Abstract
Non-small cell lung cancer (NSCLC) has a high morbidity and mortality, and it is imperative to explore the latent pathogenesis mechanism of NSCLC progression to find potential prognostic biomarkers and therapeutic targets. The present study aimed to explore the biological function of circSHKBP1 in NSCLC. circSHKBP1 was found to be upregulated in NSCLC tissues and cell lines and was enriched in exosomes derived from NSCLC cells. Exosomal circSHKBP1 enhanced the proliferation, migration, invasion, and stemness of NSCLC cells. miRNA-1294 was identified as a target for circSHKBP1, and circSHKBP1 upregulated PKM2 expression by sponging miR-1294. Exosomal circSHKBP1 regulated glycolysis through PKM2 in a HIF-1α-dependent manner in NSCLC cells and promoted M2 polarization and macrophage recruitment. Moreover, exosomal circSHKBP1 promoted NSCLC cell growth, metastasis, and M2 infiltration in vivo. Thus, exosomal circSHKBP1 participated in the progression of NSCLC via the miR-1294/PKM2 axis. circSHKBP1 may be potential biomarker for the diagnosis and treatment of NSCLC.
Keywords: non-small cell lung cancer, exosome, circRNA, glycolysis, macrophage polarization
Graphical Abstract
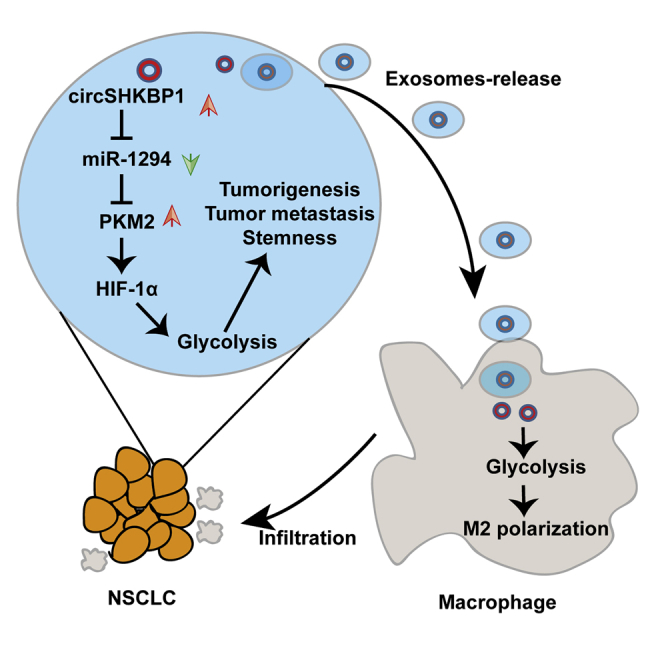
Exosomal circSHKBP1 enhances the tumorigenesis, metastasis, and stemness of NSCLC cells by sponging miR-1294 and upregulating PKM2 and promotes macrophage polarization and infiltration via glycolysis.
Introduction
Lung cancer is one of the most common malignancies and is a leading cause of cancer-related deaths worldwide.1 Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for approximately 85% of all lung cancer cases with a survival rate below 15%.2 Lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) are the most general subtypes of NSCLC. Surgical resection, chemotherapy, and radiotherapy are the main modalities for the treatment of NSCLC, with limited therapeutic effects. Although the diagnosis and treatment have improved in recent years, the prognosis for NSCLC remains poor.3 Furthermore, the potential mechanisms of NSCLC development, progression, and metastasis have not been fully elucidated. Hence, it is urgent to seek new strategies of treatment for patients with lung cancer. A better understanding of the underlying molecular mechanism and the exploration of potential prognostic biomarkers and therapeutic targets are vital to extending the survival rate of patients with NSCLC.
Exosomes are a specific subtype of extracellular vesicles with a diameter of 40–160 nm. Exosomes produced by host cells can be ingested by adjacent or distant cells and affect their biological function.4 Exosomes contain distinct biological molecules such as non-coding RNAs (ncRNAs), DNA, lipids, and proteins5 and are implicated in many diseases, including cancer.6 Exosomes containing functional ncRNAs can contribute to the progression of NSCLC. Exosomal miRNA-130a derived from cancer-associated fibroblasts confers cisplatin resistance in NSCLC cells.7 Exosomal circ-MEMO1 accelerates the progression and aerobic glycolysis of NSCLC.8 Exosome-transmitted long non-coding RNA (lncRNA) UFC1 promotes NSCLC cell proliferation, migration, and invasion by inhibiting phosphatase and tensin homolog deleted on chromosome ten (PTEN) expression.9 Therefore, it is worthwhile to further explore the role of exosomes in NSCLC progression.
Circular RNAs (circRNAs) are a category of ncRNAs with closed circular structures generated by linear RNA molecules.10 In the last few years, many circRNAs have been authenticated and found to be involved in multiple biological processes including the cell cycle, proliferation, invasion, migration, apoptosis, angiopoiesis, epithelial-mesenchymal transition, and immune escape.11, 12, 13, 14 circRNAs act as “miRNA sponges” and bind microRNAs (miRNAs) to abolish the regulation of target genes induced by miRNAs.15,16 hsa_circ_0000936, a subtype of circSHKBP1, is an oncogenic circRNA derived from the SHKBP1 gene and is reported to promote gastric cancer progression by regulating the miR-582–3p/HUR/vascular endothelial growth factor (VEGF) axis and suppressing heat shock protein 90 (HSP90) degradation.17 Moreover, hsa_circ_0000936 was also reported to regulate the angiogenesis of U87 glioma-exposed endothelial cells through the miR-544a/FOXP1 and miR-379/FOXP2 pathways.18 However, the roles of circSHKBP1 in NSCLC have rarely been reported. Hence, the present study aimed to explore the regulatory role of circSHKBP1 in NSCLC progression and further elucidate its possible mechanism in NSCLC, which may provide evidence for a promising novel diagnostic and therapeutic biomarker for NSCLC.
Results
circSHKBP1 is upregulated in NSCLC and enriched in serum exosomes
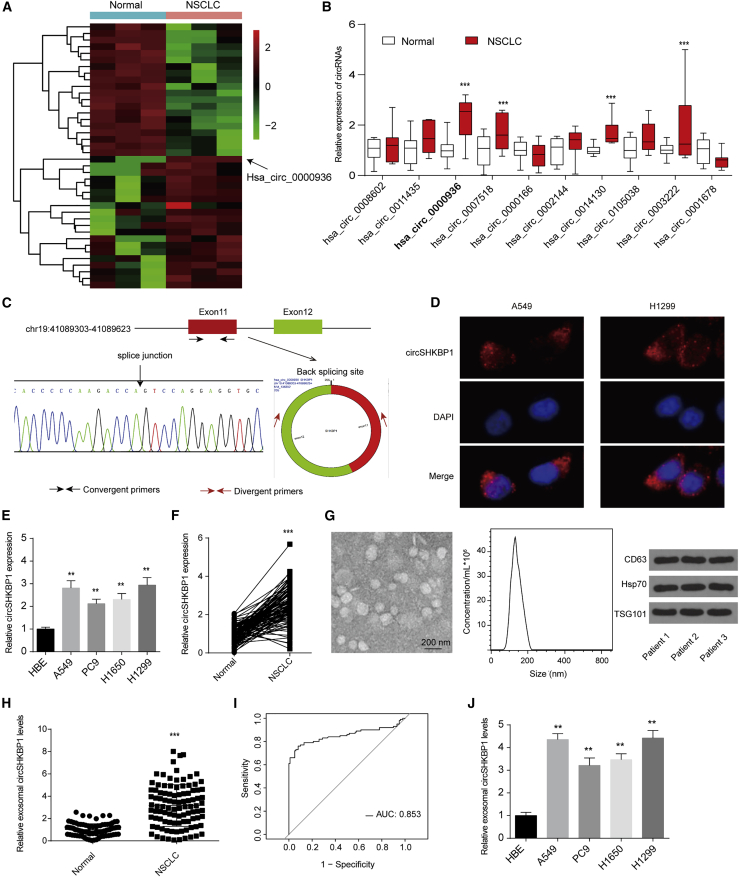
The circRNA GEO: GSE158695 dataset, comprised of three pairs of human NSCLC tissues and corresponding non-cancerous tissues, was downloaded from the GEO database. A total of 87 differentially expressed circRNAs were sorted by their fold change (|log FC| > 1, adjusted p value < 0.05). The top 40 differentially expressed genes are listed in a heatmap in Figure 1A. Then, we further analyzed the five most upregulated circRNAs by quantitative reverse transcription PCR (qRT-PCR) in 10 paired NSCLC tumors and normal tissues and found that hsa_circ_0000936, which was derived from SHKBP1 and designated circSHKBP1, was the most significantly upregulated circRNA (Figure 1B).
Figure 1.
Exosomal circSHKBP1 is overexpressed in NSCLC
(A) The heatmap shows the top 20 upregulated and downregulated circRNAs. (B) The expression of the ten most upregulated circRNAs in 10 paired NSCLC tumors and normal tissues. (C) Two exons form circSHKBP1 by back splicing from the chromosomal region, and Sanger sequencing of circSHKBP1 showed the back-splice junction. (D) A fluorescence in situ hybridization assay was conducted to determine the subcellular localization of circSHKBP1. (E) circSHKBP1 levels in four human NSCLC cell lines and one normal bronchial epithelial cell line (HBE). ∗∗p < 0.01 compared with HBE. (F) circSHKBP1 levels in 100 NSCLC tissues and matched normal tissues. ∗∗∗p < 0.01 compared with the normal group. (G) Exosomes from NSCLC tissues were identified via transmission electron microscopy (TEM), particle metrix (PMX), and western blotting. (H) Serum exosomal circSHKBP1 levels in 100 NSCLC patients. ∗∗∗p < 0.001 compared with the normal group. (I) ROC analysis was conducted to evaluate the potential diagnostic value of exosomal circSHKBP1. (J) Levels of exosomal circSHKBP1 in four human NSCLC cell lines and HBE cell culture medium. Quantitative data from three independent experiments are shown as the mean ± SD (error bars). ∗∗p < 0.01 compared with HBE.
Convergent primers were employed to detect the linear form of SHKBP1, and divergent primers containing a back-splice junction were used to amplify only the circular form of circSHKBP1. There was an evident binding around the expected size in the divergent primers for cDNA but no product for the genomic DNA (gDNA) (Figure S1A). Furthermore, Sanger sequencing confirmed the head-to-tail splicing of the amplified circSHKBP1, indicating that circSHKBP1 was derived from exons 11 and 12 of the SHKBP1 gene (Figure 1C). In addition, circSHKBP1 also showed resistance to RNase R and had a longer half-life than the linear form (Figures S1B and S1C). Importantly, we found that circSHKBP1 was largely localized in the cytoplasm of A549 and H1299 cells (Figure 1D).
Then, we analyzed the expression of circSHKBP1 in four human NSCLC cell lines and one normal bronchial epithelial cell line (HBE) as a control and found that circSHKBP1 was upregulated in NSCLC cells (Figure 1E). In addition, circSHKBP1 expression in 100 paired NSCLC tissues and normal tissues was examined, which demonstrated that circSHKBP1 was upregulated in NSCLC tissues compared with matched normal tissues (Figure 1F). We also found that SHKBP1 expression was upregulated in NSCLC tissues and was positively correlated with circSHKBP1 levels (Figures S1D and S1E). High SHKBP1 expression was associated with poor overall survival (OS) of patients with NSCLC (Figure S1F). Analysis of the clinical characteristics of circSHKBP1 suggested that circSHKBP1 expression levels were related to NSCLC lymphatic metastasis and tumor node metastasis (TNM) stage (Table S1).
Exosomes derived from the serum of patients with NSCLC were separated and identified via transmission electron microscopy (TEM), particle metrix (PMX), and western blotting (Figure 1G). Exosomes from the serum of patients with NSCLC contained high levels of circSHKBP1 (Figure 1H). The receiver operating characteristic (ROC) curve was plotted using the exosomal circSHKBP1 expression level to estimate the diagnostic value of exosomal circSHKBP1 (Figure 1I). The area under the ROC curve (AUC) was 0.853, suggesting that exosomal circSHKBP1 might serve as a diagnostic biomarker for NSCLC. Moreover, compared with the level in HBE cell culture medium, exosomal circSHKBP1 was overexpressed in 4 human NSCLC cell lines, especially A549 and H1299 (Figure 1J). Therefore, we used A549 and H1299 cells in the following analyses.
Exosomal circSHKBP1 promotes the proliferation, colony formation, migration, and invasion of NSCLC cells
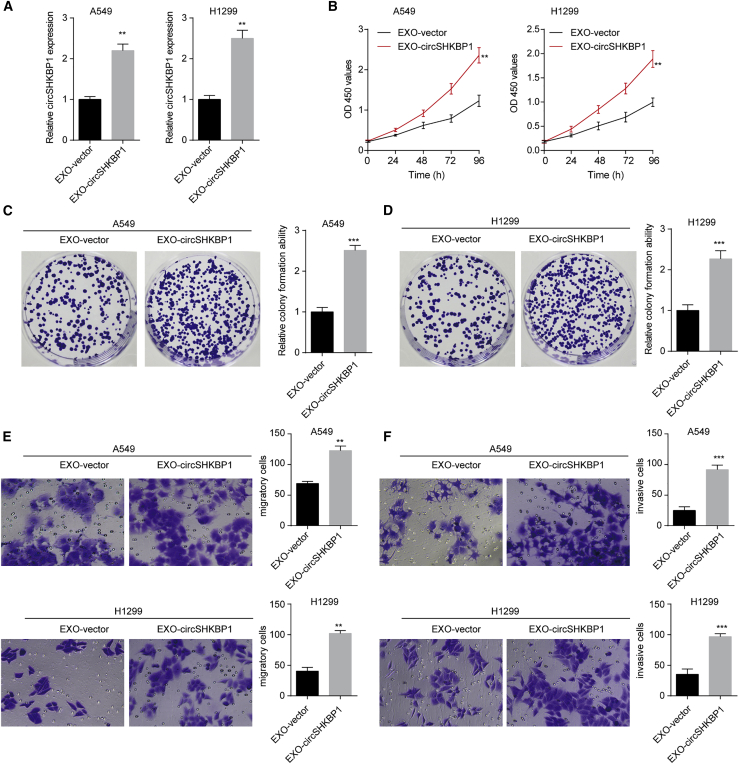
Considering the significant overexpression of exosomal circSHKBP1 in the culture medium of NSCLC cells, we further explored the role of exosomal circSHKBP1 in NSCLC cells. circSHKBP1 overexpression upregulated exosomal circSHKBP1 levels, while circSHKBP1 knockdown decreased exosomal circSHKBP1 levels in A549 and H1299 cells (Figures S2A and S2B). First, exosomes (200 μg/mL) separated from circSHKBP1-plasmid-transfected A549 and H1299 cell culture medium were used to coculture untreated A549 and H1299 cells. Exosomes from circSHKBP1-plasmid-transfected NSCLC cells (EXO-circSHKBP1) markedly enhanced circSHKBP1 levels in both A549 and H1299 cells (Figure 2A). Then, Cell Counting Kit-8 (CCK-8) assays were carried out to test the proliferation of A549 and H1299 cells, and we found that EXO-circSHKBP1 induced cell proliferation (Figure 2B). The results of colony formation assays indicated that EXO-circSHKBP1 enhanced the tumor formation ability of A549 and H1299 cells (Figures 2C and 2D). Transwell assays showed that EXO-circSHKBP1 promoted NSCLC cell migration and invasion (Figures 2E and 2F). To further confirm the effect of exosomal circSHKBP1, we also knocked down circSHKBP1 expression by specific small interfering RNA (siRNA), and then the exosomes from each group were separated. As expected, knockdown of exosomal circSHKBP1 inhibited NSCLC cell proliferation, colony formation, migration, and invasion (Figures S2C–S2G). These results suggested that circSHKBP1 could affect the biological functions of adjacent or distant NSCLC cells. Exosomal circSHKBP1 promoted NSCLC cell growth and metastasis, while the knockdown of exosomal circSHKBP1 suppressed NSCLC cell development.
Figure 2.
EXO-circSHKBP1 promotes NSCLC cell proliferation, colony formation, migration, and invasion in vitro
A549 and H1299 cells were first transfected with the circSHKBP1 plasmid or specific siRNA for circSHKBP1, and then the exosomes were isolated and designated EXO-circSHKBP1 and EXO-si-circSHKBP1, respectively. (A) The levels of circSHKBP1 in A549 and H1299 cells after coculture with EXO-circSHKBP1. (B) Assessment of the proliferation of A549 and H1299 cells after coculture with EXO-circSHKBP1 by CCK-8 assay. (C and D) Assessment of the colony formation of A549 and H1299 cells cocultured with exosomes by colony formation assay. (E and F) Assessment of the migration and invasion of A549 and H1299 cells cocultured with EXO-circSHKBP1 by Transwell assay. Quantitative data from three independent experiments are shown as the mean ± SD. ∗∗p < 0.01 and ∗∗∗p < 0.001 compared with EXO-vector.
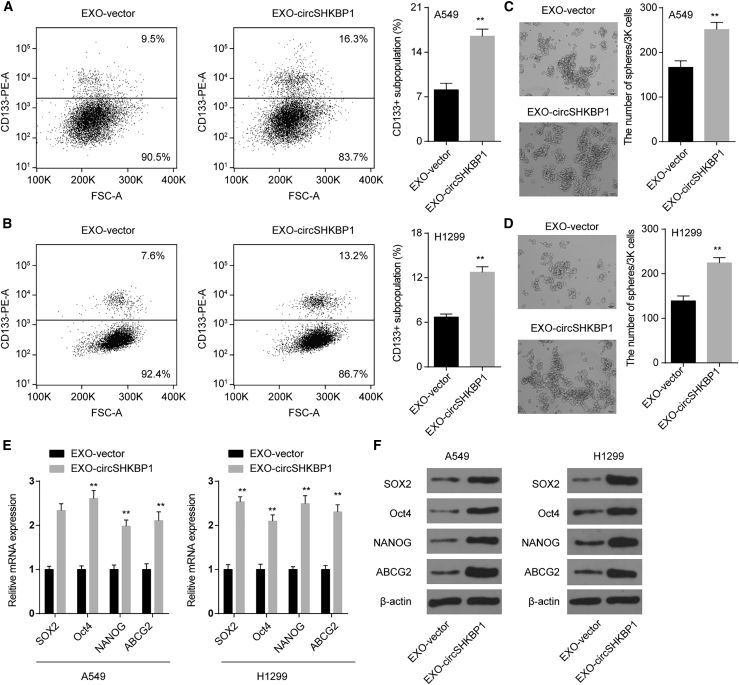
Exosomal circSHKBP1 enhances the stemness of NSCLC cells in vitro
Cancer stem cells (CSCs) are the major source of cancer initiation, relapse, and drug resistance.19 We next explored the role of exosomal circSHKBP1 on CSC properties. CD133 is a specific cell surface marker for NSCLC CSCs, and CD133+ cells show stem-like and highly tumorigenic features.20 In this study, we found that EXO-circSHKBP1 increased the percentage of CD133+ cells in both A549 and H1299 cells (Figures 3A and 3B). Moreover, sphere-formation assays revealed that EXO-circSHKBP1 promoted self-renewal ability (Figures 3C and 3D). In addition, we also showed that EXO-circSHKBP1 upregulated the mRNA and protein levels of stemness-related genes (SOX2, ABCG2, NANOG, and Oct4) (Figures 3E and 3F). All results suggested that EXO-circSHKBP1 was involved in the stemness of NSCLC cells, which further accelerated the malignant phenotype of NSCLC.
Figure 3.
EXO-circSHKBP1 enhances stemness-related gene expression and CSC properties in NSCLC cells
(A and B) The percentage of CD133+ cells was analyzed by flow cytometry. (C and D) The self-renewal ability of A549 and H1299 cells was assessed by sphere-formation analysis. (E and F) mRNA and protein levels of stemness-related genes (SOX2, ABCG2, NANOG, and Oct4) in A549 and H1299 cells after the indicated treatment.
Quantitative data from three independent experiments are shown as the mean ± SD (error bars). ∗∗p < 0.01 compared with EXO-vector.
circSHKBP1 upregulates PKM2 expression via inhibition of miR-1294
Recently, many circRNAs have been reported to function as competing endogenous RNAs (ceRNAs) in modulating the expression and biological functions of miRNAs.4 As circSHKBP1 was largely localized in the cytoplasm, we hypothesized that circSHKBP1 may act as a miRNA sponge to prevent miRNAs from binding with their target mRNAs. miR-1294 was predicted to be a cotarget for circSHKBP1 and PKM2 through an online database and article.21 miR-1294 was found to be downregulated, while PKM2 was upregulated in 30 NSCLC tissues with high levels of circSHKBP1 (Figures S3A and S3B). Furthermore, we found that the expression of miR-1294 was negatively associated with the expression of circSHKBP1, and that the expression of PKM2 was positively associated with the expression of circSHKBP1 (Figures S3C and S3D). The expression of miR-1294 was negatively associated with the expression of PKM2 (Figure S3E). Importantly, we also found that the expression of serum exosomal circSHKBP1 was negatively associated with the expression of miR-1294 and positively associated with the expression of PKM2 (Figures S3F and S3G). To further explore the clinical significance of PKM2 and miR-1294 in NSCLC, we evaluated their prognostic effects via a public database of Kaplan-Meier plotter analysis: http://www.kmplot.com. There was no predictive significance of miR-1294 in either LUAD or LUSC (Figures S3H and S3I). A higher PKM2 level was correlated with a poorer OS in LUAD, while there seemed to be no predictive significance of PKM2 in patients with LUSC (Figures S3J and S3K). These results showed that circSHKBP1, miR-1294, and PKM2 could constitute a regulatory pathway in NSCLC.
To further confirm this conjecture, luciferase assays were performed. As shown in Figure S4A, the relative firefly/Renilla luciferase activity was decreased in A549 cells cotransfected with a miR-1294 mimic in the wild-type PKM2 group or the wild-type circSHKBP1 group, while no obvious change was observed in the other groups. To further understand the mechanism between circSHKBP1 and miR-1294, radio immunoprecipitation (RIP) assays were performed. Ago2, which is the key component of the RNA-induced silencing complex, was used to verify whether circSHKBP1 serves as ceRNA to sequester miR-1294. The data suggested that circSHKBP1 was enriched in Ago2 immunoprecipitates (Figure S4B). Subsequently, RNA pull-down experiment results suggested that both circSHKBP1 and PKM2 physically bind to miR-1294, as circSHKBP1 and PKM2 were dramatically enriched in the sample pulled down by biotinylated miR-1294 (Figures S4C and S4D). We assessed the expression of miR-1294 in A549 and H1299 cells after circSHKBP1 overexpression or knockdown and found that circSHKBP1 overexpression reduced miR-1294 levels and circSHKBP1 knockdown increased miR-1294 levels (Figures S4E and S4F). Moreover, circSHKBP1 overexpression promoted PKM2 mRNA expression, and circSHKBP1 knockdown inhibited PKM2 mRNA expression (Figures S4G and S4H). The miR-1294 inhibitor upregulated PKM2 mRNA expression, and the miR-1294 mimic inhibited PKM2 mRNA expression in A549 and H1299 cells (Figures S4I and S4J). These results showed that circSHKBP1 may perform its function by upregulating PKM2 via sponging miR-1294.
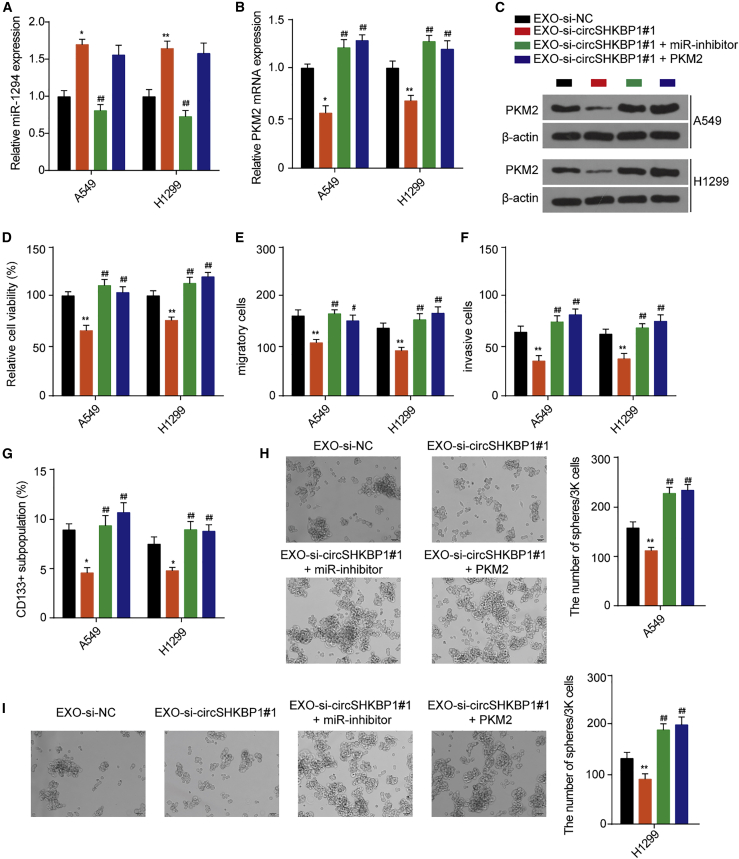
EXO-circSHKBP1 affects NSCLC cell proliferation, migration, invasion, and stemness via the miR-1294/PKM2 axis
To verify whether EXO-circSHKBP1 performs its function through the miR-1294/PKM2 axis, NSCLC cells were cotreated with EXO-si-circSHKBP1#1 and a miR-1294 inhibitor or a PKM2 overexpression vector. EXO-si-circSHKBP1#1 upregulated miR-1294 levels in A549 and H1299 cells. The miR-1294 inhibitor reversed the miR-1294 levels upregulated by EXO-si-circSHKBP1#1, while PKM2 overexpression had no significant effect on the miR-1294 levels upregulated by EXO-si-circSHKBP1#1 (Figure 4A). EXO-si-circSHKBP1#1 inhibited the mRNA and protein expression of PKM2, and both the miR-1294 inhibitor and PKM2 overexpression reversed the mRNA and protein expression of PKM2 decreased by EXO-si-circSHKBP1#1 (Figures 4B and 4C). Furthermore, EXO-si-circSHKBP1#1 inhibited NSCLC cell proliferation, migration, invasion, and stemness compared with EXO-si-NC. The inhibitory effect of EXO-si-circSHKBP1#1 on NSCLC cell proliferation, migration, invasion, and stemness was completely reversed by the miR-1294 inhibitor and PKM2 overexpression (Figures 4D and 4I). These results suggested that exosomal circSHKBP1 affected the biological functions of adjacent or distant NSCLC cells via the miR-1294/PKM2 axis.
Figure 4.
EXO-circSHKBP1 affects NSCLC cell proliferation, migration, invasion, and stemness via the miR-1294/PKM2 axis
A549 and H1299 cells cocultured with EXO-si-circSHKBP1#1 were transfected with a miR-1294 inhibitor or PKM2 overexpression vector. (A) The levels of miR-1294 in A549 and H1299 cells were assessed after the indicated treatment. (B) The levels of PKM2 mRNA in A549 and H1299 cells were assessed after the indicated treatment. (C) The levels of PKM2 protein in A549 and H1299 cells were assessed after the indicated treatment. (D) Assessment of the proliferation of A549 and H1299 cells after the indicated treatment with CCK-8. (E and F) Assessment of the migration and invasion of A549 and H1299 cells after the indicated treatment by Transwell assay. (G) The percentage of CD133+ cells was analyzed by flow cytometry. (H and I) The self-renewal abilities of A549 and H1299 cells was assessed by sphere-formation analysis. Quantitative data from three independent experiments are shown as the mean ± SD (error bars). ∗p < 0.05 and ∗∗p < 0.01 compared with EXO-si-NC; ##p < 0.01 compared with EXO-si-circSHKBP1#1.
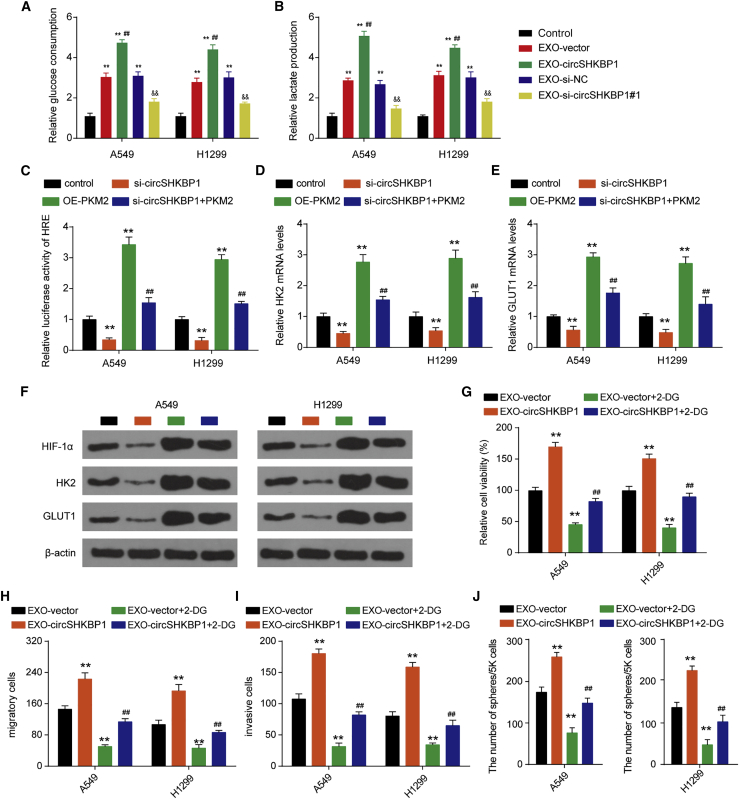
EXO-circSHKBP1 regulates glycolysis through PKM2 in a HIF-1α-dependent manner in NSCLC cells
Glycolysis is the major way by which cancer cells obtain energy, and PKM2 catalyzes the irreversible committed step of glycolysis.22 We further explored whether exosomal circSHKBP1 played a role in glycolytic metabolism. Exosomes from NSCLC cells promoted glucose uptake and lactate production in A549 and H1299 cells. EXO-circSHKBP1 promoted glucose uptake and lactate production, while EXO-si-circSHKBP1#1 inhibited the glucose uptake and lactate production in A549 and H1299 cells (Figures 5A and 5B), suggesting that circSHKBP1 plays a positive role in NSCLC glycolysis. PKM2 is known to play a crucial role in glycolysis by activating HIF-1α-dependent transcription of glycolytic enzymes.23 Luciferase assays indicated that circSHKBP1 knockdown reduced the reporter activity of HIF-1α and that PKM2 overexpression enhanced the reporter activity of HIF-1α. PKM2 overexpression reversed the inhibition of the reporter activity of HIF-1α induced by circSHKBP1 knockdown (Figure 5C). Additionally, circSHKBP1 knockdown downregulated the mRNA and protein expression of HIF-1α, HK2, and GLUT1 in A549 and H1299 cells, while PKM2 overexpression had the opposite effect. PKM2 overexpression restored the mRNA and protein expression of HIF-1α, HK2, and GLUT1 reduced by the circSHKBP1 knockdown (Figures 5D–5F). The knockdown of PKM2 in NSCLC cells abolished the exosomal circSHKBP1 effects on NSCLC cell proliferation, migration, invasion, and stemness (Figure S5). In addition, we also found that the glycolytic inhibitor 2-DG completely reversed the positive effects on NSCLC cell proliferation, migration, invasion, and stemness induced by EXO-circSHKBP1 (Figures 5G–5J). Therefore, exosomal circSHKBP1 promoted NSCLC progression by enhancing glycolysis in a PKM2/HIF-1α-dependent manner.
Figure 5.
EXO-circSHKBP1 regulates NSCLC progression through glycolysis
(A and B) A549 and H1299 cells were cocultured with EXO-circSHKBP1 or EXO-si-circSHKBP1#1 for 48 h, and then glucose consumption and lactate excretion were assessed. ∗∗p < 0.01 compared with control; ##p < 0.01 compared with EXO-vector; &&p < 0.01 compared with EXO-si-NC. (C) HIF-1α-responsive luciferase activity was examined after the indicated transfection. (D and E) The mRNA expression of HK2 and GLUT1 in A549 and H1299 cells after circSHKBP1 overexpression or knockdown. (F) The protein levels of HIF-1α, HK2, and GLUT1 in A549 and H1299 cells after circSHKBP1 overexpression or knockdown. (G) Assessment of the proliferation of A549 and H1299 cells after the indicated treatment by CCK-8. (H and I) Assessment of the migration and invasion of A549 and H1299 cells after the indicated treatment by Transwell assay. (J) The self-renewal ability of A549 and H1299 was assessed by sphere-formation analysis. ∗∗p < 0.01 compared with the control; ##p < 0.01 compared with si-circSHKBP1. Quantitative data from three independent experiments are shown as the mean ± SD (error bars).
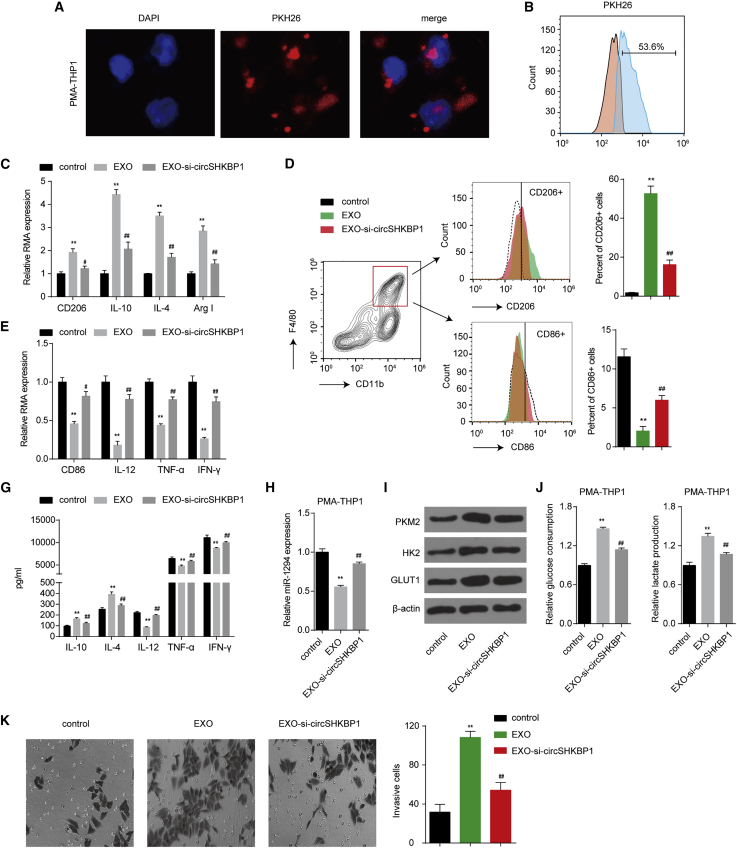
Exosomal circSHKBP1 promotes M2 polarization and macrophage recruitment
Tumor-associated macrophages (TAMs) and tumor cells are primary components of the tumor microenvironment. M2 polarization of TAMs plays a major role in NSCLC malignancy and metastasis. PKM2, a downstream effector of circSHKBP1, was also implicated in macrophage differentiation through glycolysis.22 Therefore, we next explored the function of exosomal circSHKBP1 on macrophage differentiation. A549-derived exosomes were labeled with the red fluorescent dye PKH26 and were efficiently taken up by THP-1 macrophages (Figures 6A and 6B). After coculturing A549-derived exosomes with phorbol 12-myristate 13-acetate (PMA)-induced THP-1 cells for 48 h, exosomes enhanced the mRNA expression of M2 polarization markers (CD206, interleukin [IL]-10, IL-4, and Arg1) and reduced the mRNA expression of M1 polarization markers (CD86, IL-12, tumor necrosis factor alpha [TNF-α], and interferon gamma [INF-γ]) (Figures 6C and 6D). In addition, the flow cytometry results showed an enhancement in the percentage of CD206+ cells and a decrease in the percentage of CD86+ cells after exosome treatment (Figures 6E and 6F). The cytokines of a typical M2 phenotype (IL-10 and IL-4) and M1 phenotype (IL-12, TNF-α, and IFN-γ) were investigated. THP-1 macrophages cocultured with exosomes showed increased IL-10 and IL-4 and significantly decreased IL-12, TNF-α, and IFN-γ, suggesting polarization to the M2 phenotype (Figure 6G). In terms of mechanism, we found that A549-derived exosomes inhibited miR-1294 and promoted PKM2, HK2 and GLUT1 protein expression, glucose uptake, and lactate production in macrophages (Figures 6H–6J), suggesting that circSHKBP1 also induced metabolic reprogramming that further regulated macrophage differentiation. The decrease in circSHKBP1 in exosomes partly reversed these effects (Figures 6C–6J). In short, A549-derived exosomes promoted M2 polarization and inhibited M1 polarization of THP-1 macrophages by packaging circSHKBP1 and enhancing glycolysis.
Figure 6.
Exosomal circSHKBP1 promotes M2 polarization of macrophages
(A) Representative images by confocal microscopy of the internalization of PKH67-labeled A549 exosomes (red) by THP-1 macrophages. (B) Internalization of PKH67-labeled exosomes by macrophages was assessed by flow cytometry after coculture for 24 h. (C) The mRNA expression of the M2 polarization markers CD206, IL-10, IL-4, and Arg1 in macrophages cocultured with exosomes for 48 h. (D) The mRNA expression of the M1 polarization markers CD86, IL-12, TNF-α, and INF-γ in macrophages cocultured with exosomes for 48 h. (E and F) Flow cytometry was used to explore the surface expression of CD206 and CD86 in macrophages cocultured with exosomes for 48 h. (G) The levels of the respective inflammatory cytokines in cell culture supernatants of macrophages cocultured with exosomes assessed using a ProcartaPlex combinable panel. (H) miR-1294 levels in macrophages cocultured with exosomes for 48 h. (I) Protein levels of PKM2, HK2, and GLUT1 in macrophages cocultured with exosomes for 48 h. (J) The glucose consumption and lactate excretion were detected in macrophages cocultured with exosomes for 48 h. (K) THP-1 macrophage cells were added to the upper wells of a Boyden chamber for assessment of chemotaxis toward exosomes released by A549 cells added to the lower chamber. The cells were allowed to migrate for 24 h at 37°C before staining with crystal violet and quantification. Data represent mean ± SD of triplicate experiments. ∗∗p < 0.01 compared with control; ##p < 0.01 compared with EXO.
To explore the biological function of exosomal circSHKBP1 during macrophage recruitment in NSCLC, cell migration assays were performed to assess its impact on cell motility. THP-1 macrophages exhibited greater chemotactic responses to exosome-containing media than to phosbate-buffer saline (PBS)-only control medium. The migration ability of THP-1 macrophages was significantly suppressed when the bottom chamber contained exosomes from circSHKBP1-knockdown A549 cells (Figure 6K). These data suggested that NSCLC cells exhibited a greater capacity to recruit monocytes/macrophages via exosome release into the local microenvironment.
Moreover, we next sought to validate our results with primary human macrophages derived from CD14+ monocytes isolated from the peripheral blood of healthy donors. Similarly, the migration of human macrophages was also significantly enhanced when incubated with EXO from A549 cells (Figure S6A). When compared with the EXO group, the migration of human macrophages was obviously reduced when incubated with EXO-si-circSHKBP1, indicating that exosomal circSHKBP1 promoted the migration of human macrophages. Moreover, we also found that exosomal circSHKBP1 enhanced the percentage of CD206+ cells and reduced the percentage of CD86+ cells (Figure S6B). Besides, exosomal circSHKBP1 promoted the secretion of IL-10 and IL-4 and inhibited the secretion of IL-12, TNF-α, and IFN-γ (Figure S6C). These results indicated that exosomal circSHKBP1 promoted the polarization of human macrophages to the M2 phenotype and the migration of human macrophages, which was consistent with the results of THP-1 macrophages. All of these results indicated that exosomal circSHKBP1 released by NSCLC cells promoted M2 polarization and macrophage recruitment.
Exosomal circSHKBP1 suppresses CD8+ T cell secretion of cytokines
In addition to macrophages, we also explored the role of exosomal circSHKBP1 in other immune cells, such as like CD8+ T cells. CD8+ T cells were cocultured with exosomes isolated from A549 cells with circSHKBP1 overexpression or knockdown. Then, CD3 antibodies were used to activate the T cells. The results showed that CD8+ T cells incubated with EXO-circSHKBP1 had a significantly lower expression of TNF-α, IFN-γ, granzyme-B, and perforin in the supernatant than CD8+ T cells incubated with EXO-vector. Compared with EXO-si-NC, treatment with EXO-si-circSHKBP1#1 promoted the secretion of TNF-α, IFN-γ, granzyme-B, and perforin in CD8+ T cells (Figure S7). These results indicated that exosomal circSHKBP1 suppressed CD8+ T cell secretion of cytokines, which may lead to immune escape of NSCLC.
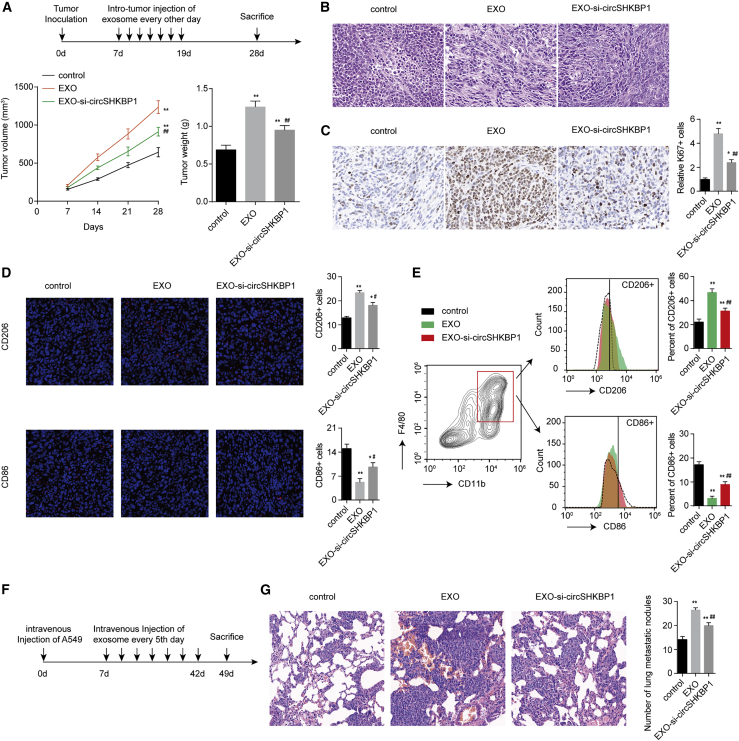
Exosomal circSHKBP1 promotes NSCLC cell growth, metastasis, and M2 infiltration in vivo
To explore the effect of exosomal circSHKBP1 on NSCLC cell growth and macrophage infiltration in vivo, a subcutaneous xenograft model of A549 cells in BALB/c nude mice was constructed. As expected, exosomes from A549 cells led to the promotion of tumor growth compared with the PBS control group. Exosomes from circSHKBP1-knockdown A549 cells suppressed tumor growth compared with the EXO group (Figure 7A). The number of Ki67+ cells was higher in the EXO group than in the control group. circSHKBP1 knockdown decreased Ki67+ cells (Figure 7B). These results showed that A549-derived exosomes promoted NSCLC cell growth in vivo partly through the packaged circSHKBP1. As exosomal circSHKBP1 has a function in macrophage polarization and recruitment, we also examined the levels of CD206+ and CD86+ macrophages through immunofluorescent staining and flow cytometry. More CD206+ cells and fewer CD86+ cells were found in the EXO group. However, more CD86+ cells and fewer CD206+ cells were found in the EXO-si-circSHKBP1 group than in the EXO group (Figures 7C and 7D). Moreover, EXO significantly enhanced the expression of circSHKBP1 and PKM2 in xenografts and decreased the expression of miR-1294 in xenografts compared with the control group (Figure S8). Compared with the EXO group, the EXO-si-circSHKBP1 group exhibited decreased expression of circSHKBP1 and PKM2 in xenografts and increased expression of miR-1294 in xenografts (Figure S8). These results suggested that exosomal circSHKBP1 also regulated miR-1294 and PKM2 expression in vivo.
Figure 7.
Exosomal circSHKBP1 promotes NSCLC cell growth and M2 infiltration in vivo
(A) Experimental scheme, tumor volume, and weights of the xenograft. (B) The proliferative status of tumors shown by Ki67 immunohistochemical staining. (C) Immunofluorescent images of CD206 (red) or CD86 (red). (D) Macrophages were separated from murine tumor tissues using a Percoll-layered liquid. Surface expression of CD206 and CD86 was assessed in macrophages using flow cytometry. (E) A549 cells were injected into nude mice via the tail vein and then treated with exosomes through intravenous injection. Autopsy of the lungs and H&E staining of the lungs in the nude mice are displayed. (F and G) Number of metastatic nodules in the lungs from the nude mice via H&E staining (five sections evaluated per lung). ∗p < 0.05 and ∗∗p < 0.01 compared with control; ##p < 0.01 compared with EXO.
Moreover, intravenous injection of exosomes promoted pulmonary metastasis in nude mice. The EXO-si-circSHKBP1 group showed less pulmonary metastasis than the EXO group but still displayed more pulmonary metastasis than the control group (Figures 7E and 7F). In conclusion, we found that exosomal circSHKBP1 promotes the growth and metastasis of NSCLC cells and CD206+ TAM infiltration in vivo.
Discussion
Emerging research has indicated that circRNAs affect the malignant biological behaviors of NSCLC.24, 25, 26 Here, we verified that circSHKBP1 functioned as an oncogene in NSCLC. circSHKBP1 was found to be improved in the tissues and serum exosomes of NSCLC patients. Exosomal circSHKBP1 affected NSCLC proliferation, migration, invasion, stemness, glycolysis, and macrophage polarization and recruitment. Furthermore, overexpression of circSHKBP1 was associated with advanced TNM stages, lymphatic metastasis, and poor prognosis. As previously reported, circSHKBP1 is plentiful in serum exosomes, and exosomal circSHKBP1 regulates the miR-582–3p/HUR/VEGF pathway, suppresses HSP90 degradation, and promotes gastric cancer progression.17 The knockdown of circ-SHKBP1 inhibits AGGF1 expression via the miR-544a/FOXP1 or the miR-379/FOXP2 pathway to further inhibit the viability, migration, and tube formation of U87 glioma-exposed endothelial cells by AGGF1 or via the PI3K/AKT and ERK1/2 pathways.18 Thus, these results suggest that circSHKBP1 may be a biomarker for NSCLC diagnosis and prognosis.
Exosomes can be secreted by all cell types and play important roles in the occurrence and development of tumors. A preponderance of evidence continues to indicate that exosomal circRNAs play an important role in the progression of NSCLC. For example, exosomal circRNA_100395 derived from adipose-derived mesenchymal stem cells was reported to inhibit NSCLC malignant transformation by sponging miR-141-3p to regulate Hippo/YAP signaling.24 circSATB2 transferred by exosomes promotes the proliferation, migration, and invasion of NSCLC cells.4 The serum exosome-based biomarker circ_0008928 has been implicated in cisplatin sensitivity, tumor progression, and glycolysis metabolism in NSCLC.27 Exosome-transmitted hsa_circ_0014235 promotes NSCLC malignant development via the miR-520a-5p/CDK4 axis.28 However, it is still not clear whether circSHKBP1 is involved in NSCLC progression. In the present study, we indicated that circSHKBP1 was enriched in exosomes and that exosomal circSHKBP1 affected NSCLC cell function and macrophage phenotype. Knockdown of circSHKBP1 in exosomes inhibited proliferation, migration, invasion, stemness, glycolysis, and macrophage polarization and recruitment in vitro and in vivo. Therefore, circSHKBP1 might serve as an oncogene in NSCLC.
PKM2 is a rate-limiting enzyme that catalyzes the progression of glycolysis and promotes tumor cells to thrive.29 PKM2 is reported to promote cancer progression by acting as a transcriptional coactivator of HIF-1α and regulating the metabolic reprogramming of NSCLC cells.23,30 PKM2 expression was upregulated in exosomes under hypoxic conditions, and exosomal PKM2 protein could be directly transferred to tumor cells and change malignant behavior.30 PKM2 may also affect the tumor microenvironment to accelerate carcinogenesis through exosomes. Exosomal PKM2 remodels the tumor microenvironment to facilitate hepatocellular carcinoma progression.22 In this study, we revealed that PKM2 was one of the downstream targets of exosomal circSHKBP1 and acted as an oncogene in NSCLC.
TAMs have been reported to promote cancer progression. Lung cancer cells can induce macrophage infiltration by upregulating CCL2 and CXCL3 production in NSCLC cells.31 Macrophages undergo polarization into M1 (classic) or M2 (alternative) subtypes in response to different stimuli.32 M1 macrophages promote inflammatory responses, while M2 macrophages inhibit inflammatory responses. TAMs, which are M2 macrophages, promote the development of several types of NSCLC tumors.33, 34, 35 In this study, we found that the exosomal circSHKBP1/miR-1294/PKM2 axis promoted the polarization of THP-1 cells from the M1 to the M2 phenotype in vitro and enhanced the inflammatory infiltration of macrophages in tumor tissue in vivo.
In summary, we demonstrate that circSHKBP1 is upregulated in patients with NSCLC and is related to TNM stages, poor prognosis, and lymphatic metastasis. Moreover, circSHKBP1 is enriched in exosomes derived from NSCLC cells, and exosomal circSHKBP1 plays an important role in the progression of NSCLC. circSHKBP1 controls PKM2 expression by sponging miR-1294 to enhance the proliferation, migration, invasion, stemness, and glycolysis of NSCLC cells as well as macrophage polarization and recruitment both in vitro and in vivo. This study has thus provided a new perspective in research on circRNAs in NSCLC and may contribute to providing potential targets for the diagnosis and treatment of NSCLC.
In conclusion, exosomal circSHKBP1 derived from NSCLC cells promotes NSCLC proliferation, migration, invasion, stemness, and glycolysis and accelerates macrophage polarization and recruitment by inhibiting miR-1294 and upregulating PKM2. circSHKBP1 can serve as a tumor promoter in NSCLC and is closely related to poor prognosis. Overall, circSHKBP1 may be a potential therapeutic biomarker for NSCLC.
Materials and methods
Cell culture
The human normal lung epithelial cell line HBE (HBE) and human lung cancer cell lines (A549, PC9, H1650, and H1299) were purchased from Guangzhou Saiku Biotechnology (Guangzhou, China). All cells were cultured in RPMI-1640 medium (HyClone, Logan, Australia) containing 10% fetal bovine serum (Clark Bioscience, Houston, TX, USA) at 37°C and 5% CO2 with a saturated humidity. Exosomes in fetal bovine serum were removed by ultracentrifugation before the experiment.
Lung cancer tissues
One hundred paired NSCLC and matched normal adjacent tissue samples were obtained from Shenzhen People's Hospital (Shenzhen, China). Surgically resected tissues were frozen in liquid nitrogen. Informed consent was obtained from each participant in the study. Tumors were histologically graded according to the Elston and Ellis method. The clinical characteristics of patients with NSCLC are shown in Table S1. To correlate serum exosomal circSHKBP1 levels with NSCLC progression, we also collected blood samples from 30 NSCLC patients and normal volunteers. Serum from lung cancer and non-cancerous donors was centrifuged, the supernatant was collected, and the exosomes were isolated using Total Exosome Isolation Reagent (from serum) (Invitrogen, Carlsbad, CA, USA). The exosomes in the cell culture medium supernatant were isolated using Total Exosome Isolation Reagent (from cell culture media) (Invitrogen). Exosome precipitates were washed with PBS for purity and then resuspended in PBS for further research. The concentration and size distribution of exosomes were examined by nanoparticle tracking analysis using a NanoSight NS300 (Malvern, UK). Hsp70 and CD63 proteins were used to further identify exosomes by western blotting. The protocols of this study were approved by the Ethics Committee of People's Hospital of Longhua.
Total RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen), and total RNA was isolated from exosomes using an Exosomal RNA and Protein Extraction Kit (101Bio, Mountain View, CA, USA). The quality and concentration of the purified total RNAs were assessed using a NanoDrop1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). A High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) was applied to measure circRNA and other linear gene expressions. For circRNA reverse transcription, the RNA sample was treated with RNase R (Epicentre, Madison, WI, USA) for 10 min at 37°C. For linear gene reverse transcription, RNase R-free water was used. For miRNA reverse transcription, TaqMan MicroRNA Reverse-Transcription kits (Applied Biosystems) were used to measure miRNA expression. qRT-PCR was carried out using TaqMan Universal Master Mix II (Applied Biosystems) according to the manufacturer's procedure. β-actin and U6 were used as internal references. The normalized method was performed using the 2−ΔΔct method. The primer sequences are listed in Table S2.
Western blotting and antibodies
Total protein was extracted from the frozen cells using RIP assay (RIPA) buffer. Thirty micrograms of protein was loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After transfer to a polyvinylidene difluoride membrane (PVDF), the proteins were blocked with 5% non-fat milk and incubated in primary antibodies at 4°C overnight. Subsequently, the membranes were incubated with a horseradish-peroxidase-conjugated secondary antibody for 2 h. The targeted proteins were detected and visualized with an enhanced chemiluminescence system (ECL, Beyotime, Shanghai, China) and X-ray film (GE Healthcare). β-actin was used as a loading control. The antibodies used (all purchased from Abcam [Cambridge, MA, USA]) are listed as follows: CD63 (ab134045), Hsp70 (ab2787), TSG101 (ab125011), SOX4 (ab70598), Oct4 (ab200834), NANOG (ab109250), ABCG2 (ab207732), β-actin (ab8227), PKM2 (ab137852), HiF-1α (ab179483), GLUT1 (ab115730), HK2 (ab209847), goat anti-rabbit immunoglobulin G (IgG) H&L (HRP) (ab6721), and rabbit anti-mouse IgG H&L (HRP) (ab6728).
RNase R treatment
Two micrograms of total RNA was incubated for 30 min at 37°C with or without 5 U/μg RNase R (Epicentre), subsequently purified by an RNeasy MinElute Cleaning Kit (Qiagen, Germantown, MD, USA), and then analyzed by qPCR.
Fluorescence in situ hybridization (FISH)
A specific circSHKBP1 FISH probe was designed and used in the experiment. Cells attached to slides were immobilized with 4% paraformaldehyde, washed with PBS, and then digested by Proteinase K (Sangon, Shanghai, China) at 37°C for 5 min. After washing with PBS, the cells were immobilized with 1% paraformaldehyde and mixed with 70%, 95%, and 100% ethanol overnight at 4°C. A fluorescently labeled probe for circRNA_09505 was applied during hybridization, and DAPI (Beyotime, Shanghai, China) was used to stain the nucleus.
Flow cytometry
Cells were collected, washed, and incubated for 30 min at 4°C with florescence-conjugated antibodies (CD133, CD206, and CD86, all obtained from Abcam). Labeled cells were analyzed using a FACSCalibur flow cytometer (BD, San Jose, CA, USA) and FlowJo software (Tree Star, Ashland, OR, USA).
Metabolic assays
Glycolysis was investigated by assessing glucose consumption and lactate production. After transfection or treatment with exosomes for 48 h, the levels of glucose and lactate in culture medium were separately measured by using a Glucose Assay Kit (Rsbio, Shanghai, China) and Lactate Assay Kit (Rsbio).
Enzyme-linked immunosorbent assay (ELISA)
ELISA was used to evaluate the IL-10, IL-12, IL-4, TNF-α, and INF-γ (Enzo Life Sciences, New York, NY, USA) contents in culture medium derived from macrophages. Following the manufacturer's instructions, each sample was evaluated in duplicate, and protein levels were normalized to the number of adherent cells.
Luciferase reporter assay
A549 cells were seeded in 96-well plates and then cotransfected with circSHKBP1/PKM2 wild-type or mutant plasmids and miR-1294 mimics or miR-NC using Lipofectamine 2000. After 48 h of incubation, the luciferase activity was measured with a kit according to the manufacturer's instructions (Promega, Madison, WI, USA). Relative luciferase activities were measured and expressed as the ratio of firefly luciferase activity to Renilla luciferase activity.
To determine the reporter activity of HIF-1α, HIF-1 luciferase reporter plasmids (YEASEN, Shanghai, China) containing multiple HIF-1 binding sites were cotransfected with PKM2-overexpression plasmids or si-circSHKBP1. Forty-eight hours after transfection, firefly and Renilla luciferase activities were measured by a dual-luciferase reporter assay system.
RIPA
RIPAs were performed using a RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) following the protocol provided by the manufacturer. Cells were lysed by hypotonic RIP buffer and mixed with magnetic beads conjugated with rabbit anti-human IgG and an anti-ago2 antibody (Abcam, Cambridge, MA, USA). The extracted RNAs were analyzed by qPCR to demonstrate the enrichment of circSHKBP1.
Pull-down assay
After cross-linking, the cells were lysed. Biotinylated miR-1294 and non-specified probes ordered from GenePharma (Shanghai, China) were added to cell lysates to pull down their targets. M-280 Streptavidin Dynabeads (Invitrogen, Carlsbad, CA, USA) were added to all samples and incubated overnight. The beads were then separated by a magnetic support, and RNAs were analyzed by qRT-PCR to demonstrate the enrichment of circSHKBP1 and PKM2.
Cell proliferation assay
Cell proliferation assays were performed with the CCK-8 (Beyotime, Shanghai, China). Cells were seeded in 96-well plates at 1 × 104 cells per well for 24 h and then cocultured with exosomes for 48 h. Ten microliters of CCK-8 solution was added to each well and incubated for 1 h at 37°C. The absorbance at 450 nm was then measured.
Colony formation assay
Cells were plated on a 6-well plate at a density of 1 × 103 cells/well and cultured for 14 days with exosome treatment. Cells were fixed with methanol and then stained with crystal violet for 20 min. The number of clones was counted.
Migration and invasion assays of NSCLC cells
Migration assays were performed using Transwell pore polycarbonate membrane inserts (Corning, NY, USA), and invasion assays were performed using Matrigel-coated invasion chambers (BD Biocoat, Corning, NY, USA). NSCLC cells, transfected or untransfected, were seeded into the upper chamber, with or without exosome treatment, and cultured in serum-free medium. The lower chamber contained medium supplemented with 20% exosome-free fetal bovine serum. After incubation for 24 h at 37°C, cells on the bottom surface of the filter were fixed, stained with 0.5% crystal violet, and counted under a microscope.
Monocyte isolation and macrophage differentiation
Buffy coats were collected from healthy donors, and peripheral blood mononuclear cells were isolated by centrifugation in Ficoll (PromoCell, Germany). Then, the CD14+ monocytes were enriched by anti-CD14 magnetic beads (Miltenyi Biotec) following the manufacturer's instructions. The percentage of CD14+ monocytes was higher than 90%. For human macrophage differentiation, CD14+ monocytes were cultured in RPMI-1640 containing 15% fetal bovine serum (FBS), 100 unit/mL streptomycin/penicillin, and PMA (10 nM) for 7 days.
THP-1 cell differentiation and exosome treatment
THP-1 monocytic cells were induced to develop a macrophage-like (M0) phenotype by 20 ng/mL PMA (Sigma-Aldrich) treatment for 48 h in 5% FBS–RPMI media and then cultured in 10% FBS–RPMI control media for 24 h. Differentiated cells were digested and seeded onto a 6-well plate. Exosomes (5 μg/mL) in 10% exosome-free FBS–RPMI media were treated for 48 h.
Macrophage invasion assay
Differentiated THP-1 cells were seeded onto Transwell inserts coated with Matrigel and cultured in a 24-well plate with 10 μg of exosome-containing medium (10% exosome-free FBS-DMEM) in the lower chamber. Cell invasion in response to exosomes was monitored for 24 h, and migrated cells on the bottom membrane were examined by crystal violet staining.
CD8+ T cell isolation and incubation
Human CD8+ T cells were isolated and purified from healthy donor peripheral blood mononuclear cells using an Easy-Sep Direct Human CD8+ T Cell Isolation Kit (STEMCELL Technologies). Anti-CD3 antibodies (BD Biosciences) were used for CD8+ T cell activation. Exosomes derived from the A549 cells were placed into 12-well plates, and then preactivated CD8+ T cells were added for incubation for 24 h. The culture medium supernatant was collected for IFN-γ, TNF-α, granzyme-B, and perforin assessment by using IFN-γ, TNF-α, granzyme-B, and perforin ELISA kits (eBioscience, San Diego, CA, USA) in accordance with the manufacturer's guidelines.
Animal models
All experiments were approved by the Ethics Committee of People's Hospital of Longhua. To establish the subcutaneous NSCLC model in female nude mice, 5 × 106 A549 cells were injected into the right posterior flanks of the mice. Intratumor injection of exosomes (10 μg per mouse) was started on day 7 after the tumor cell injection and continued every other day. The mice were sacrificed 28 days after tumor cell injection, and the tumor size and weight were calculated (tumor volume = (L × W2)/2, where L = long axis and W = short axis). Subcutaneous tumor tissues were harvested for hematoxylin and eosin (H&E) and Ki67 staining. For tissue immunofluorescence, CD206 and CD86 antibodies were used to detect macrophage recruitment in vivo. Sections stained with primary antibody were subsequently incubated with the appropriate Alexa Fluor 488- or Alexa Fluor 568-conjugated secondary antibody. Macrophages of nude mouse subcutaneous tumors were separated and obtained using Percoll (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer's instructions. Flow cytometry was performed after incubation for 30 min at 4°C with fluorescence-conjugated antibodies.
For the lung metastasis model, 1 × 107 A549 cells were injected into the tail vein. Intravenous injection of exosomes (10 μg per mouse) was started on day 7 after tumor cell injection and continued every 5 days. The mice were sacrificed 49 days after tumor-cell injection, and the tumor nodules on the surface of the lungs were counted. The lungs were fixed in formalin and embedded in paraffin, and lung sections of 5 mm thickness were then stained with H&E for histologic analysis.
Statistical analysis
The results were displayed as the mean ± SD or SEM. Two-tailed Student's t test and one-way analysis of variance (ANOVA) followed by Tukey's test were used to compare the differences between two groups and among multiple groups. Kaplan–Meier survival curves were analyzed with the log rank test. p < 0.05 was considered statistically significant.
Acknowledgments
The research was supported by China Postdoctoral Foundation (no. 2021M703372); Basic and Applied Basic Research Fund of Guangdong Province (NO. 2021A1515110967); Construction Fund of Key Medical Subject in Longhua District, Shenzhen City; National Natural Science Foundation of China (no. 82003172).
Author contributions
All authors participated in the design and interpretation of the study, analysis of the data, and review of the manuscript; W.C., D.T., S.L., and G.S. conducted the experiments; J.L., X.H., and Y.D. supplied critical reagents, W.C. and D.T. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.01.012.
Contributor Information
Guanle Shen, Email: shenguanlelh@163.com.
Yong Dai, Email: daiyong22@aliyun.com.
Supplemental information
References
- 1.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Jr., Wu Y.L., Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Yao Y., Zhou Y., Hua Q. circRNA hsa_circ_0018414 inhibits the progression of LUAD by sponging miR-6807-3p and upregulating DKK1. Mol. Ther. Nucleic Acids. 2021;23:783–796. doi: 10.1016/j.omtn.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge W.L., Chen Q., Meng L.D., Huang X.M., Shi G.D., Zong Q.Q., Shen P., Lu Y.C., Zhang Y.H., Miao Y., et al. The YY1/miR-548t-5p/CXCL11 signaling axis regulates cell proliferation and metastasis in human pancreatic cancer. Cell Death Dis. 2020;11:294. doi: 10.1038/s41419-020-2475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao L., Wang L., Dai T., Jin K., Zhang Z., Wang S., Xie F., Fang P., Yang B., Huang H., et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat. Immunol. 2018;19:233–245. doi: 10.1038/s41590-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y., Xu X., Ocansey D.K.W., Cao H., Qiu W., Tu Q., Mao F. CircRNAs as promising biomarkers of inflammatory bowel disease and its associated-colorectal cancer. Am. J. Transl. Res. 2021;13:1580–1593. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T., Zhang P., Li H.X. CAFs-derived exosomal miRNA-130a confers cisplatin resistance of NSCLC cells through PUM2-dependent packaging. Int. J. Nanomedicine. 2021;16:561–577. doi: 10.2147/IJN.S271976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding C., Xi G., Wang G., Cui D., Zhang B., Wang H., Jiang G., Song J., Xu G., Wang J. Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS Axis. Front. Genet. 2020;11:962. doi: 10.3389/fgene.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang X., Gu J., Zhang J., Shi H., Hou S., Xu X., Chen Y., Zhang Y., Mao F., Qian H., et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11:215. doi: 10.1038/s41419-020-2409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang T., Xia Y., Lv J., Li B., Li Y., Wang S., Xuan Z., Xie L., Qiu S., He Z., et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer. 2021;20:66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z., Zhu X., Li Y., Liang M., Liu M., Liu Z., Qin L., Wu X., Du K., Liu L., et al. Circ-HMGA2 (hsa_circ_0027446) promotes the metastasis and epithelial-mesenchymal transition of lung adenocarcinoma cells through the miR-1236-3p/ZEB1 axis. Cell Death Dis. 2021;12:313. doi: 10.1038/s41419-021-03601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan L., Lian W., Zhang X., Han S., Cao C., Li X., Li M. Human circular RNA0054633 regulates high glucoseinduced vascular endothelial cell dysfunction through the microRNA218/roundabout 1 and microRNA218/heme oxygenase1 axes. Int. J. Mol. Med. 2018;42:597–606. doi: 10.3892/ijmm.2018.3625. [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Jia Y., Wang B., Yang S., Du K., Luo Y., Li Y., Zhu B. Circular RNA CHST15 sponges miR-155-5p and miR-194-5p to promote the immune escape of lung cancer cells mediated by PD-L1. Front. Oncol. 2021;11:595609. doi: 10.3389/fonc.2021.595609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Panda A.C. Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 17.Xie M., Yu T., Jing X., Ma L., Fan Y., Yang F., Ma P., Jiang H., Wu X., Shu Y., et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Q., Zhao L., Liu Y., Liu X., Zheng J., Yu H., Cai H., Ma J., Liu L., Wang P., et al. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol. Ther. Nucleic Acids. 2018;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng K., Liu L., Tan X., Zhang Z., Li J., Ou Y., Wang X., Yang S., Xiang R., Sun P. WIP1 promotes cancer stem cell properties by inhibiting p38 MAPK in NSCLC. Signal Transduct. Target. Ther. 2020;5:36. doi: 10.1038/s41392-020-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao S., Wang Z., Gao X., He W., Cai Y., Chen H., Xu R. FOXC1 induces cancer stem cell-like properties through upregulation of beta-catenin in NSCLC. J. Exp. Clin. Cancer Res. 2018;37:220. doi: 10.1186/s13046-018-0894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Q., Yu H., Chen J., Song X., Sun L. Antitumor effect of miR-1294/pyruvate kinase M2 signaling cascade in osteosarcoma cells. Onco. Targets Ther. 2020;13:1637–1647. doi: 10.2147/OTT.S232718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P.P., Luo L.J., Chen H.Z., Chen Q.T., Bian X.L., Wu S.F., Zhou J.X., Zhao W.X., Liu J.M., Wang X.M., et al. Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol. Cell. 2020;78:1192–1206.e10. doi: 10.1016/j.molcel.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R.N., Pandey A., Semenza G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W., Zhang K., Li L., Xu Y., Ma K., Xie H., Zhou J., Cai L., Gong Y., Gong K. Downregulation of lncRNA ZNF582-AS1 due to DNA hypermethylation promotes clear cell renal cell carcinoma growth and metastasis by regulating the N(6)-methyladenosine modification of MT-RNR1. J. Exp. Clin. Cancer Res. 2021;40:92. doi: 10.1186/s13046-021-01889-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Tang X., Ren H., Guo M., Qian J., Yang Y., Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z., Jiang H., Fu H., Zhang Y. A circGLIS3/miR-644a/PTBP1 positive feedback loop promotes the malignant biological progressions of non-small cell lung cancer. Am. J. Cancer Res. 2021;11:108–122. [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y., Dou Y., Zhang J., Qi J., Xin Z., Zhang M., Xiao Y., Ci W. The RNA N6-Methyladenosine Methyltransferase METTL3 promotes the progression of kidney cancer via N6-Methyladenosine-dependent translational enhancement of ABCD1. Front. Cell Dev. Biol. 2021;9:737498. doi: 10.3389/fcell.2021.737498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X., Tao R., Sun L., Ji X. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 2020;20:552. doi: 10.1186/s12935-020-01642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong N., Ojo D., Yan J., Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 30.Wang D., Zhao C., Xu F., Zhang A., Jin M., Zhang K., Liu L., Hua Q., Zhao J., Liu J., et al. Cisplatin-resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics. 2021;11:2860–2875. doi: 10.7150/thno.51797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmall A., Al-Tamari H.M., Herold S., Kampschulte M., Weigert A., Wietelmann A., Vipotnik N., Grimminger F., Seeger W., Pullamsetti S.S., et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am. J. Respir. Crit. Care Med. 2015;191:437–447. doi: 10.1164/rccm.201406-1137OC. [DOI] [PubMed] [Google Scholar]
- 32.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Gao A., Zhang F., Yang Z., Wang S., Fang Y., Li J., Wang J., Shi W., Wang L., et al. ILT4 inhibition prevents TAM- and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation. Theranostics. 2021;11:3392–3416. doi: 10.7150/thno.52435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Fleur L., Botling J., He F., Pelicano C., Zhou C., He C., Palano G., Mezheyeuski A., Micke P., Ravetch J.V., et al. Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory t cells and supports cytotoxic lymphocyte function. Cancer Res. 2021;81:956–967. doi: 10.1158/0008-5472.CAN-20-1885. [DOI] [PubMed] [Google Scholar]
- 35.Hwang I., Kim J.W., Ylaya K., Chung E.J., Kitano H., Perry C., Hanaoka J., Fukuoka J., Chung J.Y., Hewitt S.M. Tumor-associated macrophage, angiogenesis and lymphangiogenesis markers predict prognosis of non-small cell lung cancer patients. J. Transl. Med. 2020;18:443. doi: 10.1186/s12967-020-02618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.