Abstract
The published genome sequence of Campylobacter jejuni strain NCTC 11168 was used to model an accurate and highly reproducible fluorescent amplified fragment length polymorphism (FAFLP) analysis. Predicted and experimentally observed amplified fragments (AFs) generated with the primer pair HindIII+A and HhaI+A were compared. All but one of the 61 predicted AFs were reproducibly detected, and no unpredicted fragments were amplified. This FAFLP analysis was used to genotype 74 C. jejuni strains belonging to the nine heat-stable (HS) serotypes most prevalent in human disease in England and Wales. The 74 C. jejuni strains exhibited 60 FAFLP profiles, and cluster analysis of them yielded a radial tree showing genetic relationships between and within 13 major clusters. Some clusters were related, and others were unrelated, to a single HS serotype. For example, all strains belonging to serotypes HS6 and HS19 grouped into corresponding single genotypic clusters, while strains of serotypes HS11 and HS18 each grouped into two genotypic clusters. Strains of HS50, the most prevalent serotype infecting humans, were found both in one large (multiserotype) cluster complex and dispersed throughout the tree. The strain genotypes within each FAFLP cluster were characterized by a particular combination of AFs, and among the cluster there were additional differential AFs. Identification of such AFs could act as a search tool to look for potential associations with disease or animal hosts, when applied to large number of human isolates. Genome-sequence based FAFLP, thus, has the potential to establish a genetic database for epidemiological investigations of Campylobacter.
Campylobacter is the commonest cause of bacterial gastroenteritis in developed countries. The incidence of human campylobacteriosis in England and Wales has risen in recent years from ∼34,000 cases reported to the Public Health Laboratory Service Communicable Disease Surveillance Centre in 1990 to ∼54,000 in 2000. It now surpasses the incidence of reported salmonella infections 3.6-fold (information found at the website http://www.phls.co.uk). Although most isolates are reported simply as Campylobacter spp., available data suggest that over 90% of campylobacters detected by culture belong to the species Campylobacter jejuni. The next commonest species is Campylobacter coli (10). Reproducible and discriminatory typing methods are needed to identify the sources of campylobacter infections, and to trace the routes of transmission of strains through the food chain.
Phenotypic discrimination between isolates of C. jejuni and C. coli uses serotyping of the heat stable (HS) (10, 24) or heat labile (16) surface-exposed antigens. The two HS schemes both use antisera raised against the same type strains but differ in their antigen detection methods. The method of Penner and Hennessy (24) uses passive hemagglutination, while the method of Frost et al. (10) employs direct whole bacterial cell agglutination. Recent studies have shown the existence of two distinct HS antigens in Campylobacter, which may account for the differences in the two schemes. These antigens are a polysaccharide capsule (3, 13) and a lipopolysaccharide and/or lipooligosaccharide component of the cell surface (17). The majority of C. jejuni isolates belong to only a limited number of HS serotypes, and 19% of isolates are nontypeable with the current antiserum panel using the direct agglutination method. Some of these limitations have been addressed by employing phage typing (9) as an adjunct to serotyping.
General drawbacks of these phenotypic typing methods are the restricted availability, cost and quality of the antiserum reagents and phage panels; cross-reactivity; and the level of nontypeability. These factors have limited epidemiological studies of Campylobacter, both nationally and globally (26). Genotyping may therefore have a particularly important role in studying the epidemiology of Campylobacter, and a number of molecular methods have been applied and evaluated. They include flagellin gene (PCR-restriction fragment length polymorphism analysis) typing, pulsed-field gel electrophoresis (PFGE), random amplified polymorphic DNA, and ribotyping. The main features of and findings by these methods have been reviewed by Wassenaar and Newell (26).
The genome-sampling technique of amplified fragment length polymorphism (AFLP) analysis has been applied in empirical fashion to Campylobacter spp., yielding promising new information based on percentage similarities of AFLP profiles, with regard to taxonomic relationships, genetic groupings, and outbreaks (7, 15, 20). Recently the complete C. jejuni genome sequence (22) has been released, enabling us to apply to C. jejuni, for the first time, predictive modeling of fluorescent AFLP (FAFLP) (1, 12).
The aim of the present study was to model FAFLP from the genome sequence of C. jejuni strain NCTC 11168 and to validate the accuracy and reproducibility of this FAFLP by comparing predicted with experimental AF sizes. We then used the standardized FAFLP analysis to examine genotypic variation in relation to HS serotype for 74 C. jejuni isolates.
MATERIALS AND METHODS
Bacterial strains, phenotypic typing, and culture conditions.
The 74 strains of C. jejuni studied were from samples of meat from retail outlets, unpasteurized milk, and human fecal specimens from sporadic campylobacter infections. They belonged to nine of the most prevalent HS serotypes isolated from humans in England and Wales during 1998 and 1999 and belonged to various phage types (PTs) (Table 1). One C. coli strain (strain 75 [Table 1]) isolated from a sample of poultry meat was also included. The genome-sequenced strain NCTC 11168 (22) and the reference strains for six serotypes were also included. All strains were HS serotyped using the direct agglutination method and phage typed according to the standard protocols of the Campylobacter Reference Unit (CRU) (Central Public Health Laboratory, London, United Kingdom). Strains were cultured microaerobically at 37°C for 48 h on blood agar plates and preserved for reference at −80°C in Microbank cryovials (Pro-Lab Diagnostics, Cheshire, United Kingdom).
TABLE 1.
Campylobacter jejuni isolate data
| Genotype group | No. | CRU no. | Yr of isolation | Source | Serotypea | Phage type | FAFLP genotypeb |
|---|---|---|---|---|---|---|---|
| Cluster A complex | 1 | NCTC 11168 | 1977 | Human | HS44 | 38 | A-2a |
| 2 | C012487 | 1998 | Chicken portion | HS50 | 35 | A-1a | |
| 3 | C011973 | 1998 | Chicken portion | HS2 | 35 | A-1b | |
| 4 | C011919 | 1998 | Chicken portion | HS2 | 35 | A-1b | |
| 5 | C011848 | 1998 | Chicken portion | HS2 | 35 | A-1c | |
| 6 | C012450 | 1998 | Chicken portion | HS50 | 1 | A-1d | |
| 7 | C013064 | 1998 | Ox liver | HS44 | 33 | A-1e | |
| 8 | C012878 | 1998 | Chicken portion | HS44 | 33 | A-1e | |
| 9 | C012026 | 1998 | Chicken portion | HS44 | 36 | A-1f | |
| 10 | C012025 | 1998 | Chicken portion | HS44 | 36 | A-1f | |
| 11 | C012567 | 1998 | Chicken portion | HS2 | 36 | A-UGc | |
| 12 | C013713 | 1998 | Ox liver | HS2 | 33 | A-2b | |
| 13 | C013082 | 1998 | Chicken portion | HS50 | 15 | A-2c | |
| 14 | C030280 | 1999 | Human | HS16 | 1 | A-3a | |
| 15 | C030328 | 1999 | Human | HS16 | 1 | A-3a | |
| 16 | C031392 | 1999 | Human | HS16 | 6 | A-3b | |
| 17 | C012912 | 1998 | Ox liver | HS50 | 33 | A-4a | |
| 18 | C013606 | 1998 | Wild strain | HS44 | 33 | A-4b | |
| 19 | C028659 | 1999 | Human | HS16 | 5 | A-5a | |
| 20 | C013040 | 1998 | Chicken portion | HS50 | 59 | A-5b | |
| 21 | C028450 | 1999 | Human | HS16 | 5 | A-5c | |
| 22 | C013634 | 1998 | Chicken portion | HS50 | 44 | A-5d | |
| 23 | C011818 | 1998 | Chicken portion | HS50 | 34 | A-5e | |
| 24 | C011856 | 1998 | Chicken portion | HS50 | 5 | A-5e | |
| 25 | C012196 | 1998 | Chicken portion | HS50 | 5 | A-UGc | |
| 26 | C013249 | 1998 | Chicken portion | HS44 | 19 | A-6a | |
| 27 | C011974 | 1998 | Chicken portion | HS44 | 2 | A-6b | |
| 28 | C012154 | 1998 | Chicken portion | HS44 | 2 | A-6b | |
| 29 | C013411 | 1998 | Chicken portion | HS44 | 14 | A-6c | |
| 30 | C026822 | 1999 | Human | HS16 | 2 | A-6d | |
| 31 | C026657 | 1999 | Human | HS16 | 2 | A-6e | |
| Cluster B | 32 | C011993 | 1998 | Chicken portion | HS11 | 44 | B1 |
| 33 | C012240 | 1998 | Chicken portion | HS11 | 44 | B2 | |
| 34 | C012234 | 1998 | Chicken portion | HS11 | 1 | B3 | |
| 35 | C012215 | 1998 | Ox liver | HS11 | 44 | B4 | |
| Cluster C | 36 | C022391 | 1999 | Human | HS18 | 2 | C1 |
| 37 | C022380 | 1999 | Human | HS18 | 2 | C2 | |
| 38 | C023887 | 1999 | Human (Cyprus) | HS18 | 1 | C3 | |
| 39 | C024010 | 1999 | Human (Spain) | HS18 | 1 | C4 | |
| Cluster D | 40 | C012238 | 1998 | Chicken portion | HS11 | 1 | D1 |
| 41 | C013100 | 1998 | Chicken portion | HS11 | 5 | D1 | |
| 42 | C013650 | 1998 | Chicken portion | HS11 | 39 | D1 | |
| 43 | C013431 | 1998 | Chicken portion | HS11 | 62 | D1 | |
| 44 | C013417 | 1998 | Chicken portion | HS11 | UT | D1 | |
| 45 | C012013 | 1998 | Chicken portion | HS11 | 1 | D2 | |
| Cluster E | 46 | C027012 | 1999 | Human | HS18 | 30 | E1 |
| 47 | NCTC 12516 | 1981 | Not known | HS18 | 9 | E2 | |
| Cluster F | 48 | C015168 | 1998 | Human | HS19 | 2 | F1 |
| 49 | C018836 | 1998 | Human GBS | HS19 | 2 | F1 | |
| 50 | C022472 | 1999 | Human | HS19 | 2 | F1 | |
| 51 | C014456 | 1998 | Human | HS19 | 2 | F2 | |
| 52 | C015207 | 1998 | Human | HS19 | 1 | F2 | |
| 53 | C014633 | 1998 | Human | HS19 | 2 | F3 | |
| 54 | C028007 | 1999 | Human | HS19 | 2 | F4 | |
| 55 | NCTC 12517 | 1981 | Not known | HS19 | 2 | F5 | |
| Cluster G | 56 | C027968 | 1999 | Human | HS16 | 20 | G1 |
| 57 | NCTC 12549 | 1982 | Not known | HS44 | 51 | G2 | |
| 58 | C011921 | 1998 | Chicken portion | HS50 | 6 | G3 | |
| 59 | C001126 | 1996 | Unpasteurized milk | HS50 | 6 | G4 | |
| Cluster H | 60 | C005281 | 1997 | Human | HS6 | 2 | H1 |
| 61 | C005477 | 1997 | Human | HS6 | 2 | H1 | |
| 62 | C026868 | 1999 | Human | HS6 | 39 | H2 | |
| 63 | NCTC 12505 | 1982 | Not known | HS6 | 2 | H3 | |
| 64 | C027635 | 1999 | Human | HS6 | 2 | H4 | |
| Unclustered (UG)d | 65 | NCTC 12504 | 1982 | Not known | HS5 | 5 | UG1 |
| 66 | C011986 | 1998 | Chicken portion | HS50 | 49 | UG2 | |
| 67 | C013423 | 1998 | Chicken portion | HS5 | 44 | UG3 | |
| 68 | NCTC 12559 | Not known | Not known | HS50 | 6 | UG4 | |
| 69 | C028937 | 1999 | Human | HS16 | 6 | UG5 | |
| 70 | C012938 | 1998 | Human | HS16 | 20 | UG6 | |
| 71 | C011839 | 1998 | Chicken portion | HS50 | 5 | UG7 | |
| 72 | C012483 | 1998 | Chicken portion | HS50 | 1 | UG8 | |
| 73 | C013039 | 1998 | Chicken portion | HS50 | 2 | UG9 | |
| 74 | C013073 | 1998 | Chicken portion | HS5 | 48 | UG10 | |
| 75e | C012586 | 1998 | Chicken portion | HS UT | UT | UG11 |
Serotype of isolates as determined by direct agglutination method (see Materials and Methods).
FAFLP genotypes were designated following cluster analysis (see Fig. 2) as cluster B to H followed by clone number or, for cluster A complex, as clusters A-1 to A-6 followed by clone type (designated by letters a to f).
A-UG designated to a characterized FAFLP genotype which is unclustered within the cluster A complex.
UG designated to a characterized FAFLP genotype which is unclustered within this study.
Indicates the C. coli strain, which exhibited a diverse and unclustered genotype.
Standard nucleic acid extraction.
Genomic DNA was extracted from 48-h Campylobacter plate cultures using the DNeasy tissue kit (Qiagen Ltd., Crawley, West Sussex, United Kingdom) according to the manufacturer's instructions. The concentration of DNA was estimated using a spectrophotometer (Beckman DU 640) by standard methods (25).
Computer methods.
The complete genome sequence of C. jejuni NCTC 11168 (accession no. AL111168) was analyzed with Lasergene (DNAStar, Madison, Wis.) and MacVector (Oxford Molecular, Oxford, United Kingdom) software. Data for the sizes and number of fragments generated by HindIII and HhaI digestion were imported into a spreadsheet. These fragment sizes were then adjusted to allow the addition of primer sequence during PCR amplification.
FAFLP.
FAFLP was performed on 500 ng of genomic DNA digested in a total volume of 30 μl, which consisted of 5 U of HindIII endonuclease (New England BioLabs [NEB], Hitchin, Hertfordshire, England), 3 μl of 10× NEB buffer 2, and 1.5 μl of DNase-free RNase A (10 μg/μl), for 1.5 h at 37°C. To this digest was then added 5 U of HhaI (NEB) and 0.3 μl of 100× bovine serum albumin (BSA), and the reactions were incubated for a further 1.5 h at 37°C. The endonucleases were inactivated (65°C for 10 min) prior to ligation.
To the double-digested DNA, 25 μl of a solution containing 0.5 μl of 2 μM HindIII adapter (Genosys Biotechnologies, Cambridge, United Kingdom), 0.5 μl of 20 μM HhaI adapter (Genosys Biotechnologies), 40 U of T4 DNA-ligase (NEB), and 5 μl of 10× T4 ligase buffer (NEB) was added. The reaction mixture was incubated at 16°C for 3 to 4 h, heated at 65°C for 10 min to inactivate the ligase, and stored at −20°C. The sequence of the HindIII adapter was 5′-CTC GTA GAC TGC GTA CC, 3′-CTG ACG CAT GGT CGA. The sequence of the HhaI adapter was 5′-GAC GAT GAG TCC TGA TCG, 3′-G CTA CTC AGG ACT A. The forward primer (HindIII adaptor specific), labeled with the blue fluorescent dye 5-carboxyfluorescein (FAM), contained an extra selective base at the 3′ end (HindIII+A, 5′-GAC TGC GTA CCA GCT TA-3′ [Genosys Biotechnologies]) (7). The reverse primer (HhaI adapter specific) also contained an extra selective base at the 3′ end (HhaI+A, 5′-GAT GAG TCC TGA TCG CA-3′ [Genosys Biotechnologies]) (7). PCRs were performed in 25-μl volumes containing 2.5 μl of ligated DNA, 2.0 μl of 1 μM FAM-labeled HindIII+A primer, 1.0 μl of 5.0 μM HhaI+A primer, 2.5 μl of 10× Taq polymerase buffer (Life Technologies, Paisley, United Kingdom), 1.25 μl of 50 mM MgCl2 (Life Technologies), 0.5 μl of 10 mM concentrations of each of the four deoxynucleoside triphosphates (dNTPs) (Life Technologies), and 1.25 U of Taq DNA polymerase (Life Technologies). Touchdown PCR cycling conditions were as described previously (4, 5). FAFLP products were separated on an ABI 377 automated DNA sequencer (Perkin-Elmer Corp., Norwalk, Conn.) using Premix Long Ranger 5% polyacrylamide gel solution (FMC BioProducts, Vallensbaek Strand, Denmark) as described previously (4, 5). Each FAFLP reaction (1 μl) was loaded with an internal size marker (GeneScan-2500 labeled with the red fluorescent dye 6-carboxy-x-rhodamine [ROX; PE Applied Biosystems, Warrington, United Kingdom]). The running buffer was 1× Tris-borate-EDTA (TBE), and the electrophoresis conditions were 2.0 kV at 51°C for 14 h. The well-to-read distance was 48 cm.
Fragment analysis.
Fluorescent amplified fragments (AFs) visualized on polyacrylamide sequencing gels were sized with GeneScan 3.1.0 software (Perkin-Elmer Corp.). Gel displays were transformed into electropherograms, and Genotyper 2.5 software (Perkin-Elmer Corp.) was used to generate a table with presence or absence of fragments. Fragment data were recorded in a binary format in Excel (version 6.0; Microsoft). Dice coefficients of similarity were calculated with an in-house program. Cluster analysis was performed by UPGMA (NEIGHBOR program of PHYLIP) (8), and a dendrogram was displayed with the TreeView program (21).
RESULTS
Predictive modeling of FAFLP for the genome sequence of strain NCTC 11168 and its experimental validation.
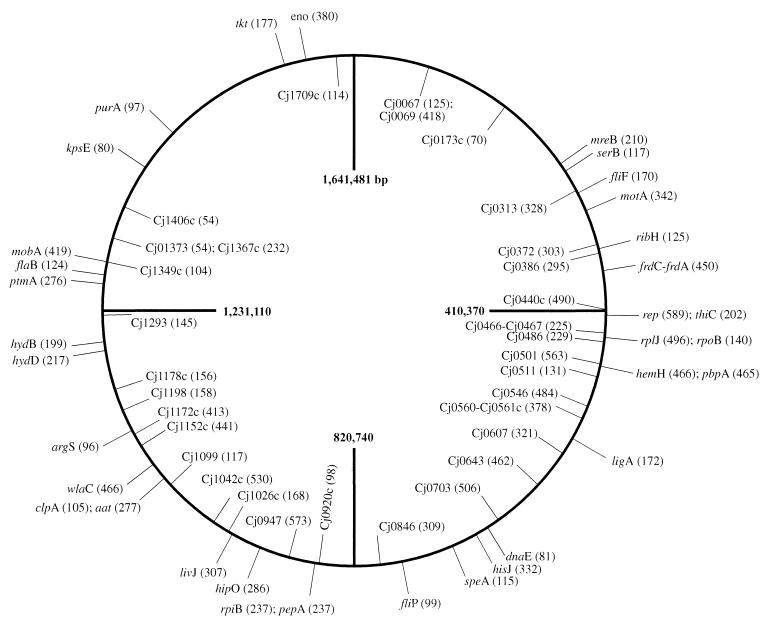
When the complete genome sequence of NCTC 11168 was analyzed (see Materials and Methods) to predict the sizes of HindIII and HhaI restriction fragments amplified by the primer pair HindIII+A and HhaI+A (7), 61 AFs were expected between 50 and 600 bp. The precision of sizing these AFs was ±0.5 bp. Their positions in relation to the genetic loci on the NCTC 11168 chromosome (22) are shown in Fig. 1. To determine the accuracy and reproducibility of FAFLP for C. jejuni, three different DNA preparations of strain NCTC 11168 were each subjected to three different FAFLP reactions and run on individual sequencing gels. When experimental data were compared with expected values, all but one (sized 462 bp) of the predicted 61 AFs were observed in all nine of the reactions, and no unpredicted fragments were amplified. Of the 60 AFs observed, 36 were in the size range 55 to 300 bp; 35 of these were within 1 bp and 1 was within 2 bp of the predicted size. Among the remaining 24 AFs in the higher size range (300 to 600 bp), 9 AFs were within 1 bp, 6 were within 2 bp, 8 were within 3 bp, and 1 was within 4 bp of the predicted size.
FIG. 1.
FAFLP genome map of C. jejuni strain NCTC 11168. Relative positions and predicted sizes (in base pairs) of AFs in the C. jejuni NCTC 11168 genome (22) modeled for the primer pair HindIII+A and HhaI+A and experimentally validated by FAFLP analysis. The genes that AFs represent are indicated on the outer circle; AFs in the inner circle represent putative genes.
Strain genotyping by FAFLP analysis.
The FAFLP profiles of the 75 strains analyzed in this study consisted of 45 to 100 AFs ranging in size from 60 to 600 bp; AFs larger than 600 bp were not included in our analysis. Sixty-one different FAFLP profiles were detected among the 75 strains, and 51 strains including NCTC 11168 and the C. coli strain had unique profiles. There were eight profiles which were shared by eight pairs of isolates each, including a pair of isolates from the same meat sample (isolates 9 and 10 [Table 1]). One profile was shared by three isolates, and another profile was shared by five isolates; these isolates sharing the same profile were from sporadic infections. Among all the 75 isolates, there were 231 polymorphic AFs (a polymorphic AF is here defined as one that is either uniquely present in a single profile or present in some profiles and absent in others). There were 182 polymorphic AFs among the 74 C. jejuni isolates.
Cluster analysis of FAFLP profiles and relationship to HS serotype.
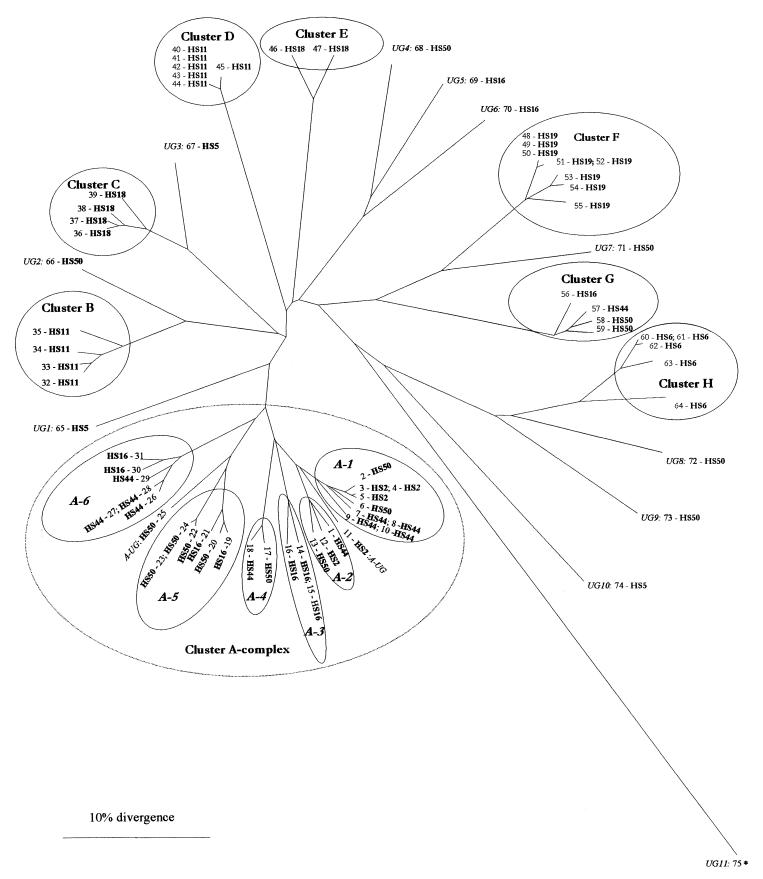
For each strain, AFs were sized (size calling tolerance was ±0.5 bp) and scored as present (as 1) or absent (as 0) in a binary matrix. Cluster analysis was performed and a radial distance tree was generated, on the basis of which FAFLP profiles were given genotype designations (Fig. 2; Table 1). The radial tree shows 13 major genotypic clusters (designated A-1 to A-6 and B to H in Fig. 2; Table 1). The isolates within all the 13 clusters showed ≤10% divergence. Cluster A complex (shown as a dotted line in Fig. 2) was defined as such to reflect the serological relatedness of its isolates (10) and constituted the genotypic clusters A-1 to A-6. The isolates within the cluster A complex exhibited ≤25% genotypic divergence.
FIG. 2.
Dendrogram showing relationship between FAFLP genotypes and prevalent HS serotypes of C. jejuni. The dendrogram was derived from FAFLP data by UPGMA and the PHYLIP and TreeView programs (21). The HS serotype of each strain is shown in boldface type following the strain number (see Table 1). Following cluster analysis, FAFLP genotypes were designated as clusters A-1 to A-6 and clusters B to H or as unclustered genotypes, shown in italics (UG1 to UG11). The isolates in all 13 of the clusters showed ≤10% divergence. The dotted line delineates cluster A complex isolates that are serologically related (8), and the isolates within this complex exhibited ≤25% divergence. Within this cluster complex, six genotypic clusters were designated A-1 to A-6 (shown in italics), and two isolates with unique genotypes were designated A-UG (see Results). An asterisk indicates a C. coli strain which was untypeable by both sero- and phage-typing and exhibited genotype UG11.
Two FAFLP clusters corresponded to specific HS serotypes (clusters F and H), while six other clusters included multiple HS serotypes. Strains belonging to two further serotypes, HS11 and HS18, grouped in two genotypic clusters each (clusters B and D and clusters C and E, respectively). All eight strains belonging to serotype HS19 grouped into cluster F, and all five HS6 strains grouped together in cluster H. Cluster A complex (Fig. 2) comprised 31 strains belonging to four serotypes: HS2, HS16, HS44, and HS50. Five of the six clusters within the cluster A complex included multiple serotypes. It also included NCTC 11168, which serotypes as HS44. Strains belonging to serotype HS50 were distributed throughout the tree. Nine were found in the multiserotype cluster A complex described above; four of these grouped into cluster A-5, two grouped into cluster A-1, and one strain each grouped in clusters A-2 and A-4, respectively. Two strains within cluster A-complex showed unclustered genotypes (designated A-UG [Table 1]). Five others were distributed throughout the tree with unclustered genotypes (designated UG), and two were found in genotypic cluster G. Of the 10 HS16 strains in this study, three grouped into the serotype-specific cluster A-3, two strains each grouped into multiserotype clusters A-5 and A-6, and one strain grouped into cluster G. The remaining two strains had unclustered genotypes (Fig. 2; Table 1). All three strains belonging to serotype HS5 had unclustered genotypes (Fig. 2, Table 1). The C. coli strain exhibited a divergent and unclustered genotype.
All genotypic clusters included isolates belonging to diverse PTs. The 31 strains in cluster A complex strains belonged to 14 PTs, including five strains of PT33; four strains each of PT2, PT5, and PT35; and three strains each of PT1 and PT36, respectively (Table 1). The remaining eight strains, however, belonged to single PTs (Table 1). All the three HS2 PT35 isolates grouped together in cluster A-1. Three of the four strains within cluster B belonged to HS11 PT44, while the six HS11 strains of cluster D belonged to five different PTs. Of the four strains in cluster C, two phenotyped as HS18 PT1 and the other two phenotyped as HS18 PT2. Seven of the eight strains in cluster F belonged to HS19 PT2.
For the 74 C. jejuni isolates, 165 of the 182 polymorphic AFs were found to combine in various ways to yield specific FAFLP genotypes. For example, cluster F strains were characterized by a combination of 55 marker AFs, and 10 other intragroup marker AFs distinguished strains within the cluster. This cluster contained clonally related strains of serotype HS19. Among strains of serotype HS11, the four strains in cluster B were differentiated from other clusters by a combination of 59 marker AFs, and cluster D was differentiated by a combination of 63 marker AFs. The two clusters, C and E, containing strains of serotype HS18 were distinguished by a combination of 59 marker AFs and a combination of 57 marker AFs, respectively. The cluster A complex, containing 41% of the strains (31 out of 75), was differentiated from the other clusters by a combination of 39 marker AFs; 57 other marker AFs distinguished the individual strains within it.
DISCUSSION
Human Campylobacter infections now greatly exceed Salmonella infections in incidence, but very few outbreaks have been linked to particular food sources (23). The frequent contamination of food by multiple strains has added to the difficulty of tracing the origins and routes of human infection and of recognizing outbreaks (14). Various phenotypic and genotypic methods have been applied in epidemiological studies of Campylobacter (18), but the question remains whether the recent large rise in C. jejuni infections reflects an increased incidence of sporadic infections or a number of outbreaks. In this study, we have standardized FAFLP to the genome sequence of C. jejuni NCTC 11168 (22) and experimentally validated this high-resolution genotyping technique using the published primer combination HindIII+A and HhaI+A, first defined in the novel empirical FAFLP studies of Duim and colleagues (7). We then investigated its use in reproducibly genotyping C. jejuni isolates.
The 60 experimentally recovered AFs (98% of the predicted AFs) derived from the NCTC 11168 strain represent approximately 1% of its total genome. The precision of sizing AFs was ±0.5 bp. Seventy-three percent of these AFs were within 1 bp of the predicted size. The accuracy of sizing was ±1 bp for AFs up to 300 bp but decreased to ±2 or 3 bp for AFs between 300 and 600 bp. AFs larger than 600 bp were not included in our analysis since the accuracy of sizing decreased further for larger fragments. The variation in sizing of >±1 bp can be attributed to the irregular spacing of bands in the ROX 2500 internal lane standard, and the use of more evenly spaced size markers would minimize this slight inaccuracy in AF sizing (data not shown). A few single base pair differences between pairs of predicted AFs (e.g., 97 and 98 bp and 104 and 105 bp) which could not be resolved by the ABI 377 sequencer presented as doublets on the electropherograms, both with higher signal strength. A few other identically sized AFs amplified from different regions of the genome (e.g., two of 117 bp and two of 237 bp) appeared either as broader peaks on the trace with a higher signal strength (e.g., the 117-bp AF) or as two AFs 1 bp apart.
Among the 74 isolates of C. jejuni examined in this study, seven AFs were common to all genotypes. This finding is being further investigated in our laboratory, with a view to sequencing AFs which have potential in molecular identification assays, e.g., in DNA arrays. The identification of AFs such as these also shows that FAFLP could act as a powerful search tool when used to screen large numbers of isolates from human disease. Clearly, there is a potential application in searching clinical microbiology FAFLP databases for as-yet-unidentified associations with host specificity and specific differential AFs.
Dingle et al. (6) examined the genetic variation present in seven housekeeping genes of C. jejuni by multilocus sequence typing (MLST). In a strain collection comprising 194 diverse C. jejuni strains from humans, animals, and the environment, they found 155 sequence types (STs) comprising 11 clone complexes and 51 unique STs. One of the largest MLST lineages, the ST-21 complex, predominantly comprised strains of the Penner HS serotypes Pen1, Pen2, and Pen4 and included NCTC 11168. The most commonly encountered HS serotypes in England and Wales include Pen1, Pen2, and Pen44 as serotyped by passive hemagglutination (24), or HS1, HS2, HS44, and HS50 as serotyped by direct agglutination (10). This group of serotypes can therefore be related to the FAFLP cluster A complex. Since our FAFLP cluster A complex included strain NCTC 11168 and comprised multiple isolates from serotypes HS2, HS16, HS44, and HS50, it is likely to correspond to or overlap with the MLST lineage ST-21. By the same reasoning, FAFLP cluster F, comprising strains of serotype HS19, can be related to MLST lineage ST-22. These conclusions are in line with the previously published congruencies of FAFLP with either MLST or multilocus enzyme electrophoresis (2, 11). Olive and Bean (19), in a recently published comparison of various DNA-based typing methods of bacterial organisms, estimate the costs for a single FAFLP reaction at $20 and that for seven-locus MLST of a single strain at $280. In our view, FAFLP exhibits superior cost-effectiveness, adaptability, discriminatory power, and ease of use. Nonetheless the two methods can be described as complementary because, both being based on the genome sequence, they are theoretically as well as experimentally congruent.
Serotype HS50 is the most prevalent serotype among C. jejuni strains isolated from humans, and it accounts for approximately 20% of all infections. The FAFLP data presented here indicate that this serotype is genetically heterogeneous. Although nine HS50 strains were found in the multiserotype cluster A complex and two strains were found in cluster G, there was no congruence between FAFLP genotypes and serotype for the remaining five HS50 strains included in the study. Strains belonging to serotypes HS11 and HS18 grouped into two separate genotypic clusters each. In contrast, all strains belonging to serotypes HS19 and HS6 grouped into serotype-specific clusters, indicating the genetic homogeneity of these two serotypes. The lack of congruence between FAFLP genotypes and certain serotypes and between FAFLP genotypes and sero-phage types of C. jejuni indicates that further investigative genotyping of this major enteropathogen will define its epidemiological clonality more accurately.
We have also demonstrated that FAFLP defines strain genotypes within and between clusters by unique combinations of precisely sized marker AFs. These AFs could serve as identification markers (“bar codes”) that define clonality and could be used as the basis of a molecular typing scheme. A continuously updated radial tree such as that shown in Fig. 2 could, in effect, serve as a genetic record of strain genotypes. The genetic data could then be linked in a comprehensive database with epidemiological information and phenotyping data, to facilitate the identification of outbreaks and sources of apparently sporadic human infection.
We conclude that FAFLP is a highly reproducible method for typing C. jejuni (as demonstrated with the NCTC 11168 strain), capable of recognizing genotypic clusters. These FAFLP genotypic clusters were not congruent with all HS serotypes. Within genotypic clusters, FAFLP could readily distinguish between individual strains. We expect FAFLP to become a powerful tool in the macro- and microepidemiological analysis of campylobacter incidence, including outbreak investigations. It could also be used to establish a genetic database of strains drawn from across the food chain and human cases.
ACKNOWLEDGMENTS
We thank Richard Thwaites and Judith Richardson for providing the strains and epidemiological data and Philip Mortimer and Henry Smith for critically reading the manuscript.
This work was partly supported by a grant (B03014) from the Food Standards Agency, London, United Kingdom.
REFERENCES
- 1.Arnold C, Metherell L, Clewley J P, Stanley J. Predictive modelling of fluorescent AFLP: a new approach to the molecular epidemiology of E. coli. Res Microbiol. 1999;150:33–44. doi: 10.1016/s0923-2508(99)80044-8. [DOI] [PubMed] [Google Scholar]
- 2.Arnold C, Metherell L, Willshaw G, Maggs A, Stanley J. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J Clin Microbiol. 1999;37:1274–1279. doi: 10.1128/jcm.37.5.1274-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chart H, Frost J A, Oza A, Thwaites R, Gillanders S, Rowe B. Heat-stable serotyping antigens expressed by strains of Campylobacter jejuni are probably capsular and not long-chain lipopolysaccharide. J Appl Bacteriol. 1996;81:635–640. doi: 10.1111/j.1365-2672.1996.tb03558.x. [DOI] [PubMed] [Google Scholar]
- 4.Desai M, Efstratiou A, George R, Stanley J. High-resolution genotyping of Streptococcus pyogenes serotype M1 isolates by fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 1999;37:1948–1952. doi: 10.1128/jcm.37.6.1948-1952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle K E, Colles F M, Wareing D R, Ure R, Fox A J, Bolton F E, Bootsma H J, Willems R J, Urwin R, Maiden M C. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duim B, Ang C W, van Belkum A, Rigter A, van Leeuwen N W J, Endtz H P, Wagenaar J A. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barré or Miller Fisher syndrome. Appl Environ Microbiol. 2000;66:3917–3923. doi: 10.1128/aem.66.9.3917-3923.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP—Phylogeny Inference Package. Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Frost J A, Kramer J M, Gillanders S A. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol Infect. 1999;123:47–55. doi: 10.1017/s095026889900254x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost J A, Oza A N, Thwaites R T, Rowe B. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J Clin Microbiol. 1998;36:335–339. doi: 10.1128/jcm.36.2.335-339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulding J N, Hookey J V, Stanley J, Olver W, Neal K R, Ala'Aldeen D A, Arnold C. Fluorescent amplified-fragment length polymorphism genotyping of Neisseria meningitidis identifies clones associated with invasive disease. J Clin Microbiol. 2000;38:4580–4585. doi: 10.1128/jcm.38.12.4580-4585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulding J N, Stanley J, Saunders N, Arnold C. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:1121–1126. doi: 10.1128/jcm.38.3.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlyshev A V, Linton D, Gregson N A, Lastovica A J, Wren B W. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- 14.Kramer J M, Frost J A, Bolton F J, Wareing D R. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J Food Protect. 2000;63:1654–1659. doi: 10.4315/0362-028x-63.12.1654. [DOI] [PubMed] [Google Scholar]
- 15.Lindstedt B A, Heir E, Vardund T, Melby K K, Kapperud G. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J Clin Microbiol. 2000;38:3379–3387. doi: 10.1128/jcm.38.9.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran A P, Penner J L. Serotyping of Campylobacter jejuni based on heat-stable antigens: relevance, molecular basis and implications in pathogenesis. J Appl Microbiol. 1999;86:361–377. doi: 10.1046/j.1365-2672.1999.00713.x. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen E M, Engberg J, Fussing V, Petersen L, Brogren C H, On S L. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol. 2000;38:3800–3810. doi: 10.1128/jcm.38.10.3800-3810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.On S L, Harrington C S. Identification of taxonomic and epidemiological relationships among campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol Lett. 2000;193:161–169. doi: 10.1111/j.1574-6968.2000.tb09419.x. [DOI] [PubMed] [Google Scholar]
- 21.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 23.Pebody R G, Ryan M J, Wall P G. Outbreaks of campylobacter infection: rare events for a common pathogen. Commun Dis Rep Rev. 1997;7:R33–R37. [PubMed] [Google Scholar]
- 24.Penner J L, Hennessy J N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. J Appl Bacteriol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]