committee of The Catalan Health Service (Ref. CEIC-1410 Hospital Arnau de Vilanova, Lleida, Spain) (ILERVAS)).
Role of funders
Funding sources played no role in study design; collection, analysis or interpretation of the data; writing of the report, or in submission of this paper for publication.
Results
Clinical characteristics of cases and controls from the PESA, AWHS and ILERVAS cohorts are depicted in Table 1. Overall, the three cohorts were constituted by low or low-to-moderate risk participants according to conventional risk scales.
Table 1.
Characteristics of the PESA, AWHS and ILERVAS populations.
| Controls | Cases | p value | |
|---|---|---|---|
| PESA population at baseline | (n = 222) | (n = 222) | |
| Age, y, mean (SD) | 48 (4) | 49 (4) | 0.023 |
| SBP, mm Hg, mean (SD) | 120 (12) | 124 (12) | 0.001 |
| DBP, mm Hg, mean (SD) | 75 (9) | 77 (9) | 0.004 |
| Fasting glucose, mg/dl, mean (SD) | 94 (11) | 96 (15) | 0.132 |
| Total cholesterol, mg/dl, mean (SD) | 201 (33) | 210 (36) | 0.012 |
| LDL-C (mg/dl), mean (SD) | 136 (30) | 143 (33) | 0.024 |
| HDL-C, mg/dl, mean (SD) | 44 (10) | 43 (10) | 0.139 |
| Triglycerides, mg/dl, mean (SD) | 106 (58) | 121 (65) | 0.009 |
| BMI, kg/m2, mean (SD) | 27.24 (3.1) | 27.5 (3.2) | 0.385 |
| Current smoking, No. (%) | 55 (25%) | 91 (41%) | <0.001 |
| Hypertension, No. (%) | 20 (9%) | 24 (11%) | 0.526 |
| Obesity, No. (%) | 39 (17.5%) | 41 (18.4%) | 0.788 |
| Dislypemia, No. (%) | 12 (5.4%) | 27 (12.2%) | 0.011 |
| History of CVD, No. (%) | 28 (12.6%) | 47 (21%) | 0.016 |
| AWHS population | (n = 175) | (n = 175) | |
| Age, y, mean (SD) | 49.6 (4.1) | 51.0 (3.6) | 0.001 |
| SBP, mm Hg, mean (SD) | 122.1 (12.0) | 125.8 (13.9) | 0.01 |
| DBP, mm Hg, mean (SD) | 81.4 (8.7) | 82.5 (8.8) | 0.243 |
| Fasting glucose, mg/dl, mean (SD) | 98.2 (17.9) | 98.3 (17.2) | 0.971 |
| Total cholesterol, mg/dl, mean (SD) | 215.4 (35.5) | 221.6 (37.3) | 0.111 |
| HDL-C, mg/dl, mean (SD) | 54.0 (11.1) | 50.4 (10.4) | 0.002 |
| BMI, kg/m2, mean (SD) | 27.21 (3) | 27.6 (3.2) | 0.369 |
| Current smoking, No. (%) | 30 (17.1%) | 77 (44%) | <0.001 |
| Hypertension, No. (%) | 42 (24%) | 50 (28.6%) | 0.379 |
| Obesity, No. (%) | 20 (11.4%) | 19 (11%) | 1 |
| Dislypemia, No. (%) | 49 (28%) | 62 (35.4%) | 0.111 |
| ILERVAS population | (n = 1404) | (n = 1595) | |
| Male, No. (%) | 489 (34.8%) | 992 (62.2%) | <0.001 |
| Age, y, mean (SD) | 56.31 (6.11) | 58.90 (6.09) | <0.001 |
| SBP, mmHg, mean (SD) | 127.91 (16.22) | 135.90 (16.69) | <0.001 |
| DBP, mmHg, mean (SD) | 80.79 (9.67) | 83.43 (9.60) | <0.001 |
| Fasting glucose, mg/dl, mean (SD) | 95.54 (14.86) | 98.81 (18.35) | <0.001 |
| Total cholesterol, mg/dl, mean (SD) | 218.02 (37.21) | 226.34 (39.96) | <0.001 |
| LDL-C, mg/dl, mean (SD) | 123.03 (30.54) | 128.18 (33.80) | <0.001 |
| HDL-C, mg/dl, mean (SD) | 66.70 (19.42) | 64.58 (17.66) | 0.002 |
| Triglycerides, mg/dl, mean (SD) | 142.73 (85.80) | 169.59 (112.84) | <0.001 |
| BMI, kg/m2, mean (SD) | 29.05 (5.12) | 29.16 (4.8) | 0.525 |
| Current smoking, No. (%) | 279 (19.8%) | 626 (39.3%) | <0.001 |
| Hypertension, No. (%) | 480 (34.2%) | 768 (48.2%) | <0.001 |
| Obesity, No. (%) | 426 (30.4%) | 497 (31.2%) | 0.63 |
| Dislypemia, No. (%) | 660 (47%) | 899 (56.4%) | <0.001 |
| History of CVD, No. (%) | 126 (9%) | 186 (11.6%) | 0.026 |
SI conversion factors: To convert glucose to mmol/L, multiply values by 0.0555; to convert cholesterol, cholesterol-low density and cholesterol-high-density to mmol/L, multiply values by 0.0259; to convert tryglicerides to mmol/L, multiply values by 0.0113.
Unbiased discovery of plasma proteins associated to subclinical atherosclerosis
Quantitative proteomics was performed on 444 plasma samples from PESA-V1 and 440 samples from PESA-V2. The complete analysis required more than 800 LC-MS runs of 5.6 h each. A mean of 1093 proteins were quantified per sample, and 470 proteins could be quantified in more than 80% of the individuals in each cohort. MS data were processed using automated statistical workflows28,31 constructed with the SanXoT package.30
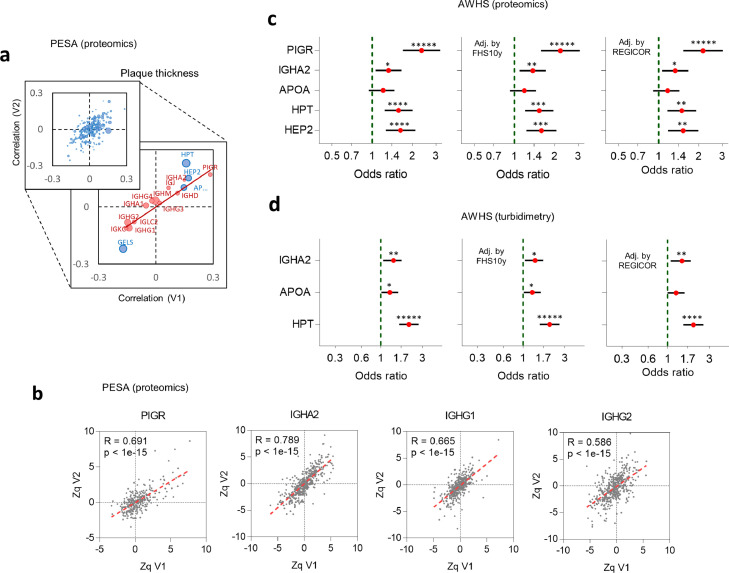
The correlation of protein levels with plaque thickness found at baseline (PESA-V1) were in general well-reproduced in the second visit (PESA-V2) (Figure 1a and Tables S4, S5). The behavior of proteins related to the humoral immune response suggested that increased plaque thickness was associated with an immunoglobulin isotype switch (Figure 1a, red circles). Protein levels in PESA-V1 individuals highly correlated with those of the same individuals in PESA-V2 (Figure 1b), suggesting that the switch remained stable along time.
Figure 1.
Selection of protein biomarkers. (a) Comparison of Pearson's correlations of relative plasma proteins levels with plaque thickness in PESA-V1 and PESA-V2 cohorts. Dot sizes are indicative of protein abundances (in number of peptides per protein). Inset: behavior of proteins related to humoral immune response (red) or yielding a significant correlation (blue). The immunoglobulin isotype switch is reflected in the increase of PIGR, IGHA2 and IGHD, and in the decrease of IGLC2, IGKC, IGHG1 and IGHG2, while, in contrast, the related isotypes IGHA1, IGHG3 and IGHG4 did not change. (b) Representative correlations between relative abundance values (expressed as standardized log2-ratios, Zq) of four representative proteins in PESA-V1 and PESA-V2 for the same individuals. A similar trend was found with the other proteins cited in the text. R indicates Pearson's correlation coefficient and p, the statistical significance of the correlation. (c and d) Validation of PESA results in the AWHS cohort. Forest plots show OR of subclinical atherosclerosis (cases vs controls) in AWHS, obtained by either proteomics (c) or turbidimetry (d). OR refer to protein values expressed in units of standard deviation, using univariate logistic regression models, or multivariate models adjusted by common Risk Scores, as indicated. Error bars indicate 95% confidence intervals of OR values.
To obtain a curated list of proteins that were independently associated with SA, we firstly selected a subset of proteins that yielded a significant correlation with plaque thickness in PESA-V1 (FDR < 5%) and showed correlation at 3-year follow-up (PESA-V2) (FDR < 15%) (Table S6). Among these, we selected IGHA2, PIGR, HEP2, HPT, APOA, GELS and IGKC as the proteins whose correlation with plaque thickness was more consistently independent from individual risk factors (Table S7). We also analyzed the correlation with coronary artery calcium score (CACS), finding that HPT, GELS and CD5L significantly correlated in PESA-V1 and V2 (Table S8 and Fig. S1a), independently of individual risk factors (Table S9). Interestingly, although HPT, considered an acute-phase protein, correlated with both plaque thickness and CACS, none of the typical acute-phase proteins showed any significant correlation (Table S10).
Secondly, among the eight proteins selected, we analyzed which ones were independently associated with the presence of SA by logistic regression analysis (Table 2). We selected PIGR, IGHA2, APOA, HPT and HEP2 since their levels were significantly associated with an increased likelihood of SA, and the associations were maintained after adjustment by FHS 10-year score37 or by the REGICOR score38,39 (Table 2A). Finally, in multivariate analysis we found that these five proteins could be combined into three-protein panels, where PIGR could be replaced by IGHA2 and HEP2 by HPT (Table 2B).
Table 2.
Logistic regression analysis of association with the presence of subclinical atherosclerosis in PESA-V1.
| A: Individual proteins |
Univariate |
Adj. by FHS 10-year |
Adj. by Regicor |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteins | p-val | OR | 95% CI | p-val | OR | 95% CI | p-val | OR | 95% CI | ||||
| Increased | Polymeric immunoglobulin receptor (PIGR) | ≤0.001 | 1.554 | 1.256 | 1.922 | 0.005 | 1.374 | 1.102 | 1.711 | 0.006 | 1.36 | 1.091 | 1.695 |
| Ig alpha-2 chain C region (IGHA2) | 0.008 | 1.295 | 1.069 | 1.567 | 0.024 | 1.259 | 1.031 | 1.538 | 0.021 | 1.265 | 1.037 | 1.544 | |
| Apolipoprotein(a) (APOA) | 0.018 | 1.261 | 1.041 | 1.527 | 0.015 | 1.279 | 1.049 | 1.56 | 0.011 | 1.293 | 1.062 | 1.575 | |
| Haptoglobin (HPT) | ≤0.001 | 1.412 | 1.157 | 1.723 | 0.05 | 1.225 | 0.994 | 1.51 | 0.05 | 1.228 | 0.998 | 1.512 | |
| Heparin cofactor 2 (HEP2) | ≤0.001 | 1.414 | 1.195 | 1.673 | 0.005 | 1.285 | 1.078 | 1.531 | 0.007 | 1.275 | 1.069 | 1.52 | |
| Decreased | Gelsolin (GELS) | 0.008 | 0.772 | 0.638 | 0.936 | 0.107 | 0.849 | 0.695 | 1.036 | 0.169 | 0.869 | 0.712 | 1.061 |
| Ig kappa chain C region (IGKC) | 0.049 | 0.833 | 0.694 | 0.999 | 0.181 | 0.879 | 0.727 | 1.062 | 0.197 | 0.883 | 0.73 | 1.067 | |
| CD5 antigen-like (CD5L) | 0.226 | 0.947 | 0.868 | 1.034 | 0.408 | 0.962 | 0.879 | 1.054 | 0.42 | 0.963 | 0.879 | 1.055 | |
| B: Protein panels | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteins | p-val | OR | 95% CI | Chi-square | df | Adj. p-val | |||||||
| Model 1 | Polymeric immunoglobulin receptor (PIGR) | 0.003 | 1.4 | 1.12 | 1.76 | 44.305 | 5 | 2.01E-08 | |||||
| Ig alpha-2 chain C region(IGHA2) | 0.016 | 1.29 | 1.05 | 1.6 | |||||||||
| Apolipoprotein(a) (APOA) | 0.004 | 1.34 | 1.1 | 1.64 | |||||||||
| Haptoglobin (HPT) | 0.434 | 1.1 | 0.87 | 1.38 | |||||||||
| Heparin cofactor 2 (HEP2) | 0.006 | 1.33 | 1.09 | 1.64 | |||||||||
| Model 2 | Polymeric immunoglobulin receptor (PIGR) | ≤0.001 | 1.49 | 1.19 | 1.85 | 37.238 | 3 | 2.05E-07 | |||||
| Apolipoprotein(a) (APOA) | 0.004 | 1.34 | 1.1 | 1.64 | |||||||||
| Heparin cofactor 2 (HEP2) | 0.001 | 1.34 | 1.12 | 1.6 | |||||||||
| Model 3 | Polymeric immunoglobulin receptor (PIGR) | ≤0.001 | 1.52 | 1.22 | 1.89 | 33.49 | 3 | 1.27E-06 | |||||
| Apolipoprotein(a) (APOA) | 0.006 | 1.32 | 1.08 | 1.61 | |||||||||
| Haptoglobin (HPT) | 0.016 | 1.28 | 1.05 | 1.58 | |||||||||
| Model 4 | Ig alpha-2 chain C region(IGHA2) | 0.001 | 1.39 | 1.14 | 1.71 | 33.267 | 3 | 1.41E-06 | |||||
| Apolipoprotein(a) (APOA) | 0.005 | 1.32 | 1.09 | 1.61 | |||||||||
| Heparin cofactor 2 (HEP2) | ≤0.001 | 1.49 | 1.25 | 1.77 | |||||||||
| Model 5 | Ig alpha-2 chain C region(IGHA2) | 0.009 | 1.3 | 1.07 | 1.58 | 24.317 | 3 | 1.05E-04 | |||||
| Apolipoprotein(a) (APOA) | 0.015 | 1.27 | 1.05 | 1.55 | |||||||||
| Haptoglobin (HPT) | 0.001 | 1.39 | 1.14 | 1.7 | |||||||||
Odds ratios (OR) refer to relative protein values determined by proteomics and expressed in units of standard deviation, using logistic regression models (univariate or bivariate models in A, or multivariate in B).
To analyze potential biomarker panels among the five proteins, several multivariate models containing different protein combinations were tested in B. Model 1 included the five proteins. Models 2–5 showed several combinations of three proteins. HPT maintained its association with SA when HEP2 was not included in the models (Models 3 and 5), so that in the practice HPT and HEP2 could be interchanged. PIGR and IGHA2 also showed a similar behavior, so that three-protein models could be constructed with either PIGR (Models 2 and 3) or IGHA2 (Models 4 and 5). P-values were adjusted by Bonferroni.
Validation of protein associations by proteomics and turbidimetry in the AWHS cohort
Quantitative proteomics using the same high-throughput approach was performed on 175 plasma samples from AWHS male participants with extensive SA and 175 from matched controls. The correlations of plasma proteins with plaque thickness found in PESA-V1 were reproduced in the AWHS cohort (Fig. S1b, see also Table S11), except for GELS. The immunoglobulin isotype switch was also reproduced in AWHS (Fig. S1b). The correlation of HPT and CD5L with CACS was lower in AWHS than in PESA but maintained the same trend (Fig. S1c, Table S11). The association with SA of PIGR, IGHA2, HPT and HEP2 was also confirmed in AWHS (Figure 1c). We also observed that the associations remained significant after adjusting by CV risk scores (Figure 1c). APOA did not achieve statistical significance in AWHS but showed the same tendency than in PESA (Figure 1c).
To validate the results by independent techniques, we selected IGHA2, APOA and HPT since these proteins formed a panel (Table 2B) that could be measured using commercially available antibody-based turbidimetry kits suitable to be used in clinical practice. Multivariate logistic regression revealed that increased plasma concentration of each one of these three proteins measured by turbidimetry in AWHS was significantly associated with an increased likelihood of SA (IGHA2: OR[95%CI]:1.37[1.08–1.74], p < .01; APOA: 1.27[1.02–1.57], p < .05; HPT: 2.09[1.62–2.70], p = 2 × 10−8, logistic regression analysis) (Figure 1d). We also observed that these associations remained significant after adjusting by FHS 10-year score37 or by the REGICOR score38,39 (Figure 1d).
Validation of protein associations by turbidimetry in the ILERVAS cohort
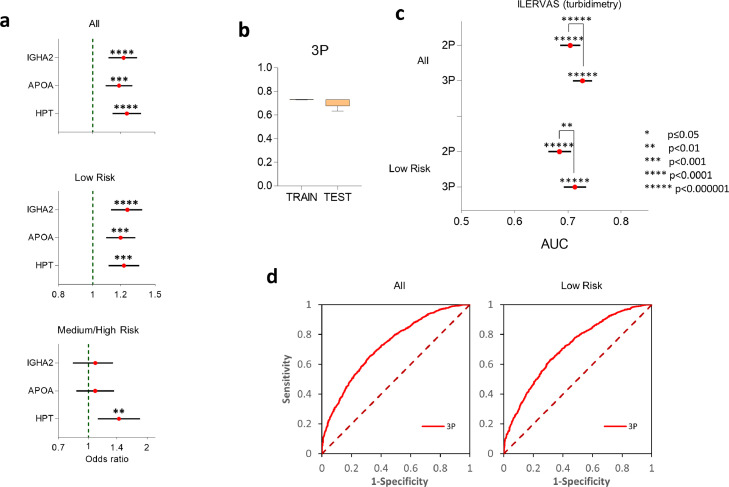
To further validate the association of IGHA2, APOA and HPT with SA using the turbidimetry kits, we analyzed blood samples from a larger cohort obtained from the ILERVAS study.21,22 This validation cohort contained individuals that had extensive SA (1.595 cases) and individuals without SA (1.404 controls), and, importantly, 51% of the subjects from this cohort were women (see Table 1). Multivariate logistic regression confirmed that the plasma concentrations of these three proteins were significantly associated with SA after adjusting by gender and conventional risk factors (IGHA2: 1.21[1.11–1.33], p = 3 × 10−5; APOA: 1.18[1.08–1.29], p = 1 × 10−4; HPT: 1.24[1.13–1.37], p = 3 × 10−5, logistic regression analysis) (Figure 2a). The independence from risk factors was further confirmed checking that the associations were maintained after stratifying the cohort into subpopulations having or not each one of the main risk factors (Table 3).
Figure 2.
Validation of biomarkers in the ILERVAS cohort. (a) Forest plots showing OR of subclinical atherosclerosis (cases vs controls) per each protein, obtained by turbidimetry in the complete ILERVAS population, or after stratifying it into low-risk (FHS 10-year score < 10%) or medium/high-risk (FHS 10-year score ≥ 10%) individuals. OR refer to protein values expressed in units of standard deviation, using multivariate logistic regression models including the three proteins, gender, smoking, obesity, hypertension, dyslipidemia,history of CV disease and body mass index. (b) 10-fold cross validation of AUC values provided by the 3P model to detect the presence of subclinical atherosclerosis in train and test populations. Data are expressed as mean ± SD. (c, d) Improvement in AUC values and in the ROC curves to detect subclinical atherosclerosis obtained by the 2P and 3P models in the complete population, or in the low-risk population (FHS 10-year score < 10%). Horizontal error bars in (c) represent 95% CI. P-values above asterisks indicate statistical significance in relation to the null hypothesis (AUC=0), calculated using the Mann-Whitney statistic; p-values from the comparative analysis between models 2P and 3P were calculated using DeLong's test.
Table 3.
Multivariate logistic regression analysis of association with the presence of subclinical atherosclerosis in ILERVAS subpopulations stratified according to main CV risk factors.
| Multivariate Adj. by Gender and all Risk Factors (RFs) |
||||||||
|---|---|---|---|---|---|---|---|---|
| p-val | OR | 95% CI | p-val | OR | 95% CI | |||
| Smoking | Non-smokers (N = 2026) | Smokers (N = 891) | ||||||
| IGHA2 | 0.002 | 1.197 | 1.07 | 1.338 | 0.057 | 1.187 | 0.995 | 1.418 |
| APOA | 0.001 | 1.188 | 1.075 | 1.313 | 0.084 | 1.182 | 0.978 | 1.428 |
| HPT | 0.358 | 1.053 | 0.944 | 1.174 | 2.206E-08 | 1.695 | 1.409 | 2.039 |
| Hypertension | Non-Hypertensive (N = 1705) | Hypertensive (N = 1212) | ||||||
| IGHA2 | 0.000378 | 1.247 | 1.104 | 1.408 | 0.083 | 1.14 | 0.983 | 1.323 |
| APOA | 0.024 | 1.139 | 1.017 | 1.276 | 0.002 | 1.254 | 1.086 | 1.446 |
| HPT | 0.000019 | 1.299 | 1.152 | 1.464 | 0.309 | 1.078 | 0.932 | 1.247 |
| Obesity | Non-Obese (N = 2021) | Obese (N = 896) | ||||||
| IGHA2 | 0.00001 | 1.288 | 1.151 | 1.442 | 0.707 | 1.034 | 0.869 | 1.23 |
| APOA | 0.001 | 1.191 | 1.073 | 1.323 | 0.037 | 1.189 | 1.01 | 1.399 |
| HPT | 0.000008 | 1.29 | 1.154 | 1.443 | 0.413 | 1.072 | 0.908 | 1.265 |
| Dyslipemia | No Dyslipemia (N = 1403) | Dyslipemia (N = 1514) | ||||||
| IGHA2 | 0.003 | 1.223 | 1.07 | 1.399 | 0.018 | 1.172 | 1.028 | 1.337 |
| APOA | 0.01 | 1.202 | 1.045 | 1.382 | 0.006 | 1.172 | 1.046 | 1.313 |
| HPT | 0.000005 | 1.359 | 1.191 | 1.55 | 0.366 | 1.062 | 0.932 | 1.21 |
| CVDH | No CVDH (N = 2607) | CVDH (N = 392) | ||||||
| IGHA2 | 0.000368 | 1.197 | 1.084 | 1.321 | 0.057 | 1.31 | 0.992 | 1.728 |
| APOA | 0.000286 | 1.189 | 1.083 | 1.305 | 0.09 | 1.225 | 0.969 | 1.549 |
| HPT | 0.000117 | 1.212 | 1.099 | 1.337 | 0.41 | 1.109 | 0.866 | 1.42 |
Multivariate logistic regression was used to determine association of protein values with the presence of subclinical atherosclerosis (SA). All the models were adjusted by risk factors including: Age, Hypertension, Obesity, Dyslipemia, Smoking, History of Cardiovascular Disease (CVDH), Systolic Blood Pressure (SBP) and Diastolic Blood Pressure (DB).
We then analyzed the behavior of these associations according to the CV risk of the individuals. Noteworthy, the associations of the three proteins with SA were maintained in the subpopulation of individuals with low CV risk (FHS 10-year score < 10%) (IGHA2: 1.24[1.12–1.38], p = 2 × 10−5; APOA: 1.19[1.09–1.31], p = 2 × 10−4; HPT: 1.22[1.11–1.35], p = 7 × 10−4, logistic regression analysis) (Figure 2a). The associations were lost in the medium/high-risk group (FHS 10-year score > 10%), except for HPT, which remained independently associated to SA (Figure 2a).
Potential to predict the presence of subclinical atherosclerosis
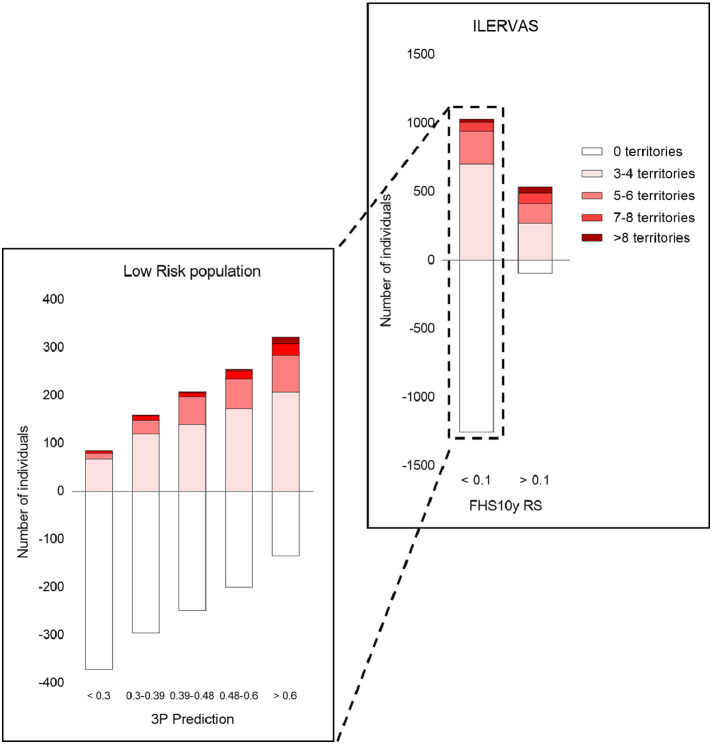
To test the potential of the three-protein panel to predict the presence of SA, we trained a machine-learning model that used as predictors the concentration of the three proteins (model 3P). Ten-fold cross validation confirmed the stability of AUC values in the testing populations with a minimal overfitting (Figure 2b). ROC analysis showed that 3P had a good performance to predict the presence of SA in the complete Ilervas population (AUC [95%CI]: 0.73 [0.70–0.74], p < 1 × 10−99 vs AUC= 0.5, Mann-Whitney statistic) (Figure 2c and d). Since APOA is a protein that is already known to associate with atherosclerosis, we also constructed a model that only considered the concentrations of IGHA2 and HPT (model 2P). While 2P maintained a reasonably good performance to predict SA (AUC [95%CI]: 0.70 [0.68–0.72], p < 1 × 10−80 vs AUC= 0.5, Mann-Whitney statistic), a comparative analysis showed that 3P was significantly better than 2P (p = 3 × 10−5, DeLong's test) (Figure 2c), suggesting that the inclusion of APOA significantly improved the performance of the biomarker panel. We further analyzed whether the performance of the panel to predict SA in the population of individuals with low CV risk. The model 3P maintained a similar performance in this subpopulation (AUC [95%CI]: 0.71 [0.69–0.73], p < 1 × 10−69 vs AUC= 0.5, Mann-Whitney statistic) (Figure 2c and d). 2P also showed a reasonable performance (AUC [95%CI]: 0.68 [0.66–0.70], p < 1 × 10−52 vs AUC= 0.5, Mann-Whitney statistic), but again 3P was still significantly better (p = .007, DeLong's test), suggesting that APOA also contributed to SA prediction in the low risk population. In the low CV risk population, the 3P model provided a means to stratify the risk of having SA (Figure 3). Thus, more than 63% of the individuals having SA in 7 or more territories were allocated to the two quintiles with highest risk according to 3P score (>0.48), increasing this proportion to 81% when individuals had more than 8 territories (Table S12). 54 to 59% of individuals with 3–6 affected territories, but only 27% of the controls, were also allocated to the two highest risk quintiles (Table S12).
Figure 3.
Risk stratification of subclinical atherosclerosis predicted by the 3P model in the low-risk ILERVAS population. Bar heights are proportional to the number of individuals in each category. In the right panel, the population was separated into two groups according to FHS 10-year CV risk score. The left panel represents inviduals with low CV risk (FHS 10-year score < 10%), stratified in quintiles according to the prediction given by 3P score. The categories in each bar represent the number of individuals according to the number of affected territories.
Finally, we also explored whether the protein panel would improve a tentative model to predict SA constructed by using conventional risk factors. As shown in Fig. S2, inclusion of the three proteins into a machine learning model constructed using risk factors significantly improved AUC values for prediction of SA as compared with a model constructed with the risk factors alone. The improvement in performance was also maintained in the subpopulation of individuals with low CV risk (Fig. S2). Taken together, these data support the notion that the three-protein biomarker panel provides useful information to predict the presence of SA, particularly in individuals that have a low risk of having CVD according to traditional risk scales.
Discussion
To our knowledge, this study is the deepest and largest unbiased mass spectrometry-based proteomics analysis to date in the search for plasma proteins related to atherosclerosis. This was possible by using well-phenotyped cohorts for SA and by combining the quantitative accuracy and robustness provided by multiplexed isobaric labeling, peptide fractionation and well-validated and fully automated quantitative statistical workflows.28,30,31 Internal validation in the 3-year follow-up PESA visit was essential to reduce error sources in the discovery phase, to discard proteins with marked biological variability, and to concentrate our efforts on a set of proteins having robust associations with SA. The selected proteins showed alterations in their plasma levels that were stable over time and were also externally validated using proteomics in an independent cohort (AWHS). Furthermore, antibody-based turbidimetry kits confirmed the association with SA of the three selected proteins in two cohorts (AWHS and ILERVAS).
Several studies have assessed potential plasma protein biomarkers of SA in large cohorts of asymptomatic individuals.40,41 In contrast with these previous analyses, which were targeted studies performed using immunoassays, here we followed a hypothesis-free approach that allowed testing the association with SA of all the plasma proteins detectable by our LC-MS setup. From more than 1.000 proteins quantified per individual, we selected five proteins (PIGR, IGHA2, APOA, HPT and HEP2) that had a significant correlation with either plaque thickness or CACS and maintained their association with an increased likelihood of SA after adjustment by risk scores in two independent cohorts (PESA and AWHS). For translational purposes, we further selected a panel composed by three proteins (APOA, HPT and IGHA2) that can be measured using standard commercial kits routinely used in clinical practice. These proteins showed independent association with SA in two separate cohorts (AWHS and ILERVAS), and our results consistently indicate that the combination of these three proteins, APOA, IGAH2 and HPT, may be a useful tool to improve prediction of atherosclerosis presence and extension in the subclinical phase, complementing the information provided by conventional risk factors.
APOA is the main component of the Lp(a) particle, which has been identified as an independent predictor of coronary artery calcification, and epidemiological studies support a strong association between elevated Lp(a) and atherosclerotic CV disease outcomes,42,43 suggesting that Lp(a) plays a causal role in the disease.44 Current guidelines on CV prevention45 recommend the assessment of Lp(a) for individuals at high risk, however our results provide novel information that suggest that Lp(a) might be also determinant for prediction of SA in individuals considered at low risk according to conventional risk factors. Our results also indicate that APOA plasma levels provide significant information, in combination with those of IGHA2 and HPT and eventually other plasma proteins, to predict the presence of SA.
Increased HPT levels have been observed in CAD patients46 and were predictive of CV events.47 In this respect, HPT could increase together with other acute phase proteins under inflammatory conditions48 and combination of HPT with other inflammatory markers (white blood cell count, CRP) has been associated to CV events.49 In contrast, we could not find any evidence of association of CRP or other inflammatory proteins with SA in our study, highlighting a promising role for HPT in the detection of the disease in its subclinical stage. Elevated levels of total serum IgA have been reported in patients with severe atherosclerosis or with previous myocardial infarction or other major ischemic events and were found to correlate with myocardial infarction and cardiac death in dyslipidemic men.50 However, the specific association between the IgA isoform IGHA2 and atherosclerosis has never been reported before. The differential behavior with plaque thickness of several kinds of immunoglobulins in PESA and AWHS cohorts support a role for the humoral immune response in the asymptomatic phases of atherosclerosis51 and may suggest the existence of Ig class switching triggered by the differential activation of specific B-cell subsets proposed to take place during atherosclerosis.51 Further supporting this idea, we have very recently demonstrated that atherosclerotic mice undergo a germinal center antibody immune response featuring increased somatic hypermutation load, producing a skewed distribution of switched antibodies with altered proportions of Ig isotypes.52
Imaging studies have revealed discordances between conventional risk scores and the presence of atherosclerosis,53,54 since a high percentage of low-risk individuals still present SA.17 Detection of SA by imaging approaches is relevant in the primary prevention field, given that it has been shown to improve risk stratification4,5 and has been related to CV events. Imaging techniques, however, require specialized personnel and are not widely available. Thus, plasma proteins whose levels associate with SA have the potential of stratifying subjects at higher risk before imaging analysis, and hence may have social and economic impact. The protein levels measured using the kits confirmed the independent association of APOA, IGAH2 and HPT with SA in the large ILERVAS cohort and were useful to predict SA in this cohort. In a previous study it was shown that the majority of individuals classified at high risk by traditional scales had SA; however SA was also present in nearly 60% of participants at low risk, suggesting an association of atherosclerosis with characteristics not considered in standard risk scales.17 In this regard, our results showed that the three-protein panel was able to predict SA in the population of ILERVAS individuals considered at low-risk of CV events, which could facilitate targeted preventive/therapeutic approaches in this group of subjects.
We should comment here that, being an observational study, our inferences did not reflect direct causality. We must recognize the potential for residual, uncontrolled confounding that might partly explain the associations. Another limitation of this study is that the discovery phase was performed in a population formed by men only. Although there is a possibility that some protein factors specific of women could have been missed, the discovered proteins were validated in ILERVAS, a cohort composed also by females, suggesting that these factors are also valid for women.
In conclusion, this high-throughput, unbiased plasma proteomics study, performed in two large cohorts, allowed the identification of a panel of biomarker proteins that associate with subclinical atherosclerosis independently of known cardiovascular risk factors. The biomarker panel was able to predict SA in a third cohort, and the performance of the prediction was maintained in a subpopulation of individuals with low cardiovascular risk. The information provided by the three biomarkers may be useful for evaluation of the risk of having subclinical atherosclerosis in individuals that otherwise would be considered at low-risk in primary cardiovascular prevention.
Declaration of interests
The authors have no relationship to disclose.
Acknowledgments
Contributors
VF, JLMV and JV conceived and designed the study. VF, LFF, JMM, JS, JMO, MBL, JCEG, BI, ELP and AFO developed the clinical cohorts, including recruitment of patients, collection of clinical and demographic data, and storing of samples. JMFA, FSC and JMGR designed the subcohorts and supported the epidemiological analysis. EN, MGS, EBK and EC conducted the plasma proteomics analyses. EN, JMR and JV performed the quantitative analysis of plasma proteomics and verified the data. DML, ICP and JLMV conducted the antibody-based analyses and verified the data. AA and JV developed the machine-learning model. EN, JLMV, JV and AFO drafted the manuscript. All authors provided important intellectual revisions to the manuscript and read and approved the final manuscript
Acknowledgments
We thank Simon Bartlett (CNIC) for English editing, Ignacio Mahillo Fernández for support in statistics and Cesar Vegas-Dominguez and Lucia Ortega-Villanueva for technical support. This study was supported by competitive grants from the Spanish Ministry of Science, Innovation and Universities (BIO2015-67580-P, PGC2018-097019-B-I00, PID2019-106814RB-I00 and SAF2016-80843-R), through the Carlos III Institute of Health-Fondo de Investigación Sanitaria grant PRB3 (IPT17/0019 - ISCIII-SGEFI / ERDF, ProteoRed), CIBERCV and CIBERDEM, the Fundació MaratóTV3 (grant 122/C/2015) and “la Caixa” Banking Foundation (project HR17-00247). The PESA study is co-funded equally by the Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid, Spain, and Banco Santander, Madrid, Spain. The ILERVAS study was funded by the Diputació de Lleida. The study also receives funding from the Instituto de Salud Carlos III (PI15/02019; PI18/00610; RD16/0009) and the FEDER funds. The CNIC is supported by the Instituto de Salud Carlos III (ISCIII), the Ministerio de Ciencia, Innovación y Universidades (MCNU) and the Pro CNIC Foundation.
Data sharing statement
All MS raw data have been deposited in Peptide Atlas (http://www.peptideatlas.org/PASS/PASS01382 and http://www.peptideatlas.org/PASS/PASS01522).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103874.
Contributor Information
José Luis Martín-Ventura, Email: jlmartin@fjd.es.
Jesús Vázquez, Email: jvazquez@cnic.es.
Appendix. Supplementary materials
References
- 1.Khot U.N., Khot M.B., Bajzer C.T., et al. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003;290(7):898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- 2.Silverman M.G., Blaha M.J., Krumholz H.M., et al. Impact of coronary artery calcium on coronary heart disease events in individuals at the extremes of traditional risk factor burden: the multi-ethnic study of atherosclerosis. Eur Heart J. 2014;35(33):2232–2241. doi: 10.1093/eurheartj/eht508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yusuf S., Rangarajan S., Teo K., et al. Cardiovascular risk and events in 17 low-, middle-, and high-income countries. N Engl J Med. 2014;371(9):818–827. doi: 10.1056/NEJMoa1311890. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Ortiz A., Jimenez-Borreguero L.J., Penalvo J.L., et al. The progression and early detection of subclinical atherosclerosis (PESA) study: rationale and design. Am Heart J. 2013;166(6):990–998. doi: 10.1016/j.ahj.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Peters S.A., den Ruijter H.M., Bots M.L., Moons K.G. Improvements in risk stratification for the occurrence of cardiovascular disease by imaging subclinical atherosclerosis: a systematic review. Heart. 2012;98(3):177–184. doi: 10.1136/heartjnl-2011-300747. [DOI] [PubMed] [Google Scholar]
- 6.Baber U., Mehran R., Sartori S., et al. Prevalence, impact, and predictive value of detecting subclinical coronary and carotid atherosclerosis in asymptomatic adults: the bioimage study. J Am Coll Cardiol. 2015;65(11):1065–1074. doi: 10.1016/j.jacc.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Martin S.S., Blaha M.J., Blankstein R., et al. Dyslipidemia, coronary artery calcium, and incident atherosclerotic cardiovascular disease: implications for statin therapy from the multi-ethnic study of atherosclerosis. Circulation. 2014;129(1):77–86. doi: 10.1161/CIRCULATIONAHA.113.003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zavodni A.E., Wasserman B.A., McClelland R.L., et al. Carotid artery plaque morphology and composition in relation to incident cardiovascular events: the multi-ethnic study of atherosclerosis (MESA) Radiology. 2014;271(2):381–389. doi: 10.1148/radiol.14131020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson A.O., Blaha M.J., Arnan M.K., et al. Coronary artery calcium and incident cerebrovascular events in an asymptomatic cohort. The MESA study. JACC Cardiovasc Imaging. 2014;7(11):1108–1115. doi: 10.1016/j.jcmg.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidsson L., Fagerberg B., Bergstrom G., Schmidt C. Ultrasound-assessed plaque occurrence in the carotid and femoral arteries are independent predictors of cardiovascular events in middle-aged men during 10 years of follow-up. Atherosclerosis. 2010;209(2):469–473. doi: 10.1016/j.atherosclerosis.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Lamina C., Meisinger C., Heid I.M., et al. Association of ankle-brachial index and plaques in the carotid and femoral arteries with cardiovascular events and total mortality in a population-based study with 13 years of follow-up. Eur Heart J. 2006;27(21):2580–2587. doi: 10.1093/eurheartj/ehl228. [DOI] [PubMed] [Google Scholar]
- 12.van der Meer I.M., Bots M.L., Hofman A., del Sol A.I., van der Kuip D.A., Witteman J.C. Predictive value of noninvasive measures of atherosclerosis for incident myocardial infarction: the rotterdam study. Circulation. 2004;109(9):1089–1094. doi: 10.1161/01.CIR.0000120708.59903.1B. [DOI] [PubMed] [Google Scholar]
- 13.Polonsky T.S., McClelland R.L., Jorgensen N.W., et al. Coronary artery calcium score and risk classification for coronary heart disease prediction. JAMA. 2010;303(16):1610–1616. doi: 10.1001/jama.2010.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visseren F.L.J., Mach F., Smulders Y.M., et al. 2021 ESC guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J. 2021;42(34):3227–3237. doi: 10.1093/eurheartj/ehab484. [DOI] [PubMed] [Google Scholar]
- 15.Casasnovas J.A., Alcaide V., Civeira F., et al. Aragon workers' health study-design and cohort description. BMC Cardiovasc Disord. 2012;12:45. doi: 10.1186/1471-2261-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betriu A., Farras C., Abajo M., et al. Randomised intervention study to assess the prevalence of subclinical vascular disease and hidden kidney disease and its impact on morbidity and mortality: the ILERVAS project. Nefrologia. 2016;36(4):389–396. doi: 10.1016/j.nefro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Friera L., Penalvo J.L., Fernandez-Ortiz A., et al. Prevalence, vascular distribution, and multiterritorial extent of subclinical atherosclerosis in a middle-aged cohort: the PESA (progression of early subclinical atherosclerosis) study. Circulation. 2015;131(24):2104–2113. doi: 10.1161/CIRCULATIONAHA.114.014310. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Friera L., Fuster V., Lopez-Melgar B., et al. Normal LDL-cholesterol levels are associated with subclinical atherosclerosis in the absence of risk factors. J Am Coll Cardiol. 2017;70(24):2979–2991. doi: 10.1016/j.jacc.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Alvira J.M., Fuster V., Dorado B., et al. Short telomere load, telomere length, and subclinical atherosclerosis: the PESA study. J Am Coll Cardiol. 2016;67(21):2467–2476. doi: 10.1016/j.jacc.2016.03.530. [DOI] [PubMed] [Google Scholar]
- 20.Laclaustra M., Casasnovas J.A., Fernandez-Ortiz A., et al. Femoral and carotid subclinical atherosclerosis association with risk factors and coronary calcium: the AWHS study. J Am Coll Cardiol. 2016;67(11):1263–1274. doi: 10.1016/j.jacc.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez E., Betriu A., Lopez-Cano C., et al. Characteristics of atheromatosis in the prediabetes stage: a cross-sectional investigation of the ILERVAS project. Cardiovasc Diabetol. 2019;18(1):154. doi: 10.1186/s12933-019-0962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermudez-Lopez M., Martinez-Alonso M., Castro-Boque E., et al. Subclinical atheromatosis localization and burden in a low-to-moderate cardiovascular risk population: the ILERVAS study. Rev Esp Cardiol (Engl Ed) 2021;74(12):1042–1053. doi: 10.1016/j.rec.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Baldan-Martin M., Lopez J.A., Corbacho-Alonso N., et al. Potential role of new molecular plasma signatures on cardiovascular risk stratification in asymptomatic individuals. Sci Rep. 2018;8(1):4802. doi: 10.1038/s41598-018-23037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagwan N., Bonzon-Kulichenko E., Calvo E., et al. Comprehensive quantification of the modified proteome reveals oxidative heart damage in mitochondrial heteroplasmy. Cell Rep. 2018;23(12):3685–3697. doi: 10.1016/j.celrep.2018.05.080. e4. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Bartolome S., Navarro P., Martin-Maroto F., et al. Properties of average score distributions of SEQUEST: the probability ratio method. Mol Cell Proteom. 2008;7(6):1135–1145. doi: 10.1074/mcp.M700239-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Navarro P., Vazquez J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J Proteome Res. 2009;8(4):1792–1796. doi: 10.1021/pr800362h. [DOI] [PubMed] [Google Scholar]
- 27.Bonzon-Kulichenko E., Garcia-Marques F., Trevisan-Herraz M., Vazquez J. Revisiting peptide identification by high-accuracy mass spectrometry: problems associated with the use of narrow mass precursor windows. J Proteome Res. 2015;14(2):700–710. doi: 10.1021/pr5007284. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Marques F., Trevisan-Herraz M., Martinez-Martinez S., et al. A novel systems-biology algorithm for the analysis of coordinated protein responses using quantitative proteomics. Mol Cell Proteom MCP. 2016;15(5):1740–1760. doi: 10.1074/mcp.M115.055905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., et al. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13(3):1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 30.Trevisan-Herraz M., Bagwan N., Garcia-Marques F., et al. SanXoT: a modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics. 2019;35(9):1594–1596. doi: 10.1093/bioinformatics/bty815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., et al. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13(3):1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 32.Yang L., Wu H., Jin X., et al. Study of cardiovascular disease prediction model based on random forest in eastern China. Sci Rep. 2020;10(1):5245. doi: 10.1038/s41598-020-62133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12(null):2825–2830. [Google Scholar]
- 34.Probst P., Wright M.N., Boulesteix A. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip Rev Data Min Knowl Discov. 2019;9(3):e1301. [Google Scholar]
- 35.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 36.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 37.Ford E.S., Giles W.H., Mokdad A.H. The distribution of 10-year risk for coronary heart disease among US adults: findings from the national health and nutrition examination survey III. J Am Coll Cardiol. 2004;43(10):1791–1796. doi: 10.1016/j.jacc.2003.11.061. [DOI] [PubMed] [Google Scholar]
- 38.Marrugat J., D’Agostino R., Sullivan L., et al. An adaptation of the Framingham coronary heart disease risk function to European mediterranean areas. J Epidemiol Commun Health. 2003;57(8):634–638. doi: 10.1136/jech.57.8.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrugat J., Solanas P., D'Agostino R., et al. [Coronary risk estimation in Spain using a calibrated Framingham function] Rev Esp Cardiol. 2003;56(3):253–261. doi: 10.1016/s0300-8932(03)76861-4. [DOI] [PubMed] [Google Scholar]
- 40.Saarikoski L.A., Huupponen R.K., Viikari J.S., et al. Adiponectin is related with carotid artery intima-media thickness and brachial flow-mediated dilatation in young adults-the cardiovascular risk in young finns study. Ann Med. 2010;42(8):603–611. doi: 10.3109/07853890.2010.514284. [DOI] [PubMed] [Google Scholar]
- 41.Oikonen M., Wendelin-Saarenhovi M., Siitonen N., et al. Tissue inhibitor of matrix metalloproteinases 4 (TIMP4) in a population of young adults: relations to cardiovascular risk markers and carotid artery intima-media thickness. The cardiovascular risk in young finns study. Scand J Clin Lab Investig. 2012;72(7):540–546. doi: 10.3109/00365513.2012.704065. [DOI] [PubMed] [Google Scholar]
- 42.Ellis K.L., Boffa M.B., Sahebkar A., Koschinsky M.L., Watts G.F. The renaissance of lipoprotein(a): brave new world for preventive cardiology? Prog Lipid Res. 2017;68:57–82. doi: 10.1016/j.plipres.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Alonso R., Andres E., Mata N., et al. Lipoprotein(a) levels in familial hypercholesterolemia: an important predictor of cardiovascular disease independent of the type of LDL receptor mutation. J Am Coll Cardiol. 2014;63(19):1982–1989. doi: 10.1016/j.jacc.2014.01.063. [DOI] [PubMed] [Google Scholar]
- 44.Ellis K.L., Watts G.F. Is lipoprotein(a) ready for prime-time use in the clinic? Cardiol Clin. 2018;36(2):287–298. doi: 10.1016/j.ccl.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Mach F., Baigent C., Catapano A.L., et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020;41(1):111–188. doi: 10.1093/eurheartj/ehz455. [DOI] [PubMed] [Google Scholar]
- 46.Lee C.W., Cheng T.M., Lin C.P., Pan J.P. Plasma haptoglobin concentrations are elevated in patients with coronary artery disease. PLoS One. 2013;8(10):e76817. doi: 10.1371/journal.pone.0076817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holme I., Aastveit A.H., Hammar N., Jungner I., Walldius G. Haptoglobin and risk of myocardial infarction, stroke, and congestive heart failure in 342,125 men and women in the apolipoprotein mortality risk study (AMORIS) Ann Med. 2009;41(7):522–532. doi: 10.1080/07853890903089453. [DOI] [PubMed] [Google Scholar]
- 48.Graves K.L., Vigerust D.J. Hp: an inflammatory indicator in cardiovascular disease. Future Cardiol. 2016;12(4):471–481. doi: 10.2217/fca-2016-0008. [DOI] [PubMed] [Google Scholar]
- 49.Holme I., Aastveit A.H., Hammar N., Jungner I., Walldius G. Inflammatory markers, lipoprotein components and risk of major cardiovascular events in 65,005 men and women in the apolipoprotein mortality risk study (AMORIS) Atherosclerosis. 2010;213(1):299–305. doi: 10.1016/j.atherosclerosis.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 50.Kovanen P.T., Manttari M., Palosuo T., Manninen V., Aho K. Prediction of myocardial infarction in dyslipidemic men by elevated levels of immunoglobulin classes A, E, and G, but not M. Arch Intern Med. 1998;158(13):1434–1439. doi: 10.1001/archinte.158.13.1434. [DOI] [PubMed] [Google Scholar]
- 51.Tsiantoulas D., Diehl C.J., Witztum J.L., Binder C.J. B cells and humoral immunity in atherosclerosis. Circ Res. 2014;114(11):1743–1756. doi: 10.1161/CIRCRESAHA.113.301145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorenzo C., Delgado P., Busse C.E., et al. ALDH4A1 is an atherosclerosis auto-antigen targeted by protective antibodies. Nature. 2021;589(7841):287–292. doi: 10.1038/s41586-020-2993-2. [DOI] [PubMed] [Google Scholar]
- 53.Naqvi T.Z., Mendoza F., Rafii F., et al. High prevalence of ultrasound detected carotid atherosclerosis in subjects with low Framingham risk score: potential implications for screening for subclinical atherosclerosis. J Am Soc Echocardiogr. 2010;23(8):809–815. doi: 10.1016/j.echo.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Pen A., Yam Y., Chen L., Dennie C., McPherson R., Chow B.J. Discordance between Framingham risk score and atherosclerotic plaque burden. Eur Heart J. 2013;34(14):1075–1082. doi: 10.1093/eurheartj/ehs473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.