Abstract
Single cell RNA-Seq technology enables the assessment of RNA expression in individual cells. This makes it popular in experimental biology for gleaning specifications of novel cell types as well as inferring heterogeneity. Experimental data conventionally contains zero counts or dropout events for many single cell transcripts. Such missing data hampers the accurate analysis using standard workflows, designed for massive RNA-Seq datasets. Imputation for single cell datasets is done to infer the missing values. This was traditionally done with ad-hoc code but later customized pipelines, workflows and specialized software appeared for this purpose. This made it easy to benchmark and cluster things in an organized manner. In this review, we have assembled a catalog of available RNA-Seq single cell imputation algorithms/workflows and associated softwares for the scientific community performing single-cell RNA-Seq data analysis. Continued development of imputation methods, especially using deep learning approaches, would be necessary for eradicating associated pitfalls and addressing challenges associated with future large scale and heterogeneous datasets.
Keywords: Single cell, RNA-Seq, imputation, algorithms, heterogeneity, analysis
1. BACKGROUND
Single cell RNA sequencing (RNA-Seq) is a cutting-edge technique, introduced in 2009, that can dissect the cellular heterogeneity of a plethora of cells [1]. Single cell RNA-Seq plays a phenomenal role in the identification of specific markers of same cell type, fluctuating states of same phenotypic cells, intra-population heterogeneity at microscopic resolution, transcript dynamicity and cell to cell variability of transcriptome [2, 3]. It has facilitated the construction of an extensive atlas of phenotypically similar human cells [4] and paved the way for researchers to initiate the “The Human Cell Atlas” project [5, 6]. It aims to map and quantify all cell types in the body, which would be useful for diagnosis and disease treatment. Above all, single-cell study supports unbiasedness in diverse research areas, treatment of many diseases by unmasking the presence of rare sub-populations of cells (i.e. cancer stem cells), underlying mechanisms in common diseases (i.e. kidney diseases) [3], reconstruction of genetic lineage trajectories, embryonic development [7], evolution and genomic diversity of bacterial ecosystem [8], etc.
All this is not without problems and single-cell sequencing has to deal with several challenges such as drop-out events and high level of noise because small amounts of RNA from a single-cell requires amplification, which is susceptible to damage, contamination and distortion [9]. Minimal expression may be read as a zero by computer and hence, loss of information impedes proper downstream analysis. To deal with this problem, computer programs based on logical and coherent algorithms are required for replacing missing or negligible values with substitute values, derived using certain formulas (either based on prior information or trained on dataset under study). This derivation of missing values and associated information is called imputation and is a critical component of single-cell data analysis.
An algorithm is a defined set of clear and implementable instructions on a computer. Usually, it addresses a problem and pertains to providing a solution through computation. With an avalanche of data from sequencing platforms, algorithms and programs to address machine derived biological data challenges and solve problems in computational biology have been in the limelight. Single cell technology produces a bulk of data but the issue of missing data is there which obstructs accurate transcriptomic studies. Algorithms have been designed to address this shortfall and impute missing or drop out values. We aim to provide an overview of such algorithms, which could be useful for scientists working with single cell RNA-Seq.
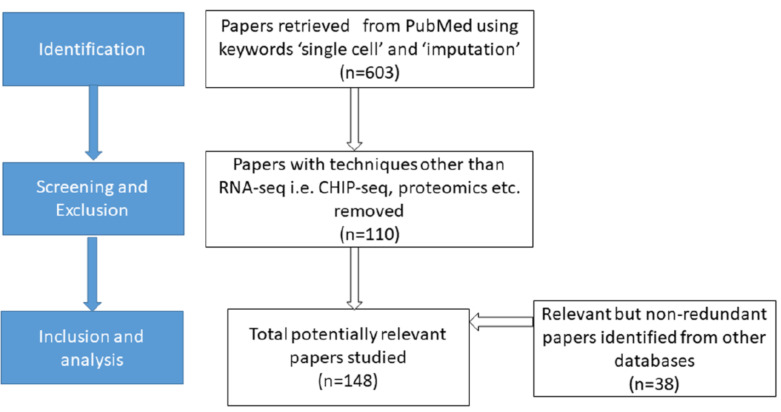
2. LITERATURE SEARCH AND CONSPECTUS
We searched for ‘single cell’ and ‘imputation’ in Pubmed (dated 22 April, 2020). Inclusion and exclusion criteria are mentioned in (Fig. 1). Imputation methods for inferring proteins from RNA-Seq and phylogenetic coupled genotype analysis were also eliminated from the study. Only the ones with imputation analysis specific to RNA-Seq data were retained and categorized into three major types according to Lahnemann et al. [10], (1) model based, (2) data smoothing and (3) data reconstruction (low-ranked matrix-based or deep learning) methods. Algorithms integrating several approaches and falling under more than one category (such as Seurat falling under random forest-based machine learning or low-ranked matrix-based method), were listed only once. Programs such as EnImpute [11], using an ensemble of methods for an output combined from several software was not listed. For chosen approaches, the full text was downloaded for each algorithm/tool and programming language, operating system (Windows/Unix), working link information was obtained. Method and implementation of the workflow was taken into account and acquired information was summarized. To the best of the authors knowledge, this is the first comprehensive review of single cell RNA-Seq software.
Fig. (1).
An outline of literature search for this review. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3. ALGORITHMS EMPLOYING MODEL-BASED APPROACH
The primary group of algorithms enforce model-based approach for inferring the data sparsity and hence, imputation. Such probabilistic models may or may not be able to differentiate amid technical and biological zeros. Usually, gene expression is imputed for technical ones if they are able to separate both. Eight such algorithms were identified listed in (Table 1). The first model-based method specific to single cell RNA-Seq data was presented in a JMLR workshop in 2016 by the name of BISCUIT [12](Bayesian Inference for Single-cell ClUstering and ImpuTing). It is an implementation of Dirchlet process mixture model, to iteratively normalize and cluster imputation expression. This was the initial, wholly Bayesian model for grouping, normalizing and imputing single-cell expression data. Biological and technical variation was resolved without spike-in and gaussian was implemented for gene-cell distribution. Imputation was inferred using Gibbs sampling. Following pursuit, SAVER [13](Single-cell Analysis Via Expression Recovery) was reported, which coalesces information across genes to infer transcript counts. Adaptive shrinkage using a Poisson-Gamma or negative binomial model is used for the purpose. SAVER-X [14] (Single-cell Analysis Via Expression Recovery via harnessing eXternal data) is an extension of the program and uses a Bayesian approach coupled to an autoencoder. This makes learned analyses from UMI counts possible. Gene-gene relation information is transferred across heterogeneous data (varying conditions, species etc) to impute a new dataset. It gives uncertainty co-efficient but associated computational intensity makes it less useful for large datasets. It is now implemented as a web app as well. ScImpute [2] is a scalable method which performs imputation only on dropout entries by probability calculation of specified gene in similar cells. This is done by fitting a Gamma-Gaussian mixture model on cell clusters. It can analyze heterogeneous datasets and is robust but does not provide uncertainty quantification values and may oversmooth the data. The package utilizing this algorithm is Granatum [15]. scRecover [16], uses zero-inflated negative binomial (ZINB) regression for maximum likelihood-based expression imputation. It docks values with ScImpute and other algorithms (SAVER, MAGIC) for final imputation. scUnif [17] is a supervised learning method, employing Bayesian approach with expectation–maximization algorithm, coupled with Gibbs sampling technique. It analyzes single as well as bulk data. Dropout inference and deconvolution are concurrent in bulk data. VIPER [18] accomplishes iterative inference of imputation using scant set of neighboring cells. A nonnegative sparse regression model is used for the estimate of expression. It is computationally efficient but does not provide uncertainty co-efficients for imputation values. scGAIN [19] applies adversarial learning to construct generative network model for imputing. Generator and the discriminator networks are trained on batches of 128 cells in each round, followed by mask matrix formation. It identifies right entries for imputation and spawned data points, with characteristics analogous to existing data help infer data distribution. Mean expression determines zero values of single-cell data. bayNorm [20] applies a novel Bayesian approach for standardizing data features and deducing expression features. Informed data structures consolidate accuracy and sensitivity in differential expression study. It can be used for UMI and non-UMI based data. A likelihood function coupled with binomial model of mRNA transcript capture is utilized after scaling, enabling it to capture mean-variance and mean-dropout relationship. Generated transcript distributions (2D using point estimate from posterior or 3D using posterior distribution) resemble fluorescence in situ hybridization (FISH) detection of single molecules. It exhibits high scalability coupled with computational efficiency. It is also useful for heterogeneous data.
Table 1.
Features of methods employing model-based approach.
| Serial No. | Software/ Method | OS | Interface |
Programming
Language |
Link |
|---|---|---|---|---|---|
| 1 | BISCUIT | Windows, Linux | Commandline | R | https://github.com/sandhya212/BISCUIT_SingleCell_IMM_ICML_2016 |
| 2 | SAVER | Windows/Linux | Commandline | R | https://github.com/mohuangx/SAVER |
| 3 | SAVER-X | Windows/Linux and web app | Commandline | R | https://github.com/jingshuw/SAVERX, https://singlecell.wharton.upenn.edu/saver-x/ |
| 4 | ScImpute | Windows, Linux, web server GRANATUM | Commandline as well as web application | R, shiny for web server |
https://github.com/ Vivianstats/scImpute, http://garmiregroup.org/granatum/app |
| 5 | scRecover | Windows, Linux | Commandline | R | https://miaozhun.github.io/scRecover/ |
| 6 | scUnif | Windows, Linux | Commandline | Python, R | https://github.com/lingxuez/URSM |
| 7 | VIPER | Windows, Linux | Commandline | R | https://github.com/ChenMengjie/VIPER |
| 8 | scGAIN | Windows, Linux | Commandline | python | https://github.com/mgunady/scGAIN |
| 9 | bayNorm | Windows/Linux | Commandline | R | https://github.com/WT215/bayNorm |
4. ALGORITHMS EMPLOYING DATA SMOOTHING APPROACH
Data smoothing is a technique to eradicate noise and filter out important patterns from a data set. Different models employ random, exponential smoothing or variants of these approaches. For single cell data imputation, smoothing is achieved through the identification of the nearest neighbors of a cell. The second class of algos for single cell data imputation employs this approach. Seven different algorithms and, if present, associated softwares were identified for imputation of single cell RNA-Seq data, using the smoothing approach listed in (Table 2). In KNN-smoothing algorithm [21], transcript counts are unified and imputation is conducted via discreet smoothing or variance-stabilization of the expression profiles. It is scalable and applicable to heterogeneous datasets. For large datasets, having a higher number of similar cells, a modified version of the approach called KNN-smoothing 2 is implemented. In this method, slightly smoothed data from nearby cells are projected onto first principal components enabling the differentiation of heterogeneous data. DrImpute [22] estimates dropout events using a hot deck, matrix construction method. To swiftly process large datasets, it does not compute large cell-cell distance matrices but instead uses sampling-based algorithm. The CellBench software (available at: https://github.com/shians/ cellbench) [23] implements KNN-smooth and DrImpute. Output is delivered in tabular form. MAGIC / Markov Affinity-based Graph Imputation of Cells [24] has the capability to impute complex and non-linear contacts of neighboring cells while retaining clusters and data structure. This augments group interactions of cells and genes (2D as well as 3D interactions). This method is computationally efficient, however, it does not provide uncertainty measurement and projection of data on low dimensional space, causing it to lose variability across cells. Moussa and Mandiou [25], later introduced an iterative algorithm, called LSImpute, based on previous algorithms. Instead of keeping a fixed quantity of nearest cells for imputation, numbers are altered based on least similarity threshold. Clusters of cells were formed based on median and mean values of neighbouring cells (n=1-10 cells per round). Clusters are then assembled in corresponding centroids and these are added to the previous unaccounted cells. The procedure is repeated using Cosine similarity metric of Hornik et al. [26] or Jaccard (available at http://cnv1.engr.uconn.edu:3838/LSImpute/) for each iteration, with a set high or low threshold (0.65-0.95). Similar results have been obtained for both metrics. This
Table 2.
Features of methods employing data smoothing approach.
| Serial No. | Algorithm/Method | Interface | OS | Programming Language | Link |
|---|---|---|---|---|---|
| 1 | DrImpute | Command line, CellBench | Windows/Linux | R | https://github.com/ikwak2/DrImpute |
| 2 | MAGIC / Markov Affinity-based Graph Imputation of Cells | Command line | Windows/Linux | Python, Matlab, R | https://github.com/KrishnaswamyLab/MAGIC |
| 3 | KNN-smoothing | Command line, CellBench | Windows/Linux | Python, Matlab, R | https://github.com/yanailab/knn-smoothing |
| 4 | LSImpute | Web application | Web application | Java script, Shiny | http://cnv1.engr.uconn.edu:3838/LSImpute |
| 5 | 2Dimpute | Command line | Windows/Linux | R |
https://github.com/ zky0708/2Dimpute |
| 6 | scNPF | Command line | Windows/Linux | R |
https://github.com/ BMILAB/scNPF. |
| 7 | netImpute | Command line | Linux | Python | http://www.cs.utsa.edu/~software/netImpute/ |
| 8 | G2S3 | Command line | Windows, Linux | Matlab, R | https://github.com/ZWang-Lab/G2S3 |
demonstrates that median imputation is disposed to a conformist approach and provides improved performance by minimizing dropout effects, decreasing data sparsity, reducing spurious expression and overimputation. 2DImpute [27] is another workflow that detects co-expression signatures by means of unsupervised ‘attractor metagene’ algorithm [28] i.e. it does not require knowledge of the preceding number of cell subpopulations. It also does not make random assumptions of statistical methods for inferring expression. Spurious or dropout-suspected events are distinguished from true biological zeros using Jaccard distance matrix. Imputation is done by leveraging correlation among gene-gene and cell-cell (Inter or intra-cell) relationship. scNPF [29] takes into account cell-cell and gene-gene interactions through a network-based propagation and fusion approach. Previous knowledge is combined with topology of network (through random walk simulation). Initial expression signal is smoothed and diffused through network, denser propagated matrix and better values are obtained for expression. Two modes of network propagation based on Random Walk with Restart (RWR), including the priori mode (using public molecular networks as base and retaining top 10% interactions) and the context mode (utilizing WCGNA package) [30] are utilized. Context mode relies solely on available RNA-Seq data and no priori interaction network is employed. Multiple networks are then fused to obtain a useful expression network based on shared and complementary network knowledge. netImpute [31] utilizes RWR method to fine-tune the gene expression of a specified cell, using gene-gene, protein-protein and cell-cell interaction network for imputing expression. Although this method has similar roots as other smoothing algos, network selection and diffusion methods differ, which lead to variation in performance. Application of log transformation (with added pseudo count value to avoid infinite values) minimizes the impact of a very large values in data. Another recently developed G2S3 method [32], infers scant signals and builds gene graph network for imputation. Expression levels are smoothed using non-linear correlation and graph is optimized. After this, random walk aimed transition matrix is generated and gene expression level is imputed through weighed average expression levels of gene network in the graph.
5. ALGORITHMS EMPLOYING DATA RECONSTRUCTION APPROACH
The third algorithmic approach initially pinpoints a latent space rendering of the cells, by capturing linear associations (low-rank matrix-based methods) or non-sequential relationships (deep-learning methods). Expression matrix is then reconstructed from the low-rank or predicted latent spaces, which then cease to be insignificant. Seven low rank matrix-based algorithms were identified (Table 3).
Table 3.
Features of methods employing low-ranked matrix-based approach.
| Serial No. | Algorithm/Method | OS | Interface | Programming Language | Link |
|---|---|---|---|---|---|
| 1 | ALRA | Windows, Linux, Seurat webserver | Commandline and implemented in SeuratWeb | R | https://github.com/nasqar/SeuratWizard/, http://nasqar.abudhabi.nyu.edu/SeuratWizard |
| 2 | mcImpute | Windows, Linux | Commandline | Matlab | https://github.com/aanchalMongia/McImpute_scRNAseq |
| 3 | PBLR | Windows, Linux | Commandline | Matlab | http://page.amss.ac.cn/shihua.zhang/software.html |
| 4 | scRMD | Windows, Linux | Commandline | R | https://github.com/XiDsLab/scRMD |
| 5 | scHinter | Windows, Linux | Commandline | Matlab | https://github.com/BMILAB/scHinter |
| 6 | CMF-Impute | Windows, Linux | Commandline | Matlab | https://github.com/xujunlin123/CMFImpute |
| 7 | netNMF-sc | Windows, Linux | Commandline | Python | https://github.com/raphael-group/netNMF-sc |
5.1. Low-Ranked Matrix-based Methods
Among these, Adaptive-threshold Low-Rank Approximation (ALRA) by Linderman and Kluger [33], is a scalable process for retrieval of single cell RNA-Seq expression. Selective imputation of technical zeroes is done through a non-negative and correlatingly structured expression matrix. Matrix is approximated via a singular vector decomposition method, followed by a thresholding. PBLR [34] employs incomplete or non-negative matrix factorization (NMF), to create a concurrent matrix. Cell-cell distances are calculated using Spearman, Pearson and Cosin metrics. Matrices are transformed to affinity matrices, with 20 rounds of NMF application on each matrix. Imputed matrices are merged to get a consolidated one and then fed as the input of hierarchical clustering. Optimization is done via Alternating Direction Method of Multipliers (ADMM) algorithm [35, 36] and submatrices/sub-populations are inferred. mcImpute [37] is a matrix completion focused workslow and imputes dropouts from single cell expression values through iterative thresholding. Raw reads are standardized by library size, sieved for expression, pseudo-count of one is added and Log2 transformed expression matrix is fed to Nuclear-norm minimization algorithm. The expression is recovered through convex optimization and distribution is not taken into account. Synthetic or planted drop-outs in the expression matrix can be retrieved through this approach. It can handle heterogeneous data. scRMD [38] utilizes matrix decomposition for imputation. Nominal assumptions (i.e. low-rankness and the sparsity) guided by random matrix theory are accounted for and scRMD can resolve dropouts with expression matric values of zero > 80%. scHinter [39] is tailored for imputation on limited sample size data. A ranked ensemble distance technique (with consensus distance from Euclidean, Manhattan, Cosine, Pearson, Spearman metrics) and synthetic minority oversampling method (SMOTE) for aleatory or hierarchal interpolation are utilized. Iteration or multi-layer random interpolation improves the accuracy of results. CMF-Impute [40] uses collaborative matrix factorization for imputation. Distance (Euclidean, Chebyshev) and correlation (Pearson’s correlation) matrices are used for finding cell-cell and gene-gene similarity. Two feature matrices are obtained from matrix decomposition algorithms and consistency is quantified. netNMF-sc [41] uses the network as well as transcript count information for making low dimensional cell and gene matrix. A network-regularized NMF is combined with a graph Laplacian for treating excess zeros in transcript count matrices having a dropout rate above 60%. Value for each entry is imputed rather than just considering values for null entries and it is adept to gather information from any gene-gene interaction network, instead of inferring parameters from a trained protein-protein interaction. A low-dimensional transcript count matrix is obtained that can be used for grouping discrete cells or imputing gene clusters with zero and non-zero values. It has been observed that cumulating representative networks, boosts performance of imputation algorithm.
5.2. Deep Learning Methods
In the case of deep-learning algorithms (e.g. ones employing variational autoencoders), the imputed data (i.e. reconstructed expression matrix) along with predicted latent space can be used for further analyses, but it is typical to only use imputed data for downstream processing. Nine algorithms employing deep learning methodology were identified from the literature (Table 4). Among these, AutoImpute [42] applies a state-of-the-art deep learning technique and imputes expression using sparse gene expression matrix. A latent factor model based on over complete autoencoders (type of neural network) is employed. Autoencoder entails a coder (which inputs the value with sigmoid activation function) and decoder (which outputs expression), with values regularized to avoid overfitting. A decreased loss and insensitivity to the peripheral gene expression distribution is characteristic of this method. The network is trained by means of gradient descent with minimal cost. Iterations are carried out and convergent imputation values are obtained at the end. ScVI [43] is a scalable method utilizing probability to impute expression of drop outs. scVI amasses information across similar cells and genes via stochastic optimization coupled with deep neural networks. Distribution values behind the observed expression are approximated and expression imputation is inferred. Even though the initial objective of ScVI was not imputation but gene filtering (~ top 700 variable genes) also facilitated accurate imputation. It is computationally efficient but more suited to homogeneous datasets. DCA [44] is an abbreviation of deep count autoencoder. It is another workflow that uses neural networks to denoise single cell RNA-Seq data. DCA accounts for data sparsity, count distribution and overdispersion using a ZINB model. Non sequential gene-gene interactions are deduced and the process scales linearly with the quantity of cells with or without zeros inflation. DCA can handle heterogeneous datasets but a limitation is that it is computationally intensive. DeepImpute [45] is a scalable method which uses sub-neural networks with correlated genes as input layer. Specific target genes are not used as direct input, to reduce overfitting. A dense layer consisting of 256 neurons is the primary hidden layer followed by a dropout layer with 20% dropout rate for misfits. Output layer is composed of to-be imputed target genes and their subsets (default N = 512). This method is computationally efficient. Deconvolution using saliency maps [46] is a method which uses autoencoder neural networks to count single cell RNA-Seq expression. This method detects the expression signal with perturbed or zeroed out input. Four layers, with dimensions 128, 64 and 128 were used for training autoencoders. Two layers are specified for encoding and two for decoding. Xavier initialization for initial weighing is followed by Poisson negative log-likelihood loss function for training the neural network. Captured information is deconvoluted through saliency maps. SAUCIE/ Sparse Autoencoder for Unsupervised Clustering, Imputation, and Embedding [47] is a scalable technique based on different layers and extracts structure from single-cell RNA-Sequencing data. Autoencoder neural network for unsupervised learning is employed and latent layer assigns digital codes, clusters input, processes near-binary inactivated values using dimensionality reduction. Denoised data is regularized and outer layer yields encoded cluster identifications. scScope [48] is a scalable, deep learning method with a self-correcting capability. It obtains imputations for zeroed entries of single cell RNA-Seq data. Iterations are performed using multilayered neural networks for imputing zero-valued entries of input single cell RNA-Seq data. Phenograph (https://github.com/jacoblevine/ PhenoGraph) is used for subpopulation discovery. deepMc or deep Matrix completion [49] is grounded on deep matrix factorization and deep dictionary learning methods. It does not account for distribution for gene expression. Gene detected with more than 3 reads (in at least 3 cells) is considered expressed. Inferred matrices are normalized and 1000 genes having high-dispersion coefficient of variance are reserved for imputation followed by log2 transformation of expression data. LATE/TRANSLATE [50] is a parametric deep learning method for imputation. Arbitrary starting values of the parameters are used for training autoencoder (LATE algorithm) while extension of the method is TRANSLATE (TRANSfer learning with LATE), which utilizes gene expression (reference data set) for training autoencoder. Input is a sequencing read count data matrix (cell IDs=row names; gene IDs=column names) in .csv, .tsv or .h5 format. Output is in .hd5 format, with the same layout as input. These algorithms are extremely scalable (can process >1 million cells in a few hours) on a graphics processing unit.
Table 4.
Features of methods employing deep learning approach.
| Serial No. | Algorithm/Method | Interface | Programming Language | Link |
|---|---|---|---|---|
| 1 | AutoImpute | Linux | Python, R |
https://github.com/divyanshu-talwar/ AutoImpute |
| 2 | ScVI | Linux | Python | https://github.com/YosefLab/scVI |
| DCA | Linux | Python |
https://github.com/theislab/ dca |
|
| 3 | DeepImpute | Linux | Python | https://github.com/lanagarmire/DeepImpute |
| 4 | SAUCIE | Linux | Python | https://github.com/KrishnaswamyLab/SAUCIE/ |
| 5 | scScope | Linux | Python | https://github.com/AltschulerWu-Lab/scScope |
| 7 | deepMc | Linux, Windows | Matlab | https://drive.google.com/drive/folders/1TMD8sjPXlpe5V-3EAi38aFHQoy1gXd6h |
| 8 | Deconvolution through saliency maps | Linux | Python | https://gitlab.com/cphgeno/expression_saliency |
| 9 | LATE/TRANSLATE | Linux | Python | https://github.com/audreyqyfu/LATE |
6. PERSPECTIVE
For majority of transcripts, single cell RNA-Seq data often contains a large fraction of zero counts due to drop out events. The term “dropout” is often used to denote observed zero values in single cell RNA-Seq data. Dropout typically integrates two different types of zero values i.e. false and true. False one is due to methodological noise i.e. there is an expression of gene, which is undetectable by the sequencing technology because of insufficient depth and low capture rate. True drop outs are due to lack of gene expression [10]. The frequency of zero counts depends on which sequencing protocol has been used and also on the depth of sequencing. For example, Microfluidic single cell RNA-Seq technologies, like inDrops, Drop-Seq, and 10x Genomics Chromium platform have 90% dropout rate as these sequence thousands of the cells with low coverage (1K-200K reads/cell). Cell-capture technologies, like Fluidigm C1 has 20-40% dropout rate as it sequences hundreds of the cells with high coverage (1-2 million reads/cell) [41]. These zero counts or dropout events increase the complexity of single cell RNA-Seq data and hinder the accurate quantitative data analysis. In single cell RNA-Seq studies, it is, therefore crucial to impute the zero values in order to facilitate exact quantification of transcriptome at the single-cell level [18]. Since the first single cell imputation method presented in 2016, several methods/workflows have been developed for the purpose. In the text, we provide a short overview of different approaches for the imputation of single cell RNA-Seq data. We have categorized these methods into three categories, where the first category includes imputation methods that use probabilistic models to directly represent sparsity. Biological and technical zeroes may not be distinguished and usually only technical ones are accounted for, in the imputation function. Such methods produce less false-positives but this rests on data homogeneity or heterogeneity. Second category includes methods that smooth or adjust zero and non-zero values by averaging expression values or their diffusion. This approach is useful for reducing noise but many false positives may be generated. It is interesting to note that first category methods may outperform algorithms of second category, in datasets having genes with small effect size [51]. Third category entails data reconstruction, either through a low-ranked matrix-based method or deep learning neural network-based approach. Low-rank matrix-based methods capture linear while deep learning methods process non-linear relationships. Denser information matrix is obtained for downstream processing. Although sparsity and scalability have been resolved by numerous methods and benchmarking has revealed the algorithms suited to heterogenous and homogenous datasets, discrete expression inference has been the hallmark of all these algorithms but trajectory-based interpretation of imputation is suggested for the future. Most methods are computationally efficient, scalable and applicable to heterogeneous datasets. Circularity issue has been addressed in several algorithms, with random input instead of specified data values. Overimputation and overfitting have also been addressed by several methods, with better results. Users can implement a statistical method of choice, depending on their requirements. We also suggest that statistical tests applied to imputed data should be treated with care and filtering by effect size as well as testing with at least one algorithm from each category should be done to eliminate errors and reduce false-positives. Benchmarking of all these methods on small and large datasets of homogeneous and heterogeneous nature should also be attempted to make a better comparison.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 2.Li W.V., Li J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018;9(1):997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner A., Regev A., Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 2016;34(11):1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozenblatt-Rosen O., Stubbington M.J.T., Regev A., Teichmann S.A. The Human Cell Atlas: from vision to reality. Nature. 2017;550(7677):451–453. doi: 10.1038/550451a. [DOI] [PubMed] [Google Scholar]
- 6.Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., Teichmann S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358(6359):58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kester L., van Oudenaarden A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell. 2018;23(2):166–179. doi: 10.1016/j.stem.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods. 2014;11(1):22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 9.Poirion O.B., Zhu X., Ching T., Garmire L. Single-cell transcriptomics bioinformatics and computational challenges. Front. Genet. 2016;7:163. doi: 10.3389/fgene.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lähnemann D., Köster J., Szczurek E., McCarthy D.J., Hicks S.C., Robinson M.D., Vallejos C.A., Campbell K.R., Beerenwinkel N., Mahfouz A., Pinello L., Skums P., Stamatakis A., Attolini C.S., Aparicio S., Baaijens J., Balvert M., Barbanson B., Cappuccio A., Corleone G., Dutilh B.E., Florescu M., Guryev V., Holmer R., Jahn K., Lobo T.J., Keizer E.M., Khatri I., Kielbasa S.M., Korbel J.O., Kozlov A.M., Kuo T.H., Lelieveldt B.P.F., Mandoiu I.I., Marioni J.C., Marschall T., Mölder F., Niknejad A., Raczkowski L., Reinders M., Ridder J., Saliba A.E., Somarakis A., Stegle O., Theis F.J., Yang H., Zelikovsky A., McHardy A.C., Raphael B.J., Shah S.P., Schönhuth A. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21(1):31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X.F., Ou-Yang L., Yang S., Zhao X.M., Hu X., Yan H. EnImpute: imputing dropout events in single-cell RNA-sequencing data via ensemble learning. Bioinformatics. 2019;35(22):4827–4829. doi: 10.1093/bioinformatics/btz435. [DOI] [PubMed] [Google Scholar]
- 12.Prabhakaran S., Azizi E., Carr A., Pe’er D. Dirichlet process mixture model for correcting technical variation in single-cell gene expression data. JMLR Workshop Conf. Proc. 2016;48:1070–1079. [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M., Wang J., Torre E., Dueck H., Shaffer S., Bonasio R., Murray J.I., Raj A., Li M., Zhang N.R. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods. 2018;15(7):539–542. doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., Agarwal D., Huang M., Hu G., Zhou Z., Ye C., Zhang N.R. Data denoising with transfer learning in single-cell transcriptomics. Nat. Methods. 2019;16(9):875–878. doi: 10.1038/s41592-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X., Wolfgruber T.K., Tasato A., Arisdakessian C., Garmire D.G., Garmire L.X. Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists. Genome Med. 2017;9(1):108. doi: 10.1186/s13073-017-0492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao Z., Li J., Zhang S. scRecover: Discriminating true and false zeros in single-cell RNA-Seq data for imputation. bioRxiv. 20192019
- 17.Zhu L., Lei J., Devlin B., Roeder K. A unified statistical framework for single cell and bulk RNA sequencing data. Ann. Appl. Stat. 2018;12(1):609–632. doi: 10.1214/17-AOAS1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M., Zhou X. VIPER: variability-preserving imputation for accurate gene expression recovery in single-cell RNA sequencing studies. Genome Biol. 2018;19(1):196. doi: 10.1186/s13059-018-1575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunady M.K., Kancherla J., Bravo H.C., Feizi S. scGAIN: Single cell RNA-seq data imputation using generative adversarial networks. bioRxiv. 20192019
- 20.Tang W., Bertaux F., Thomas P., Stefanelli C., Saint M., Marguerat S., Shahrezaei V. bayNorm: Bayesian gene expression recovery, imputation and normalization for single-cell RNA-sequencing data. Bioinformatics. 2020;36(4):1174–1181. doi: 10.1093/bioinformatics/btz726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner F., Yan Y., Yanai I. K-nearest neighbor smoothing for high-throughput single-cell RNA-Seq data. bioRxiv. 20172017
- 22.Gong W., Kwak I.Y., Pota P., Koyano-Nakagawa N., Garry D.J. DrImpute: imputing dropout events in single cell RNA sequencing data. BMC Bioinformatics. 2018;19(1):220. doi: 10.1186/s12859-018-2226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su S., Tian L., Dong X., Hickey P.F., Freytag S., Ritchie M.E. CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods. Bioinformatics. 2020;36(7):2288–2290. doi: 10.1093/bioinformatics/btz889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D., Bierie B., Mazutis L., Wolf G., Krishnaswamy S., Pe’er D. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174(3):716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moussa M., Măndoiu I.I. Locality Sensitive Imputation for Single Cell RNA-Seq Data. J. Comput. Biol. 2019;26(8):822–835. doi: 10.1089/cmb.2018.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornik K., Feinerer I., Kober M., Buchta C. Spherical k-means clustering. J. Stat. Softw. 2013;50:1–22. [Google Scholar]
- 27.Zhu K., Anastassiou D. 2DImpute: Imputation in single cell RNA-seq data from correlations in two dimensions. Bioinformatics. 2020;36(11):3588–3589. doi: 10.1093/bioinformatics/btaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng W.Y., Yang O. T.H.; Anastassiou, D. Biomolecular events in cancer revealed by attractor metagenes. PLOS Comput. Biol. 2013;9(2):e1002920. doi: 10.1371/journal.pcbi.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye W., Ji G., Ye P., Long Y., Xiao X., Li S., Su Y., Wu X. scNPF: an integrative framework assisted by network propagation and network fusion for preprocessing of single-cell RNA-seq data. BMC Genomics. 2019;20(1):347. doi: 10.1186/s12864-019-5747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zand M., Ruan J. Network-based single-cell RNA-seq data imputation enhances cell type identification. Genes (Basel) 2020;11(4):E377. doi: 10.3390/genes11040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W., Dai Q., Liu Y., Yan X., Wang Z. 2020.
- 33.Linderman G. C., Zhao J., Kluger Y. Zero-preserving imputation of scRNA-seq data using low-rank approximation. 2018.
- 34.Zhang L., Zhang S. PBLR: an accurate single cell RNA-Seq data imputation tool considering cell heterogeneity and prior expression level of dropouts. bioRxiv. 2018
- 35.Gabay D., Mercier B. A dual algorithm for the solution of nonlinear variational problems via finite element approximation. Comput. Math. Appl. 1976;2:17–40. doi: 10.1016/0898-1221(76)90003-1. [DOI] [Google Scholar]
- 36.Chen C.H., He B.S., Yuan X.M. Matrix completion via an alternating direction method. IMA J. Numer. Anal. 2012;32:227–245. doi: 10.1093/imanum/drq039. [DOI] [Google Scholar]
- 37.Mongia A., Sengupta D., Majumdar A. McImpute: Matrix completion based imputation for single cell RNA-seq data. Front. Genet. 2019;10:9. doi: 10.3389/fgene.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C., Wu C., Wu L., Wang X., Deng M., Xi R. scRMD: imputation for single cell RNA-seq data via robust matrix decomposition. Bioinformatics. 2020;36(10):3156–3161. doi: 10.1093/bioinformatics/btaa139. [DOI] [PubMed] [Google Scholar]
- 39.Ye P., Ye W., Ye C., Li S., Ye L., Ji G., Wu X. scHinter: imputing dropout events for single-cell RNA-seq data with limited sample size. Bioinformatics. 2020;36(3):789–797. doi: 10.1093/bioinformatics/btz627. [DOI] [PubMed] [Google Scholar]
- 40.Xu J., Cai L., Liao B., Zhu W., Yang J. CMF-Impute: an accurate imputation tool for single-cell RNA-seq data. Bioinformatics. 2020;36(10):3139–3147. doi: 10.1093/bioinformatics/btaa109. [DOI] [PubMed] [Google Scholar]
- 41.Elyanow R., Dumitrascu B., Engelhardt B.E., Raphael B.J. netNMF-sc: leveraging gene-gene interactions for imputation and dimensionality reduction in single-cell expression analysis. Genome Res. 2020;30(2):195–204. doi: 10.1101/gr.251603.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talwar D., Mongia A., Sengupta D., Majumdar A. AutoImpute: Autoencoder based imputation of single-cell RNA-seq data. Sci. Rep. 2018;8(1):16329. doi: 10.1038/s41598-018-34688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez R., Regier J., Cole M.B., Jordan M.I., Yosef N. Deep generative modeling for single-cell transcriptomics. Nat. Methods. 2018;15(12):1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eraslan G., Simon L.M., Mircea M., Mueller N.S., Theis F.J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 2019;10(1):390. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arisdakessian C., Poirion O., Yunits B., Zhu X., Garmire L.X. DeepImpute: an accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biol. 2019;20(1):211. doi: 10.1186/s13059-019-1837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinalis S., Nielsen F.C., Winther O., Bagger F.O. Deconvolution of autoencoders to learn biological regulatory modules from single cell mRNA sequencing data. BMC Bioinformatics. 2019;20(1):379. doi: 10.1186/s12859-019-2952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amodio M., van Dijk D., Srinivasan K., Chen W.S., Mohsen H., Moon K.R., Campbell A., Zhao Y., Wang X., Venkataswamy M., Desai A., Ravi V., Kumar P., Montgomery R., Wolf G., Krishnaswamy S. Exploring single-cell data with deep multitasking neural networks. Nat. Methods. 2019;16(11):1139–1145. doi: 10.1038/s41592-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng Y., Bao F., Dai Q., Wu L. F., Altschuler S. J. Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. 2019. [DOI] [PMC free article] [PubMed]
- 49.Mongia A., Sengupta D., Majumdar A. deepMc: Deep matrix completion for imputation of single-cell RNA-seq data. J. Comput. Biol. 2020;••• doi: 10.1089/cmb.2019.0278. [DOI] [PubMed] [Google Scholar]
- 50.Badsha M.B., Li R., Liu B., Li Y.I., Xian M., Banovich N.E., Fu A.Q. Imputation of single-cell gene expression with an autoencoder neural network. Quant. Biol. 2020;8(1):78–94. doi: 10.1007/s40484-019-0192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews T.S., Hemberg M. False signals induced by single-cell imputation. F1000 Res. 2018;7:1740. doi: 10.12688/f1000research.16613.1. [DOI] [PMC free article] [PubMed] [Google Scholar]