SUMMARY
Differentiation of intestinal T helper 17 (Th17) cells, which contribute to mucosal barrier protection from invasive pathogens, is dependent on colonization with distinct commensal bacteria. Segmented filamentous bacteria (SFB) are sufficient to support Th17 cell differentiation in mouse, but the molecular and cellular requirements for this process remain incompletely characterized. Here we show that intestine-draining mesenteric lymph nodes (MLN), not intestine proper, are the dominant site of SFB-induced intestinal Th17 cell differentiation. Subsequent migration of these cells to the intestinal lamina propria is dependent on their up-regulation of integrin β7. Stat3-dependent induction of RORγt, the Th17 cell-specifying transcription factor, largely depends on IL-6, but signaling through the receptors for IL-21 and IL-23 can compensate for absence of IL-6 to promote SFB-directed Th17 cell differentiation. These results indicate that redundant cytokine signals guide commensal microbe-dependent Th17 cell differentiation in the MLN and accumulation of the cells in the lamina propria.
Keywords: mucosal immunology, ileum, T cell activation, nuclear receptors, lymphocyte homing, cytokine receptors, CD4 T cells, Peyer’s patches, homeostatic Th17 cells, IL-17A
INTRODUCTION
CD4+ T helper cells play crucial roles in vertebrate adaptive immune responses against potentially pathogenic microbes. CD4+ T cells additionally respond to antigens encoded by commensal microbes, and thereby contribute to the diversity of the T cell antigen receptor (TCR) repertoire resident within tissues. Differentiation of T cells into effector cells with diverse programs requires their activation through the TCR and CD28, the co-stimulatory receptor, as well as signaling through a multitude of cytokine receptors. Cytokines in the microenvironment, produced in large part by myeloid cells, direct the different programs of differentiation, which are readily distinguished by the induction of subset-specific transcription factors. Thus, interferon-γ (IFNγ)-producing Th1 cells express Tbet (Tbx21) (Szabo et al., 2002), Th2 cells that secrete interleukin 4 (IL-4), IL-5, and IL-13 express GATA3 (Zhang et al., 1997) (Zheng and Flavell, 1997) and Th17 cells characterized by the production of IL-17A, IL-17F, and IL-22 express RORγt+ (Ivanov et al., 2006). In addition, induced regulatory T cells (iTreg) upregulate Foxp3 (and often other lineage-defining transcription factors, e.g. RORγt) and can produce IL-10 (Sefik et al., 2015) (Ohnmacht et al., 2015) (Xu et al., 2018).
Th17 cells are critical for epithelial homeostasis and host defense against bacteria and fungi at diverse mucosal sites (Korn et al., 2009). Dysregulated Th17 cell responses, however, promote several chronic inflammatory and autoimmune diseases such as inflammatory bowel disease (IBD), psoriasis, and various forms of arthritis (Maddur et al., 2012). Therefore, manipulation of Th17 cell responses has the potential to provide effective therapies for multiple immune system-based diseases. Developing therapies that do not hinder host-protective functions requires a thorough understanding of the molecular and cellular mechanisms underlying Th17 cell regulation during both homeostasis and chronic inflammation. In the past decade, the cytokine requirements for Th17 cell differentiation have been investigated in vitro and in vivo (Korn et al., 2009). In vitro differentiation of mouse Th17 cells from naïve CD4+ T cells can be achieved by culturing TCR/CD28-stimulated T cells with either TGF-β and IL-6 (Bettelli et al., 2006) (Mangan et al., 2006) (Veldhoen et al., 2006), with IL-6, IL-1β, and IL-23 (Ghoreschi et al., 2010), or with IL-6 and serum amyloid A (SAA) proteins (Lee et al., 2020). These conditions induce the expression of IL-23R and RORγt in a STAT3-dependent manner (Zhou et al., 2007). Th17 cell differentiation from naïve human cord blood CD4+ T cells can similarly be achieved with a combination of TGF-β, IL-1β, and IL-6, IL-21, or IL-23 (Manel et al., 2008). These cytokines have also been reported to be involved in pathogenic Th17 cell functions in the context of inflammatory conditions such as experimental autoimmune encephalomyelitis (EAE), mouse models of colitis, and IBD in humans (Bettelli et al., 2006) (Korn et al., 2007) (Langrish et al., 2005) (Nurieva et al., 2007) (Kamimura et al., 2003) (Cua et al., 2003) (Sanchez-Munoz et al., 2008) (Perrier and Rutgeerts, 2011) (Lee et al., 2020). IL-23, in particular, is critical for conferring pathogenicity in several animal models, but the transcriptional and metabolic features that distinguish homeostatic from pathogenic Th17 cells remain poorly understood.
The vertebrate gastrointestinal (GI) tract is colonized by hundreds of species of bacteria, which outnumber total host cells. Commensal microbes contribute not only to the activation of innate immune responses mediated by way of multiple sensing pathways, but also to the establishment of the peripheral adaptive immune repertoire and the acquisition of diverse T lymphocyte effector functions (Hooper et al., 2012). The generation of gut Th17 cells is especially dependent on commensal bacteria (Honda and Littman, 2016). While germ-free mice have few, if any, Th17 cells in their gut lamina propria (LP), specific pathogen-free (SPF) mice, which harbor many commensal species, have abundant intestinal Th17 cells that help maintain gut homeostasis (Ivanov et al., 2008) (Honda and Littman, 2016) (Belkaid and Harrison, 2017). We previously showed that Il-6 deficient mice have fewer IL-17A-producing CD4+ T cells in the intestine compared to Il-6 sufficient mice (Ivanov et al., 2006). However, it was also reported that IL-6 is dispensable and, instead, microbiota-induced IL-1β is essential for the development of steady-state Th17 cells in the murine intestinal LP (Shaw et al., 2012). These studies did not account for immune responses that can vary according to which bacteria colonize the host. It is known that mice from different animal facilities and vendors have different intestinal microbiota (Chudnovskiy et al., 2016). The diversity of commensal communities in different vivaria likely accounts for conflicting results in inflammation-related studies, and may explain the different reported cytokine requirements for Th17 cell differentiation in vivo. We previously demonstrated that mono-colonization of germ-free mice with commensal segmented filamentous bacteria (SFB) is sufficient to induce Th17 cells that are specific for SFB antigens (Ivanov et al., 2009) (Yang et al., 2014). Recently, we described a two-step process for the functional differentiation of homeostatic Th17 cells in response to SFB colonization in the gut of mice with intact specific pathogen-free microbiota (Sano et al., 2015). In the first step, SFB antigen-specific CD4+ T cells are primed and polarized such that they are in a poised state, marked by the expression of RORγt in Th17 cells in the draining lymph nodes, recently shown to be primarily the ileum- and cecum-draining lymph nodes (Esterhazy et al., 2019). In the second step, T cells undergo functional maturation in the intestinal LP, with triggering of Th17 signature cytokine gene expression programs by epithelial cell-derived factors. SAA1 and SAA2, produced by the epithelial cells in the ileum in response to SFB colonization, were shown to act on the poised RORγt+ Th17 cells, resulting in production of the effector cytokines IL-17A and IL-17F (Sano et al., 2015). Although the intestine-draining mesenteric lymph nodes (MLN) were described as sites for Th17 cell priming, other studies indicated that priming of naïve SFB-specific T cells can occur in Peyer’s patches (PP) (Lecuyer et al., 2014) (Teng et al., 2016) or in the small intestine lamina propria (SILP) (Goto et al., 2014) (Geem et al., 2014). In the current study, we have investigated the roles of cytokines and inductive sites in the discrete steps of homeostatic Th17 cell differentiation.
To explore the molecular mechanisms of Th17 cell induction, especially the expression of RORγt in vivo, we adoptively transferred naïve T cells from SFB-specific TCR transgenic mice into SFB-colonized hosts, allowing us to track SFB-specific Th17 cell expansion and differentiation. Here, we focused on characterizing the mechanisms by which SFB induces Th17 cells in the intestine-draining MLN and the SILP. We found that SFB-induced expression of RORγt and proliferation of naïve T cells occurred in the MLN, but not in PPs or other inductive site in the intestinal mucosa. The RORγt+ Th17 cells migrated from the MLN to the SILP and PPs in an integrin β7-dependent manner. Although IL-6 was required for early RORγt expression by SFB-specific T cells in the MLN and SILP, IL-21 or IL-23, but not IL-1β, compensated in its absence. However, the cytokines had distinct roles in inducing production of effector cytokines by RORγt+ Th17 cells. These results indicate that Th17 cell differentiation occurs in discrete steps at different locations, and that several back-up systems exist to insure induction of the RORγt-directed program.
RESULTS
Sites and kinetics of SFB-specific Th17 cell induction and proliferation
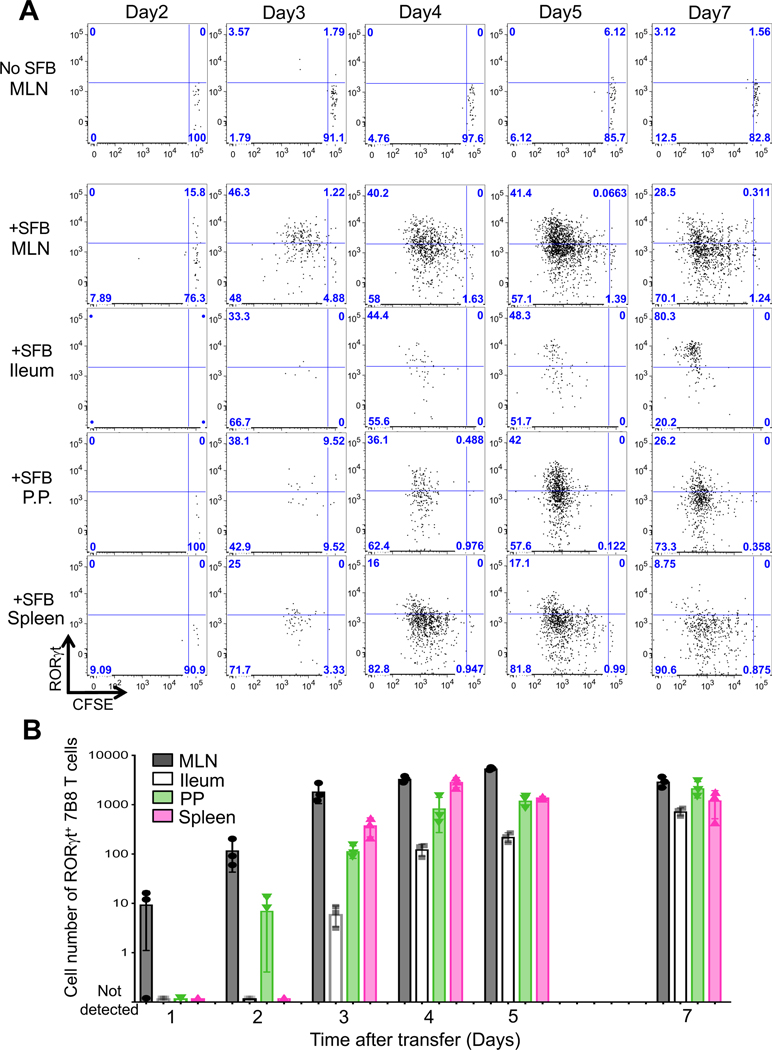
To explore the molecular and cellular mechanisms of Th17 cell induction in vivo, we first tracked SFB-induced Th17 cell expansion and differentiation based on cell division and RORγt expression, respectively. To monitor when, where, and how naïve T cells differentiate into Th17 cells upon SFB colonization, we adoptively transferred carboxyfluorescein succinimidyl ester (CFSE)-labeled naïve T cells from SFB-specific TCR transgenic (7B8) mice into SFB-gavaged host mice (Yang et al., 2014). Two days following the transfer of 50,000 naïve 7B8 T cells, some of the transferred cells began to express CD25 (data not shown) and RORγt in the intestine-draining mesenteric lymph nodes (MLN). Notably, at this time point, there was no proliferation of these cells, and CD25 (data not shown) and RORγt+ 7B8 T cells were undetectable in the Ileum, PP, or spleen (Fig. 1A). By day 3, 7B8 T cells within the MLN had undergone several rounds of proliferation and many expressed RORγt (Fig. 1A) but not Foxp3 (data not shown). Donor-derived 7B8 T cells only began to accumulate in the ileal LP at 4 or 5 days following T cell transfer (Fig. 1A and B). By 7 days post-transfer, most 7B8 T cells in the intestinal LP expressed RORγt (Fig. 1A). We also checked other tissues to determine where SFB-specific Th17 cells differentiate. We found some proliferating RORγt+ 7B8 T cells in the spleen and Peyer’s patches (PP) before we observed RORγt+ 7B8 T cells in the intestinal LP (Fig. 1A). Overall, expanded 7B8 T cells were detected in the MLN before any of the other tissues tested (Fig. 1A and 1B).
Figure 1. Kinetics of SFB-specific Th17 cell differentiation in tissues of SFB-colonized mice.
(A) Cell expansion, monitored by CFSE dilution, and RORγt expression in donor-derived 7B8 T cells from diverse tissues of SFB-gavaged mice. 50,000 naïve 7B8 TCR transgenic T cells (Cd45.1/Cd45.2) were transferred into SFB gavaged mice (Cd45.2/ Cd45.2). SFB-specific 7B8 T cells were defined as CD4+, TCRβ+, TCRVβ14+, and CD45.1+ cells. As a negative control, SFB-free SPF B6 mice were used. The time course experiment was performed once, with 3 mice per time point. Repeats with specific time points (Day2, Day3, Day4, Day5, and Day7) were performed multiple times using biological duplicates, with similar results. (B) The number of SFB-specific Th17 cells in the MLN, Ileum, PPs, and spleens of SFB-gavaged C57BL/6 mice. SFB-specific Th17 cells were defined as Foxp3− RORγt+ 7B8 T cells described in Figure 1A. These data were generated from one time course experiment and represent the mean from 3 or 4 mice +/− SD.
To test our model under conditions that better match the physiological frequency of antigen-specific T cells within a host, we transferred only 5000 naïve 7B8 T cells into SFB-gavaged mice. We again first detected Th17 cell expansion and differentiation in the MLNs and, at later times, in the intestinal LP (Fig. S1A and S1B), suggesting that they had migrated from sites of induction. SFB colonize and adhere to epithelial cells in the terminal ileum (Koopman et al., 1987), but not in the duodenum and colon (Sano et al., 2015). Nevertheless, we observed similar numbers of RORγt+ 7B8 T cells in the duodenum and colon when compared to the ileum (Fig.S1A).
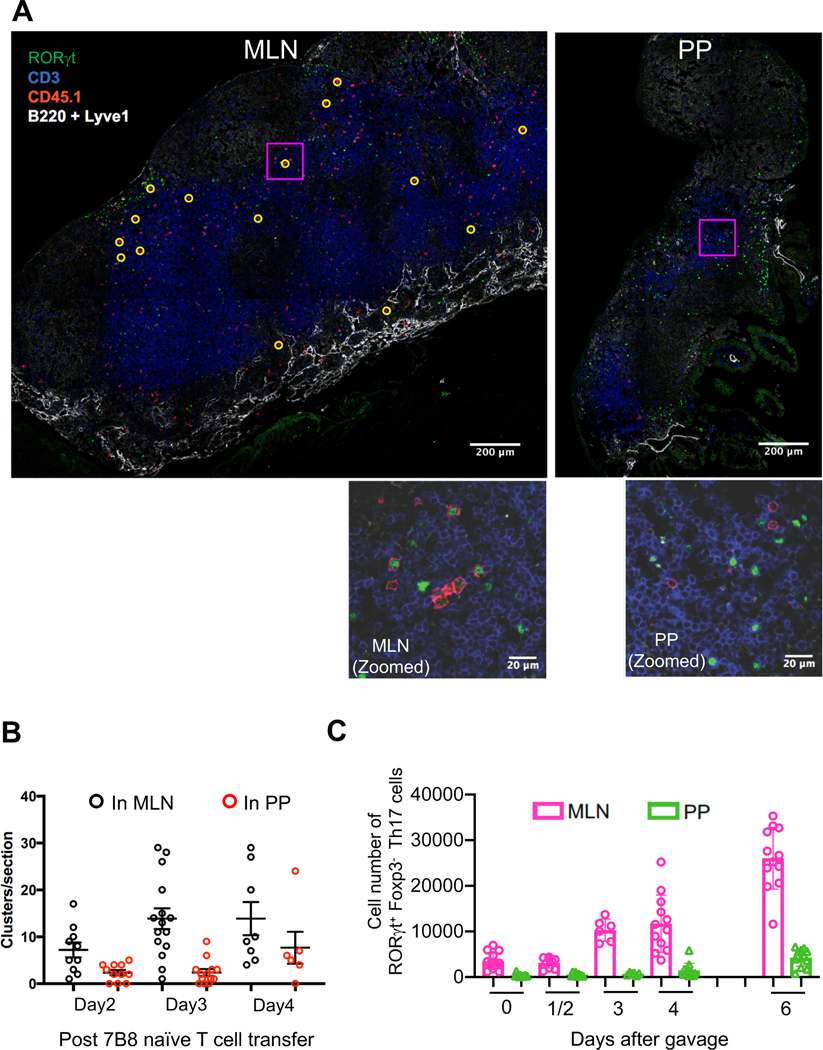
It has been proposed that PPs are the primary site of SFB-specific Th17 cell induction (Lecuyer et al., 2014). In addition, PP dendritic cells were shown to endow CD8+ T cells with small intestine-homing capability, through induction of α4β7 and CCR9 (Mora et al. 2003). We therefore compared PP and MLN tissue sections to identify clusters of SFB-specific Th17 cells at different times after naïve 7B8 T cell transfer. In the absence of SFB, there were no donor-derived RORγt+ Th17 cells in either MLN or PPs (data not shown). In SFB-gavaged mice, however, we were able to detect clusters of SFB-specific T cells in the MLNs and PPs. Many of these clustered cells expressed RORγt (Fig. 2A) and stained positive for the proliferation marker Ki67 (Fig. S2A), suggesting that these were SFB-specific Th17 cells undergoing active differentiation. Notably, these clusters were detected only in MLNs at 2 and 3 days following transfer, and only on day 4 were some clusters detected in PPs (Fig. 2B).
Figure 2. Quantification of Th17 cells in the MLN and PPs following SFB gavage.
(A) Th17 cell clusters in MLN and PPs 3 days following transfer of 50,000 naïve 7B8 T cells. Sections were stained with anti-RORγt, anti-CD3, anti-CD45.1, anti-B220, and anti-Lyve-1 and were analyzed by confocal microscopy. B220 and Lyve1 were both stained with Pacific Blue-conjugated antibodies, but can be distinguished by cell morphology and stain intensity. Yellow circles indicate clusters of CD45.1+ cells, defined as 2 or more CD45.1+ cells in contact. Representative of 5 mice from 2 experiments. The scale bars in the above photomicrographs correspond to 200 μm. Insets are enlargements of the boxed areas. The scale bars in the enlarged images correspond to 20 μm. (B) Quantification of clustering in MLN and PPs imaged as in (A). Numbers of clusters per tissue section are shown. Representative of two experiments, each with 2–3 mice per time point. Each symbol represents a single tissue slice, and the bar indicates average +/− SD. (C) Kinetics of Th17 cell accumulation in the MLN and PPs following SFB gavage. The cell numbers were calculated based on total cells in the MLN and PPs. Experiments were repeated twice with similar results and results were all combined (SFB gavaged mice, Day0: n=13, Day1/2: n=9 (Day1: n=3, Day2: n=6), Day3: n=6, Day4: n=12, Day6: n=12). Data are represented as mean +/− SD.
We also quantified the number of endogenous Th17 cells in the MLN and PP of mice following gavage with SFB. We found that while the number of RORγt+ Th17 cells began to increase in PPs at 6-days post SFB gavage, those in the MLN began to expand within 3 days following SFB inoculation (Fig. 2C). To evaluate whether APCs from MLN carry SFB antigens, we collected APCs from MLN following SFB gavage and co-cultured with SFB-specific naïve T cells in vitro. Naïve 7B8 T cells expressed the T cell activation markers CD25 and CD69, and down-regulated CD62L, when they were co-cultured with APCs isolated from MLN at least 4 days after gavage, but not with APCs from ungavaged mice (Fig. S2B and S2C).
We next investigated whether Th17 cells can accumulate in the intestinal LP in response to SFB gavage if the mice lack PPs. PP development was abrogated by injection of LTβR-Ig fusion protein into dams at E16.5 and E18.5 of fetal development (Rennert et al., 1996). Although PPs were absent in treated mice (Fig. S2D), there was no effect on the number of SFB-specific Th17 cells in the mouse gut at 7 and 10 days following SFB gavage (Fig. S2E and S2F). Importantly, MLNs were intact and had similar numbers of RORγt+ Th17 cells in mice depleted of PPs and control antibody-injected mice (data not shown). These results indicate that PPs are dispensable for the induction of Th17 cells and for their expansion in the SILP in response to SFB gavage.
Integrin β7 is essential for the migration of Th17 cells from the MLN to the intestine
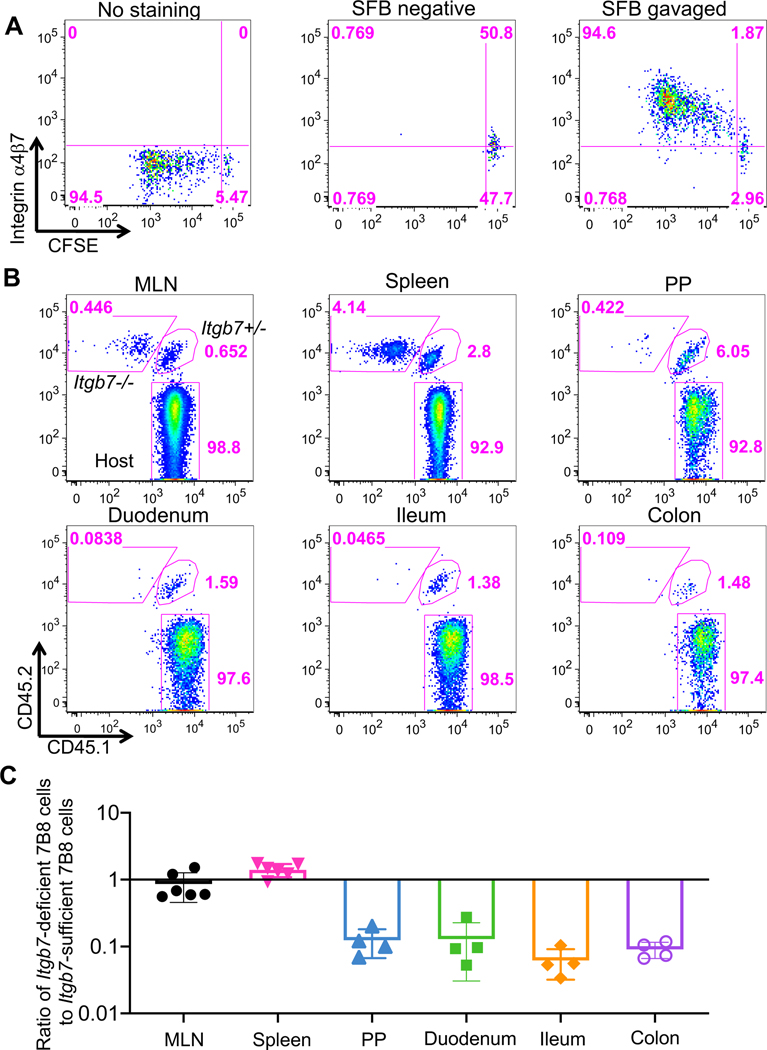
Integrins α4 and β7 are induced on T cells in gut-draining lymph nodes and have a key role in homing of the T cells to intestinal LP (Schweighoffer et al., 1993) (Erle et al., 1994) (Johansson-Lindbom and Agace, 2007). Indeed, naïve 7B8 T cells transferred into SFB-colonized mice upregulated the expression of integrin α4 and β7 heterodimers on their surface following proliferation in response to antigen (Fig. 3A). To confirm that Th17 cells migrate to the intestine after expansion in MLN (Fig. 1 and 2), we co-transferred naïve CFSE-labeled Itgb7+/− (Cd45.1/ Cd45.2) and Itgb7−/− (Cd45.2/Cd45.2) 7B8 T cells into SFB-colonized hosts (Cd45.1/Cd45.1), and tracked the isotype-marked cells in various tissues at several time-points. Both Itgb7−/− 7B8 and Itgb7+/− 7B8 T cells expanded in the MLN and spleen at 4-days post co-transfer (Fig. 3B and 3C). However, Itgb7−/− 7B8 T cells did not accumulate in the intestinal LP compared to Itgb7+/− 7B8 T cells at 4, 7, and 10 days after transfer, but had increased accumulation in spleen (Fig. 3B, 3C, S3A and S3B). Taken together, we conclude that initial expansion of Th17 cells occurs primarily in the MLN, and SFB-specific Th17 cells subsequently migrate into the intestinal LP and PPs in an integrin β7-dependent manner. These data thus imply that APCs present SFB antigens and prime and polarize Th17 cells in the MLN.
Figure 3. The role of α4β7 integrin in migration of SFB-specific Th17 cells from MLN to local tissues.
(A) Expression of α4β7 integrin heterodimer in primed SFB-specific T cells in the MLN. 50,000 CFSE-labeled naïve T cells from SFB-specific TCR transgenic (7B8) mice were adoptively transferred into SFB-colonized mice. No staining represents the sample with isotype control IgG. Similar results were observed in multiple mice (n=4) at day 4 post adoptive transfer. (B) Localization of integrin β7-sufficient and -deficient SFB-specific T cells following adoptive transfer. 25,000 naïve T cells each from Itgb7 heterozygous (Cd45.1/Cd45.2) and homozygous mutant (Cd45.2/Cd45.2) 7B8 mice were co-transferred into SFB-gavaged C57BL/6 (Cd45.1/Cd45.1) mice. Four days following transfer, isotype-marked 7B8 T cells from multiple tissues were analyzed. (C) Ratios of Itgb7-deficient to Itgb7-sufficient (heterozygous) 7B8 T cells from multiple tissues in the same host mice at 4 days post transfer. Each symbol represents a single mouse, and the color bar indicates average +/− SD. Experiments were repeated twice and results were combined. n= 4 (PPs. Duodenum, Ileum, and colon); n= 6 (MLN and spleen).
Cytokine requirements for SFB-induced Th17 cell differentiation in vivo
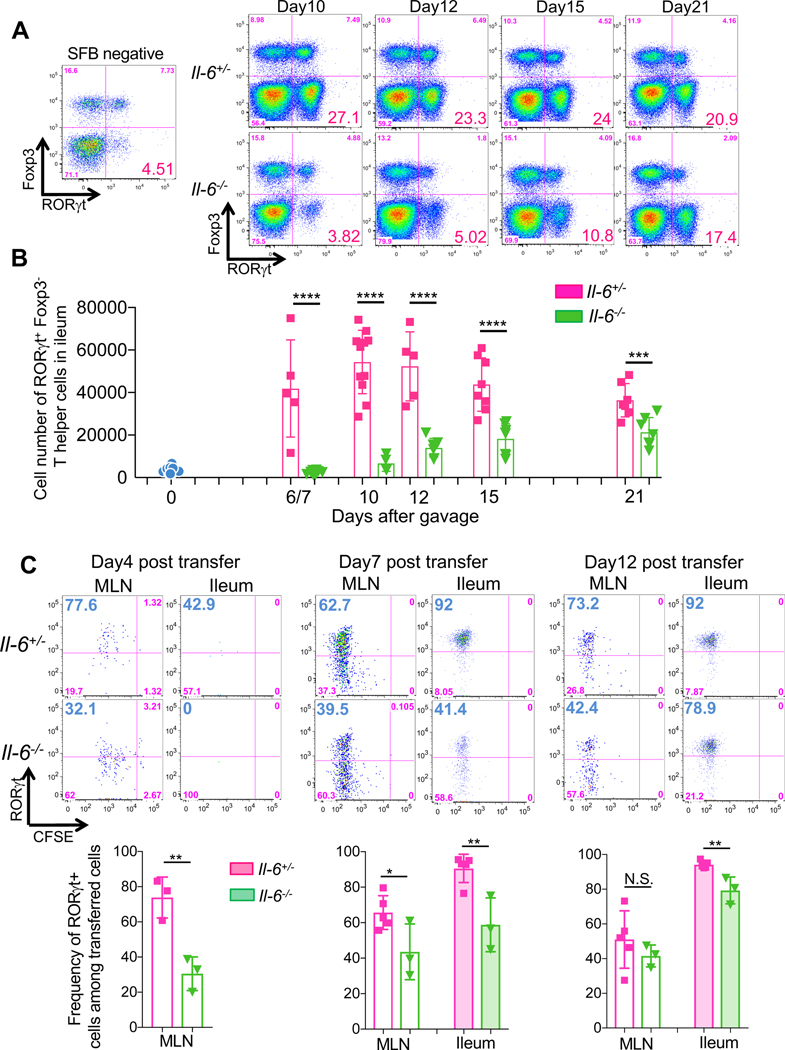
Antigen presentation is the initial step in the priming of antigen-specific T cells. Simultaneously, innate immune cell-derived cytokines direct differentiation of the T cells towards diverse effector programs marked by expression of the signature transcription factors. Thus, most of the 7B8 T cells located in the ileum in SFB-colonized mice expressed RORγt (Sano et al., 2015), but proliferating 7B8 T cells in the spleen, which may have migrated from the MLN, expressed little or no RORγt (Fig. S1C). We hence hypothesized that APCs in the MLN provide Th17-inducing cytokines, but continued RORγt expression in differentiated Th17 cells may require maintenance by cytokines in tissue microenvironments. Therefore, we next focused on cytokine requirements for Th17 cells in the MLN and intestinal LP. In vitro studies revealed that the combination of several cytokines including TGF-β, IL-1β, IL-6, IL-21, and IL-23 can induce RORγt expression and IL-17A production in CD4+ T cells (Acosta-Rodriguez et al., 2007; Bettelli et al., 2006; Manel et al., 2008; Mangan et al., 2006; Veldhoen et al., 2006; Wilson et al., 2007; Yang et al., 2007; Zhou et al., 2007). However, the identity of cytokines involved in RORγt expression in vivo remains unresolved. For example, whereas we described a requirement for IL-6 in the differentiation of intestinal CD4+ T cells that express RORγt, IL-23R, IL-17A and IL-17F (Ivanov et al., 2006), Nunez and colleagues demonstrated that IL-1β, but not IL-6, was required for steady state Th17 cell differentiation in gut (Shaw et al., 2012). To elucidate which cytokines are important for RORγt expression in SFB-induced Th17 cells, we orally introduced SFB-enriched fecal contents into SFB-free Il-21r, Il-23 p19, and Il1r1 deficient mice. Seven or 10-days post SFB gavage, the numbers and proportions of RORγt+ Th17 cells in the ileum of these cytokine signaling-deficient mice were similar to those of littermate control mice (Fig. S4). In contrast, there were markedly fewer RORγt+ Th17 cells in the ileum of SFB-colonized Il-6−/− mice compared to IL-6-sufficient littermate controls (Fig. 4A and 4B). Although this defect was pronounced during the first week after SFB gavage, there was a gradual increase in ileal RORγt+ Th17 cells in Il-6−/− mice during the subsequent two weeks (Fig. 4A and 4B).
Figure 4. Early requirement for IL-6 in SFB-induced Th17 cell differentiation.
(A and B) IL-6 requirement for Th17 cell differentiation in response to SFB gavage. IL-6-sufficient (heterozygous) and -deficient mice were gavaged with SFB, and RORγt+ Foxp3− Th17 cells in the ileal LP were assessed by FACS at different times after gavage. Representative FACS panels (A) and cell numbers (B). Results were combined from two experiments with littermates. Dots indicate individual mice and the color bars indicate mean values +/− SD. (C) Requirement for IL-6 in SFB-specific Th17 cell differentiation in the MLN and ileum. 50,000 (for Day 4) or 5,000 (for Day 7 and Day 12) naïve T cells from SFB Tg (7B8) mice (Cd45.1/Cd45.2) were adoptively transferred into SFB-gavaged Il-6 sufficient and deficient mice (Cd45.2/Cd45.2). Donor-derived T cells in the MLN and ileum were analyzed for RORγt expression on the indicated days. Representative FACS profiles in top panels, aggregate data in bottom panels. Each symbol represents a single mouse, and the color bar indicates average +/− SD. p-values were determined by two-tailed unpaired Student’s t-test. *p<0.05, ** p <0.001, *** p<0.001, and **** p<0.0001.
To confirm these results with SFB-induced antigen-specific T cells, we transferred CFSE-labeled 7B8 naïve T cells into SFB-gavaged Il-6+/− and Il-6−/− mice. Four days following transfer, proliferation of 7B8 T cells in MLN of Il-6−/− mice was similar to that in Il-6+/− mice based on CFSE dilution (Fig. 4C). However, there was substantial reduction of RORγt-expressing 7B8 T cells in the MLN of Il-6−/− compared to Il-6+/− mice (Fig. 4C). Similarly to the T cells in the MLN, there were fewer RORγt-expressing SFB-specific T cells in the ileum of Il-6−/− hosts at 7 days following transfer (Fig. 4C). However, expression of RORγt in transferred 7B8 T cells was similar in Il-6−/− and Il-6+/− mice at the later time points (Fig. 4C). To confirm IL-6 dependency using different mouse models, we also quantified the number of Th17 cells in Il-6+/− and Il-6−/− mice that had been stably colonized with SFB and various other commensals in a SPF room of the animal facility at the New York University Skirball Institute (NYUSK). Long-term SFB-colonized Il-6−/− mice in the NYUSK facility had a moderate, but statistically significant, reduction in Th17 cells compared to littermate Il-6+/− mice (Fig. S5A and S5B). We also tested other microbial communities that can induce Th17 cells in the mouse gut. Mice from Jackson Laboratory (JAX), which have a microbiota lacking SFB, consistently contain a small population of intestinal RORγt+ Th17 cells (Ivanov et al., 2008). We quantified the number of RORγt+ Th17 cells in IL-6-sufficient and -deficient littermates bearing JAX flora, and found a similar reduction in RORγt+ Th17 cells in the MLN and ileum of the homozygous mutant mice (Fig. S5C and S5D). Taken together, these results indicate that, following colonization with Th17-inducing bacteria, IL-6 is a strong early inducer of RORγt expression in responsive T cells in the MLN. However, it is not absolutely required for Th17 cell differentiation, suggesting that there are other cytokines capable of inducing RORγt in intestinal T cells.
Stat3 is required for Th17 cell differentiation in response to SFB colonization
Because at least some RORγt+ Th17 cell differentiation proceeds in the absence of IL-6 (Fig. 4 and S5), we speculated that other cytokines may compensate. Multiple cytokines, including IL-1β, IL-6, IL-21, and IL-23, have been reported to induce RORγt expression during in vitro Th17 cell differentiation, and several of these function through stimulation of Stat3 phosphorylation. Moreover, it was reported that Stat3 signaling is required in IL-17-producing Th17 cells in multiple tissues including MLN and intestinal LP (Harris et al., 2007). To investigate whether Stat3-stimulating cytokines compensate for loss of IL-6 in microbiota-dependent induction of RORγt expression in CD4+ T cells in vivo, we next examined whether SFB-dependent Th17 cell differentiation can occur in the absence of Stat3. We quantified RORγt-expressing Th17 cells in mice that were stably colonized with SFB and lacked expression of Stat3 in T cells (Cd4cre/+ Stat3flox/flox). As previously reported (Harris et al., 2007) (Ohnmacht et al., 2015), Stat3 deficiency in CD4+ T cells led to a substantial reduction in both frequency and numbers of RORγt+ Th17 cells in the MLN, to almost complete absence of such cells in the ileum, and to loss of RORγt+ induced Treg cells (Fig. S6A and S6B). The reduction in Th17 cells was much more marked, particularly in the ileum, in the absence of Stat3 as compared to the absence of IL-6. This result suggested that Stat3-stimulating cytokines other than IL-6, particularly IL-21 and IL-23, may contribute to RORγt expression in Th17 cell differentiation in vivo.
IL-21 and IL-23, but not IL-1β, compensate for the lack of IL-6 in SFB-dependent Th17 cell differentiation
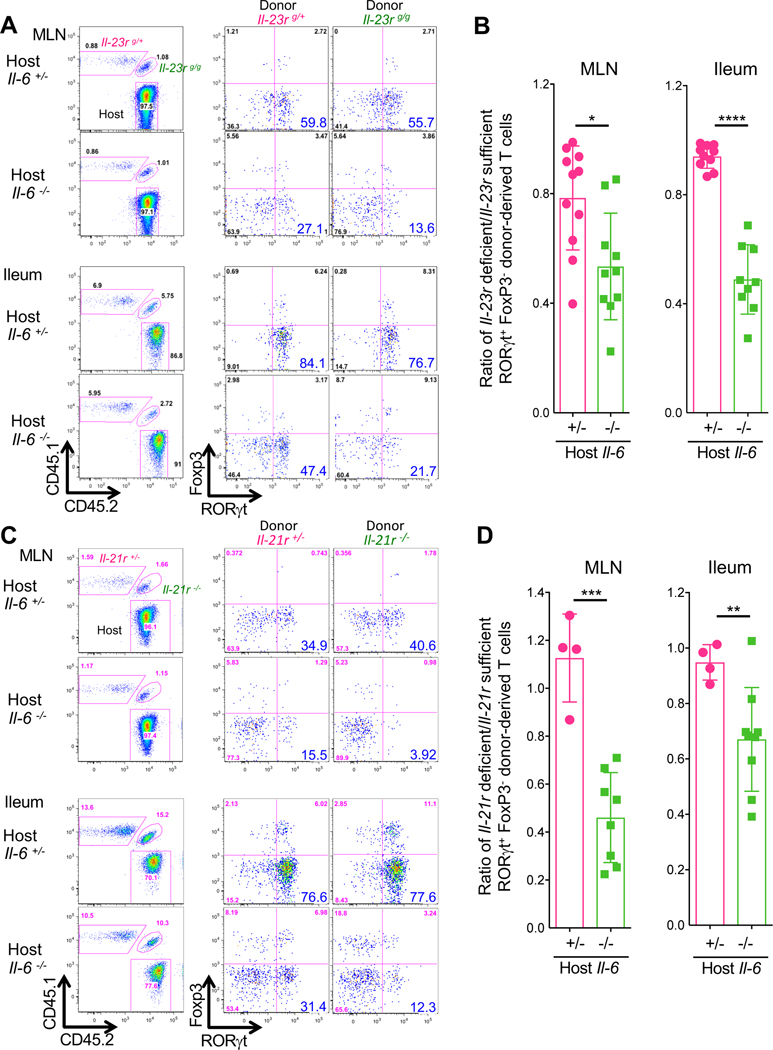
To determine which cytokines contribute to Th17 cell differentiation in the absence of IL-6 in vivo, we prepared isotype-marked 7B8 TCR transgenic mice deficient in Il-1r1, Il-21r, or Il-23r. We first determined whether IL-23R is expressed on 7B8 T cells in the Il-6−/− host colonized with SFB. Indeed, there was reduced GFP expression in the SFB-specific T cells from Il-23rgfp/+ reporter mice in Il-6−/− compared to Il-6+/− recipient mice, consistent with partial dependency of IL-23R expression on IL-6 (Zhou et al., 2007) (Fig. S6C). We then co-transferred Il-23rgfp/+ (Cd45.1/Cd45.1) and Il-23rgfp/gfp (Cd45.1/Cd45.2) naïve 7B8 T cells into stably SFB-colonized Il-6+/− and Il-6−/− hosts (Cd45.2/Cd45.2). In Il-6+/− hosts, there was little difference in RORγt+ induction between Il-23rgfp/+ and Il-23rgfp/gfp 7B8 T cells in the MLN and the ileum. In Il-6−/− hosts, however, there was substantial reduction in RORγt expression among Il-23r-deficient 7B8 T cells compared to Il-23r -sufficient cells in MLN and the ileum (Fig. 5A and 5B). Transferred Il-21r−/− 7B8 T cells displayed similarly impaired expression of RORγt in the MLN and ileum in Il-6−/− hosts but differentiated normally into Th17 cells in Il-6-sufficient mice (Fig. 5C and 5D). Notably, Il-1r1 deficiency did not affect RORγt expression even in the absence of IL-6 signaling in vivo (Fig. S6D and S6E).
Figure 5. Cytokine redundancy in the differentiation of SFB-specific Th17 cells in the MLN and ileum.
(A-D) Requirement for IL-23R (A and B) and IL-21R (C and D) signaling in SFB-specific Th17 cell differentiation in Il-6-sufficient and -deficient mice. 5,000 naïve T cells from cytokine receptor sufficient and deficient isotype-marked 7B8 TCR transgenic mice were co-transferred into SFB-gavaged Il-6 sufficient and deficient mice (Cd45.2/Cd45.2). Representative FACS plots of RORγt and Foxp3 expression in ileal 7B8 T cells with homozygous or heterozygous Il-23r and Il-21r mutations at 7 days post transfer into Il-6-sufficient and -deficient mice (A and C). Panels at left indicate gating for donor-derived versus host T cells. Ratios of RORγt+ Foxp3− Th17 cells (cytokine receptor sufficient vs deficient T cells) in Il-6 sufficient and deficient mice (B and D). Experiments were performed twice with littermate controls. All results were combined. Dots indicate individual mice and color bars indicate average +/− SD. For IL-23R signaling (Il6+/−: n=9, Il6−/−: n=10). For IL-21R signaling (Il6+/−: n=4, Il6−/−: n=9). p-values were determined by two-tailed unpaired Student’s t-test. *p<0.05, ** p <0.001, *** p<0.001, and **** p<0.0001.
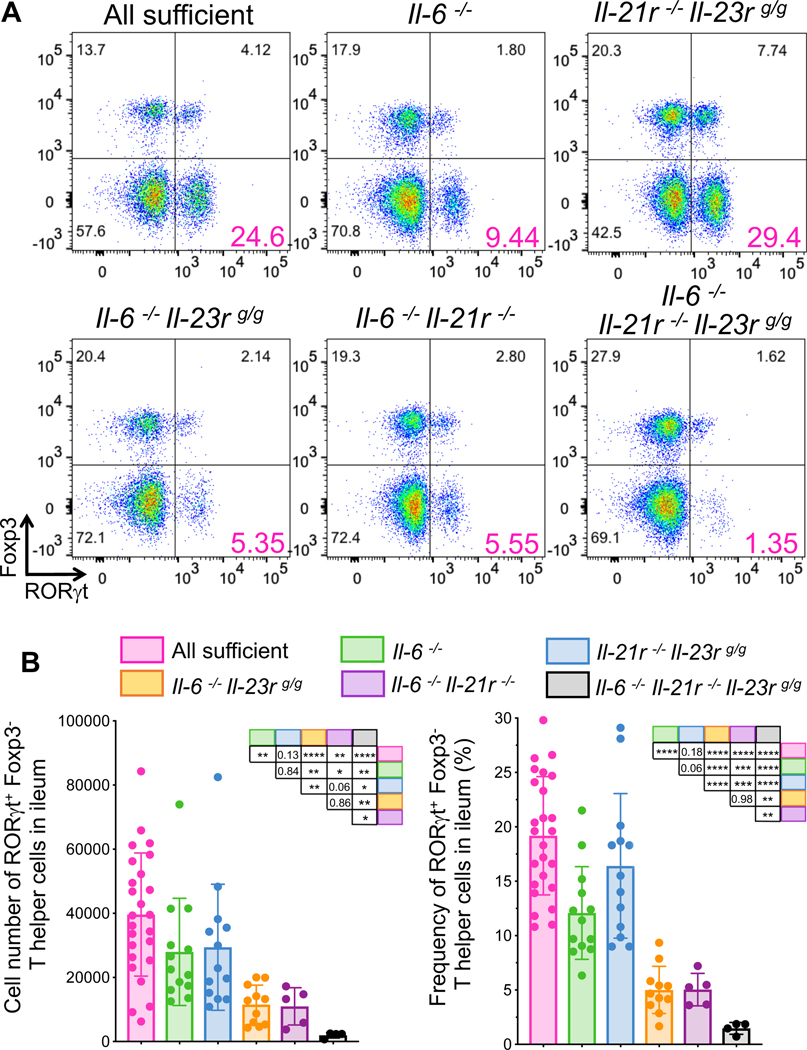
We next compared Th17 cell differentiation in mice deficient for different combinations of the Stat3-signaling cytokines. In IL-6, IL-21R, and IL-23R triple-deficient mice, RORγt-expressing Th17 were barely detectable in the ileum of SFB-colonized mice at steady state (Fig 6A and B). These animals were housed in the vivarium at the University of Illinois at Chicago (UIC), and control, single-, or double-deficient mice recapitulated the partial loss of Th17 cells observed in the NYUSK mice. The number of RORγt-expressing Th17 cells in the ileum of IL-21R/IL-23R double-deficient mice was similar to that in the sufficient mice (Fig 6A and B), and deficiency of IL-6 was required to observe defective Th17 cell differentiation. The results thus indicate that signaling through receptors for IL-21 and IL-23, but not IL-1β, can partially compensate for the loss of IL-6 in differentiation of SFB-specific Th17 cells in the mouse intestine.
Figure 6. Contribution of multiple cytokines to Stat3-dependent signaling redundancy in SFB-induced Th17 cell differentiation.
(A and B) Representative FACS plots of ileal Th17 cells (RORγt+Foxp3−) in stably SFB-colonized mice with mutations in cytokines or their receptors (A). CD4+ T cells were gated for MHCII−, TCRβ+, CD4+, and CD8−. Cell numbers (B, left) and frequencies (B, right) of Th17 cells in the ileum. Experiments were repeated with indicated genotypes at least three times and results are combined. Littermates were used in most cases. Mice were confirmed as SFB+ by qPCR or microscopic observation. p-values for differences between each group of mice are shown in the tables. Dots represent individual mice and color bars indicate average +/− SD. p-values were determined by two-tailed unpaired Student’s t-test. *p<0.05, ** p <0.001, *** p<0.001, and **** p<0.0001.
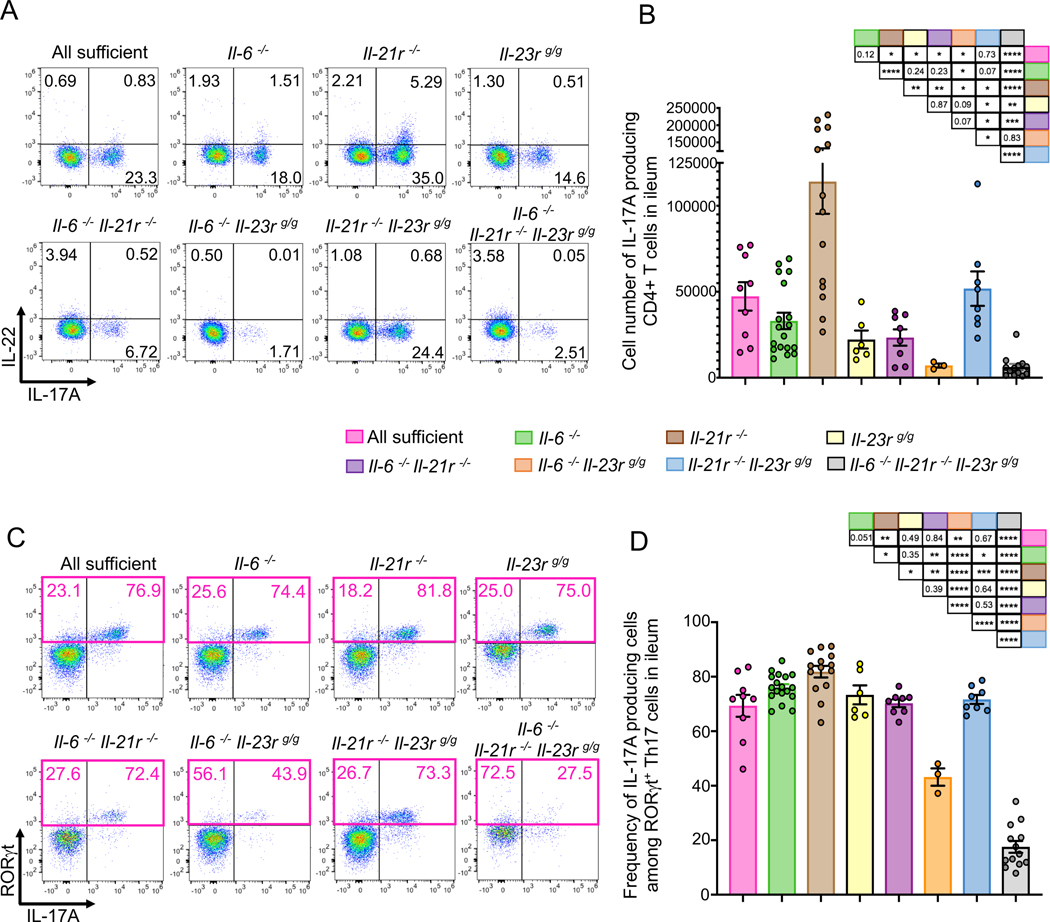
We also investigated how deficiencies of the Stat3-activating cytokines and receptors affect the production of effector cytokines from Th17 cells in SFB-colonized ileum. Il-21r deficiency resulted in an increased number of IL-17A-producing Th17 cells, consistent with a previous study (Cho et al., 2019), but the number of IL-17A-producing CD4+ T cells was significantly reduced in Il-23r deficient mice (Fig. 7A and B). In addition, IL-23R-deficient mice tended to lose IL-22-producing cells, but loss of IL-21R resulted in an increase in such cells (Fig. 7A, S7A and S7B). Despite having a substantial reduction in RORγt+ Th17 cells, 70% of the residual RORγt+ T cells in IL-6/IL21R double deficient mice produced IL-17A (Fig. 7C and D). In contrast, IL-6/IL23R double deficient and IL6/IL21R/IL23R triple deficient mice, which had similar decreases in ileal Th17 cells to IL-6/IL21R double deficient mice, displayed loss of IL-17A production in the remaining RORγt+ Th17 cells (Fig.7C and D). Deficiency in IL-6 or IL-23R, and, to a lesser extent, in IL-21R, resulted in reduced TNFα production by RORγt+ Th17 cells, but not other T cells (Fig. S7B and C). Taken together, our results indicate that IL-6, IL-21R, and IL-23R possess redundant roles in RORγt expression in T cells, but have distinct roles in regulating production of cytokines by Th17 cells.
Figure 7. Contribution of IL-6, IL21R, and IL-23R to cytokine production by ileal Th17 cells.
(A) Representative FACS plots of ileal Th17 cell cytokines (IL17A and IL-22) in SFB-colonized mice with mutations in cytokines or their receptors. Ileal CD4+ T cells were gated for MHCII−, TCRβ+, CD4+, and CD8−. (B) Cell numbers of IL-17A-producing CD4+ T cells in the ileum of wild-type and mutant mice stably colonized with SFB. (C) Representative FACS plots for RORγt and IL-17A in Foxp3− ileal CD4+ T cells. (D) Frequency of IL-17A producing cells among RORγt+ Th17 cells shown in (C). Experiments were repeated with indicated genotypes at least three times and results are combined. Littermates were used in most cases. p-values for differences between each group of mice are shown in the tables. Dots represent individual mice and color bars indicate average +/− SEM. p-values were determined by two-tailed unpaired Student’s t-test. *p<0.05, ** p <0.001, *** p<0.001, and **** p<0.0001.
DISCUSSION
We showed in this report that Th17 cell differentiation in response to colonization with the commensal microbe SFB is primarily initiated in the gut-draining MLN and that the requisite T cell intrinsic Stat3 signaling can be achieved through the action of cytokines that can compensate in part for each other’s absence. We found that IL-6 is a potent early inducer of RORγt in SFB-specific Th17 cells in the draining lymph nodes, but, in its absence, IL-21 and IL-23 can compensate to activate the Th17 program. Our study also shows that, after initial differentiation in the MLN, SFB-specific Th17 cells migrate to the intestinal LP in an integrin β7-dependent manner. Finally, RORγt+ Th17 cells that have migrated from the MLN accumulate in the intestinal LP and are further stimulated by the tissue microenvironment to produce effector cytokines (Sano et al., 2015). RORγt+ Th17 cells localized at mucosal sites likely serve as a reservoir of microbiota-induced cells that can be reactivated in response to tissue damage and thus maintain homeostasis (Honda and Littman, 2016). However, Th17 cells also contribute to autoimmune disease and inflammatory pathology in humans, and IL-1β, IL-6, and IL-23 are required in Th17-dependent mouse models of autoimmunity (Kamimura et al., 2003) (Chen et al., 2006). Furthermore, antibodies targeting these cytokines or their receptors have been clinically effective for treating rheumatoid arthritis and inflammatory bowel disease in humans (Jones et al., 2011) (Gaffen et al., 2014) (Tanaka and Kishimoto, 2012). Our results suggest that such treatments may not fully block homeostatic Th17 cell differentiation, as IL-6 and IL-23 signaling are partially redundant for RORγt induction and Th17 cell differentiation.
Microbiota-dependent Th17 cell priming and differentiation occurs in MLNs
It has been reported that SFB induces bacterial antigen-specific IgA and Th17 cell responses in secondary and tertiary lymphoid tissues, including Peyer’s patches and isolated lymphoid follicles (ILFs) (Lecuyer et al., 2014) (Teng et al., 2016). On the other hand, it has also been suggested that Th17 cells are generated even in mice lacking secondary lymphoid organs (Goto et al., 2014) (Geem et al., 2014). Here, we have demonstrated that the MLNs are the initial site for RORγt+ Th17 cell differentiation. SFB-specific RORγt+ Th17 cells were detected in similar numbers not only in SFB-colonized ileum but also in the lamina propria of duodenum and colon, which are not colonized with SFB (Sano et al., 2015). Moreover, RORγt+ Th17 cells in the intestinal LP were detected much later than those in the MLN and PPs. These results indicated that the intestinal LP and ILFs are not the predominant nor initial sites of SFB-specific Th17 cell differentiation. We therefore investigated the other candidate sites for Th17 cell priming and differentiation, the MLN and PPs. RORγt and CD25 were upregulated early in MLN, but not PPs, spleen or intestinal LP, after transfer of SFB-specific 7B8 cells. In addition, clusters of SFB-specific RORγt+ Th17 cells were observed in the MLN, but not in the PPs, at early time points. Moreover, even in mice depleted of PPs, RORγt+ Th17 cells were detected in the MLN and ileum LP with similar kinetics and abundance to control mice. The integrin α4β7, which mediates lymphocyte migration to intestinal lamina propria, was critical for accumulation of SFB-specific Th17 cells in the small intestine, but not for their induction in the MLN. Finally, APCs from MLN stimulated SFB-specific naïve T cells in vitro early after SFB colonization. Based on these results, we conclude that the MLN, rather than PPs or intestinal LP, is the principal site for early SFB-specific Th17 cell differentiation. However, our results do not rule out the possibility that Th17 cells can be also generated in the PPs and gut associated lymphoid tissue (GALT). Th17 cells were induced in the intestine of LTα-deficient mice, which lack secondary and tertiary lymphoid tissues, but the kinetics of induction were not compared to those in control mice (Goto et al., 2014). In the absence of secondary lymphoid tissues, APCs carrying SFB antigens may prime T cells locally in addition to maintaining homeostatic Th17 cells in the ileal LP. In germ-free mice, there are more naïve T cells in the intestinal LP than in SPF mice. Under these non-physiological conditions, commensal-specific T cells may thus be primed ectopically.
Cytokine requirements and redundancy during commensal-induced Th17 cell differentiation
Studies on conditions required for in vitro Th17 cell differentiation have reported the importance of several cytokines, including TGF-β, IL-1β, IL-6, IL-21, and IL-23 (Korn et al., 2009). Here, we showed that IL-6 was required in vivo for early induction of RORγt expression in SFB-specific Th17 cells. However, in its absence, other Stat3-stimulating cytokines, IL-21 and IL-23, provided the necessary stimuli for the Th17 cell differentiation program in mouse intestine. IL-21 is known to be produced by activated CD4+ T cells following IL-6 stimulation (Korn et al., 2007) (Zhou et al., 2007) (Nurieva et al., 2007). IL-6 and IL-21 induce IL-23R expression on activated T cells (Zhou et al., 2007). Induced RORγt itself can also enhance the expression of IL-23R on Th17 cells (Wang et al., 2015). These features may provide positive feedback and backup systems to maintain Th17 cells in the gastrointestinal tract.
Unlike IL-21 and IL-23, IL-1β failed to compensate for the loss of IL-6 in rescuing SFB-dependent Th17 cell differentiation. However, IL-1β has been shown to be required for pathogenic functions of Th17 cells, e.g. in EAE. In addition, IL-1β, but not IL-6, was reported to be required for microbiota-induced gut Th17 cell differentiation in mice that were colonized with SFB (Shaw et al., 2012). A parsimonious explanation for these disparate findings is that SFB induces a different set of cytokines in the context of the microbiota in different vivaria. The frequency of SILP-localized RORγt+ Th17 cells reported by Shaw et al. in IL-1R1 sufficient mice was considerably lower than what we observed, consistent with this possibility.
We found divergent roles of IL-21 and IL-23 in the induction of Th17-associated cytokines. CD4+ T cells producing IL-17A and IL-22 were increased in the IL-21R deficient ileal T cells, but severely reduced in the absence of IL-23R. IL-6/IL-21R double deficient mice lost a significant proportion of IL-17A producing cells compared to IL-6 deficient mice, yet 70% of the remaining RORγt+ Th17 cells produced Il-17A. In contrast, IL-6/IL23R double deficient and IL6/IL21R/IL23R triple deficient mice had both reduced numbers of RORγt+ Th17 cells in their ileum, and reduced IL-17A in the remaining cells. These data suggest that IL-21 restrains Th17 cells under some circumstances, but also compensates for loss of IL-6 together with IL-23. Moreover, IL-6 or IL-23R, but not IL-21R deficiency, resulted in reduced TNFα from RORγt+ Th17 cells. Taken together, we conclude that the STAT3-dependent cytokines have both redundant and distinct roles in regulating Th17 cytokine production in the SFB-colonized ileum. Because the mutations of Il21r and Il23r are in the germ line, we cannot rule out the possibility that their divergent effects on cytokine-producing Th17 cells may be indirect, by way of non-T cells that respond to IL-21 and IL-23.
Although our results indicate that IL-21 and IL-23 can compensate for the absence of IL-6 in supporting Th17 cell differentiation in the ileum, it is less clear that these cytokines provide a similar function in the MLNs, the sites of priming and early differentiation of the microbiota-specific T cells. Additional studies are needed to determine if these cytokines have regionally selective functions due to differences in their production by cells in distinct regions of the intestinal lamina propria and in different MLNs.
Microbiota-induced Th17 cells in autoimmune disease
The SFB colonization model that we and others study is considered to result in differentiation of non-pathogenic or homeostatic Th17 cells. However, under some circumstances, SFB can contribute to systemic autoimmune disease. Thus, SFB mono-colonized mice had more severe disease in the K/BxN autoimmune arthritis model (Wu et al., 2010), and IL-17A-producing SFB-specific Th17 cells were detected in the spleen and lung of K/BxN mice (Bradley et al., 2017). SFB-specific T cells activated in pregnant female mice following poly(I:C) injection can also contribute to IL-17A-mediated neurodevelopmental abnormalities in mouse offspring (Kim et al., 2017). Whether the same or different cytokine requirements apply to SFB-induced Th17 cells in the ileum at steady state and at distal sites during inflammation remains to be determined. It is still unclear whether humans harbor SFB, although a recent report provides evidence that some infants may have small amounts of a related species (Jonsson et al., 2020). It will be of considerable interest to determine if such bacteria in humans influence Th17 cell differentiation, activation, and trafficking.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resourses and reagents should be directed to and will be fulfilled by the lead contact, Teruyuki Sano (tsano1@uic.edu).
Materials Availability
This study did not generate unique reagents.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Strains and Vivarium Housing
C57BL/6J mice were obtained from the Jackson Laboratory as SFB-negative mice. All transgenic animals were bred and maintained in the animal facility of the Skirball Institute (NYU School of Medicine) and Bio Research Laboratory of University of Illinois at Chicago (UIC) under specific-pathogen-free (SPF) conditions. To maintain SFB-free mice, autoclaved cages, autoclaved acidified water, and fresh chow were used and all cages were changed weekly by investigators. Il-23rgfp mice (Awasthi et al., 2009) were provided by M. Oukka and maintained in Rag2−/− (JAX) background. SFB-specific Th17-TCRTg (7B8) mice were previously described (Yang et al., 2014) and maintained on the Ly5.1 (Cd45.1) background (JAX; B6. SJL-Ptprca Pepcb/BoyJ). Itgb7 mice were purchased from JAX and maintained on the Ly5.1 (Cd45.1) or Ly5.2 (Cd45.2) background. Il-23 p19−/− mice were obtained from DNAX (Lieberman et al., 2004) and maintained in SFB-free condition by treatment with ampicillin using autoclaved cages for 2 weeks. Il-1r1 (JAX), Il-6 (JAX), and Il-21r (JAX) mice were maintained with Jackson flora (SFB negative). Six- to 16-week old animals (both males and females) were used for in vivo experiments. All animal procedures were in accordance with protocols approved by the Institutional Animal Care and Use Committee of the NYU School of Medicine and UIC College of Medicine. All phenotypes were compared using littermate controls and the results were combined in most of the experiments.
METHODS DETAILS
Acute SFB Colonization
Fecal pellets were collected from SFB-enriched Il-23r Rag2 DKO mice (SFB), or SFB-free JAX B6 mice (Jax). Fresh fecal pellets were homogenized in ice-cold PBS, passed through a 100 μm filter, pelleted at 3400 rpm at 4°C for 10 minutes, and re-suspended in ice-cold PBS. Each SFB-free animal was administered 1/4 pellet by oral gavage. Before mice were used for the gavage experiments, we confirmed that they were SFB negative by qPCR, as described in (Sano et al., 2015). The state of SFB colonization in gavaged mice was confirmed by microscopic observation of the disrupted epithelial fraction during gut preparation or by qPCR. Littermates were cohoused from their birth or after SFB gavage in most of the experiments.
SFB-specific Transgenic T Cell Transfer
SFB-enriched fecal extracts were administered by oral gavage to SFB-free mice 3 or 4 days before T cell transfer. Spleen and lymph nodes (except MLNs) were collected from 7B8 TCR Tg mice. Cells from these tissues were dissociated using 100 μm cell strainers and then red blood cells were lysed by ACK Lysing Buffer. 7B8 naïve T cells were purified as DAPI−, CD4+, CD62Lhi, CD44low, CD25−, TCRVβ14+ T cells from spleen and lymph nodes (except MLN) of 7B8 TCR Tg mice using the Aria II cell sorter (BD). To monitor proliferation, sorted 7B8 naïve T cells were stained with CellTrace CFSE Cell Proliferation kit (Thermo Fisher Scientific). After cell numbers were calculated, 5,000 or 50,000 naïve 7B8 T cells were administered to recipient mice by intravenous retro-orbital injection. Animals were sacrificed and analyzed at multiple time points following naïve T cell transfer. Mononuclear cells from multiple tissues such as MLN, PP, spleen, duodenum, ileum, and colon were harvested as described (Yang et al., 2014).
Depletion of PPs from Mouse Small Intestine
To abrogate development of gut-associated PPs, we intravenously injected SFB-free pregnant C57BL/6 mice with 150 μg of LTβR-Ig on gestational days 16.5 and 18.5, as described (Rennert et al., 1996). As an experimental negative control, control isotype-matched IgG was intravenously injected. During the gut prep, the number of PPs and MLNs were isolated and counted.
Isolation of Lamina Propria Lymphocytes
Mesenteric fat tissue and Peyer’s patches were carefully removed from intestinal tissues. The proximal and distal 1/3 of small intestine were designated as duodenum and ileum, respectively. Tissues were incubated in 5 mM EDTA in PBS containing freshly prepared 1 mM of DTT for 20 min at 37°C with rotation (200–250 rpm). During the step, we confirmed that the treated tissues were not stacked at the bottom of the tubes. After this 1st wash step, SFB attaching to epithelial cells can be detected by microscopic observation of the supernatant (epithelial rich fraction). Remaining tissues were incubated in a second EDTA wash (5 mM EDTA in PBS) for 10 min at 37°C with rotation (200–250 rpm). After tissues were washed with RPMI 10% FCS medium to remove EDTA, tissues were then further digested (RPMI media containing 10% fetal calf serum (FCS), 1.0 mg/ml each of Collagenase D (Roche) and 100 μg/ml DNase I (Sigma), and 0.1 U/ml Dispase (Worthington)) at 37°C for 30 min (duodenum and ileum) or 45 min (colon) with rotation (125–250 rpm). During the step, we confirmed that the treated tissues were not stacked at the bottom of the tubes. After the tissues were shaken vigorously in 50 ml tubes by hand for 30 seconds, 14 ml of ice cold RPMI 10% FCS was added to the tube. The digested tissues were then passed through a 70μm cell strainer. Mononuclear cells were enriched on 40:80 Percoll gradients by centrifugation (2200 rpm) for 22 min at room temperature. Lamina propria (LP) lymphocytes were collected from the Percoll gradient interphase. The recovered LP lymphocytes were washed with ice cold RPMI 10% FCS and the total cell numbers were counted by Vi-Cell XR Cell Viability Analyzer (Beckman Coulter) or Countess™ II automated Cell Counter (ThermoFisher).
Cell Staining for Flow Cytometry
For analysis of T helper cell lineages, live cells were stained with anti-CD3, anti-TCRβ, anti-CD4, and anti-MHCII with control IgG and Fc block. To distinguish transferred 7B8 T cells from the host cells, live cells were stained with anti-TCRVβ14, anti-CD45.1, and anti-CD45.2. Live-dead labeling with DAPI (for live cell analysis) was performed and only live cells were analyzed. For intracellular protein staining, extracellular proteins were first labeled with antibodies (anti-CD3, anti-TCRβ, anti-CD4, and anti-MHC-II), control IgG, and Fc-blocker for 30 min at 4 oC. After the cells were stained with live-dead labeling Aqua (Thermo Fisher Scientific), they were fixed using fixation/permeabilization buffer (Thermo Fisher Scientific) at room temperature for 30 min or at 4 oC for over night, re-suspended in permeabilization buffer (Thermo Fisher Scientific) and further stained with anti-Foxp3 and anti-RORγt using the Foxp3 staining buffer set (Thermo Fisher Scientific). To detect both cytokines and transcription factors together, one million LP mononuclear cells were stimulated with 50 ng/ml phorbol myristate acetate (PMA) (Sigma) and 500 ng/ml Ionomycin, in the presence of GolgiPlug (BD Biosciences) at 37 oC for 4 hours in a 5% CO2 incubator. The cells were then incubated with control IgG and Fc-blocker for 15 min at 4 oC, and stained with anti-TCRβ, anti-CD4, anti-CD8a, anti-MHC-II for 30 min at 4 oC. Dead cells were further stained with Aqua. Following with the fixation and permeabilization, as described above, cells were stained with intracellular antibodies (Anti-IL-17A, anti-IL-22, anti-TNFα, anti-RORγt, and anti-Foxp3) for 30 min at room temperature. Non-specific or Fc-receptor binding were blocked with control IgG and Fc-blocker during staining. Flow cytometric analysis was performed on a LSRII (BD Biosciences), a FACS Aria (BD Biosciences), or Attune (Thermo Fisher Scientific). All data were re-analyzed using FlowJo (Tree Star).
Confocal microscopy
Mesenteric lymph nodes and Peyer’s patches were harvested and fixed in 4% PFA for 1.5 hours at 22–25°C with gentle shaking. Organs were then dehydrated by sucrose gradient (15% sucrose in PBS for 1 hour at 4°C, then 30% sucrose at 4°C overnight), embedded in OCT (Sakura), and snap-frozen in dry ice-cooled 2-methylbutane. 8- to 10-μm sections were cut and air-dried at least 30 minutes. All staining was performed at 22–25°C. Sections were permeabilized for 30 min with 0.5% Triton X-100 in PBS, then incubated for 10–30 min in block buffer consisting of 0.1% Triton X-100 in PBS with 1% BSA (mass/vol) and 10% normal donkey serum. An Avidin/Biotin blocking kit (Vector Laboratories) was used with biotinylated antibodies. Stains were done with antibodies diluted in the block buffer. The following fluorochrome-conjugated antibodies were used: anti-CD45.1 (A20), anti-CD3 (17A2) anti-B220/CD45RO (RA3–6B2), and streptavidin (all from BioLegend), Biotin-conjugated anti-Lyve1 (ALY7, 2.5 μg/ml) was from Thermo Fisher Scientific.
For RORγt detection, primary anti-RORγt antibodies (1:30, clone AFKJS-9, Thermo Fisher Scientific 14–6988) were detected with secondary FITC Goat Anti-Rat Ig (1:500, BD Biosciences 554016), then tertiary Alexa Fluor® 488 donkey anti-goat (1:500, Thermo Fisher Scientific A11055). 10 μg/ml anti-CD16/32 (BioLegend, clone 93) was used in the buffer for tertiary staining. Slides were mounted with G-Fluoromount (Southern Biotech) and visualized using a Zeiss 710 inverted confocal microscope with a 25× or 40× oil-immersion objective and ZEN 2010 software. For all direct comparisons, samples were stained on the same day and imaged with the same settings.
Ex-vivo priming assay
Cd45.1/Cd45.1 WT B6 mice (8 week old females) were gavaged with SFB for two sequential days, and MLN (L1: second closest from cecum: highly associated to ileum (Esterhazy et al., 2019)) and PPs were isolated at several time points following the first gavage. Cd45.2/Cd45.2 WT B6 mice were also prepared as SFB-negative control and their MLN and PPs were collected in the same way. Collected tissues from SFB negative and positive mice were combined and digested together with RPMI 10% FCS containing 100 μg/ml DNase I (Sigma), 1.0 mg/ml of Collagenase D (Roche) and 0. 1 U/ml of Dispase (Worthington)) at 37oC for 30 min by gently shaking. Digested tissues were passed through 100 μm filter then stained for cell sorting. APCs were sorted following the gating. APCs from SFB-gavaged: DAPI-, Dump− (CD45R−, TCRβ−, NK1.1− Ly6G−), CD45.1+, and CD11b+ or CD11c+ singlet cells, APCs from SFB negative mice: Dump− (CD45R−, TCRβ−, NK1.1− Ly6G−), CD45.2+, and CD11b+ or CD11c+ singlet cells. APCs were sorted from indicated whole tissues and incubated with naïve 7B8 T cells with IMDM and 10% FCS for 24 h. T cell activation was assessed by flow cytometry after staining for TCRvβ14, CD4, CD25, CD69, and CD62L.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
Two-tailed unpaired Student’s t-tests were performed to compare the results using Excel and Prism. We treated less than 0.05 of P-value as significant differences. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001. N.S.: No Significant. All of the statistical details of experiments can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Flow Cytometry: anti-mouse CD4 (RM4-5), eFluor450 | Thermo Fisher Scientific | Cat#48-0042-82; RRID: AB_1272194 |
| Flow Cytometry: anti-mouse CD4 (RM4-5), Alexa Fluor 700 | Thermo Fisher Scientific | Cat#56-0042-82; RRID: AB_494000 |
| Flow Cytometry: anti-mouse CD4 (GK1.5), APC-Cy7 | BD Biosciences | Cat#552051; RRID: AB_394331 |
| Flow Cytometry: anti-mouse CD4 (RM4-5), APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-0042-82; RRID: AB_1272183 |
| Flow Cytometry: anti-mouse CD4 (GK1.5), APC-eFluor 780 | BD Biosciences | Cat#55205; RRID: AB_394331 |
| Flow Cytometry: anti-mouse CD45.1 (A20), APC | BioLegend | Cat#110714; RRID: AB_313503 |
| Flow Cytometry: anti-mouse CD45.1 (A20), APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-0453-82; RRID: AB_1582228 |
| Flow Cytometry: anti-mouse CD45.1 (A20), eFluor450 | Thermo Fisher Scientific | Cat#48-0453-82; RRID: AB_1272189 |
| Flow Cytometry: anti-mouse CD45.2 (104), APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-0454-82; RRID: AB_1272175 |
| Flow Cytometry: anti-mouse CD45.2 (104), PerCP-cy5.5 | Thermo Fisher Scientific | Cat#45-0454-82; RRID: AB_953590 |
| Flow Cytometry: anti-mouse I-A/I-E (M5/114.15.2), Alexa Fluor 700 | Thermo Fisher Scientific | Cat#56-5321-82; RRID: AB_494009 |
| Flow Cytometry: anti-mouse I-A/I-E (M5/114.15.2), BV510 | BD Biosciences | Cat#742893; RRID: AB_2741133 |
| Flow Cytometry: anti-mouse Integrin alpha 4 beta 7 (DATK32), PE | Thermo Fisher Scientific | Cat#14-5887-82; RRID: AB_529605 |
| Flow Cytometry: anti-mouse TCR β (H57-597), Alexa Fluor 700 | BD Biosciences | Cat#560705; RRID: AB_1727573 |
| Flow Cytometry: anti-mouse TCR β (H57-597), APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-5961-82; RRID: AB_1272173 |
| Flow Cytometry: anti-mouse Vβ14 TCR (14-2), biotin | BD Biosciences | Cat#553257; RRID: AB_394737 |
| Flow Cytometry: anti-mouse Vβ14 TCR (14-2), FITC | BD Biosciences | Cat#553258; RRID: AB_394738 |
| Flow Cytometry: anti-mouse FoxP3 (FJK-16s), APC | Thermo Fisher Scientific | Cat#17-5773-82; RRID: AB_469457 |
| Flow Cytometry: anti-mouse FoxP3 (FJK-16s), FITC | Thermo Fisher Scientific | Cat#11-5773-82; RRID: AB_465243 |
| Flow Cytometry: anti-mouse FoxP3 (FJK-16s), PE | Thermo Fisher Scientific | Cat#12-4777-42; RRID: AB_1944444 |
| Flow Cytometry: anti-mouse RORγt (B2D), APC | Thermo Fisher Scientific | Cat#17-6981-82; RRID: AB_2573254 |
| Flow Cytometry: anti-mouse RORγt (B2D), PE | Thermo Fisher Scientific | Cat#12-6981-82; RRID: AB_10807092 |
| Flow Cytometry: anti-mouse RORγt (Q31-378), BV421 | BD Biosciences | Cat#562894; RRID: AB_2687545 |
| Flow Cytometry: anti-mouse CD16/CD32 (2.4G2) | Tonbo Biosciences | Cat#70-0161; RRID: AB_2621487 |
| Flow Cytometry: anti-armenian Hamster IgG (H+L) | Jackson ImmunoResearch Labs | Cat#127-005-099; RRID: AB_2338971 |
| Flow Cytometry: Streptavidin, APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-4317-82; RRID: AB_10366688 |
| Flow Cytometry (TF and cytokines): anti-mouse Foxp3 (FJK-16s), FITC | Thermo Fisher Scientific | Cat#11-5773-82; RRID: AB_465243 |
| Flow Cytometry (TF and cytokines): anti-mouse RORγt (Q31-378), BV421 | BD Biosciences | Cat#562894; RRID: AB_2687545 |
| Flow Cytometry (TF and cytokines): anti-mouse IL-17A (TC11-18H10.1), AF700 | BioLegend | Cat#506914; RRID: AB_536016 |
| Flow Cytometry (TF and cytokines): anti-mouse IL-22 (IL-22JOP), APC | Thermo Fisher Scientific | Cat#17-7222-82; RRID: AB_10597583 |
| Flow Cytometry (TF and cytokines): anti-mouse TNF alpha (MP6-XT22), PE | Thermo Fisher Scientific | Cat#12-7321-82; RRID: AB_466199 |
| Flow Cytometry (TF and cytokines): anti-mouse CD4 (RM4-5), BV605 | BioLegend | Cat#100548; RRID: AB_2563054 |
| Flow Cytometry (TF and cytokines): anti-mouse CD8a (53-6.7), Super Bright 702 | Thermo Fisher Scientific | Cat#12-7321-82; RRID: AB_466199 |
| Flow Cytometry (TF and cytokines): anti-mouse TCRβ (H57-597), APC-eFluor 780 | Thermo Fisher Scientific | Cat#47-5961-82; RRID: AB_1272173 |
| Naïve T cell sorting: anti-mouse CD4 (RM4-5), eFluor450 | Thermo Fisher Scientific | Cat#48-0042-82; RRID: AB_1272194 |
| Naïve T cell sorting: anti-mouse CD25 (PC61.5), APC | Thermo Fisher Scientific | Cat#17-0251-82; RRID: AB_469366 |
| Naïve T cell sorting: anti-mouse CD44 (IM7), PerCP-cy5.5 | Thermo Fisher Scientific | Cat#45-0441-82; RRID: AB_925746 |
| Naïve T cell sorting: anti-mouse CD62L (MEL-14), PE | Thermo Fisher Scientific | Cat#12-0621-81; RRID: AB_465720 |
| Naïve T cell sorting: anti-mouse Vβ14 TCR (14-2), biotin | BD Biosciences | Cat#553257; RRID: AB_394737 |
| Naïve T cell sorting: anti-mouse Vβ14 TCR (14-2), FITC | BD Biosciences | Cat#553258; RRID: AB_394738 |
| Naïve T cell sorting: Streptavidin PE-Cy7 Conjugate | Thermo Fisher Scientific | Cat#25-4317-82; RRID: AB_10116480 |
| In-vivo injection: LTβR-Ig | Gift from Biogen | https://rupress.org/jem/article-lookup/doi/10.1084/jem.184.5.1999 |
| In-vivo injection: Control Ig | Gift from Biogen | https://rupress.org/jem/article-lookup/doi/10.1084/jem.184.5.1999 |
| Immunohistchemistry: Purified anti-mouse CD16/32 antibody | BioLegend | Cat#101302; RRID: AB_312801 |
| Immunohistchemistry: anti-goat IgG (H+L) Cross-Adsorbed secondary antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11055; RRID: AB_2534102 |
| Immunohistchemistry: anti-rat Ig (Polyclonal), FITC | BD Biosciences | Cat# 554016; RRID: AB_395210 |
| Immunohistchemistry: RORγt monoclonal antibody (AFKJS-9) | Thermo Fisher Scientific | Cat#14-6988-82; RRID: AB_1834475 |
| Immunohistchemistry: Lyve1 monoclonal antibody (ALY7), biotin | Thermo Fisher Scientific | Cat#13-0443-82; RRID: AB_1724157 |
| Immunohistchemistry: anti-B220/CD45RO (RA3-6B2), BV421 | BioLegend | Cat#103239; RRID: AB_10933424 |
| Immunohistchemistry: anti-CD3 (17A2), APC | BioLegend | Cat#100236; RRID: AB_2561456 |
| Immunohistchemistry: anti-CD45.1 (A20), PE | BioLegend | Cat#110707; RRID: AB_313496 |
| Immunohistchemistry: Streptavidin, BV421 | BioLegend | Cat#405226 |
| Bacterial and Virus Strains | ||
| None | ||
| Biological Samples | ||
| None | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DPBS | HyClone | Cat#SH30028.FS |
| PBS 10X Solution | Fisher BioReagents | Cat#BP39920 |
| RPMI 1640 | Gibco | Cat#11875119 |
| Fetal Bovine Serum | Gibco | Cat#16140071 |
| HEPES Solution | HyClone | Cat#SH30237.01 |
| MEM Amino Acids solution | HyClone | Cat#SH30598.01 |
| Penicillin Streptomycin 100X Solution | HyClone | Cat#SV30010 |
| Sodium Pyruvate Solution | HyClone | Cat#SH30239.01 |
| EDTA | Thermo Fisher Scientific | Cat#AM9262 |
| Collagenase D | Roche | Cat#11088882001 |
| Dispase | Worthington Biochemical Corporation | Cat#LS02104 |
| DNase 1 | Sigma Aldrich | Cat#DN25 |
| DTT | Sigma Aldrich | Cat# D9779 |
| Percoll | GE Healthcare | Cat#17089101 |
| DAPI | Sigma Aldrich | Cat#D9542 |
| G-Fluoromount | Southern Biotech | Cat#0100-01 |
| Tissue-Tek O.C.T. Compound | Sakura | Cat#4583 |
| PMA | Sigma | Cat#P1585 |
| Ionomycin calcium salt from Streptomyces conglobatus | Sigma | Cat#I0634 |
| BD GolgiPlug™ Protein Transport Inhibitor (containing Brefeldin A) | BD Biosciences | Cat#555029 |
| ACK Lysing Buffer | Gibco | Cat#A1049201 |
| Critical Commercial Assays | ||
| FoxP3/Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat#00-5523-00 |
| CellTrace CFSE Cell Proliferation Kit | Thermo Fisher Scientific | Cat#C34554 |
| Avidin/Biotin Blocking Kit | Vector Laboratories | Cat#SP-2001 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34961 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34966 |
| Deposited Data | ||
| None | ||
| Experimental Models: Cell Lines | ||
| None | ||
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | The Jackson Laboratory | JAX:000664 |
| B6. SJL Ptprca Pepcb /BoyJ | The Jackson Laboratory | JAX:002014 |
| B6.129S7-Il1r1tm1Imx/J | The Jackson Laboratory | JAX:003245 |
| B6.129S2-Il6tm1Kopf/J | The Jackson Laboratory | JAX:002650 |
| B6N.129-Il21rtm1Kopf/J | The Jackson Laboratory | JAX:019115 |
| C57BL/6-Tg(Tcra,Tcrb)2Litt/J | The Jackson Laboratory | JAX:027230 |
| B6(Cg)-Rag2tm1.1Cgn/J | The Jackson Laboratory | JAX:008449 |
| B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ | The Jackson Laboratory | JAX:022071 |
| B6.129S1-Stat3tm1Xyfu/J | The Jackson Laboratory | JAX:016923 |
| IL23R-gfp knock-in-knock-out reporter mice | Gift of the Mohamed Oukka Laboratory | http://www.jimmunol.org/content/182/10/5904.long |
| Il-23 p19 mice | Gift of DNAX | https://www.jimmunol.org/content/173/3/1887 |
| Oligonucleotides | ||
| None | ||
| Recombinant DNA | ||
| None | ||
| Software and Algorithms | ||
| FlowJo | Tree Star, Inc. | N/A |
| GraphPadPrism | GraphPad Software, Inc. | https://www.graphpad.com/scientificsoftware/prism/ |
| ZEN 2010 software | Zeiss | N/A |
| Other | ||
| Attune NxT Flow Cytometer | Thermo Fisher Scientific | N/A |
| BD FACSAria II | BD Biosciences | N/A |
| BD LSR II | BD Biosciences | N/A |
| Zeiss 710 inverted confocal microscope | Zeiss | N/A |
| Vi-Cell XR Cell Viability Analyzer | Beckman Coulter | N/A |
| Countess II automated Cell Counter | Thermo Fisher Scientific | N/A |
ACKNOWLEDGEMENTS
We thank Ken Cadwell, Sang V. Kim, Jason A. Hall, and Lin Wu for valuable discussion, and Biogen for providing control-Ig and LTβR-Ig. This work was supported by fellowships from the TOYOBO Bioscience foundation (T.S.), Human Frontier Science Program (T.S.), UIC start-up funds (T.S.), Schweppe Award in Translational Research (T.S.), the Helen and Martin Kimmel Center for Biology and Medicine (D.R.L.), The Judith and Stewart Colton Center for Autoimmunity (D.R.L), NYSTEM award (D.R.L), National Institutes of Health grant R01DK103358 (D.R.L.), and by the Howard Hughes Medical Institute (D.R.L.)
Footnotes
Declaration of Interests
D.R.L consults for and has equity interest in Chemocentryx, Vedanta Biosciences, Vor Biopharma, and Immunai, and is a Director of Pfizer, Inc.
REFERENCES
- Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, and Sallusto F. (2007). Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol 8, 942–949. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, Bettelli E, Kuchroo VK, and Oukka M. (2009). Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol 182, 5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Harrison OJ (2017). Homeostatic Immunity and the Microbiota. Immunity 46, 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. [DOI] [PubMed] [Google Scholar]
- Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, Huang H, Knox KS, Littman DR, and Wu HJ (2017). Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 22, 697–704 e694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Langrish CL, McKenzie B, Joyce-Shaikh B, Stumhofer JS, McClanahan T, Blumenschein W, Churakovsa T, Low J, Presta L, et al. (2006). Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest 116, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Jaime H, de Oliveira RP, Kang B, Spolski R, Vaziri T, Myers TG, Thovarai V, Shen Z, Fox JG, et al. (2019). Defective IgA response to atypical intestinal commensals in IL-21 receptor deficiency reshapes immune cell homeostasis and mucosal immunity. Mucosal Immunol 12, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, et al. (2016). Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167, 444–456 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. (2003). Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748. [DOI] [PubMed] [Google Scholar]
- Erle DJ, Briskin MJ, Butcher EC, Garcia-Pardo A, Lazarovits AI, and Tidswell M. (1994). Expression and function of the MAdCAM-1 receptor, integrin alpha 4 beta 7, on human leukocytes. J Immunol 153, 517–528. [PubMed] [Google Scholar]
- Esterhazy D, Canesso MCC, Mesin L, Muller PA, de Castro TBR, Lockhart A, ElJalby M, Faria AMC, and Mucida D. (2019). Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL, Jain R, Garg AV, and Cua DJ (2014). The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol 14, 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geem D, Medina-Contreras O, McBride M, Newberry RD, Koni PA, and Denning TL (2014). Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol 193, 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. (2010). Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, and Ivanov II (2014). Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, et al. (2007). Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol 179, 4313–4317. [DOI] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, and Littman DR (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, and Littman DR (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Johansson-Lindbom B, and Agace WW (2007). Generation of gut-homing T cells and their localization to the small intestinal mucosa. Immunol Rev 215, 226–242. [DOI] [PubMed] [Google Scholar]
- Jones SA, Scheller J, and Rose-John S. (2011). Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest 121, 3375–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson H, Hugerth LW, Sundh J, Lundin E, and Andersson AF (2020). Genome sequence of segmented filamentous bacteria present in the human intestine. Commun Biol 3, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura D, Ishihara K, and Hirano T. (2003). IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev Physiol Biochem Pharmacol 149, 1–38. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, and Huh JR (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JP, Stadhouders AM, Kennis HM, and De Boer H. (1987). The attachment of filamentous segmented micro-organisms to the distal ileum wall of the mouse: a scanning and transmission electron microscopy study. Lab Anim 21, 48–52. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, and Kuchroo VK (2007). IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 448, 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, and Kuchroo VK (2009). IL-17 and Th17 Cells. Annu Rev Immunol 27, 485–517. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, and Cua DJ (2005). IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 201, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, et al. (2014). Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620. [DOI] [PubMed] [Google Scholar]
- Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al. (2020). Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 180, 79–91 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman LA, Cardillo F, Owyang AM, Rennick DM, Cua DJ, Kastelein RA, and Hunter CA (2004). IL-23 provides a limited mechanism of resistance to acute toxoplasmosis in the absence of IL-12. J Immunol 173, 1887–1893. [DOI] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. (2013). Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39, 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddur MS, Miossec P, Kaveri SV, and Bayry J. (2012). Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am J Pathol 181, 8–18. [DOI] [PubMed] [Google Scholar]
- Manel N, Unutmaz D, and Littman DR (2008). The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol 9, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, and Weaver CT (2006). Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, and Dong C. (2007). Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483. [DOI] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. (2015). MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 349, 989–993. [DOI] [PubMed] [Google Scholar]
- Perrier C, and Rutgeerts P. (2011). Cytokine blockade in inflammatory bowel diseases. Immunotherapy 3, 1341–1352. [DOI] [PubMed] [Google Scholar]
- Rennert PD, Browning JL, Mebius R, Mackay F, and Hochman PS (1996). Surface lymphotoxin alpha/beta complex is required for the development of peripheral lymphoid organs. J Exp Med 184, 1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Munoz F, Dominguez-Lopez A, and Yamamoto-Furusho JK (2008). Role of cytokines in inflammatory bowel disease. World J Gastroenterol 14, 4280–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighoffer T, Tanaka Y, Tidswell M, Erle DJ, Horgan KJ, Luce GE, Lazarovits AI, Buck D, and Shaw S. (1993). Selective expression of integrin alpha 4 beta 7 on a subset of human CD4+ memory T cells with Hallmarks of gut-trophism. J Immunol 151, 717–729. [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim YG, and Nunez G. (2012). Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med 209, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, and Glimcher LH (2002). Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295, 338–342. [DOI] [PubMed] [Google Scholar]
- Tanaka T, and Kishimoto T. (2012). Targeting interleukin-6: all the way to treat autoimmune and inflammatory diseases. Int J Biol Sci 8, 1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, and Wu HJ (2016). Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity 44, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, and Stockinger B. (2006). TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189. [DOI] [PubMed] [Google Scholar]
- Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Meyer Zu Horste G, Pawlak M, et al. (2015). CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell 163, 1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, et al. (2007). Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8, 950–957. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, and Mathis D. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, and Dong C. (2007). STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 282, 9358–9363. [DOI] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, and Ray A. (1997). Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem 272, 21597–21603. [DOI] [PubMed] [Google Scholar]
- Zheng W, and Flavell RA (1997). The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, and Littman DR (2007). IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 8, 967–974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.