Letter to the Editor
SMARCE1 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1) is a constant component of the SWI/SNF (SWItch/Sucrose Non-Fermentable) chromatin remodeling complexes. While heterozygous missense variants are responsible for the rare genotype of Coffin-Siris syndrome, heterozygous truncating variants predispose individuals to an increased risk of clear cell meningiomas (CCM) that occur from childhood to adulthood. Here we report an undescribed supra-tentorial intra-parenchymal malignant tumor with an inherited heterozygous truncating variant of SMARCE1, and thereby expand the spectrum of SMARCE1-related cancer predisposition syndrome.
A 4-year-old child without congenital abnormality and normal development was addressed for seizures. MRI revealed a 40 × 33 × 30 mm right temporal tumor. There was no family history of tumor, especially no brain tumor or meningioma. The tumor was not dura matter based on imaging and on per-operative findings. Complete resection was performed. The patient was treated by radiotherapy and temozolomide. After 30 months of follow-up the tumor did not relapse.
Histopathological findings showed a highly cellular tumor devoid of specific pattern. Tumor cells were small to medium sized with moderate amount of cytoplasm. There was no clear cell morphology neither rhabdoid cells. More than 20 mitoses/2mm2, endothelial proliferation and necrosis were observed. Automated immunohistochemistry demonstrated labeling of tumor cells by Olig2 (AF2418, R1D Systems) stained most of tumor cells, GFAP (6F2, Dako), synaptophysin (DAK-SYNAP, Dako), EMA (E29, Dako) in 10% of tumor cells, alpha smooth actin (1A4, Dako) stained most of tumor cells, and p53 (DO-7, Dako) diffusely stained tumor cells, Vimentin (V9, Dako) diffusely stained tumor cells. There was no staining by SSTR2A (ab134152, Abcam), IDH1R132H (H09, Dianova), H3K27M (K27M, Sigma-Aldrich), LIN28(A177, Cell Signaling), NFKappaB (D14E12, Cell Signaling), Internexine Alpha (BB300-140, Novus), AE1AE3 (AE1/AE3, Dako-Agilent), FGFR3 (SC-13121, Santa Cruz), CD34 (QBEnd 10, Beckman Coulter) antibodies. There was no loss of ATRX (poly, Sigma-Aldrich) H3K27me3 (H3K27Me3, Sigma-Aldrich), BAF47 (25/BAF47, BD Biosciences), BRG1 (ab110641, Abcam). Ki67 (MIB1, Dako) stained 80% of tumor cells.
Given the absence of definite diagnosis, we next investigated whether molecular characterization could help classify the tumor. Methylation profiling (Illumina EPIC Human Methylation microarray) failed to classify the tumor in any known subgroup: the DNA methylation classifier V11b4 gave as prediction class “atypical teratoid and rhabdoid tumor (ATRT)” with a low calibration score of 0.67. Its latest version V12.3 assessed the lesion to “GBM pedMYCN” with a score of 0.17 while “ATRT-SHH subtype” had a score of 0.13. On a t-SNE plot perfomed on v11b4 version of the DNA methylation classifier, the tumor was closely related to ATRT, and clearly distinct from clear cell meningioma.
Then, a targeted NGS custom panel of 571 genes involved in oncogenesis was performed on the tumor DNA; it surprisingly revealed a c.460G > T / p.(Glu154*) pathogenic variation (PV) in SMARCE1. The inactivation of SMARCE1 was confirmed at the protein level with anti-BAF57 (poly, Sigma-Aldrich). No other variant was found in the SWI/SNF complex genes; noticeably, NGS also revealed a c.1009C > T / p.(Arg337Cys) PV in TP53. The final retained diagnosis was a malignant tumor with SMARCE1 inactivation, unclassifiable in the current World Health Organization Classification.
The chromosome CNV profile showed a complex profile unlikely found in ATRT or clear cell meningioma. It also revealed an isodisomy of the whole chromosome 17 where SMARCE1 and TP53 genes are located. Besides, the relatively complex genomic profile and the mutation in TP53 are not consistent with typical ATRT since those tumors most generally harbor a very simple profile and no PV except SMARCB1 or SMARCA4 alterations. Targeted mRNA sequencing (FusionPLex Comprehensive Panel, ArcherDx) on formalin-fixed, paraffin embedded material did not show any fusion gene.
The presence of a SMARCE1 biallelic PV in a brain tumor occurring at 4 years prompted us to test the germline DNA, which confirmed that the child was bearing the heterozygous PV in the lymphocytes. The familial screening revealed that the PV was inherited from the child’s unaffected father (54 years old at diagnosis).
Truncating germline variants of SMARCE1 are so far restrictedly associated to CCM inferring a central role of this gene to CCM oncogenesis [4]. To the best of our knowledge, the only other brain tumor reported in the context of SMARCE1 PV was an anaplastic astrocytoma occurring in a child with a Coffin-Siris syndrome and demonstrating a missense heterozygous PV [3]. The biallelic hit resulting in the loss of protein expression [5] further argues in favor of a driver role of SMARCE1 in our case. Thus, the tumor we report enlarges the spectrum of SMARCE1-related brain tumors to a so far undescribed high-grade neoplasm. Indeed, a CCM could be formally ruled out because the tumor was not dura matter based, microscopic findings are not consistent with clear cell meningioma, immunohistochemical features were those of a tumor without SSTR2A expression and methylation profiling was clearly in disfavor of this diagnosis. Furthermore complex CNV profile was found whereas recurrent chromosomal aberrations in CCM are chromosome 17q (segmental) loss, chromosome 6q loss and chromosome 22q loss. Interestingly, the methylation profiling was the most related to ATRT, which may indicate some kindship with this family of tumors due to SWI/SNF deficiency [2]. Besides, the relatively complex genomic profile and the mutation in TP53 are not consistent with typical ATRT, those tumors most generally harboring a very simple profile and no PV other than SMARCB1 or SMARCA4 alterations. Nevertheless, given the association of mutations in SWI/SNF proteins with ATRT and the close clustering of the tumor with ATRTs in methylation analysis, it is not possible to formally exclude that this tumor could be an ATRT with a SMARCE1 mutation (Fig. 1).
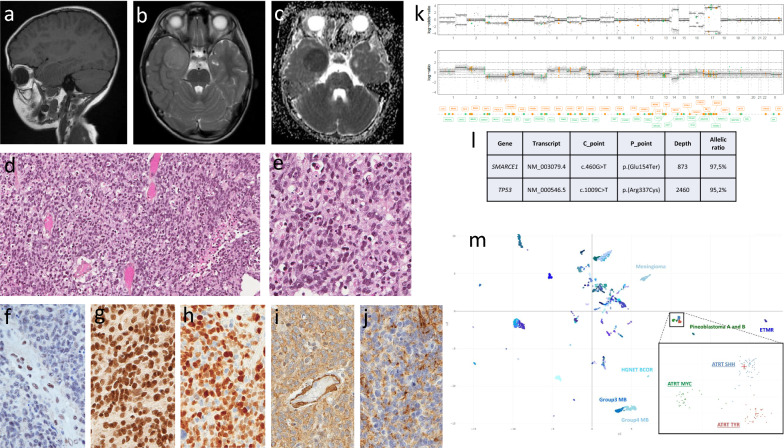
Fig. 1.
a MRI sagittal plane, T1 sequence without gadolinium injection showing a hypointense right temporal tumor. b MRI axial plane, T2 sequence showing a hyperintense right temporal tumor. c MRI axial plane, apparent diffusion coefficient (ADC) values showing a low ADC in the right temporal tumor. d, e Hematoxylin and Eosin (H&E) stain × 200 (d) and × 400 (e) showing a hypercellular tumor with pleomorphic cells harboring numerous mitoses. f SMARCE1 immunohistochemistry, × 400, showing a lack of expression in tumor cells with positive cells in a vessel as an internal control. g BAF47 immunohistochemistry, × 400, showing a maintained nuclear expression in tumor cells. h olig2 immunohistochemistry, × 400, showing a diffuse nuclear expression in tumor cells. i α-smooth actin immunohistochemistry, × 400, showing a cytoplasmic expression in tumor cells. j EMA immunohistochemistry, × 400, showing a patchy expression in tumor cells. k Copy number variation profile showing a complex profile. l Details of SMARCE1 and TP53 pathogenic variants. m UMAP showing the close localization of the tumor with ATRT
Altogether, this case expands the spectrum of SWI/SNF altered related tumors and of SMARCE1-related predisposition syndrome. Whether this risk should impact genetic counseling and tumor surveillance will depend on potential further reports of similar findings in individuals with SMARCE1 germline truncating variants.
Acknowledgements
The authors would like to thank the child’s parent for giving consent for publication. Theraprint (CLARA), RENOCLIP-LOC network.
Authors' contributions
FF and CG performed the histological examination. JMP and CG performed molecular analysis. FBV provided critical reagents and performed immunohistochemical analysis. FP, FV and CB provided clinical data and medical history. CB and CB provided imaging data and reviewed them. FF, JMP, FB and CG wrote the manuscript. All authors critically reviewed the manuscript. FF and JMP contributed equally and share a co-first author position. FB and CG contributed equally share a co-last author position. All authors read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not needed.
Consent for publication
Consent for publication was given by the child’s parents.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fabien Forest and Julien Masliah Planchon contributed equally and share a co-first author position
Franck Bourdeaut and Catherine Godfraind contributed equally share a co-last author position
References
- 1.Ho B, Johann PD, Grabovska Y, De DieuAndrianteranagna MJ, Yao F, Frühwald M, Hasselblatt M, Bourdeaut F, Williamson D, Huang A, Kool M. Molecular subgrouping of atypical teratoid/rhabdoid tumors—a reinvestigation and current consensus. Neuro Oncol. 2020;22:613–624. doi: 10.1093/neuonc/noz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holdhof D, Johann PD, Spohn M, Bockmayr M, Safaei S, Joshi P, Masliah-Planchon J, Ho B, Andrianteranagna M, Bourdeaut F, Huang A, Kool M, Upadhyaya SA, Bendel AE, Indenbirken D, Foulkes WD, Bush JW, Creytens D, Kordes U, Frühwald MC, Hasselblatt M, Schüller U. Atypical teratoid/rhabdoid tumors (ATRTs) with SMARCA4 mutation are molecularly distinct from SMARCB1-deficient cases. Acta Neuropathol. 2021;141:291–301. doi: 10.1007/s00401-020-02250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin B, Kesserwan C, Quinn EA, Einhaus SL, Wright KD, Azzato EM, Orr BA, Upadhyaya SA. Anaplastic Astrocytoma in a Child With Coffin-Siris Syndrome and a Germline SMARCE1 Mutation: A Case Report. J Pediatr Hematol Oncol. 2020;42:e177–e180. doi: 10.1097/MPH.0000000000001361. [DOI] [PubMed] [Google Scholar]
- 4.Sievers P, Sill M, Blume C, Tauziede-Espariat A, Schrimpf D, Stichel D, Reuss DE, Dogan H, Hartmann C, Mawrin C, Hasselblatt M, Stummer W, Schick U, Hench J, Frank S, Ketter R, Schweizer L, Schittenhelm J, Puget S, Brandner S, Jaunmuktane Z, Küsters B, Abdullaev Z, Pekmezci M, Snuderl M, Ratliff M, Herold-Mende C, Unterberg A, Aldape K, Ellison DW, Wesseling P, Reifenberger G, Wick W, Perry A, Varlet P, Pfister SM, Jones DTW, von Deimling A, Sahm F. Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1. Acta Neuropathol. 2021;141:281–290. doi: 10.1007/s00401-020-02247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith MJ, O’Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, Sharif S, Eccles D, Fitzpatrick D, Rawluk D, du Plessis D, Newman WG, Evans DG. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.