Abstract
PCR analysis of tRNA intergenic spacer (tDNA-PCR) and of the 16S-23S internal transcribed spacer (ITS-PCR) and random amplified polymorphic DNA (RAPD) analysis were evaluated for their usefulness in characterization of Enterobacter cloacae strains isolated from both clinical origins and vaccine microbial contamination. tDNA-PCR presented specific and reproducible patterns for Enterobacter sakazakii ATCC 29004, Enterobacter aerogenes ATCC 13048, and Enterobacter cloacae ATCC 13047 and 23355 that presented the same profile for all 16 E. cloacae isolates, offering an alternative tool for species-level identification. ITS-PCR and RAPD analysis yielded completely different banding patterns for the 20 strains studied, except for E. cloacae strains isolated from different batches of vaccine that exhibited a unique pattern, suggesting contamination by the same strain. The combined use of tDNA-PCR and ITS-PCR in a one-step protocol allows accurate identification and typing of E. cloacae strains a few hours after the colony has been isolated.
Medical microbiology is extremely reliant on the culture of bacteria from clinical specimens and their subsequent identification for the diagnosis of disease (17). Determining the relatedness of isolates of microorganisms has become increasingly important as the number and spectrum of nosocomial pathogens continue to expand (16). Generally, the etiologic agent causing an outbreak of infection is derived from a single cell whose progeny are genetically identical or closely related to the source organism (3). Enterobacter cloacae, found in the normal flora of the human gastrointestinal tract, has emerged as an important nosocomial pathogen (9), and cases of infection in surgical wards (29) and burn units (26) and of neonatal sepsis have frequently been reported.
The vaccines and sera destined for human use must be free from microbial contamination, which may originate from various sources during the manufacturing process. In order to verify the contamination-free status, the articles are subjected in batches to a sterility test (37), which must be conducted in a controlled environment to avoid accidental contamination (7, 30). An accurate identification to the genus and species level of the bacterial isolates provides invaluable information concerning the source of contamination (1). Traditional methods used for identification of bacterial contaminants are often based on phenotypic characteristics, including colonial morphology and biochemical reactions. However, most of these techniques are affected by physiological factors or are not sufficiently sensitive to distinguish between strains (29).
Recently, DNA analysis has become the preferred method of identification, since it provides a more stable determination of isolate identity (20). PCR-based fingerprinting has been used for assessing the genetic diversity of many microorganisms. Depending on the primers and amplification conditions employed, the results allow discrimination between organisms at the level of genera, species, or strains.
The genes for the three rRNA molecules (16S, 23S, and 5S) found in the ribosome are generally linked together and cotranscribed in a single operon in prokaryotes. The length of these genes and their sizes as well as sequences are conserved between different prokaryotic species (24). However, the number of operons for a given species largely depends on its growth rate (15) and can range from 1 to 11, generally dispersed throughout the prokaryotic genome (15). The 16S and 23S genes are separated by internal spacer regions (ITS), which exhibit a large degree of sequence and length variation at the levels of genus and species. The size of the spacer may vary considerably for different species, and even among different operons within a single cell in the case of multiple operons (11). The variation in length is mainly due to the presence of several functional units within them, such as tRNA genes; sequences for enzyme recognition, such as RNase III, involved in the process of splicing to yield the mature ribosome (6); and boxA, which has an antiterminator role during transcription (19). The rest of this region consists of nonessential sequences subject to frequent insertion-deletion events, such as rsl in some Escherichia coli operons (11). The tRNA genes occur in multiple copies dispersed throughout the genome in most species. The shared sequence motifs of tRNA genes imply that the use of primers that contain consensus tRNA sequences in the PCR are likely to result in a number of characteristic PCR products (34). Analysis of tRNA intergenic spacer (tDNA-PCR), based on PCR amplification of spacers between tRNA genes, was proposed by McClelland et al. (27), to distinguish at the species level streptococcal strains of groups A, B, and G and Streptococcus mutans. This technique has also been applied successfully for the identification of Staphylococcus (25, 34, 35), Acinetobacter (14), Listeria (31), and Streptococcus viridans (13) strains.
Another important PCR-based fingerprinting technique used for typing of a wide range of bacteria is the random amplified polymorphic DNA (RAPD) analysis developed in 1990 by Williams et al. and Welsh and McClelland (33, 36). It has been widely adopted in gene mapping (22), phylogenetic analyses (21), population studies (10), and molecular typing of various microorganisms (12, 28, 38). Several investigators found poor reproducibility with this method; however, it is reliable if the PCR conditions are optimized (4) and can potentially be used to screen for genetic similarities and differences in whole genomes (23).
MATERIALS AND METHODS
Bacterial strains.
All the strains used in this study are listed in Table 1. The reference strains were obtained from the American Type Culture Collection (ATCC). Clinical samples were isolated from patients of the Federal University of Rio de Janeiro (HU-UFRJ) and Adolpho Lutz Institute (IAL), São Paulo, Brazil. The five contaminant strains of E. cloacae were isolated from three batches of the same vaccine at the National Quality Control Institute, Rio de Janeiro, Brazil. Contaminant 1 was isolated from a batch produced in 1998. Contaminants 2, 3, and 4 were isolated from another batch produced in 1998, and contaminant 5 was from a batch produced in 1999.
TABLE 1.
tDNA-PCR, ITS-PCR, and RAPD patterns of the 20 strains analyzed
| Species | Strain or strain origin | Pattern
|
||
|---|---|---|---|---|
| tDNA-PCR | ITS-PCR | RAPD (primer 1) | ||
| E. cloacae | Type strain ATCC 13047 | 1 | A | I |
| E. cloacae | Reference strain ATCC 23355 | 1 | B | II |
| E. sakazakii | Reference strain ATCC 29004 | 2 | C | III |
| E. aerogenes | Type strain ATCC 13048 | 3 | D | IV |
| E. cloacae | Clinical strain IAL-125 | 1 | E | V |
| E. cloacae | Clinical strain IAL-131 | 1 | F | VI |
| E. cloacae | Clinical strain IAL-134 | 1 | G | VII |
| E. cloacae | Clinical strain IAL-127 | 1 | H | VIII |
| E. cloacae | Clinical strain IAL-124 | 1 | I | IX |
| E. cloacae | Clinical strain IAL-132 | 1 | J | X |
| E. cloacae | Clinical strain HU-UFRJ 10281 | 1 | K | XI |
| E. cloacae | Clinical strain HU-UFRJ 4807 | 1 | L | XII |
| E. cloacae | Clinical strain HU-UFRJ 10232 | 1 | M | XIII |
| E. cloacae | Clinical strain HU-UFRJ 2303 | 1 | N | XIV |
| E. cloacae | Clinical strain HU-UFRJ 1135 | 1 | O | XIV |
| E. cloacae | Vaccine contaminant 1 | 1 | P | XV |
| E. cloacae | Vaccine contaminant 2 | 1 | Q | XVI |
| E. cloacae | Vaccine contaminant 3 | 1 | Q | XVI |
| E. cloacae | Vaccine contaminant 4 | 1 | Q | XVI |
| E. cloacae | Vaccine contaminant 5 | 1 | Q | XVI |
The sterility tests were performed in class 100 laminar airflow cabinets by different technicians on different dates by the direct inoculation method. The strains were identified by conventional biochemical tests (8) and the Vitek 32 system (bioMérieux Vitek, Inc.), and accompanying software (version 5.1) was used according to the manufacturer's instructions.
DNA extraction.
All strains were grown on nutrient broth (Difco, Detroit, Mich.) for 24 h at 30°C and checked for purity on nutrient agar plates. Approximately two loops worth of biomass were scraped off the agar plates, suspended in 100 μl of sterile distilled water, and boiled for 10 min. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatants were recovered and 5 μl was directly used as the template for PCR.
tDNA-PCR and ITS-PCR.
For tDNA-PCR the reaction was carried out with the outwardly directed consensus primers T5A (5′-AGTCCGGTGCTCTAACCAACTGAG-3′) and T3B (5′-AGGTCGCGGGTTCGAATCC-3′) described by Welsh and McClelland (34), which result in the amplification of regions between adjacent tRNA genes. ITS-PCR was carried out with primers L1 (5′-CAAGGCATCCACCGT-3′) and G1 (5′-GAAGTCGTAACAAGG-3′). These sequences are complementary to conserved regions in the 16S and 23S rRNA genes and result in the amplification of the variable spacer regions between these genes. The primers were obtained from DNAgency (Malvern, Pa.), and the amplifications were done on a Perkin-Elmer thermocycler in 25-μl reaction mixtures consisting of 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 3 mM MgCl2; a 200 μM concentration (each) of dATP, dCTP, dGTP, and dTTP (Amersham-Pharmacia Biotech); a 0.5 μM concentration of each primer; 1 U of Taq DNA polymerase (Gibco-BRL); and 5 μl of DNA. The program consisted of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min for 30 cycles, with an additional extension at 72°C for 10 min.
RAPD analysis.
For RAPD analysis we used arbitrarily chosen primers, which are able to anneal at multiple sites throughout the genome, producing a spectrum of amplified products characteristic of each template DNA. The primers used were primer 1 (5′-d[GGTGCGGGAA]-3′), primer 2 (5′-d[GTTTCGCTCC]-3′), primer 3 (5′-d[GTAGACCCGT]-3′), primer 4 (5′-d[AAGAGCCCGT]-3′), and primer 5 (5′-d[AACGCGCAAC]-3′) obtained from the RAPD Analysis primer set (Amersham Pharmacia Biotech). Amplification was done in 25-μl reaction mixtures consisting of 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 3 mM MgCl2; a 200 μM concentration (each) of dATP, dCTP, dGTP, and dTTP (Amersham-Pharmacia Biotech); 25 pmol of each primer; 2 U of Taq DNA polymerase (Gibco-BRL); and 5 μl of DNA. The program consisted of denaturation at 94°C for 1 min, annealing at 36°C for 1 min, and extension at 72°C for 2 min for 45 cycles, with an additional extension at 72°C for 7 min.
Agarose gel electrophoresis.
A sample of 10 μl of the PCR mixture was loaded onto a 2% (wt/vol) agarose gel, and the PCR products were separated by electrophoresis at 50 V for 3 h in 0.5× Tris-borate-EDTA (pH 8.0) buffer with φX174 RF DNA/HaeIII fragment size markers (GIBCO BRL) or a 100-bp DNA ladder; the gels were stained with ethidium bromide, and the gel images were digitized with the Video Documentation System and analyzed with ImageMaster software (Amersham Pharmacia Biotech).
Reproducibility evaluation.
The experiments were evaluated in triplicate with at least two bacterial lysates of each strain, and DNAs were coamplified in separate PCRs. The banding patterns were highly reproducible after visual and automated analysis.
Phylogenetic analysis.
A phylogenetic tree was generated using NTSYSpc 2.02 h software. Similarity was determined on the basis of the simple matching coefficient, and the generated matrix was subjected to clustering by the unweighted pair group method with arithmetic average algorithm.
RESULTS
Phenotypic characteristics.
All isolates were gram-negative rods, oxidase-negative, glucose fermenters with production of acid and gas, and no spores were observed. The strains were identified as Enterobacter cloacae after being subjected to standard conventional biochemical tests (8) and the Vitek System (bioMérieux Vitek).
tDNA-PCR.
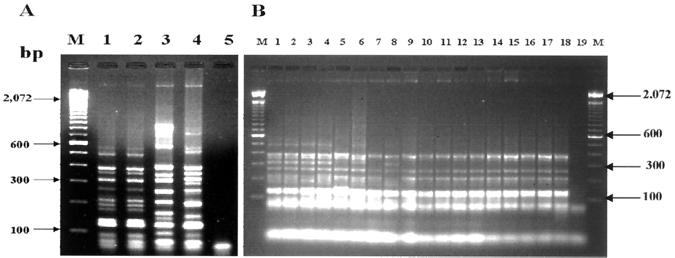
The tDNA-PCR profiles of Enterobacter species showed patterns of 9 to 12 DNA fragments ranging in size from 70 to 1,000 bp that distinguished the species by a number of specific DNA fragments (Fig. 1A). The two reference strains of E. cloacae showed the same set of fragments (Fig. 1A, lanes 1 and 2), whereas Enterobacter aerogenes and Enterobacter sakazakii exhibited distinct profiles (Fig. 1A, lanes 3 and 4). As indicated in Fig. 1B, the 11 clinical (lanes 3 to 13) and 5 vaccine (lanes 14 to 18) isolates showed the same pattern obtained by E. cloacae reference strains (lanes 1 and 2).
FIG. 1.
tDNA-PCR patterns of reference, clinical, and vaccine samples of Enterobacter species by electrophoresis in a 2% agarose gel. Lanes M, molecular size marker (100 bp). (A) Lane 1, E. cloacae ATCC 13047; lane 2, E. cloacae ATCC 23355; lane 3, E. sakazakii ATCC 29004; lane 4, E. aerogenes ATCC 13048; lane 5, negative control with no added template DNA. (B) Lane 1, E. cloacae ATCC 13047; lane 2, E. cloacae ATCC 23355; lanes 3 to 8, clinical isolates of E. cloacae from IAL; lanes 9 to 13, clinical isolates of E. cloacae from HU-UFRJ; lanes 14 to 18, vaccine isolates of E. cloacae; lane 19, negative control with no added template DNA.
ITS-PCR.
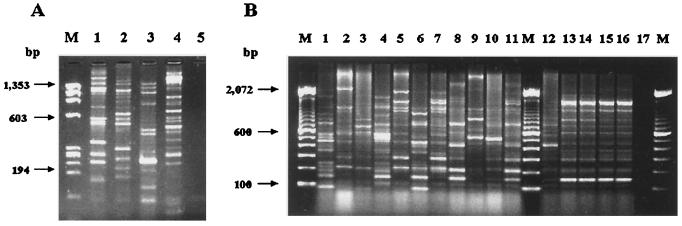
As seen in Fig. 2, an ITS-PCR of each reference strain of Enterobacter showed each strain's own banding pattern, including those of the two strains of E. cloacae (lanes 1 to 2). All clinical samples (Fig. 2B, lanes 1 to 11) showed distinct patterns, indicating a very significant degree of intraspecies variation. However, the four E. cloacae strains isolated from two different batches of vaccine produced an indistinguishable banding pattern (Fig. 2B, lanes 13, 14, 15, and 16). Considerable variation was observed in E. cloacae isolated from another batch (Fig. 2B, lane 12).
FIG. 2.
ITS-PCR patterns of reference, clinical, and vaccine samples of Enterobacter species by electrophoresis in a 2% agarose gel. (A) Lane M, molecular size marker (φX174 RF DNA/HaeIII); lane 1, E. cloacae ATCC 13047; lane 2, E. cloacae ATCC 23355; lane 3, E. sakazakii ATCC 29004; lane 4, E. aerogenes ATCC 13048; lane 5, negative control with no added template DNA. (B) Lanes M, molecular size markers (100-bp DNA ladder); lanes 1 to 6, clinical isolates of E. cloacae from IAL; lanes 7 to 11, clinical isolates of E. cloacae from HU-UFRJ; lanes 12 to 16, vaccine isolates; lane 17, negative control with no added template DNA.
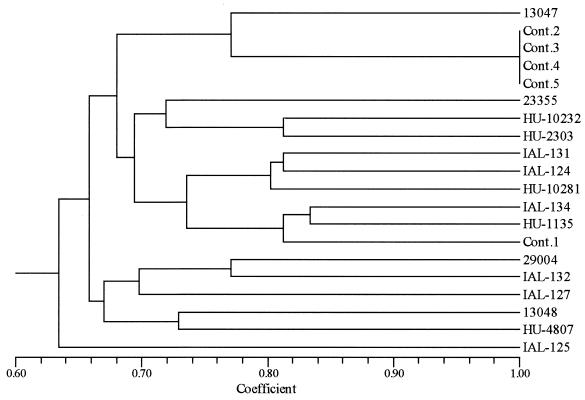
These results can be also observed by the dendrogram of Fig. 3, which was generated from ITS-PCR analysis, showing the genetic diversity of the E. cloacae strains analyzed. The four vaccine contaminant strains (contaminants 2, 3, 4, and 5) showed the same pattern, clustering at 100% similarity, and clustered at 77 and 72% with E. cloacae ATCC 13074 and ATCC 23055, respectively. Meanwhile, the vaccine contaminant (contaminant 1) clustered at 68% with the other contaminants. The group formed by the vaccine contaminant and the ATCC type strain clustered at 64% with the group formed by all the clinical samples and the two other ATCC species of Enterobacter. This dendrogram showed once more the high similarity (100%) of the contaminant strains, suggesting they belong to the same strain isolated from different batches of the same vaccine.
FIG. 3.
Phenetic relationship of 20 strains of Enterobacter sp. by ITS-PCR. The dendrogram was constructed by unweighted pair group method with arithmetic average. The numbers 13047, 23355, 29004, and 13048 are the ATCC strains; IAL and HU strains are E. cloacae strains from clinical samples; and Cont. 1 to 5 are E. cloacae strains from vaccine contaminants (Table 1).
RAPD analysis.
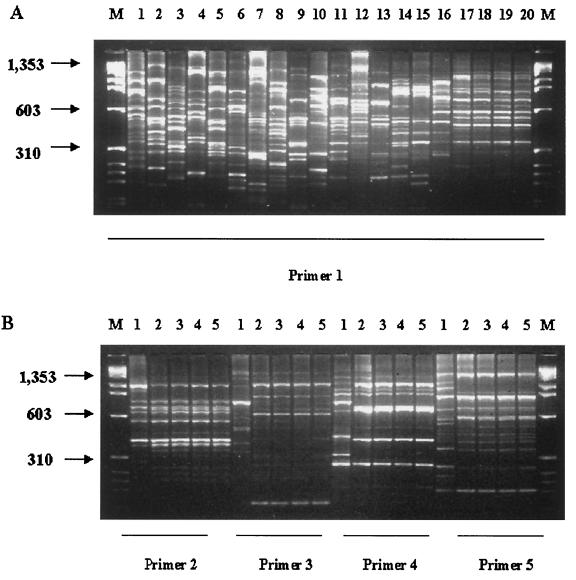
RAPD analysis with primer 1 demonstrated distinct fingerprints of Enterobacter reference strains (Fig. 4A, lanes 1 to 4) as well as all clinical samples of E. cloacae (Fig. 4A, lanes 5 to 15). It is noteworthy that the four vaccine isolates showed identical banding patterns after RAPD analysis (Fig. 4A, lanes 17, 18, 19, and 20); a different banding pattern was observed with the E. cloacae isolate from another batch of the vaccine (Fig. 4A, lane 16). The amplifications with RAPD primers 2, 3, 4, and 5 presented the same results described with primer 1 (Fig. 4B)
FIG. 4.
RAPD patterns of reference, clinical, and vaccine samples of Enterobacter species by electrophoresis in a 2% agarose gel. Lanes M, molecular size marker (φX174 RF DNA/HaeIII). (A) Lane 1, E. sakazakii ATCC 29004; lane 2, E. aerogenes ATCC 13048; lane 3, E. cloacae ATCC 13047; lane 4, E. cloacae ATCC 23355; lanes 5 to 10, clinical isolates of E. cloacae from IAL; lanes 11 to 15, clinical isolates of E. cloacae from HU-UFRJ; lanes 16 to 20, vaccine isolates. (B) Lanes 1 to 5, vaccine isolates analyzed with RAPD primers 2, 3, 4, and 5.
DISCUSSION
The most striking data presented in this study were that the use of tDNA-PCR and ITS-PCR together offered a hierarchical system for E. cloacae strain differentiation; tDNA-PCR resolved the cultures at the species level (Fig. 1), and ITS-PCR differentiated them at the intraspecies level (Fig. 2). The presence of microbial contamination in a sterility test of vaccine is a sporadic and random event requiring the analysis of large sample numbers and demands highly sensitive procedures due to a heterogeneous distribution of microorganisms in a given biological production batch (30). For analyzing sterility test results it is necessary to have an accurate identification and differentiation between the isolates of one species, in order to make a statement about the source and ways of contamination for effective quality control (1). In order to fulfill these requirements we checked the efficiency of the combined methods by using 18 samples of E. cloacae. In the tDNA-PCR 11 clinical samples and five vaccine isolates showed the same pattern exhibited by two E. cloacae reference strains (Fig. 1B), and ITS-PCR yielded distinct patterns for the strains, except for the four E. cloacae strains isolated from different batches of vaccine that presented a unique single pattern (Fig. 2B). With the aim of evaluating if the genetic variability of E. cloacae is confined only to the more variable part of the genome or is scattered over the whole genome, we compared DNA fingerprinting obtained with ITS-PCR and RAPD analyses. We observed an expressive agreement between ITS-PCR results and those of RAPD analysis with five different random primers (Fig. 2 and 4). Our results permitted us to conclude that all clinical and vaccine isolates were E. cloacae and the four vaccine isolates were actually the same strain, probably originating from a common source during the manufacturing process, instead of an accidental contamination during the sterility assays. These findings were essential for our quality control laboratory to perform a proper interpretation of sterility tests. It is important to point out that three isolates (Fig. 2B, lanes 13, 14, and 15) belonged to the same batch of the vaccine produced in 1998, for which a sterility test and retests were performed on different dates by different technicians; the other isolate (lane 16) was from a vaccine batch produced in 1999, and all of them showed the same set of fragments.
Recently, a number of molecular biology-based approaches directed at species identification have been described. Ribotyping (5) and pulsed-field gel electrophoresis have been used for typing of E. cloacae, with high discriminatory potential and good reproducibility (18); however, they are labor-intensive and time-consuming (29). Another method reported to discriminate bacteria of different genera to the species level is amplified ribosomal DNA restriction analysis (32). It is very sensitive, but compared to tDNA-PCR analysis several restrictions may be needed to obtain a final discrimination between species.
tDNA-PCR and ITS-PCR have been used successfully in our laboratory to identify and determine the genetic relationships between Staphylococcus aureus and Bacillus sp. strains isolated from different sources (data not shown). The assays offer potential usefulness in the clinical laboratory and can be performed independent of concentration of DNA, genome size, or sequencing when, for example, rapid identification and typing with a high degree of specificity of clinical isolates are desired for choice of antibiotic therapy (2) or even for surveillance of outbreaks (5).
ACKNOWLEDGMENTS
We thank C. F. A. Pereira from the HU-UFRJ and L. T. Bastos from the IAL for the clinical samples kindly provided. We are also grateful to R. M. Albano for critical review of the manuscript.
This work was supported by INCQS/ANVISA, CNPq (OBM), PADCT III, and grant 4196092800 from FINEP/BID.
REFERENCES
- 1.Akers M J. Parenteral quality control: sterility, pyrogen, particulate, and package integrity testing. 2nd ed. New York, N.Y: Marcel Dekker; 1994. [Google Scholar]
- 2.Baele M, Baele P, Vaneechoutte M, Storms V, Butaye P, Devriese L, Verschraegen G, Gillis M, Haesebrouck F. Application of tRNA intergenic spacer for identification of Enterococcus species. J Clin Microbiol. 2000;38:4201–4207. doi: 10.1128/jcm.38.11.4201-4207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean P, Olive D M. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benter T, Papadopoulos S, Pape M, Manns M, Poliwada H. Optimization and reproducibility of random amplified DNA in human. Anal Biochem. 1995;230:92–100. doi: 10.1006/abio.1995.1442. [DOI] [PubMed] [Google Scholar]
- 5.Bingen E, Denamur E, Lambert-Zechovsky N, Brahimi N, El Lakami M, Elion J. Rapid genotyping shows the absence of cross contamination in Enterobacter cloacae nosocomial infections. J Hosp Infect. 1992;21:95–101. doi: 10.1016/0195-6701(92)90028-k. [DOI] [PubMed] [Google Scholar]
- 6.Bram R J, Young R A, Steitz J A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of Escherichia coli. Cell. 1980;19:393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- 7.Brazilian Pharmacopeial Convention. Brazilian Pharmacopeia. 4th ed. São Paulo, Brazil: Atheneu; 1988. Biological methods. Biological safety tests: sterility, p. v.5.1.1.1.–v.5.1.1.6. [Google Scholar]
- 8.Brenner D J, Farmer III J J, Frederikesen W, Le Minor L, Sakazaki R. Facultatively anaerobic gram-negative rods. In: Murray R G E, Brenner D J, Bryant M P, Holt J G, Krieg N R, Moulder J W, Pfennig N, Sneath P H A, Staley J T, editors. Bergey's manual of systematic bacteriology. 1st ed. Baltimore, Md: Williams and Wilkins; 1984. pp. 408–516. [Google Scholar]
- 9.Burchard K W, Barral D T, Reed M, Stoman G J. Enterobacter bacteremia in surgical patients. Surgery. 1986;100:857–862. [PubMed] [Google Scholar]
- 10.Chen K H, Chen T A. A novel method for cloning DNA of plant-pathogenic mycoplasmalike organisms. Can J Microbiol. 1995;41:753–757. doi: 10.1139/m95-104. [DOI] [PubMed] [Google Scholar]
- 11.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debast S B, Melchers W J, Voss A, Hoogkamp-Korstanje J A, Meis J F. Epidemiological survey of an outbreak of multi-resistant Serratia marcescens by PCR-fingerprinting. Infection. 1995;23:267–271. doi: 10.1007/BF01716283. [DOI] [PubMed] [Google Scholar]
- 13.De Gheldre Y, Vandamme P, Goosses H, Struelens M J. Identification of clinically relevant viridans streptococci by analysis of transfer DNA intergenic spacer length polymorphism. Int J Syst Bacteriol. 1999;49:1591–1598. doi: 10.1099/00207713-49-4-1591. [DOI] [PubMed] [Google Scholar]
- 14.Ehrenstein B, Bernards A T, Dijkshoorn L, Gerner-Smidt P, Towner K J, Bouvet P J, Daschner F D, Grundmann H. Acinetobacter species identification by using tRNA spacer fingerprinting. J Clin Microbiobiol. 1996;34:2414–2420. doi: 10.1128/jcm.34.10.2414-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Martínez J, Acinas S G, Antón A I, Rodríguez-Valera F. Use of the ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999;36:55–64. doi: 10.1016/s0167-7012(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 16.Gaston M A. Enterobacter: an emerging nosocomial pathogen. J Hosp Infect. 1988;11:197–208. doi: 10.1016/0195-6701(88)90098-9. [DOI] [PubMed] [Google Scholar]
- 17.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 18.Haerti R, Bandlow G. Epidemiological fingerprinting of Enterobacter cloacae by small fragment restriction endonuclease analysis and pulsed-field gel electrophoresis of genomic restriction fragments. J Clin Microbiol. 1993;31:128–133. doi: 10.1128/jcm.31.1.128-133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey S, Hill C W, Squires C, Squires C L. Loss of the spacer loop sequence from the rrnB operon in the Escherichia coli K-12 subline that bears the relA1 mutation. J Bacteriol. 1988;170:1235–1238. doi: 10.1128/jb.170.3.1235-1238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurata N, Nakamura K, Yamamoto K, et al. A300 kilobase interval genetic map of rice including 283 expressed sequences. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence L M, Harvey J, Gilmour A. Development of a random amplification of polymorphic DNA typing method for Listeria monocytogenes. Appl Environ Microbiol. 1993;59:3117–3119. doi: 10.1128/aem.59.9.3117-3119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leelayuwat C, Romphruk A, Lulitanond A S. Genotype analysis of Burkholderia pseudomallei using randomly amplified polymorphic DNA (RAPD): indicative of genetic difference amongst environmental and clinical isolates. Acta Trop. 2000;77:229–237. doi: 10.1016/s0001-706x(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 24.Liao D. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J Mol Evol. 2000;51:305–317. doi: 10.1007/s002390010093. [DOI] [PubMed] [Google Scholar]
- 25.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markowitz S M, Smith S M, Williams D S. Retrospective analysis of plasmid patterns in a study of burn unit outbreaks of infection due to Enterobacter cloacae. J Infect Dis. 1983;148:18–23. doi: 10.1093/infdis/148.1.18. [DOI] [PubMed] [Google Scholar]
- 27.McClelland M, Petersen C, Welsh J. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J Clin Microbiol. 1992;30:1499–1504. doi: 10.1128/jcm.30.6.1499-1504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monna L, Miyao A, Zhong H S, Sasaki T, Minobe Y. Screening of RAPD markers linked to the photoperiod-sensitive gene in rice chromosome 6 using bulked segregant analysis. DNA Res. 1995;2:101–106. doi: 10.1093/dnares/2.3.101. [DOI] [PubMed] [Google Scholar]
- 29.Shi Z-Y, Liu P Y-F, Lau Y-J, Hu B-S. Epidemiological typing of isolates from an outbreak of infection with multidrug-resistant Enterobacter cloacae by repetitive extragenic palindromic unit b1-primed PCR and pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2784–2790. doi: 10.1128/jcm.34.11.2784-2790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.U.S. Pharmacopoeial Convention. Microbiological tests. Sterility tests. In: Pharmacopeia US, editor. vol. 23. Rockville, Md: U.S. Pharmacopeial Convention; 1995. pp. 1483–1488. [Google Scholar]
- 31.Vaneechoutte M, Boerlin P, Tichy H-V, Bannerman E, Jäger B, Bille J. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int J Syst Bacteriol. 1998;48:127–139. doi: 10.1099/00207713-48-1-127. [DOI] [PubMed] [Google Scholar]
- 32.Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, De Vos P, Claeys G, Verschraegen G. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1995;33:11–15. doi: 10.1128/jcm.33.1.11-15.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welsh J, McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991;19:861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welsh J, McClelland M. PCR-amplified length polymorphisms in tRNA intergenic spacers for categorizing staphylococci. Mol Microbiol. 1992;6:1673–1680. doi: 10.1111/j.1365-2958.1992.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 36.Williams J, G K, Kubelik A R, Livak K J, Rafalski J A, Tingly S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. Requirements for biological substances. 6. General requirements for the sterility of biological substances. Technical report series no. 530. Geneva, Switzerland: World Health Organization; 1973. [Google Scholar]
- 38.Yamamoto Y, Kohmo S, Koga H, et al. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol. 1995;33:3328–3332. doi: 10.1128/jcm.33.12.3328-3332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]