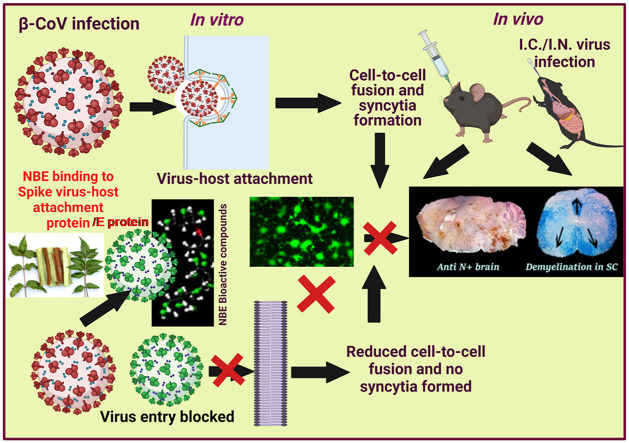
Abstract
Emerging mutations in the SARS-CoV-2 genome pose a challenge for vaccine development and antiviral therapy. The antiviral efficacy of Azadirachta indica bark extract (NBE) was assessed against SARS-CoV-2 and m-CoV-RSA59 infection. Effects of in vivo intranasal or oral NBE administration on viral load, inflammatory response, and histopathological changes were assessed in m-CoV-RSA59-infection. NBE administered inhibits SARS-CoV-2 and m-CoV-RSA59 infection and replication in vitro, reducing Envelope and Nucleocapsid gene expression. NBE ameliorates neuroinflammation and hepatitis in vivo by restricting viral replication and spread. Isolated fractions of NBE enriched in Nimbin isomers shows potent inhibition of m-CoV-RSA59 infection in vitro. In silico studies revealed that NBE could target Spike and RdRp of m-CoV and SARS-CoV-2 with high affinity. NBE has a triterpenoids origin that may allow them to competitively target panoply of viral proteins to inhibit mouse and different strains of human coronavirus infections, suggesting its potential as an antiviral against pan-β-Coronaviruses.
Keywords: Beta-coronavirus, SARS-CoV-2, m-CoV-MHV-A59/RSA59, Azadirachtaindica A. Juss (Neem bark extract), Antiviral, Inhibitor of viral entry and spread, Virus spike protein, RdRp (RNA dependant RNA polymerase), Epinimbin/Nimbin
Graphical abstract
Abbreviations
- ACE2
Angiotensin-Converting Enzyme 2
- ATCC
American Type Culture Collection
- B.W,
Bodyweight
- BIMP
Bioactivity of Indian Medicinal Plants
- CC
Column chromatography
- CEACAM1
Carcinoembryonic antigen-related cell adhesion molecule 1
- COVID-19
Coronavirus disease 2019
- CPE
Cytopathic effects
- DCM
Dichloromethane
- DMSO
Dimethyl sulfoxide;
- dpi
Days post-infection
- E
Envelope gene
- EC
Effective concentration
- EGFP
Enhanced green fluorescence protein
- ESI-MS
Electrospray Mass Spectrometry
- F
Fraction
- HCoV
Human Coronaviruses
- I.C.
Intracranial
- IHC
Immunohistochemistry
- LC50
Lethal concentration 50% of maximum response
- m-CoV-MHV
Murine β-Coronavirus Mouse hepatitis virus
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MIC
Minimum inhibitory concentration
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide;
- N
MHV Nucleocapsid gene
- N1
SARS-CoV-2 Nucleocapsid gene
- NBE
Neem bark extract
- ND
Neem database
- NeemDB
Neem Metabolites Structure Database
- PFU
Plaque forming unit
- P.I.
Post-infection
- RdRp,
RNA-dependant RNA Polymerase
- RM
Repeated measures
- ROS
Reactive oxygen species
- SARS-CoV
Severe acute respiratory syndrome Coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome Coronavirus 2
- S
Viral Spike gene
- T
NBE-treated
- TLC
Thin-layer chromatography
- DMSO
(vehicle, non-NBE)-treated + infected (NT + I), NBE-treated + infected (T + I)
1. Introduction
COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), created a worldwide crisis (Yang et al., 2020; Zhou et al., 2020). Recurrent outbreaks of SARS-CoV-2 and other human Coronaviruses (HCoVs), and the potential for new HCoVs to emerge, make finding pan-coronavirus antiviral therapies critical (Tay et al., 2020; Wang et al., 2020). While antibodies (Jiang et al., 2020), repurposed drugs (Hossein-Khannazer et al., 2020), anti-convalescent plasma therapy (Muecksch et al., 2021), and advances in diagnosis and vaccine development (Padron-Regalado, 2020) helped manage this pandemic (Rabaan et al., 2020), no single antiviral drug has shown pan-anti-coronaviral activity irrespective of zoonotic potential and host targets. Understanding Coronaviruses' properties, including genomic control of pathogenesis, host cell entry, and cell-to-cell fusion may guide new therapeutic efforts (Lu et al., 2020; V'Kovski et al., 2021).
Mechanistic studies of SARS-CoV-2 pathogenesis are difficult in patients, and experimental animal models of HCoVs are limited in mimicking human disease. Related murine-β-Coronaviruses (m-β-CoVs) facilitate studying conserved mechanisms of COVID pathobiology. MHV-1 produces a SARS-CoV-like lethal disease (De Albuquerque et al., 2006; Hua et al., 2018), and neurotropic m-CoV-MHV-A59 or its spike protein targeted recombinant strain RSA59 infection inC57BL/6 mice initiates a biphasic disorder from acute meningoencephalomyelitis to chronic demyelination (Chakravarty and Das Sarma, 2021; Das Sarma, 2010, 2014). The consequences of SARS-CoV-2 neuro-infection as reported may be inferred from m-CoV-RSA59-induced neuro-COVID (Chakravarty and Das Sarma, 2021; Mao et al., 2020).
m-CoV-MHV Spike glycoprotein is a major determinant of viral entry, virus-host interaction, infection-initiation, viral antigen spread, and consecutive pathogenesis. MHV Spike protein can initiate viral entry, fusion and subsequent pathogenesis without the CEACAM1 receptor, due to immune activation (Sadasivan et al., 2017; Singh et al., 2019). Replacement of a single amino acid in the Spike fusion domain alters pathology (Singh et al., 2019). Fusion depends on conformational transition of the six-helix bundle viral fusion core driven by hydrophobic interactions between Heptad repeats HR1 and HR2 of the S2 domain (Hoffmann et al., 2020), providing a target for mimetic peptide design. Thus, identifying anti-fusogenic properties of bark extract of the ethnomedicinal plant Azadirachta indica A. Juss (Neem) may reveal novel therapeutics.
Neem bark extract (NBE) restricts viral-host attachment, cell-to-cell fusion, viral spread, viral replication, and viral-induced demyelination induced by m-CoV-RSA59 (Sarkar et al., 2020). NBE also inhibits Herpes simplex virus type-1 glycoprotein mediated cell-to-cell fusion and polykaryocyte formation (Tiwari et al., 2010). Moreover, NBE blocks in vitro virus-free cell-to-cell fusion induced by cells expressing the m-CoV-MHV-A59 spike glycoprotein (Sarkar et al., 2020).
Viral Membrane (M), Envelope (E), and RNA-dependant RNA polymerase (RdRp) proteins also contribute to infectivity (Harrison et al., 2020) and may be affected by NBE. NBE boosts host immunity and metabolism (Alzohairy, 2016). Its antibacterial, anti-inflammatory, anti-cancer, anti-allergic, anti-parasitic, and antifungal activities support repurposing of this drug to combat COVID-19 (Lim et al., 2021). In silico studies suggest that Neem components Nimbolin-A, Nimocin, and Cycloartanols can bind to SARS-CoV-2 E and M proteins resulting in inhibition of their function (Borkotoky and Banerjee, 2020). Desacetylgedunin (DCG), a limonoid, bindstoSARS-CoV-2 papain-like protease (PLpro) (Baildya et al., 2021), and Azadirachtin, Nimbolinin, Nimbolide, Quercetin, and β-sitosterol have therapeutic benefits in pulmonary fibrosis and inflammation models (Prashanth Goud, Bale, Pulivendala and Godugu, 2019; Thota et al., 2020).
Here, we have tested the efficacy of NBE against SARS-CoV-2 infection in Vero E6 and A549-ACE2 cells, and effects of NBE administered intranasally or orally in m-CoV-RSA59-infectedC57BL/6 mice. Dichloromethane fractionation of NBE and in silico studies identified key Nimbin isomers that bind to Spike proteins of m-CoV-MHV-RSA59 and SARS-CoV-2. These data advance our understanding of COVID biology and support using NBE as a pan-CoV antiviral therapy.
2. Materials and methods
2.1. Chemicals and reagents
MTT reagent Thiazolyl Blue Tetrazolium Bromide (Sigma Aldrich), All cell culture dishes (Nunc), TRIzol (Ambion), DEPC, Diethyl pyrocarbonate (Ambion), High-Capacity Reverse Transcription Kit (Applied Biosciences, Inc. Foster, CA), DyNAmo ColorFlash SYBR Green qPCR kit (Thermo Scientific), EDTA-free Protease-cocktail inhibitor (Roche Mannheim Germany), Agarose (Invitrogen by Life Technologies), Crystal violet (Sigma-Aldrich), Gelatin (Merck), PFA, Paraformaldehyde (Merck), Mouse monoclonal Anti-N, Nucleocapsid protein of MHV-JHM (monoclonal clone 1-16-1, kindly provided by Julian Leibowitz, Texas A&M, College Station, TX), Anti-CD11b (Abcam; catalog no. ab133357), Anti-Iba1 (Wako, Richmond, VA, USA, Cat no. 019-19741, RRID:AB_839504) antibody, Avidin-biotin immunoperoxidase technique (Vector Laboratories), Refrax mounting medium (Anatech Ltd., MI, USA), Direct-Zol RNA MiniPrep (Zymo Research), Turbo DNA-Free Kit (Life Technology), High Fidelity cDNA Synthesis Kit (Roche), Fast Start Universal Probe Master (Rox) (Roche), Viral-ToxGlo assay (Promega), Prime-direct probe RT-qPCR mix (Takara).
2.2. Preparation of A. indica A. Juss (neem) bark extract
Dr. Bhargav's Neem Bark Powder (Bhargav's Enclave, Haryana, India, 100 gm) was suspended in 300 mL methanol by maceration for one week of vigorous shaking at 25 °C for 24 h. The extract was collected by filtering through Grade 1 Whatmann™ filter paper and the extract was dried using a rotary vacuum evaporator at 55 °C. The lyophilized fine brown powder (crude bark extract) was dissolved in Dimethyl sulfoxide (DMSO; cell-culture grade) at a concentration of 100 mg/mL followed by filtration through a 0.22 μm membrane filter and stored in the freezer at −20 °C (Sarkar et al., 2020).
2.3. The voucher specimen/plant authentication for A. indica A. Juss
Azadirachta indica A. Juss (Neem tree) was authenticated by India's “Central National Herbarium, Botanical Survey of India, Ministry of Environment, Forest and Climate Change” Government of India. Specimen number is IISER/JDS_01 (Azadirachta indica A. Juss) family Meliaceae. Dr. R.K. Gupta, Scientist -E and Head of Office, Central National Herbarium, Botanical Survey of India, Howrah, India, provided the voucher specimen. The specimen is kept in the Central National Herbarium, Botanical Survey of India, Howrah, India.
2.4. Cell lines
Vero E6 cells (ATCC CRL-1586) were cultured in DMEM, 10% FBS, 1% penicillin-streptomycin (P/S), 10 mM HEPES, 1% Glutamax, 1% non-essential amino acids (NEAA). A549 cells stably overexpressing human ACE2 (A549-ACE2) were cultured in Han's F12 media, 10% FBS, 1% P/S, 0.5 μg/mL Puromycin.
Murine Neuro-2A neuroblastoma cells (kind gift from Dr. Anirban Basu NBRC, Haryana, India, ATCC, and RRID: CVCL_0470) were maintained in MEM media supplemented with 10% FBS and 1% Penicillin (10,000 m/mL)-Streptomycin (100 mg/mL) antibiotic cocktail. DBT astrocytoma cells used for titre estimation were maintained in DMEM supplemented with 10% FBS and 1% P/S, 10 mM HEPES, 7.5% NaHCO3 and 0.1% l-glutamine.
2.5. Viruses
SARS-CoV-2 Swedish strain (SWE/01/2020, SS) was isolated from nasopharyngeal sample of a patient in Sweden, confirmed by sequencing (Gene bank accession number MT093571). SARS-CoV-2 WT virus (U.S.-WA1/2020, Bei Resources, and NR-52284) was a clinical isolate in the U.S. SS and U.S.-WA1 strains were used for infection at MOI 0.1 & 0.01.
Dual hepatotropic and neurotropic EGFP-expressing targeted spike protein recombinant demyelinating strain of m-CoV-MHV-A59, m-CoV-RSA59, was used to infect mice at half of the LD50 dose (20000 & 10^6 PFU) and cells at MOI 0.01, 0.1 & 1.0. M-CoV-RSA59 (isogenic recombinant, demyelinating strain of dual tropic murine β Coronavirus MHV-A59) viruses infect mouse liver and central nervous system (CNS). The gene 4a and part of 4B of MHV-A59 are replaced by Enhanced green fluorescence protein (EGFP), which is useful to trace viral entry and dissemination through cells and tissues. Intracranial inoculation of MHV-A59 in C57BL/6 mice induces an acute neuroinflammatory and chronic demyelinating stages, which mimics the common pathological hallmarks of human neurological disorder multiple sclerosis (MS) (Das Sarma, 2010; Das Sarma, Fu, Tsai, Weiss and Lavi, 2000; Das Sarma, Scheen, Seo, Koval and Weiss, 2002).
2.6. Cytotoxicity, EC determination, NBE pre-and post-treatment, viral infectivity in vitro
NBE cytotoxicity was tested in Vero E6 and A549-ACE2 cell cultures by ToxGlo™ assay (Saccon et al., 2021) or MTT assays. For the Swedish SS strain, Vero E6 cells were treated at 1 h 30 min post-adsorption of virus (MOI 0.1) or pre-incubated at MOI 0.1 of SARS-CoV-2 SS with 200 μg/mL of NBE for 15 min at 4 °C, and the cells were then infected with the preincubated virus. During the infection all incubations were performed in DMEM supplemented with 5% heat-inactivated FBS. Similarly, Vero E6 and A549-ACE2 cells were either post-infection-treated or treated with NBE (150–200 μg/mL) preincubated U.S. strain at MOI 0.01.
2.7. Viral growth-kinetics assay in Neuro-2A cells
The effect of NBE in restricting viral replication was determined by routine plaque assay for titer estimation. After the infection with the preincubated m-CoV-RSA59 virus, the cell culture plates were transferred to −80 °C at 0–24 h p.i. The cells were freeze-thawed three times, and after a final round of thawing, dislodged cells with culture supernatant were centrifuged at 3000 rpm for 15 min at 4 °C. The viral supernatants were collected from V 200 (infected + DMSO) and T 200 (NBE-treated). A monolayer of L2 cells was infected with V 200 and T 200 samples and incubated at 37 °C in 5% CO2. The titer value was calculated according to routine procedures and plotted against the corresponding time post-infection.
2.8. RNA extraction, reverse transcription, and RT-PCR analysis in vitro
To determine viral replication and estimate the expression of viral E (Appelberg et al., 2020; Corman et al., 2020) gene in the cell culture supernatant, real-time PCR assay targeting the E-gene of the virus using Prime-direct probe RT-qPCR mix was performed at 48 h post-infection (p.i.) as described previously (Chen et al., 2021). Alternatively, cell culture supernatants plus scraped cellular samples were collected at 72 h p.i. and centrifuged for 30 min at 16,000 g. Following removal of supernatant 400 μl of TRIzol reagent was added for 10 min at room temperature. RNA was isolated using Direct-Zol RNA MiniPrep, DNA removed using Turbo DNA-Free Kit, RNA normalized, and cDNA generated using Roche Transcription High Fidelity cDNA Synthesis Kit using random hexamer, all following manufactures’ protocols. Real-time qPCR was performed using E gene and N1 gene primers, Fast Start Universal Probe Master (Rox), and sub-genomic N1 SARS-2 primers and probes (Supplementary Table_S1, S2).
2.9. Facilities
All experiments on SARS-CoV-2 were done in BSL-3 containment laboratories either at Karolinska Institutet, Sweden, or University of Colorado Anschutz Medical Campus, USA following all safety regulation guidelines until the infectious virus was deactivated with TRIzol reagent.
2.10. Animal ethics statements
Use of C57BL/6 male mice (Jackson Laboratory, USA) and all experimental procedures were reviewed following good animal ethics approved by the IAEC committee at IISER Kolkata, India. Animal protocols adhered to the guidelines of the CPCSEA, India (Ref IISERK/IAEC/2020/003, Protocol Name- Understanding the ameliorative role of Neem (Azadirachta Indica) bark extract in viral-induced acute and chronic neuroinflammation), originally approved on July 13, 2020 and successively renewed (Ref IISERK/IAEC/2021/015) on July 08, 2021.
2.11. m-CoV-RSA59 inoculation in vivo
Four-week-old C57BL/6 mice were anesthetized using 2–2.5% vaporized inhaled isoflurane (Fuentes et al., 2006) and infected either intranasally (10^6 PFU) or intracranially (20000 PFU, half of LD50 dose) by m-CoV-RSA59 in 20–30 μL of endotoxin-free, filter-sterilized PBS-0.75% BSA (Das Sarma, 2010; Das Sarma et al., 2002; Singh et al., 2018).
2.12. NBE preparation and in vivo treatment groups
We have determined the doses of NBE for intranasal and oral administration in C57BL/6 mice based on a thorough in vivo pharmacological literature study (Anisuzzman et al., 2017; Bandyopadhyay et al., 2002; Beuth et al., 2006; Haasbach et al., 2014; Habluetzel et al., 2019; He et al., 2016; Ngo et al., 2017; Othman et al., 2011; Seddiek et al., 2013; Youan et al., 1997). We have chosen 100 mg/kg B.W and 500 mg/kg B.W of NBE as the non-lethal LD50, which showed potent antiviral activity and appeared to be the effective dose for intranasal and oral NBE treatment upon m-CoV-RSA59 infection, respectively.
Stock NBE was prepared in DMSO (100 mg/mL). For intranasal treatment, 30 μL NBE at 100 mg/kg in PBS was administered one day before intranasal inoculation of m-CoV-RSA59 in 4–5 mice named the intranasal treated group (T). Age-matched non-treatment group (NT), 4-5 mice, were sham-treated with only DMSO in PBS one day before infection (10^6 PFU). All mice treated with NBE/sham DMSO and infected intranasally had treatment repeated every alternate day after infection until the peak of inflammation (day 5–6 p.i.) and are grouped as intranasal NBE treated + infected (T + I) or intranasal-DMSO (no-NBE) treated + infected (NT + I).
For oral administration, mice received NBE (500 mg/kg) through drinking water (Bandyopadhyay et al., 2002) for 11 days prior to intracranial inoculation with 20000 PFU. NBE treatment continued for NBE-treated + infected group (T); whereas, non-NBE treated + infected group was called as (NT + I).
Experimental mice were observed daily for clinical signs and symptoms and sacrificed at day 5–6 p.i. Brain, liver, and spinal cords were harvested for RNA extraction or histopathological and immunohistochemical analyses.
2.13. Viral titer estimation
Virus titers in brain and liver were determined (Das Sarma et al., 2000) by homogenizing tissues in PBS and centrifuging at 400 g for 7 min at 4 °C. Supernatants were stored at −80 °C until thawed for plaque assay on DBT astrocytoma cell monolayers.
2.14. RNA extraction, reverse transcription, and real-time PCR
RNA was extracted from brain, liver, and spinal cords on day 6 p.i. using TRIzol reagent per the manufacturer's instruction. Nucleocapsid (N), Spike (S), and pro-inflammatory chemokine CCL5 gene expression was estimated by qRT-PCR. 1 μg of RNA was used for cDNA preparation using a High-Capacity Reverse Transcription Kit protocol. qRT-PCR analysis was done using the DyNAmo ColorFlash SYBR Green qPCR kit in a Step One plus Real-time PCR system (Sarkar et al., 2020) using enlisted primer sets ( SupplementaryTable_S3 ). mRNA expression levels of target genes were normalized with GAPDH and expressed as relative fold change (2-ΔΔCt).
2.15. Histopathology and immunohistochemistry
On day 5–6 p.i., mice were perfused transcardially with PBS. Brain, liver, and spinal cord tissues were collected, post-fixed in 4% Paraformaldehyde (PFA) overnight, and embedded in paraffin (Das Sarma et al., 2008). Five micron thin sagittal sections were prepared and stained with Hematoxylin/eosin (H&E) or stained using the avidin-biotin-immunoperoxidase technique with 3, 3-diaminobenzidine as substrate and anti-Iba1 (1:500), anti-N (1:20), and anti-CD11b (1:2000) primary antibodies ( SupplementaryTable_S4). Staining in different neuro-anatomic regions (McGavern et al., 1999) was quantified (Sarkar et al., 2020) and analyzed by Fiji (ImageJ 1.52g) software.
2.16. Identification of NBE compounds
Dr. ‘Bhargav's Neem bark was extracted in low-polar solvent Dichloromethane (CH2Cl2/DCM), dried, and fractionated by thin-layer chromatography followed by open column chromatography using silica gel G (Hay et al., 2007) ( Supplementary Scheme_S1 ). Analytical grade solvents were used after passing through hot Na2SO4. Fractions were eluted using DCM and tested in cell culture for viral-induced Cytopathic effect (CPE, by quantification of syncytia) and viral replication (viral titer assay) to assess antiviral activity (Sarkar et al., 2020). Mass spectra were acquired in positive mode electrospray ionization using a Bruker Maxis IITM mass spectrometer. Molecular information on the neem compounds was obtained from NeemDB (http://www.vmsrfdatabase.org/), BIMP (http://www.scfbio-iitd.res.in/plants_scf/search_neem1.html), ND (https://sites.google.com/site/neemdatabase1/). A tolerance value of ±0.0001 from observed and expected masses of molecular ion peaks were used to identify molecules. DCM extracts post chromatographic separation were labeled DCM. F [1 … 6], corresponding to six fractions.
2.17. Quantification of syncytia (cell-to-cell fusion)
Syncytia formation was monitored with a Nikon eclipse Ti2 microscope. Images were blindly analyzed with ImageJ software (Schneider et al., 2012). Percent area of syncytia was plotted in GraphPad software 9.0.
2.18. Statistics
Student's unpaired two-tailed t-test was used to identify significant differences in two-group comparisons. Multiple comparisons were achieved by RM one-way and two-way ANOVA tests followed by Tukey's and Sidak's multiple comparison tests, respectively. All statistical analyses were performed using GraphPad Prism 9.0 software, P-value of <0.05.
2.19. Molecular docking of active neem compounds linked to fraction 1 of chromatographically separated NBE
A) Preparation of the neem compound receptors. The Spike glycoprotein, human ACE2, and the RNA-dependent RNA polymerase (RdRp) protein were downloaded from the Protein Data Bank (PDB) (http://www.rcsb.org) and used as receptors to dock the neem compounds listed in Table S4. We used Spike proteins from mCoV strain A59 and SARS-COV-2 with PDB ID: 3JCL and 6VXX, respectively. The Spike protein is a trimer with structural coordinates of lengths 1118, 1147 residues for the monomeric m-CoV and SARS-CoV-2, respectively. The human ACE2 receptor coordinates were obtained from the PDB ID: 1R42, for the extracellular domain of 615 residues. The RdRp protein coordinates were only available for SARS-CoV-2 from PDB ID: 7AAP. The model for m-CoV strain A59 was obtained using homology modeling with the software Modeller (https://salilab.org/modeller/). All the receptors were prepared by attaching hydrogen and assigning Amber force field charges to the atoms using the AutoDock MGL Tools and saved in the PDBQT format.
B) Neem compounds. The coordinate information on the neem compounds was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). If in case a given neem compound did not have any entry in the PubChem database it was obtained from the NeemDB database (http://www.vmsrfdatabase.org/index.php). All files were formatted in the PDBQT format for use.
a) C) Structure-based screening. The Neem compounds were screened for high-affinity binding against the receptor proteins using the AutoDock Vina (http://vina.scripps.edu/) software. Scanning was done in triplicate using exhaustiveness values of 100, 200, and 300. Each of the proteins was placed in a box covering all the protein dimensions for the scan. The top 10 binding sites of a protein provided by the software for a given compound were chosen for further analysis. Among these, the one with the lowest binding energy was noted along with the presence of another site with low root-mean-square-deviation to it. All nonhydrogen atoms of the docked ligands were screened for finding nearest contact with the protein atoms using a threshold distance of <4 Å. The residue with the closest contact with each ligand was noted as the site residues. The top binding sites were noted in each run along with their site residues. In case the site was common, evidenced by the presence of shared site residues, then an average of the ligand binding energies was calculated for further analysis. The AutoDock Vina program was selected for in silico screening due to its unique scoring function that emphasizes non-polar interactions and docking multiplicity at a given site.
Molecular docking analysis was performed using 21 Neem compounds identified from mass spectrometry analysis of DCM.F1 fraction, which showed maximum inhibition of m-CoV-RSA59. Docked compounds were used to find high-affinity binding sites and binding energy to spike glycoprotein (Huang et al., 2020) and RdRp (Shang et al., 2020) from m-CoV-MHV-A59/RSA59 and SARS-CoV-2 viruses. Coordinates of MHV-A59 RdRp were modeled from the SARS-CoV-2 RdRp structure. The hACE2 receptor (Du et al., 2009) was also analyzed. The docking scan was done using the AutoDock Vina and MGL Tools software (Xia et al., 2020). Complexes were structurally analyzed, and binding energy was tabulated.
3. Results
3.1. NBE shows inhibitory effect on SARS-CoV-2 (SS)-induced cell toxicity and susceptibility
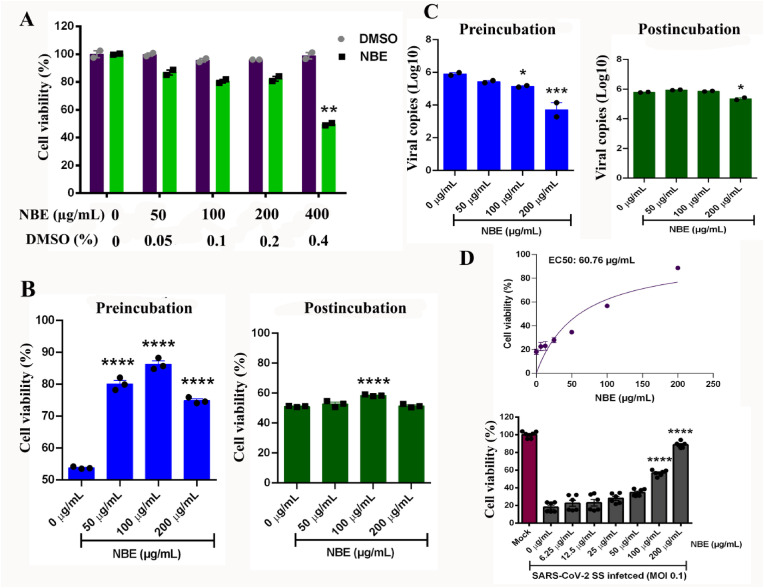
NBE cytotoxicity in Vero-E6 cells was determined by viral ToxGlo™ assay at a concentration range of 50–400 μg/mL for 48 h compared to DMSO treated controls. At 200 μg/mL NBE, Vero-E6 cells rendered 82% viability, whereas 400 μg/mL NBE was toxic, showing only 50% cell viability (considered lethal concentration, LC) (Fig. 1 A) [Biological replicates, n = 2, technical replicates = 3]. Fifty percent of the LC (200 μg/mL) was considered effective concentration (EC) with minimal cell death.
Fig. 1.
NBE shows an inhibitory effect in SARS-CoV-2 (SS) susceptibility in Vero E6 cells. (A) The cytotoxic effect of NBE at indicated concentrations (0–400 μg/mL) and their corresponding DMSO concentrations is represented as bar-scatter graphs as a measure of % cell viability. The cell viability was measured by detecting ATP using ToxGlo assay, and the % cell viability was calculated for the respective agents compared to untreated cells. The data is representative of technical duplicates and presented as mean ± SEM. Statistical significance compared to the DMSO control is shown. (B) NBE (0–200 μg/mL) was either preincubated with SARS-CoV-2 SS (MOI 0.1) prior to infection of Vero E6 cells or added to cell cultures immediately after SARS-CoV-2 SS (MOI 0.1) infection. Preincubation of the virus with NBE (50–200 μg/mL) significantly reduced virus-induced cellular cytotoxicity but not post-incubation. The data is represented as % cell viability in comparison to untreated and mock-infected cells. The bar-scatter graph is representative of technical triplicate and presented as mean ± SEM. Statistical significance compared to the NBE-untreated is shown. (C) NBE preincubation significantly reduced virus production in the cell culture supernatant (E gene expression) at 100–200 μg/mL. Post-infection NBE treatment showed a low but significant reduction of viral replication (E gene expression) only at 200 μg/mL. The bar-scatter graph is representative of technical duplicates and presented as mean ± SEM. Statistical significance compared to the NBE-untreated is shown. (D) The EC50 concentration of NBE was determined by pre-incubating the virus with NBE concentrations ranging from 6.25 to 200 μg/mL as indicated. The cellular cytotoxicity at 48 h after SARS-CoV-2 SS infection (MOI 0.1) was determined by viral ToxGlo™ assay (Luminescence). The values were normalized as % viability to the Mock-infected and untreated cells. The EC50 was calculated using GraphPad Prism. The bar-scatter graphs show the calculated % viability (mean ± SEM) and represent biological duplicates with technical triplicates in each. The significance level between treatment groups and respective controls was calculated by unpaired student's t-test and RM two-way ANOVA test followed by Sidak's multiple comparison test, *p < 0.05, **p < 0.01; ***p < 0.001; ****p < 0.0001.
Next, alteration of SARS-CoV-2 SS mediated Vero E6 cellular toxicity by NBE was measured by the Viral ToxGlo™ assay. With NBE preincubation the virus induced cytotoxicity was significantly reversed and the cell viability increased in a NBE dose-dependent manner (Fig. 1B, left panel). However, No significant alteration in virus induced cytotoxicity was observed when the cells were treated with NBE post-infection (Fig. 1B, right panel). The effect of NBE in susceptibility of Vero E6 cells to SARS-CoV-2 SS was also determined by measuring the virus production in the cell culture supernatant using RT-PCR targeting E-gene. In concordance to the cytotoxicity assay NBE preincubation (100–200 μg/mL) (Fig. 1C, left panel) significantly reduced viral E expression at 100 μg/mL (>0.75 log10copies/mL) and at 200 μg/mL (>2 log10copies/mL) in the culture supernatant and post-treatment showed marginal inhibition in virus production at 200 μg/mL (Fig. 1C, right panel). The EC50 concentration of NBE was determined by pre-incubating the virus with NBE concentrations ranging from 6.25 to 200 μg/mL as indicated in Fig. 1D, upper panel and EC50 value for the pre-incubated NBE was determined as 60.76 μg/mL (Fig. 1D, upper panel); [Biological replicates, n = 2, technical replicates = 3]. The cellular cytotoxicity at 48 h after SARS-CoV-2 SS infection (MOI 0.1) was determined by viral ToxGlo™ assay (Luminescence). The values were normalized as % viability to the Mock-infected and untreated cells (Fig. 1D, lower panel). These data suggest that NBE may have the ability to inhibit the early steps of the SARS-CoV-2 SS infection.
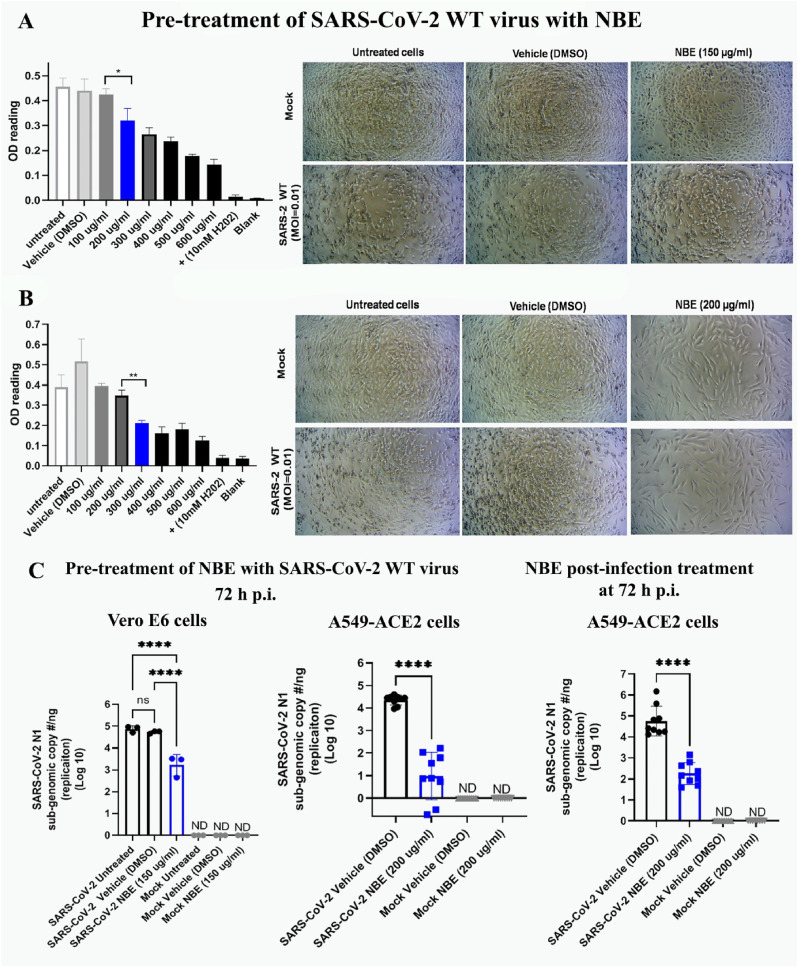
3.2. NBE impairs SARS-CoV-2 WT (U.S.-WA1) -induced cytopathy and viral replication
Vero E6 and A549-ACE2 cells were infected with SARS-CoV-2 that was pre-treated with NBE, or cells were post-infection-treated with NBE (100–600 μg/mL) for 72 h.
MTT cytotoxicity assay estimated trivial toxicity at 200 μg/mL of NBE in Vero E6 cells; EC of NBE was 150 μg/mL. SARS-CoV-2 WT infected (MOI 0.01) untreated and DMSO treated Vero E6 cells had characteristic CPE (few rounded cells) at 3 days p.i. NBE preincubated SARS-CoV-2 WT infected cells showed little CPE (Fig. 2 A). In A549-ACE2 cells, the LC of NBE was between 300 and 600 μg/mL (based on CPE and MTT) with a significant difference between 200 and 300 μg/ml. EC of NBE was 200 μg/mL. NBE (200 μg/mL) pre-treated SARS-CoV-2 WT infection had significantly less CPE (slightly elongated in morphology) like mock + NBE samples, suggesting NBE has potential antiviral properties (Fig. 2B). RT-qPCR sub-genomic analysis following pre-incubation of SARS-CoV-2 WT with NBE at 150 and 200 μg/mL and infection at 3 days p.i. revealed 20% and 75% downregulation in replication of SARS-CoV-2 Nucleocapsid N1 gene upon NBE preincubation than DMSO)/untreated Vero E6 (Fig. 2C, left panel) and A549-ACE2 cells, respectively (Fig. 2C, middle panel). No significant changes were observed between SARS-CoV-2 untreated and DMSO treated (Supplementary Figure_ S1) sets. Moreover, NBE-treated (200 μg/mL) A549-ACE2 cells showed a significant decrease in virus replication upon SARS-CoV-2 WT infection (MOI 0.01) for 2 h (Fig. 2C, right panel); [n = 3].
Fig. 2.
NBE restricts SARS-CoV-2 WT (U.S.-WA1/2020)-induced Cytopathy (CPE) and replication. (A) Vero E6 cells were treated with a range of NBE concentrations (100–600 μg/mL) for 72 h and analyzed utilizing an MTT toxicity assay. Ten mM H2O2 was used as a positive control, which causes cell death at 3 days post-infection (dpi). SARS-CoV-2 WT infected (MOI 0.01) untreated and DMSO treated Vero E6 cells revealed characteristic CPE (few rounded cells) at 72 h p.i. Vero E6 cells were sensitive to NBE at 200 μg/mL; hence the MTT assay estimated the EC value of NBE is 150 μg/mL, which had significantly less CPE on Vero E6 cells upon preincubation of SARS-CoV-2 with NBE. (B) A549-ACE2 cells were sensitive to NBE at 300 μg/mL, and 200 μg/mL as EC value was used for subsequent analyses. There was significantly less CPE upon 200 μg/mL NBE preincubated SARS-CoV-2 infection in A549-ACE2 cells. NBE (200 μg/mL) pre-treated SARS-CoV-2 WT infection had significantly less CPE (slightly elongated in morphology) like mock + NBE samples. (C) SARS-CoV-2 N1 gene replication was measured at 3 dpi from RNA harvested from cells infected with SARS-CoV-2-WT that was preincubated with NBE (150 μg/mL) for Vero E6 cells; 200 μg/mL for A549-ACE2 cells, and from A549-ACE2 cells treated with NBE (200 μg/mL) beginning immediately after infection. Data represent mean ± SEM; level of significance was calculated using unpaired student's t-test, and RM one-way ANOVA test followed by Tukey's multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.0001, ****p < 0.0001; ND = not detected, LOD = limit of detection); [n = 3].
Parallelly, NBE pre-incubation with m-CoV-RSA59 significantly reduces infectivity and replication even at MOI 0.1& MOI 0.01 like MOI 1(Supplementary Figure_S2); [n = 5]. Our data cumulatively imply that NBE significantly restricts SARS-CoV-2 replication independent of the isolate type.
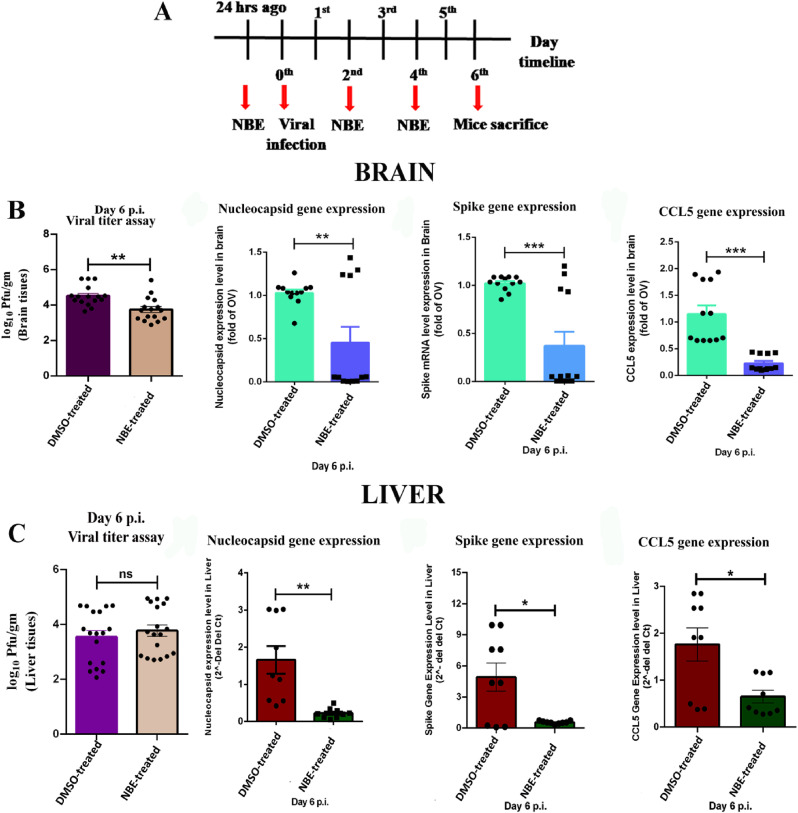
3.3. NBE treatment significantly restricts m-CoV-RSA59 replication and proinflammatory chemokine expression in vivo
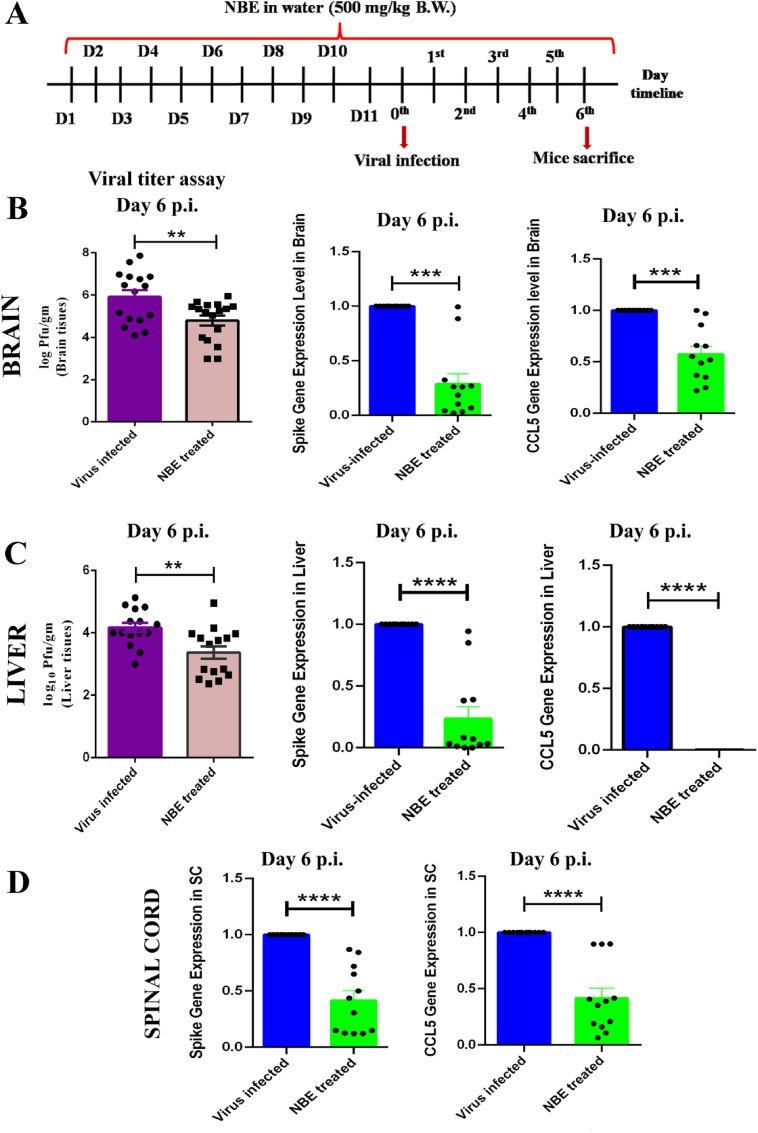
The line-graphs represent the timeline for intranasal (Fig. 3 A) and oral (Fig. 4 A) NBE administration, m-CoV-RSA59 inoculation, and euthanasia for tissue harvesting. Intranasal (100 mg/kg) NBE treatment (Fig. 3B and C) significantly reduced viral infectivity by plaque assay, and reduced Nucleocapsid (N), Spike (S) and CCL5 gene expression in brain, liver tissues. Similarly, oral NBE treatment (500 mg/kg) (Fig. 4B–D) significantly reduced viral infectivity by plaque assay, and reduced Nucleocapsid (N), Spike (S) and CCL5 gene expression in brain, liver and spinal cord in NBE-treated + infected (T + I) mice compared to non-treated, infected mice (NT + I); [n = 4–5].
Fig. 3.
Intranasal NBE administration followed by intranasal m-CoV-RSA59 inoculation significantly restricts viral replication and pro-inflammatory chemokine expression in vivo. (A) Mice were treated with NBE (100 mg/kg B.W) or DMSO intranasally 24 h before intranasal infection of m-CoV-RSA59 (10^6 PFU) and subjected for NBE/DMSO treatment every alternate day until day 6 p.i. The schematic represents the timeline for NBE administration, m-CoV-RSA59 inoculation, and euthanasia for tissue harvesting. (B) Viral burden and mRNA level expression of viral N, S, and CCL5 from brains of DMSO treated and NBE-treated infected mice. (C) Viral burden and mRNA level expression of viral N, S, and CCL5 from livers of DMSO treated and NBE-treated infected mice. Brain and liver tissue homogenates were analyzed by routine plaque assay and represented as PFU/gm of brain or liver from individual mice. Data represent mean ± SEM and statistical significance was determined by unpaired student's t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, significantly different from DMSO treated sets, [n = 4–5].
Fig. 4.
Oral NBE administration through drinking water before and after intracranial m-CoV-RSA59 infection significantly reduces viral S gene and pro-inflammatory chemokine CCL5 expression. (A) The schematic represents the timeline of NBE treatment and m-CoV-RSA59 infection. 2.5-week-old C57BL/6 mice were bottle-fed with NBE (500 mg/kg B.W) in drinking water for 11 days prior to infection, and then both NBE-treated and age-matched untreated mice were inoculated intracranially with m-CoV-RSA59 (half of the LD50 dose, 20, 000 PFU) at age 4-weeks old. At day 6 p.i., viral load and mRNA level expression of viral S and CCL5 from untreated and NBE-treated infected mouse brain (B) and liver tissues (C) shows that NBE treated mice had significantly lower viral titers (based on routine plaque assay of tissue homogenates and represented as PFU/gm) and reduced expression of viral S and CCL5. (D) mRNA level expression of viral S and CCL5 from spinal cords of untreated and NBE-treated infected mice shows reduced expression in NBE-treated mice. Data represent mean ± SEM and statistical significance was determined by unpaired student's t-test and RM two-way ANOVA and Tukey's multiple comparison test analyses; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, significantly different from untreated sets, [n = 4–5].
3.4. NBE treatment reduces meningitis, encephalitis, myelitis and hepatitis
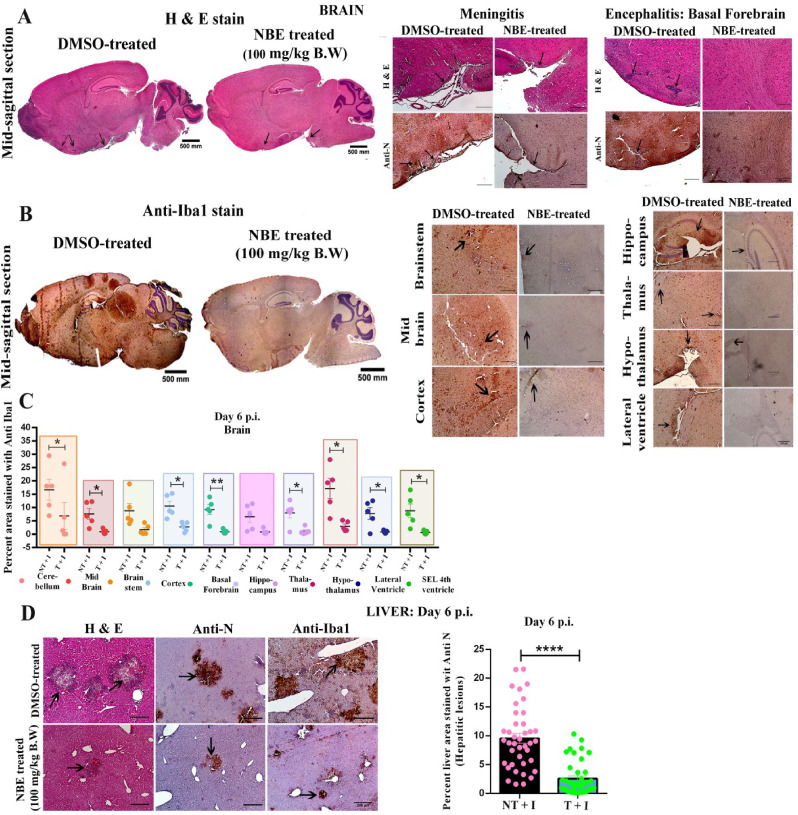
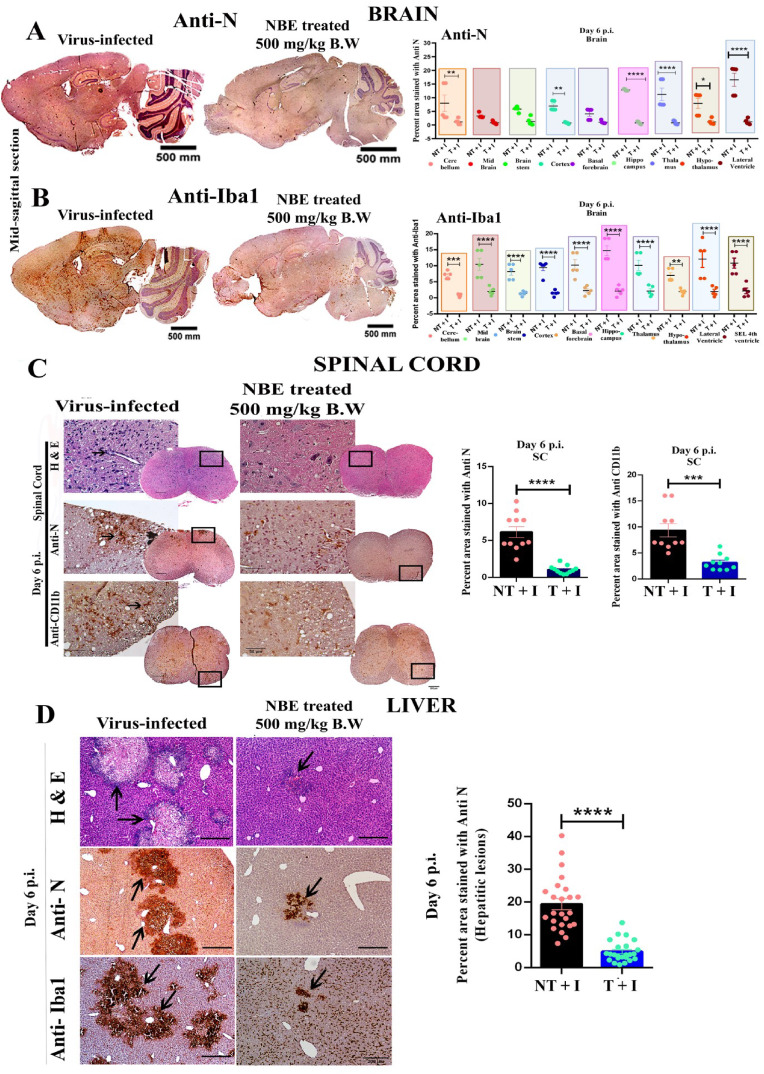
Mice were treated with DMSO as vehicle control (considered as untreated) or treated with NBE (100 mg/kg B.W) through intranasal route at 24 h before intranasal infection of m-CoV-RSA59 (10^6 PFU) and subjected to DMSO or NBE treatment every alternate day until day 6 p.i. when brain and liver tissues were harvested, as shown in Fig. 3A. For oral NBE administration, 2.5-week-old C57BL/6 mice were bottle-fed with NBE (500 mg/kg B.W) in drinking water for 11 days prior to infection, and then both NBE-treated and age-matched untreated mice were inoculated intracranially with m-CoV-RSA59 (half of the LD50 dose, 20, 000 PFU) at age 4-weeks old as shown in Fig. 4A. H&E, viral antigen (anti-Nucleocapsid), and anti-Iba1 (microglia/macrophage marker) stained m-CoV-RSA59 infected, untreated mouse brains showed characteristic perivascular cuffing and microglial nodules on day 5–6 p.i. In contrast, brains from both intranasal (Fig. 5 A–C) and oral (Fig. 6 A and B) NBE treatment groups showed much less anti-N staining throughout meninges and brain parenchyma, and reduced perivascular cuffing with minimal Iba-1 staining; [n = 4–5].
Fig. 5.
Intranasal NBE treatment reduces m-CoV-RSA59-induced neuroinflammatory pathology and hepatitis. (A) Mice were treated with intranasal NBE(100 mg/kg B.W) or DMSO (control) 24 h before intranasal infection of m-CoV-RSA59 (10^6 PFU) and subjected to NBE/DMSO treatment every alternate day until day 6 p.i. when tissues were harvested as depicted in Fig. 3A. Five μm thin paraffin-embedded serial sections of m-CoV-RSA59 infected + DMSO treated (no-NBE), and NBE-treated brain tissues were processed for H & E, anti-Nucleocapsid (A) and Iba1 staining (B). (A) Representative images of the whole brain with selected neuroanatomic regions are shown for DMSO treated mice with meningeal infiltration and encephalitis characterized by perivascular cuffing much more prominent in untreated control mice than in NBE-treated mice. (B) Iba1 staining in different neuroanatomic regions like cerebellum, midbrain, cortex, ventral striatum/basal forebrain, thalamus, hypothalamus, lateral ventricle, and the sub-ependymal layer of 4th ventricle is shown for DMSO treated mice brains. NBE-treated mice showed significantly reduced inflammation determined by no perivascular cuffing and less scattered Iba-1+ cells without apparent nodule formation. (C) Quantification of the intensity of Iba-1 staining representing microglial activation in different neuroanatomic regions of DMSO treated and NBE-treated mice brain sections were plotted in the graph and showed significantly reduced inflammation in most brain regions in NBE-treated mice. (D) Serial sections from liver tissues of the same DMSO treated and NBE-treated mice were stained with H&E or immunostained with anti-N and anti-Iba1. DMSO treated mice liver sections show a characteristic hepatic lesion with profuse anti-N and Iba-1+ cells. In contrast, NBE-treated mice showed comparatively much less anti-N and anti-Iba1 staining. The graph plotted the quantification of the intensity of anti-N staining in hepatic lesions in DMSO treated and NBE-treated liver sections. Arrows indicate the presence of viral antigen and Iba1+ cells in hepatic lesions. Data represent mean ± SEM and statistical significance was determined by unpaired student's t-test, and RM two-way ANOVA and Tukey's multiple comparison tests; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, significantly different from untreated sets; T + I = NBE treated + infected; NT + I = DMSO treated + infected [n = 4–5].
Fig. 6.
Oral NBE treatment through drinking water reduces m-CoV-RSA59-induced meningitis, encephalitis, myelitis, and hepatitis. 2.5-week-old age mice received oral NBE (500 mg/kg B.W) treatment through drinking water for 11 days and were infected intracranially with m-CoV-RSA59 (20000 PFU) at 4-weeks old, as depicted in Fig. 4A. NBE treatment continued until day 6 p.i. when the brain, spinal cord, and liver tissues were harvested for histopathological and immunohistochemical analyses. (A–B) Five μm thin serial brain sections from NBE-treated and age-matched untreated infected mice were processed for anti-Nucleocapsid and anti-Iba1 staining. (A) Representative images of anti-N stained brain sections. Untreated mouse brains revealed meningeal infiltration and encephalitis characterized by perivascular cuffing and well-disseminated viral antigen throughout the brain, including cortex, hippocampus, thalamus, hypothalamus, lateral ventricle, and brain stem. In contrast, NBE-treated mice showed significantly restricted viral antigens with less accumulation. The viral antigen distribution of both mice groups was quantified and compared. (B) Representative Iba1 staining throughout the brain parenchyma, including cortex, hippocampus, thalamus, hypothalamus, lateral ventricle, brain stem, and the sub-ependymal layer of the 4th ventricle shows significantly reduced neuroinflammation and few resident microglia/macrophages without nodule formation in NBE-treated mice as compared with untreated mice. Quantification of the intensity of Iba-1 staining in different neuroanatomic regions of untreated and NBE-treated mouse brains is shown.(C) Serial cross-sections of the spinal cords from the same untreated and NBE-treated mice at day 6 p.i. were stained with H & E, anti-N, and anti-CD11b (microglia/macrophages) antibodies. Untreated mice showed significantly more viral antigen distributed in the white matter and the gray-white matter junction than the NBE-treated group. Iba1+ cells were well-disseminated, and a characteristic microglial nodule was formed in untreated spinal cord sections, whereas the NBE-treated group showed restricted viral antigen dissemination and microglial-mediated inflammation. The total anti-N+ areas and Iba1+ areas were quantified and plotted.(D) H&E stained liver sections from untreated mice show more severe necrotizing hepatitis with profuse viral antigen and Iba-1+ monocytes/macrophages than treated mice. NBE treatment significantly restricted viral antigen (anti-N) and accumulation of Iba1+ cells in hepatic lesions at day 6 p.i.; confirmed by quantitative analysis. Arrows indicate the presence of viral antigen and Iba1+ cells in hepatic lesions. Data represent mean ± SEM and statistical significance was determined by unpaired student's t-test and RM two-way ANOVA and Tukey's multiple comparison test analyses; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, significantly different from DMSO treated sets, T + I = NBE treated + infected; NT + I = untreated + infected [n = 4–5].
5 μm thin longitudinal liver sections from intranasally (Fig. 5D) or orally (Fig. 6D) administered NBE-treated and non-NBE treated infected mice were stained with H&E, anti-N and anti-Iba1. Non-NBE treated liver sections showed moderate hepatitis with profuse viral antigen spread followed by accumulation of large number of Iba1+ cells, In contrast, infected NBE-treated liver sections showed very few necrotic loci with restricted viral antigen spread and fewer number of Iba1+ cells. The differential quantification of viral N gene staining between non-NBE treated and NBE-treated liver sections was depicted in scatter diagram [n = 4–5].
Spinal cords from the oral NBE treatment group showed minimal myelitis, whereas untreated infected mice showed characteristic myelitis with profuse distribution of viral antigen. Anti-N and anti-CD11b (microglia/macrophage/monocytes marker) staining showed a significant reduction of viral antigen spread with restricted number of CD11b+ cells in treated mice compared to untreated mice (Fig. 6C, left panel); [n = 4–5]. The differential quantification of viral antigen and CD11b staining of treated and untreated mice were plotted as scatter diagram (Fig. 6C, middle and right panels).
3.5. Identification of bioactive NBE components
Dr. Bhargav's neem bark was extracted in DCM (low polar) following published methodology (Hay et al., 2007).
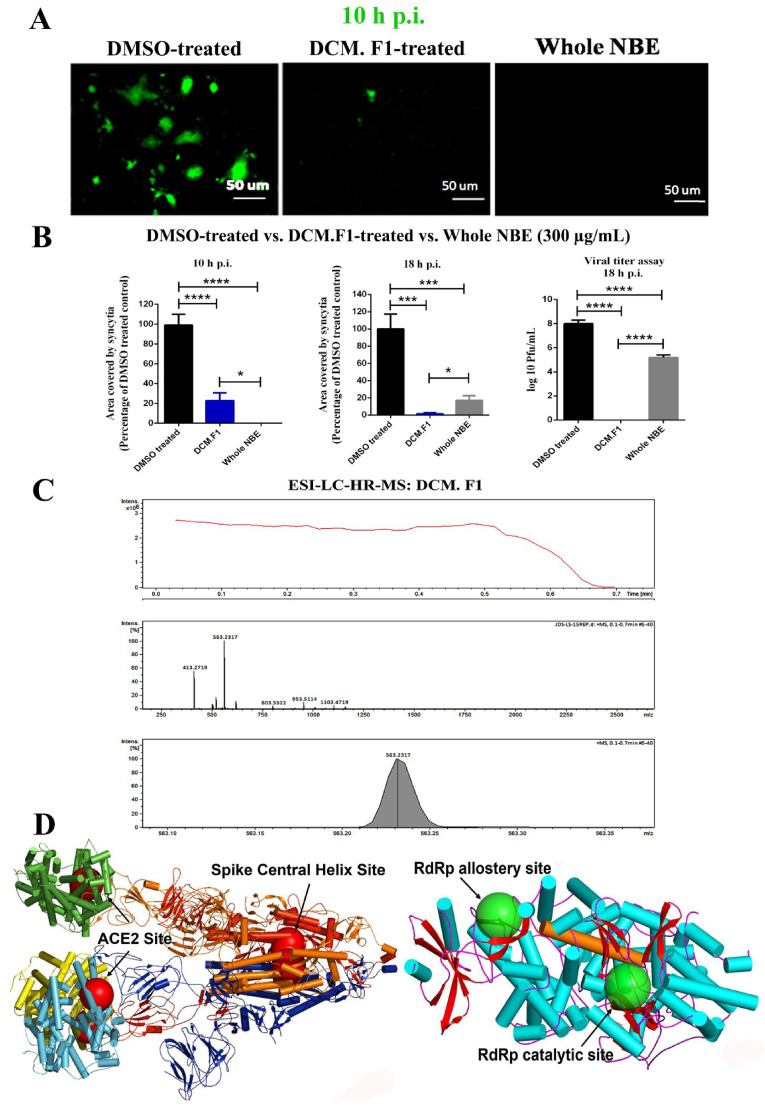
Electrospray ionization Liquid Chromatography High-resolution mass spectrometry of NBE fractions shows mostly either [M + H] + or [M + Na] + ion peak. The first elute (DCM.F1) mapped21 molecules, of which 4-Epinimbin/Nimbin, Nimbocetin, and Nimbidic acid were the three most abundant peaks (Supplementary Table_S5). Except for Nimbocetin, all compounds are of the triterpenoids class. Several compounds with identical names map to different PubChem IDs, representing different isomers of the same compound. DCM.F1 inhibited cell-cell fusion and viral-induced syncytia formation at 10 (Fig. 7 A, left panel) and 18 h post m-CoV-RSA59 infection (MOI 1) (Fig. 7B, middle panel). DCM.F1 pre-treatment with m-CoV-RSA59 also reduced viral replication determined by routine plaque assay in Neuro-2A cells at 18 h p.i. (Fig. 7B, right panel); [n = 3]. The peak value of Nimbin/4-Epinimbin and area under the curve show it contributes almost 50% of all molecules present in DCM.F1 (Fig. 7C), suggesting it may be responsible for its inhibitory activity. The molecular formula, ion fragmentation, and m/z value data of all molecules in DCM.F1 are presented in Supplementary Table_S5.
Fig. 7.
Identification of bioactive NBE components and a cartoon diagram for SARS-CoV-2 proteins showing dominant binding sites for bioactive compounds. The DCM.F1 (300 μg/mL) fraction of NBE was analyzed to test the potential antiviral efficiency against m-CoV-RSA59 infection by preincubating it with the virus at MOI 1, followed by infection in Neuro-2A cells. Effects of DCM.F1 were compared to the effects of preincubation of the virus with NBE (300 μg/mL) prior to infection. (A) Both DCM.F1 and whole NBE show reduced syncytia formation at 10 h p.i. (B) Virus-induced cell-to-cell fusogenicity was significantly reduced to a similar degree in cultures infected with both DCM.F1 pre-treated and whole NBE pre-treated m-CoV-RSA59, as compared with cultures infected with the untreated virus at 10 h and 18 h p.i. Viral titer assay of cell culture supernatants revealed a significant reduction in viral replication upon preincubating the virus with DCM.F1 at 18 h p.i., indicating that DCM.F1 can inhibit viral cytopathy as well as viral replication effectively. Whole NBE also reduced viral replication but was less effective than DCM.F1. Data represent mean ± SEM. Level of significance was calculated using student's t-test and RM one-way ANOVA followed by Sidak's multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.0001, ****p < 0.0001); [n = 3]. (C) Mass spectrometry data corresponding to DCM.F1. The top panel shows the total ion chromatogram obtained from the elution of DCM.F1. The extracted ions from the total ion chromatogram are shown in the middle panel. The area under the curve (AUC) for the base peak is shown in the bottom-most panel. The peaks map to neem compounds Nimbin and 4- Epinimbin. (D) A cartoon diagram of Spike from SARS-CoV-2 complex with ACE2 protein is drawn using coordinates from PDB ID: 7A98. The top-ranked docking sites of the neem compounds from NBE fraction DCM.F1 are shown as large transparent spheres in red. These are anchored around residue L1024 in the Central helix for all three trimers of Spike. For ACE2, the top-ranked docking sites for a majority of neem compounds are around residue F40 located close to the ACE2-Spike interaction site. A cartoon diagram of RdRp protein is drawn using coordinates from the PDB ID: 7AAP. The top-ranked docking sites of the neem compounds are shown in large green transparent spheres. Of the two sites, one is the RdRp-associated nucleotidyl transferase catalytic site and may influence the binding at the main catalytic site that is bridged by a long helix (686-709) shown in orange. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Top-ranked docking hits of neem compounds on m-CoV/SARS-CoV-2 Spike, hACE2, modeled MHV RdRp and SARS-CoV-2 RdRp as receptor are summarized in Supplementary Table_S6. Except for Nimbocetin, all compounds show high binding energy between −8.5 and −10.5 kcal/mol for MHV Spike and −8.4 to −10.5 kcal/mol for SARS-CoV-2 Spike, centered on a common binding location hemmed by residues D1041, N1023, L1024 within the evolutionarily conserved central helix of the Spike skeleton (Fig. 7D). Most compounds accrue their high binding affinity from the triterpenoids structure, in contrast to the non-triterpenoids compound Nimbocetin. The trend is identical for top-ranked hits from m-CoV Spike, where the common binding site is hemmed by central helix residues A1020, D1024 (Supplementary figure_S3).
Top-ranked docking hits of neem compounds on hACE2 receptor returned comparatively lower binding energies (−6.8 to −9.6 kcal/mol) and are not relevant because they locate away from the Spike-hACE2 interaction surface (Fig. 7D). Only Nimbin, 4-Epinimbin, Azadiradionol, Azadironol, Azadirachtin I, Gedunin, Nimocinol, and O-methylazadironolide returned hits that bound close to the Spike-hACE2 interaction site. The binding interface of Spike and ACE2 is formed by two N-terminal helices spanning residues 22-53, 55-82. Neem compounds bind on one side of these helices, while Spike-ACE2 interacts on the opposite surface.
Top-ranked docking hits for RdRp were consistent in replicate runs, but only in 50% were sites common between MHV and SARS-CoV-2 (Supplementary Table_S6), and binding energy values (−6.7 to −9.4 kcal/mol) were lower than Spike or ACE2. A majority of docked neem compounds have contact with residue I37, in the RdRp pyrophosphate binding location (PDB ID: 7AAP; Fig. 7D), possibly important as the same site binds rNTP in the influenza RNA polymerase (Xia et al., 2020a, Xia et al., 2020b). This site may have an allosteric effect that regulates binding in the catalytic site through a long helix (686–709) that tethers the two locations. Only Nimbin, 1β,2β epoxy Nimbin, Diepoxyazadiradione, Nimolicinol, O-methylazadironolide bind to both sites(Supplementary Table_S6), which may elicit higher inhibition of RdRp activity despite marginally low RdRp binding affinity compared to Spike or ACE2.
4. Discussion and conclusions
In this study, NBE was found to inhibit the deleterious effects of three distinct Coronaviruses in diverse infection models, including in vivo and in vitro systems, suggesting that NBE and NBE-derived compounds have promise as a pan-β-CoV therapeutic.
The respiratory tract is an early target for SARS-CoV-2 infection which then induces a wide range of complications from mild cough and congestion to acute respiratory distress syndrome (ARDS). Clinically, ARDS is also a characteristic feature of patients with SARS and MERS, in addition to COVID-19, whereas HCoVs- 229E, -OC43, - NL63, and -HKU1 trigger mild common seasonal cold symptoms. New CoVs continue to emerge periodically in humans, likely due to the high prevalence, vast genetic diversity, and frequent recombination of their genomes, and the ever-increasing human-animal interface. The diversity of symptoms associated with different Coronaviruses underscores the need to identify therapeutic agents with the capacity to attenuate disease severity irrespective of virus identity or tropism.
The in vivo model used was m-CoV-RSA59 infected mice, which targets the CNS and liver, two clinically relevant organ targets which are associated with severe COVID-19 (Chakravarty and Das Sarma, 2021; Mao et al., 2020) (Saviano et al., 2021). Numerous neurologic manifestations of COVID-19 have been documented, including encephalopathy, encephalitis, acute disseminated encephalomyelitis, meningitis, ischemic and hemorrhagic stroke, venous sinus thrombosis, loss of smell (anosmia) and taste (ageusia), difficulty concentrating, altered consciousness, confusion, neuropsychiatric disorders, focal seizures, and Guillain-Barre syndrome. SARS-CoV-2 also can infect other organs, including the kidneys, intestines, heart, testis, and blood vessels.
Although we used the in vivo system as a model for Coronavirus infection in the CNS and liver, one limitation is that m-CoV-RSA59 does not infect the respiratory system, which is a major target of SARS-CoV-2. Nonetheless, NBE applied intranasally and orally showed significant systemic efficacy in preventing CNS and liver injury. Moreover, NBE has the ability to inhibit the pathologic effects of SARS-CoV-2 infection using a human lung cell model in vitro. Although not directly tested here, we anticipate that NBE and NBE-derived compounds will have efficacy in preventing SARS-CoV-2 infection of nasal and lung tissue in vivo as well. Consistent with this, a recent clinical trial suggests that subject's prophylactically taking neem capsules had a reduced risk of infection resulting in COVID-19 (Nesari et al., 2021). Moreover, neem leaf extract has been shown to inhibit inflammation associated with endotoxin-induced lung injury in mice (Lee et al., 2017), indicating that NBE may have a comparable effect in ameliorating the impact of SARS-CoV-2 on the respiratory system.
Our data suggest a potential mechanism of action for the protective effects of NBE. Decade-long studies on different β-CoVs and associated disease suggest Spike is a major antigenic determinant and thus a valuable target in antiviral therapies (Huang et al., 2020; J. Shang et al., 2020). Spike-induced cell-to-cell fusion, viral spread, and host immune responses drive immunopathogenesis (Du et al., 2009; Tay et al., 2020). The receptor-binding domain of Spike has received great attention as it initiates host attachment. However, the fusion peptide, part of the fusion core complex assembled by Heptad repeats 1 and 2 has therapeutic potential (Xia et al., 2020; Xia et al., 2020). Blocking β-CoV receptor attachment, entry, and inhibiting fusion by NBE or its bioactive compounds holds therapeutic promise.
While antibodies, vaccines, and repurposed drugs can combat the SARS-CoV-2 outbreak (Xia et al., 2020; Xia et al., 2020), no therapeutic tools are equally effective against all strains (Kozlov, 2021), and may not affect new human CoVs that emerge in the future (Sanders et al., 2020). Thus, pan-β-CoV antivirals are needed. Significant inhibitory effects of NBE against two different isolates of SARS-CoV-2 and m-CoV present a promising candidate for designing pan-antivirals against β-CoV infection. NBE significantly inhibited viral replication, spread, and pathogenesis. Furthermore, various routes of NBE delivery can be explored in an in vivo m-CoV-RSA59-induced neuroinflammatory model to maximize therapeutic efficacy and minimize side effects (Wen et al., 2015). Current results of intranasal and oral NBE treatment were consistent with previous studies showing effects of m-CoV preincubation with NBE prior to intracranial inoculation, and intraperitoneal NBE treatment (Sarkar et al., 2020). Together, results support consistent and conserved antiviral effects of NBE on mouse and human β-CoVs.
The triterpenoids group of compounds isolated from the DCM.F1 fraction of NBE significantly inhibited m-CoV infectivity, suggesting these compounds are key mediators of NBE-induced antiviral properties. 4-epinimbin or its stereoisomers are most abundant in this fraction, and together with other triterpenoids modeling results show their high affinity binding is centered on a common binding location within the central helix of the Spike skeleton in both h-CoV and m-CoV. This region is evolutionarily more conserved than other regions, suggesting functional importance. The high-affinity binding site at an equivalent position in two Spike proteins is remarkable as the two proteins share an overall amino acid identity of <30%. Binding of neem compounds is expected to stabilize the Spike protein to prevent the conformational transition necessary to trigger virus fusion.
Overall, the current findings of the antiviral properties of NBE both in vitro and in vivo against β-CoV offer a new premise for restricting viral spread, replication, and fusion associated with pathogenesis. Our studies can guide new antiviral therapeutic efforts to combat the ongoing COVID-19 pandemic and hold promise for treating the future emergence of new coronavirus strains.
Funding statement
This work is endorsed by Indo-U.S. Science & Technology Forum (IUSSTF) Virtual Networks for COVID-19 (Ref: IUSSTF/VN-COVID/107/2020), India. The Swedish part of the study is supported by the Swedish Research Council (2017-01330 to UN and 2018-06156). SG acknowledges the grant received from Karolinska Institute Stiftelser och Fonder (2020-02153) and Åke Wibergs Stiftelse (M20-0220).
Declaration of competing interest
The authors declare that they have no conflict of interest with the contents of this article.
Animal ethics approval statement
Use of C57BL/6 male mice (Jackson Laboratory, USA) and all experimental procedures were reviewed following ethical animal protocols approved by the IAEC committee at IISER Kolkata, India. Animal protocols adhered to the guidelines of the CPCSEA, India (Ref IISERK/IAEC/2020/003, Protocol Name- Understanding the ameliorative role of Neem (Azadirachta Indica) bark extract in viral-induced acute and chronic neuroinflammation), originally approved on July 13, 2020 and successively renewed (Ref IISERK/IAEC/2021/015) on July 08, 2021.
Lay summary (Word count: 158)
The COVID-19 pandemic is an urgent global challenge, demanding treatments that can broadly treat viral infection. Several traditional phytochemicals have been used to treat many diseases and have been confirmed to inhibit viral replication and transcription. Azadirachta indica A. Juss (Neem), a traditional ethnomedicinal plant, is known for its antimicrobial, anti-oncogenic, anti-inflammatory, and antioxidant properties. Here we demonstrated that Neem bark extract (NBE) effectively restricted the pathological effects of multiple Coronaviruses, including Swedish SARS-CoV-2 (SS) strain, SARS-CoV-2 (U.S.-WA1) 2020 strain, and prototype murine β-Coronavirus (m-CoV)-MHV-A59/RSA59. Critically, both intranasal and oral administration of NBE to mice successfully restricted the spread of m-CoV and inflammation of the brain, spinal cord, and liver. We identified Nimbin/4-Epinimbin compounds as an active component of NBE that inhibits m-CoV infection. Molecular modeling confirmed that Nimbin/4-Epinimbin binds to SARS-CoV-2 Spike protein and RNA-dependent RNA polymerase with high affinity. Our study suggests that NBE has significant potential as a broad-spectrum antiviral targeting multiple Coronaviruses.
Author contribution statement
J. D. S.: originally conceptualized and designed the experimental protocol. provided the day-to-day supervision of the project, analyzed data with L. S. in a double-blinded manner, helped in critically reviewing and re-writing the manuscript. All the authors have gone through the final manuscript and approved the submission. L.S.: performed all the in vitro and in vivo lab experiments, analyzed data, prepared the figures, and drafted the manuscript. L. O.: performed all the in vitro and in vivo lab experiments, analyzed data, prepared the figures, and drafted the manuscript. S. G.: performed all the in vitro and in vivo lab experiments, analyzed data, prepared the figures, and drafted the manuscript. D. P: performed in silico analyses, prepared the in silico figure, and wrote the in silico part of the manuscript. B. D.: assisted in NBE fractionation work. P. G.: assisted in NBE fractionation work. A. N. B: assisted in in vitro and in silico work. A. A. S: assisted in in vitro and in silico work. C. S.: assisted in in vitro and in silico work. A. B.: helped in critical reviewing of the work and the editing of the manuscript. D. B.: helped in critical reviewing of the work and the editing of the manuscript. R. M.: helped in critical reviewing of the work and the editing of the manuscript. U. N.: helped in critical reviewing of the work and the editing of the manuscript. R. J. C: helped in critical reviewing of the work and the editing of the manuscript. M. N.: helped in critical reviewing of the work and the editing of the manuscript. K. S. S.: helped design experiments, performed a thorough revision and editing of the manuscript. M. K.: helped design experiments, performed a thorough revision and editing of the manuscript.
Declaration of transparency and scientific rigor
This declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigor of preclinical research, as stated in the Virology guidelines for Design & Analysis, Immunoblotting, and Immunochemistry and Animal Experimentation, and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Acknowledgments
These studies utilized the RT-qPCR protocol generated by Dr. Thomas E. Morrison at the University of Colorado Anschutz Medical Campus for the sub-genomic N1 SARS-2 to identify SARS-2 replication. These studies also utilized the stable A549-ACE2 cell generated by Dr. Mario Santiago at the University of Colorado Anschutz Medical Campus. The authors would also like to acknowledge the support from Division of Clinical Microbiology, Karolinska University Hospital for access to BSL-3 laboratory, and Prof. Ali Mirazimi to provide the first Swedish SARS-CoV-2 virus.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.01.002.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Supplementary materials are available at ‘Virology’ online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Data availability
Data will be made available on request.
References
- Alzohairy M.A. Therapeutics role of Azadirachta indica (neem) and their active constituents in diseases prevention and treatment. Evid. Based Complement. Alternat. Med. 2016:7382506. doi: 10.1155/2016/7382506. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisuzzman M., Hasan M.M., Acharzo A.K., Das A.K., Rahman S. In vivo and in vitro evaluation of pharmacological potentials of secondary bioactive Metabolites of Dalbergia candenatensis leaves. Evid. Based Complement. Alternat. Med. 2017:5034827. doi: 10.1155/2017/5034827. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelberg S., Gupta S., Svensson Akusjarvi S., Ambikan A.T., Mikaeloff F., Saccon E.…Neogi U. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microb. Infect. 2020;9(1):1748–1760. doi: 10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baildya N., Khan A.A., Ghosh N.N., Dutta T., Chattopadhyay A.P. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: an insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021;1227:129390. doi: 10.1016/j.molstruc.2020.129390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U., Biswas K., Chatterjee R., Bandyopadhyay D., Chattopadhyay I., Ganguly C.K.…Banerjee R.K. Gastroprotective effect of Neem (Azadirachta indica) bark extract: possible involvement of H(+)-K(+)-ATPase inhibition and scavenging of hydroxyl radical. Life Sci. 2002;71(24):2845–2865. doi: 10.1016/s0024-3205(02)02143-4. [DOI] [PubMed] [Google Scholar]
- Beuth J., Schneider H., Ko H.L. Enhancement of immune responses to neem leaf extract (Azadirachta indica) correlates with antineoplastic activity in BALB/c-mice. In Vivo. 2006;20:247–251. 2. [PubMed] [Google Scholar]
- Borkotoky S., Banerjee M. A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem) J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1774419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D., Das Sarma J. Murine-beta-coronavirus-induced neuropathogenesis sheds light on CNS pathobiology of SARS-CoV2. J. Neurovirol. 2021;27(2):197–216. doi: 10.1007/s13365-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Saccon E., Appelberg K.S., Mikaeloff F., Rodriguez J.E., Vinhas B.S., et al. Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells. Cell Death Dis. 2021;7(1):114. doi: 10.1038/s41420-021-00487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J. A mechanism of virus-induced demyelination. Interdiscip. Perspect. Infect. Dis. 2010:109239. doi: 10.1155/2010/109239. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J. Microglia-mediated neuroinflammation is an amplifier of virus-induced neuropathology. J. Neurovirol. 2014;20(2):122–136. doi: 10.1007/s13365-013-0188-4. [DOI] [PubMed] [Google Scholar]
- Das Sarma J., Fu L., Tsai J.C., Weiss S.R., Lavi E. Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J. Virol. 2000;74(19):9206–9213. doi: 10.1128/jvi.74.19.9206-9213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Iacono K., Gard L., Marek R., Kenyon L.C., Koval M., Weiss S.R. Demyelinating and nondemyelinating strains of mouse hepatitis virus differ in their neural cell tropism. J. Virol. 2008;82(11):5519–5526. doi: 10.1128/JVI.01488-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Scheen E., Seo S.H., Koval M., Weiss S.R. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 2002;8(5):381–391. doi: 10.1080/13550280260422686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Albuquerque N., Baig E., Ma X., Zhang J., He W., Rowe A., et al. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J. Virol. 2006;80(21):10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes J.M., Talamini M.A., Fulton W.B., Hanly E.J., Aurora A.R., De Maio A. General anesthesia delays the inflammatory response and increases survival for mice with endotoxic shock. Clin. Vaccine Immunol. 2006;13(2):281–288. doi: 10.1128/CVI.13.2.281-288.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasbach E., Hartmayer C., Hettler A., Sarnecka A., Wulle U., Ehrhardt C., et al. Antiviral activity of Ladania067, an extract from wild black currant leaves against influenza A virus in vitro and in vivo. Front. Microbiol. 2014;5:171. doi: 10.3389/fmicb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habluetzel A., Pinto B., Tapanelli S., Nkouangang J., Saviozzi M., Chianese G., et al. Effects of Azadirachta indica seed kernel extracts on early erythrocytic schizogony of Plasmodium berghei and pro-inflammatory response in inbred mice. Malar. J. 2019;18(1):35. doi: 10.1186/s12936-019-2671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A.E., Ioset J.R., Ahua K.M., Diallo D., Brun R., Hostettmann K. Limonoid orthoacetates and antiprotozoal compounds from the roots of Pseudocedrela kotschyi. J. Nat. Prod. 2007;70(1):9–13. doi: 10.1021/np0680230. [DOI] [PubMed] [Google Scholar]
- He Z., Jiang C., Zhang J., Yin Z., Yin Z., Zhu Y., Fu J. Neem tree (Azadirachta indica) extract specifically suppresses the growth of tumors in H22-bearing Kunming mice. Z. Naturforsch. C J. Biosci. 2016;71(7–8):201–208. doi: 10.1515/znc-2014-4210. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossein-Khannazer N., Shokoohian B., Shpichka A., Aghdaei H.A., Timashev P., Vosough M. Novel therapeutic approaches for treatment of COVID-19. J. Mol. Med. (Berl.) 2020;98(6):789–803. doi: 10.1007/s00109-020-01927-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Vijay R., Channappanavar R., Athmer J., Meyerholz D.K., Pagedar N.…Perlman S. Nasal priming by a murine coronavirus provides protective immunity against lethal heterologous virus pneumonia. JCI Insight. 2018;3(11) doi: 10.1172/jci.insight.99025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human Coronaviruses. Trends Immunol. 2020;41(5):355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M. Omicron overpowers key COVID antibody treatments in early tests. Nature. 2021 doi: 10.1038/d41586-021-03829-0. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Ryu H.W., Park S.Y., Park H.A., Kwon O.K., Yuk H.J., et al. Protective effects of neem (Azadirachta indica A. Juss.) leaf extract against cigarette smoke- and lipopolysaccharide-induced pulmonary inflammation. Int. J. Mol. Med. 2017;40(6):1932–1940. doi: 10.3892/ijmm.2017.3178. [DOI] [PubMed] [Google Scholar]
- Lim X.Y., Teh B.P., Tan T.Y.C. Medicinal plants in COVID-19: potential and limitations. Front. Pharmacol. 2021;12:611408. doi: 10.3389/fphar.2021.611408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavern D.B., Murray P.D., Rodriguez M. Quantitation of spinal cord demyelination, remyelination, atrophy, and axonal loss in a model of progressive neurologic injury. J. Neurosci. Res. 1999;58(4):492–504. doi: 10.1002/(sici)1097-4547(19991115)58:4<492::aid-jnr3>3.0.co;2-p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F., Wise H., Batchelor B., Squires M., Semple E., Richardson C., et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J. Infect. Dis. 2021;223(3):389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesari T.M., Bhardwaj A., ShriKrishna R., Ruknuddin G., Ghildiyal S., Das A., et al. Neem (Azadirachta indica A. Juss) capsules for prophylaxis of COVID-19 infection: a pilot, double-blind, randomized controlled trial. Alternative Ther. Health Med. 2021;27(S1):196–203. [PubMed] [Google Scholar]
- Ngo H.T., Hwang E., Seo S.A., Park B., Sun Z.W., Zhang M., et al. Topical application of neem leaves prevents wrinkles formation in UVB-exposed hairless mice. J. Photochem. Photobiol., B. 2017;169:161–170. doi: 10.1016/j.jphotobiol.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Othman F., Motalleb G., Lam Tsuey Peng S., Rahmat A., Fakurazi S., Pei Pei C. Extract of Azadirachta indica (neem) leaf induces apoptosis in 4T1 breast cancer BALB/c mice. Cell J. 2011;13(2):107–116. [PMC free article] [PubMed] [Google Scholar]
- Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Ther. 2020:1–20. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashanth Goud M., Bale S., Pulivendala G., Godugu C. Therapeutic effects of Nimbolide, an autophagy regulator, in ameliorating pulmonary fibrosis through attenuation of TGF-beta1 driven epithelial-to-mesenchymal transition. Int. Immunopharm. 2019;75:105755. doi: 10.1016/j.intimp.2019.105755. [DOI] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Sah R., Tiwari R., Yatoo M.I., Patel S.K., et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic. Ann. Clin. Microbiol. Antimicrob. 2020;19(1):40. doi: 10.1186/s12941-020-00384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccon E., Chen X., Mikaeloff F., Rodriguez J.E., Szekely L., Vinhas B.S., et al. Cell-type-resolved quantitative proteomics map of interferon response against SARS-CoV-2. iScience. 2021;24(5):102420. doi: 10.1016/j.isci.2021.102420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivan J., Singh M., Sarma J.D. Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals. J. Biosci. 2017;42(2):231–244. doi: 10.1007/s12038-017-9676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Sarkar L., Putchala R.K., Safiriyu A.A., Das Sarma J. Azadirachta indica A. Juss ameliorates mouse hepatitis virus-induced neuroinflammatory demyelination by modulating cell-to-cell fusion in an experimental animal model of multiple sclerosis. Front. Cell. Neurosci. 2020;14:116. doi: 10.3389/fncel.2020.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviano A., Wrensch F., Ghany M.G., Baumert T.F. Liver disease and coronavirus disease 2019: from pathogenesis to clinical care. Hepatology. 2021;74(2):1088–1100. doi: 10.1002/hep.31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddiek S.A., Khater H.F., El-Shorbagy M.M., Ali A.M. The acaricidal efficacy of aqueous neem extract and ivermectin against Sarcoptes scabiei var. cuniculi in experimentally infested rabbits. Parasitol. Res. 2013;112(6):2319–2330. doi: 10.1007/s00436-013-3395-2. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Khan R.S., Dine K., Das Sarma J., Shindler K.S. Intracranial inoculation is more potent than intranasal inoculation for inducing optic neuritis in the mouse hepatitis virus-induced model of multiple sclerosis. Front. Cell. Infect. Microbiol. 2018;8:311. doi: 10.3389/fcimb.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Kishore A., Maity D., Sunanda P., Krishnarjuna B., Vappala S., et al. A proline insertion-deletion in the spike glycoprotein fusion peptide of mouse hepatitis virus strongly alters neuropathology. J. Biol. Chem. 2019;294(20):8064–8087. doi: 10.1074/jbc.RA118.004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota S.M., Balan V., Sivaramakrishnan V. Natural products as home-based prophylactic and symptom management agents in the setting of COVID-19. Phytother Res. 2020;34(12):3148–3167. doi: 10.1002/ptr.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Darmani N.A., Yue B.Y., Shukla D. In vitro antiviral activity of neem (Azardirachta indica L.) bark extract against herpes simplex virus type-1 infection. Phytother Res. 2010;24(8):1132–1140. doi: 10.1002/ptr.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H., Jung H., Li X. Drug delivery approaches in addressing clinical pharmacology-related issues: opportunities and challenges. AAPS J. 2015;17(6):1327–1340. doi: 10.1208/s12248-015-9814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youan B.B., Coulibaly S., Miezan T.B., Doua F., Bamba M. In vivo evaluation of sixteen plant extracts on mice inoculated with Trypanosoma brucei gambiense. Bull. World Health Organ. 1997;75(4):343–348. [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.