Abstract
The utility of luciferase reporter mycobacteriophages (LRPs) for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis was prospectively evaluated in a clinical microbiology laboratory in Mexico City, Mexico. Five hundred twenty-three consecutive sputum samples submitted to the laboratory during a 5-month period were included in this study. These specimens were cultivated in Middlebrook 7H9 (MADC), MGIT, and Löwenstein-Jensen (LJ) media. Of the 71 mycobacterial isolates recovered with any of the three media, 76% were detected with the LRPs, 97% were detected with the MGIT 960 method, and 90% were detected with LJ medium. When contaminated specimens were excluded from the analysis, the LRPs detected 92% (54 of 59) of the cultures. The median time to detection of bacteria was 7 days with both the LRPs and the MGIT 960 method. LRP detection of growth in the presence of p-nitro-α-acetylamino-β-hydroxypropiophenone (NAP) was used for selective identification of M. tuberculosis complex (MTC) and compared to identification with BACTEC 460. Using the LRP NAP test, 47 (94%) out of 50 isolates were correctly identified as tuberculosis complex. The accuracy and speed of LRP antibiotic susceptibility testing with rifampin, streptomycin, isoniazid, and ethambutol were compared to those of the BACTEC 460 method, and discrepant results were checked by the conventional proportion method. In total, 50 MTC isolates were tested. The overall agreement between the LRP and BACTEC 460 results was 98.5%. The median LRP-based susceptibility turnaround time was 2 days (range, 2 to 4 days) compared to 10.5 days (range, 7 to 16 days) by the BACTEC 460 method. Phage resistance was not detected in any of the 243 MTC isolates tested. Mycobacteriophage-based approaches to tuberculosis diagnostics can be implemented in clinical laboratories with sensitivity, specificity, and rapidity that compare favorably with those of the MGIT 960 and BACTEC 460 methods. The phages currently provide the fastest phenotypic assay for susceptibility testing.
The microscopic examination of sputum to detect the presence of acid-fast bacilli (AFB) (AFB smear) has served the world well for the past 100 years and is the diagnostic basis of directly observed therapy. However, it fails to identify 30 to 50% of active cases of tuberculosis and cannot identify the increasing numbers of persons diseased by drug-resistant Mycobacterium tuberculosis (7, 10). A reliance solely upon the limited AFB smear is one of the many factors that contribute to poor tuberculosis control in resource-poor countries (1). Although available, conventional mycobacteriological techniques for culture isolation and antibiotic susceptibility testing (AST) are slow, and the more recently developed rapid methods such as the BACTEC and MGIT systems are expensive and require elaborate technology (3, 13, 15). Thus, there is an urgent need for a rapid and affordable method to detect mycobacteria in clinical specimens, to identify whether they are M. tuberculosis, and to determine their antimicrobial susceptibility.
Over the past decade, luciferase reporter mycobacteriophages (LRPs) have been developed that show great promise for diagnostic microbiology (4). This novel approach utilizes a genetically engineered reporter phage to detect viable mycobacteria, which upon LRP infection produces quantifiable light. In the presence of antibiotics, bacilli that are drug resistant and retain their viability undergo phage infection and also produce light. In this way, quantification of photons with a luminometer can be used to reveal the susceptibility profile of each isolate. Unlike in the radiometric BACTEC system, the phages do not have any requirement for radioactive isotopes. When mycobacteria are grown in media containing agents such as p-nitro-α-acetylamino-β-hydroxypropiophenone (NAP), selective inhibition of M. tuberculosis complex (MTC) organisms allows differentiation from nontuberculous mycobacteria (NTM) (11).
To date, the performance of the LRPs for mycobacterial diagnostics has only been tested against a limited number of laboratory strains in a research laboratory (4, 12). In this study we prospectively evaluated the diagnostic utility of the LRPs in a centralized diagnostic laboratory in Mexico City, Mexico. We showed (i) that LRP assay can rapidly and sensitively detect growth of MTC from sputum cultures, (ii) that the LRP NAP test can rapidly and specifically distinguish M. tuberculosis complex from other mycobacteria, (iii) that LRPs can rapidly and accurately perform AST with the four first-line drugs, and (iv) that phage-resistant MTC isolates were not detected in Mexico.
MATERIALS AND METHODS
Sputum specimens included in study.
From February to June of 2000, all 487 sputum samples collected from 213 patients suspected of having pulmonary tuberculosis in the Huauchinango Health Jurisdiction, Huauchinango, Puebla, were sent to the reference laboratory of clinical microbiology of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City (INCMNSZ). Concurrently, 36 additional sputum samples were obtained from 21 patients hospitalized at the same institution.
LRPs.
Phage phAE142, a second generation of TM4-derived mycobacteriophage, was constructed in the laboratory of W. R. Jacobs (S. Bardarov, Jr., H. Dou, K. Eisenach, N. Banaiee, S. Ya, J. Chan, W. R. Jacobs, Jr., and P. F. Riska, submitted for publication) and used in this study. High-titer phage stocks were prepared as previously described (4) with the following modifications: Mycobacterium smegmatis mc2-4502 cells were used as the propagating strain and grown in the presence of hygromycin at 150 μg/ml.
Antibiotics.
Lyophilized antibiotics (Becton Dickinson, Sparks, Md.) were dissolved in sterile water to make the following 200× stock concentrations: isoniazid (INH), 40 μg/ml; rifampin (RIF), 400 μg/ml; streptomycin (STR), 80 μg/ml; and ethambutol (EMB), 1,000 μg/ml. Working stocks (20×) were prepared with sterile water. p-Nitro-α-acetylamino-β-hydroxypropiophenone (NAP) stock solutions (11) were diluted with sterile water to prepare 20× (150-μg/ml) working stocks. All stocks were stored at −40°C.
Mycobacteriologic procedures.
Specimens were decontaminated and digested with an equal volume of 4% NaOH–0.5% N-acetylcysteine solution according to standard methods (8). Samples were neutralized with 45 ml of 0.067 M phosphate buffer (pH 6.8) and centrifuged at 3,164 × g for 20 min. A small aliquot of the sediment was used to prepare smears for auramine-rhodamine and Ziehl-Neelsen staining. Sediments were resuspended in approximately 3 ml of Middlebrook 7H9 broth (Difco, Detroit, Mich.) supplemented with 1% (vol/vol) glycerol and 10% (vol/vol) albumin-dextrose-catalase (MADC) (Difco). An aliquot of 0.5 ml was used to prepare MGIT 960 tubes according to the manufacturer's standard protocol (Becton Dickinson Diagnostic Instrument Systems). An additional aliquot of 0.5 ml was inoculated onto the surface of Löwenstein-Jensen (LJ) slants. The remaining 2 ml was cultured and subsequently referred to as MADC culture. To each MADC tube 40 μl of MGIT PANTA, with a final concentration similar to the one added to MGIT tubes, was added. Each culture was screened for contamination on sheep blood agar, and contaminated cultures were eliminated. All cultures were incubated at 37°C until found positive or for up to 8 weeks. Throughout the course of incubation, breakthrough contaminations were discarded.
Growth detection. (i) MGIT 960 system.
The MGIT 960 automated instrument read each tube hourly and triggered an alarm when growth was detected. Positive cultures were checked for the presence of mycobacteria with an AFB smear and subculture on sheep blood agar. Cultures free of contaminants were advanced for identification and susceptibility testing by the BACTEC 460 system. Time to detection (TTD) was based on the date of earliest instrument positivity for contamination-free cultures.
(ii) LJ method.
LJ slants were incubated in an atmosphere of 5 to 10% CO2. Growth on LJ medium was checked visually every 7 days and considered positive upon appearance of colonies. TTD was based on the earliest date of detection of colonies.
(iii) Phage assay.
Cultures were checked for mycobacterial growth on post-incubation days 1, 3, 5, 7, 11, 15, 19, 23, 27, 41, and 55. On each designated day, 80 μl of each culture was infected with 8 μl of LRP phAE142 in a Falcon 96-well plate (Becton Dickinson Labware, Lincoln Park, N.J). The plate was covered with a lid, sealed with Parafilm, and incubated at 37°C. Three hours after the phage infection 20-μl aliquots were transferred to disposable cuvettes for quantitative luciferase assay with a TD-20/20 luminometer (5-s integration, 1-s pause; Turner Design, Mountain View, Calif.). Upon autoinjection of 100 μl of 0.33 mM d-luciferin solution (Sigma, St. Louis, Mo.) into each cuvette, light production was quantified and expressed in relative light units (RLU). The value from a blank read was automatically subtracted from each reading. Samples with ≥0.5 RLU were considered positive, and those with <0.1 RLU were considered negative. Samples with <0.5 and ≥0.1 RLU were considered equivocal and were rechecked at 6 h post-phage infection. All positives were confirmed with a duplicate read. Samples with a negative 3-h read or discrepant 3- and 6-h reads were considered negative for that day. The TTD was based on the earliest date of LRP assay positivity. Samples with negative reads on day 55 were reported as negative cultures.
Culture titration.
Bacillus concentrations were quantified on the days cultures were detected by the LRP assay. Serial 10-fold dilutions were plated onto 7H10 agar supplemented with OADC (Difco) and PANTA (Becton Dickinson). Plates were incubated at 37°C, and CFU were counted at 3 to 6 weeks.
NAP test and susceptibility testing. (i) LRP method.
Identification and susceptibility testing were performed simultaneously with the LRPs. Drug concentrations for RIF, STR, INH, and EMB were determined from antibiotic concentration curves with M. tuberculosis ATCC 35801, 35820, 35822, 35837, and 35838. Test concentrations of the following drugs were as indicated: RIF, 2 μg/ml; INH, 0.2 μg/ml; STR, 0.4 μg/ml; and EMB, 5 μg/ml. The NAP concentration was 7.5 μg/ml (11). Sterile water (5 μl) and 5 μl of each of the 20× antibiotic stocks and 20× NAP were placed in separate wells of a sterile Falcon 96-well plate (Becton Dickinson Labware). Subsequently, 95 μl of culture was added to each well, and the plate was covered with a sealing membrane and incubated at 37°C for 40 h. After incubation, each well was infected with 10 μl of phage phAE142, and the plate was returned to the incubator. At 3 and 6 h post-phage infection, 20-μl aliquots of each well were transferred to disposable cuvettes for quantitative luciferase assay in the luminometer. Inhibition indices [(RLUantibiotic)/(RLUcontrol)×100] were calculated and interpreted as follows: for antibiotics, <10% was considered to indicate susceptibility and ≥10% was considered to represent resistance (12); for the NAP test, ≤25% was interpreted as representing an MTC isolate and >25% was interpreted as representing an NTM isolate (11). In the case of resistant, borderline (inhibition index of >5% for AST and >15% for identification), or inconclusive (<1 RLU for control, discrepant duplicate reads, and discrepant 3- and 6-h inhibition index) results, AST and NAP tests were repeated.
(ii) Radiometric method.
All MGIT cultures underwent identification with the BACTEC 460 NAP differentiation test (Becton Dickinson Diagnostic Instrument Systems). Those identified as MTC were advanced for AST with the BACTEC 460 protocol using standardized antibiotic concentrations and cutoff points (Becton Dickinson Diagnostic Instrument Systems). Final concentrations of the following antibiotics were as indicated: RIF, 2 μg/ml; INH, 0.1 μg/ml; STR, 2 μg/ml; and EMB, 7.5 μg/ml.
Phage infectibility.
Mexican MTC isolates from our library collection were cultured on solid or liquid media. From solid media, several colonies were scraped and transferred to a 15-ml glass tube containing sterile 4-mm-diameter glass beads and 1.5 ml of MADC. After vortexing for 20 s, large clumps of cells were allowed to settle and 1 ml of supernatant was removed for phage infection. From liquid cultures, 0.5 ml of cells was washed twice with MADC and suspended in an equal volume of MADC. In both cases, the cells were incubated overnight at 37°C and infected on the following day as described above.
Statistical analysis.
The sensitivity, specificity, and accuracy of the LRP AST and NAP tests were calculated. Differences in proportions were evaluated by the chi-square test.
RESULTS
Growth detection.
From February to June of 2000, 523 sputum samples (82 smear positive) from 234 patients were cultured in MADC, MGIT, and LJ media. Contamination losses accounted for 142 (27.2%) of the MADC, 35 (6.7%) of the MGIT, and 56 (10.7%) of the LJ cultures. Sensitivity and speed of the LRP assay for detection of primary mycobacterial cultures were compared to those of the MGIT 960 and LJ assays. Of the 71 cultures (66 smear positive) recovered by any of the three culture media, 54 (76.1%) were detected with the LRPs, 69 (97.2%) were detected by the MGIT 960 method, and 64 (90.1%) were detected by the LJ method. Distribution of these cultures is shown in Table 1. When cultures recovered with the liquid media were combined with those detected with the solid medium, the LRPs plus LJ detected 69 (97.2%) cultures compared to 70 (98.6%) detected by the MGIT 960 method plus LJ. Of the 68 MTC cultures recovered by all three methods, 51 (75%) isolates were detected with the LRPs, 66 (97.1%) were detected by the MGIT 960 assay, and 61 (89.7%) were detected with LJ medium. For the MTC cultures, when the liquid media were paired with solid medium, the LRPs plus LJ detected 66 (97.1%) of the cultures compared to 67 (98.5%) with the MGIT 960 assay plus LJ. There were 12 cultures for which breakthrough contaminations were found in the MADC culturing tubes while mycobacterial growth was detected in their cohorts. With contaminated cultures and their cohorts excluded from the analysis, the rate of culture detection with the LRPs improved to 91.5% (54 of 59) but remained unchanged for the other two methods.
TABLE 1.
Distribution of bacterial isolates recovered by the LRP, MGIT 960, and LJ systems
| Culture system | No. of isolates recovered/total no. of isolates (%)
|
||
|---|---|---|---|
| MTC | NTM | Total | |
| LRP | 51/68 (75.0) | 3/3 | 54/71 (76.1) |
| LRPa | 51/56 (91.1) | 3/3 | 54/59 (91.5) |
| MGIT | 66/68 (97.1) | 3/3 | 69/71 (97.2) |
| LJ | 61/68 (89.7) | 3/3 | 64/71 (90.1) |
| LRP + LJ | 66/68 (97.1) | 3/3 | 69/71 (97.2) |
| MGIT + LJ | 67/68 (98.5) | 3/3 | 70/71 (98.6) |
Contaminated cultures and their cohorts were excluded from the analysis.
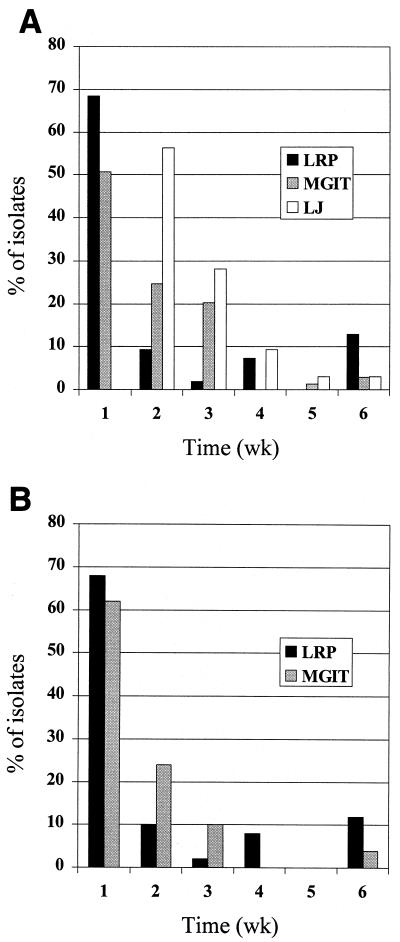
The TTD of primary mycobacterial isolates with the three culture systems is shown in Table 2. The median TTD for all mycobacterial isolates was 6 days (range, 1 to 41 days) with the LRPs, 7 days (range, 4 to 42 days) by the MGIT 960 system, and 14 days (range, 8 to 42 days) by the LJ method. Growth was detected within 7 days for 37 out of 54 (68.5%) cultures with the LRPs in contrast to 35 out of 69 (50.7%) by the MGIT 960 system (Fig. 1A). The median TTD for MTC isolates was 7, 7.5, and 14 days for the three methods, respectively. When TTD was analyzed for 50 MTC isolates that were recovered by both the LRPs and the MGIT 960 system, the median TTD was 7 days for both methods. Within this group of isolates, the LRPs detected 34 (68%) cultures within 7 days compared to 31 (62%) cultures by the MGIT 960 system (Fig. 1B). The sensitivity of the LRP assay was also determined in terms of minimal bacillus concentrations needed to detect positive cultures. Among the 37 MADC cultures quantified on the days growth was detected, titers ranged from 103 to 4.3 × 107 CFU/ml, with a median of 106 CFU/ml.
TABLE 2.
TTD of primary bacterial isolates by the LRP, MGIT 960, and LJ systems
| Culture system | Median TTD (days) (range, no. of isolates)
|
|||
|---|---|---|---|---|
| M. tuberculosis complex
|
NTM | All cultures | ||
| Nonpaired | Paireda | |||
| LRP | 7 (1–41, 51) | 7 (1–41, 50) | 3 (3, 3) | 6 (1–41, 54) |
| MGIT | 7.5 (4–42, 66) | 7 (4–42, 50) | 7 (6–10, 3) | 7 (4–42, 69) |
| LJ | 14 (8–42, 51) | 20 (20, 3) | 14 (8–42, 64) | |
Isolates recovered by the LRP and MGIT 960 systems.
FIG. 1.
Percentage of mycobacterial isolates recovered versus time. (A) Mycobacterial isolates detected per week by the LRPs (n = 54), the MGIT 960 system (n = 69), and the LJ method (n = 64). (B) Paired MTC isolates (n = 50) detected per week by the LRP and MGIT 960 methods.
Identification.
NAP, a compound which selectively inhibits the growth of MTC but not that of NTM, was used to tentatively identify the MTC cultures. LRP and BACTEC 460 NAP tests were performed on 53 MADC cultures and their MGIT cohorts, respectively, and the results were compared. Overall agreement between the LRPs and the BACTEC 460 method was found in 50 (94.3%) out of 53 tests. A total of 47 (94%) of 50 cultures were correctly identified as MTC. The three cultures with discrepant results were falsely resistant to NAP and incorrectly identified as NTM. When we screened these cultures for microbial contaminants, all three cultures harbored non-acid-fast organisms. The sensitivity of the LRP NAP test, or the ability to detect MTC strains, was 94%. Specificity, defined as the ability to detect NTM isolates, was 100%. There was no statistically significant difference between the NAP test results obtained by the LRPs and the BACTEC 460 method (P = 0.586). The turnaround times for the identification of 50 primary MTC cultures ranged from 2 to 4 days (median, 2 days) with the LRPs and 2 to 4 days (median, 3 days) by the BACTEC 460 method. With the LRPs, 47 (94%) of these cultures were identified in 2 days compared to 15 (30%) by the BACTEC 460 method.
Susceptibility testing.
Accuracy and reproducibility of the LRPs for susceptibility testing with the first-line drugs were evaluated and compared to those for the BACTEC 460 method. Fifty MADC M. tuberculosis complex cultures and their MGIT cohorts were tested, and their susceptibility patterns are shown in Table 3. Overall agreement for all four drugs was found in 197 (98.5%) out of 200 tests. Three discordant results were found between the LRP and BACTEC 460 results.
TABLE 3.
Susceptibility of MTC isolates to four first-line drugs as determined by the LRP and BACTEC 460 systems
| Druga | Total no. of isolates tested | No. of strains with following test resultsb:
|
|||
|---|---|---|---|---|---|
| Both S | BACTEC, S; LRP, R | BACTEC, R; LRP, S | Both R | ||
| RIF | 50 | 47 | 3 | ||
| STR | 50 | 48 | 2 | ||
| INH | 50 | 41 | 2 | 1 | 6 |
| EMB | 50 | 47 | 3 | ||
The following drugs were used at the indicated concentrations (in micrograms per milliliter): RMP, 2 (both systems); SM, 0.4 (LRP) and 2 (BACTEC); INH, 0.2 (LRP) and 0.1 (BACTEC); EMB, 5 (LRP) and 7.5 (BACTEC).
Abbreviations: S, susceptible; R, resistant.
Of the 50 LRP susceptibility tests performed with RIF, STR, and EMB, 100% agreement was found with BACTEC 460 results. Three resistant isolates were identified for RIF and EMB, and two were identified for STR. With INH, 47 (94.1%) out of 50 (43 susceptible and 7 resistant) isolates gave test results that were in agreement between the two methods. Among the three isolates with discrepant INH results, one was susceptible by the LRP method but resistant by the BACTEC method, and two were resistant by the LRP method but susceptible by the BACTEC method. Upon retesting by the conventional agar-based proportion method on Middlebrook 7H11 (National Jewish Medical and Research Center) (3), all three isolates gave results in agreement with the BACTEC 460 results. However, in the case of the strain shown by BACTEC to be resistant and shown by LRP to be susceptible, it was shown to be only moderately resistant to INH as it was 100% resistant at an INH concentration of 0.2 μg/ml but completely susceptible at 1.0 μg/ml. In order to assess the reproducibility of AST results with the LRPs, we blindly repeated LRP AST on 24 MGIT cohort cultures. Under these conditions, all 24 isolates (22 pan-susceptible and 2 STR resistant) gave results 100% (96 of 96) in agreement with the susceptibility results as previously determined by the LRPs and BACTEC 460 (data not shown). The sensitivity and specificity of the LRP AST were determined for each drug. Sensitivity, or the ability to detect drug-resistant isolates, was 100% for RIF, STR, and EMB and 85.7% for INH. The specificity, or ability to detect susceptible isolates, was 100% for RIF, STR, and EMB and 95.3% for INH. There was no statistically significant difference between the two AST methods for INH (P = 0.377).
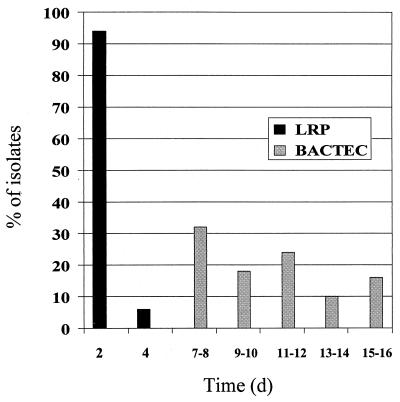
The amount of time taken to complete susceptibility testing with the LRPs and by the BACTEC 460 method was evaluated. For the 50 MADC MTC cultures and their MGIT cohorts, the turnaround times ranged from 2 to 4 days (median, 2 days) with the LRPs and 7 to 16 days (median, 10.5 days) by the BACTEC 460 method, respectively. With the LRPs, 94% of the AST results were completed in 2 days, while BACTEC 460 results did not become available until day 7 (Fig. 2).
FIG. 2.
Percentage of isolates with AST results available versus time. Susceptibility of MTC cultures to first-line drugs was determined by the LRP and BACTEC 460 methods, and turnaround times were recorded accordingly (n = 50).
Phage resistance.
In order to show that phage-resistant MTC isolates do not exist in Southern Mexico, we performed the LRP assay on 243 Mexican MTC isolates from 131 patients. These strains included the culture-positive cohorts of culture-negative MADC cultures. Among the 243 MTC isolates tested with LRP phAE142, 100% infectivity was observed. In each case, adequate light production was noted after phage infection (data not shown). We did not evaluate the efficiency of infection given that the luminometer settings and bacterial loads were different for each isolate.
DISCUSSION
Previous studies have shown that under research conditions LRP-based assays can rapidly identify mycobacteria and perform susceptibility tests. In this study we have demonstrated that it is feasible to perform LRP-based assays in a clinical laboratory and compared their abilities to detect, identify, and determine the antibiotic susceptibility of clinical isolates with those of established methods. Because the majority of these results were obtained from sequential specimens from a single geographical area, we believe these encouraging results are unbiased and accurately predict the results likely to be obtained when these tests are used in other reference laboratories in other developing countries.
As in other studies, no single method was able to recover all positive cultures (2, 5) (Table 1). Of the 71 mycobacterium isolates recovered with any of the three media, 76% were detected by the LRP assay, 97% were detected by the MGIT 960 method, and 90% were detected with LJ medium. The apparent sensitivity of the LRPs is diminished primarily because there were 12 samples for which MADC tubes were contaminated while MTC growth was detected for these sputa using MGIT and LJ. Suboptimal PANTA concentration was the likely cause of contaminations in MADC cultures, though the use of original sputum-containing tubes for MADC cultures exacerbated the problem. In this study, we underestimated the final volume of the sputum sediments, and this in turn resulted in suboptimal PANTA concentrations and an opportunity for the growth of nonmycobacterial contaminants. Because this contamination was unrelated to the LRP assay and can be easily reduced by using a fixed sediment volume and sterile tubes, a more accurate assessment is likely provided by excluding these sputa from the analysis. Analyzed in this manner, the sensitivity of LRPs increases to 92% (54 of 59). Regardless of how one analyzes the contaminated samples, the yield of culture is greatest if one follows the recommendations of the Centers for Disease Control and Prevention to use a combination of solid and liquid media for mycobacterial isolation (6). The use of LJ with the MGIT 960 system or LRPs recovered 98 and 97% of positive cultures, respectively. Unfortunately, because virtually all culture-positive specimens were also AFB positive regardless of the detection method, we could not evaluate the sensitivity and speed of the LRPs for detection of smear-negative culture-positive isolates. In order to make such an assessment, the LRPs must be evaluated in a laboratory with adequate rates of recovery for smear-negative cultures.
Reports of phage-resistant NTM (11, 14) raise the concern that some MTC strains will be resistant to phage and thus reduce the sensitivity of detection by this system. Over the course of this work we tested MTC bacteria isolated from over 130 different patients and never encountered a phage-resistant strain. While this does not exclude their existence, it does suggest that, at least in southern Mexico, this is not likely to be a clinically significant phenomenon.
The TTD of MTC bacteria from sputum samples was statistically equivalent between the LRP and MGIT 960 methods. The median TTD of MTC isolates was 7 days for both methods (Table 2). Although similar TTD values have been reported for MGIT 960 system in other studies (2, 5), the design of our study was biased towards slower TTD by the MGIT 960 system and LJ compared to the LRPs. This was due to the fact that sputum sediments were resuspended in 3 ml, a volume larger than that recommended by the Centers for Disease Control and Prevention, and resulted in smaller starting inoculum sizes for MGIT and LJ cultures. Furthermore, the reading schedule for the three detection systems also varied from one another and thus biased the TTD. The reading schedule favored more rapid detection by the MGIT 960 system given that this system was on a continuous (hourly) reading schedule while the LRPs had fixed schedules ranging from every 2 days to every 14 days.
By combining an agent that selectively inhibits the growth of tuberculous mycobacteria with the LRP assay it is possible to rapidly and specifically identify MTC. Among the 50 specimens tested, the sensitivity of this approach was 94%, and results were available in a time frame comparable to that of the BACTEC 460 system. In three instances the system misidentified MTC as NTM due to contaminations with non-AFB. Erroneous NAP test results due to contamination have been reported in other studies (9). This problem could be reduced by excluding contaminated specimens (as determined by AFB smears and blood agar cultures) prior to conducting NAP tests. Despite the accurate performance of the NAP test, it fails to identify isolates beyond MTC and NTM, a feature that may be necessary in today's diagnostic laboratories. Thus, the LRP NAP test can only be used to provide preliminary results while awaiting further species determination by biochemical and genotypic assays.
The greatest advantage of the LRP system is its ability to rapidly and accurately perform AST with the four first-line drugs. Ninety-eight percent of the 200 LRP susceptibility tests performed with all four drugs gave identical results to those from the BACTEC 460 system. There were only three discrepant results, two in which mycobacteria were falsely assessed as resistant and one in which an isolate was falsely identified as susceptible to INH. Furthermore, results were 100% reproducible when 24 MGIT cohorts were retested with the LRPs in a blinded fashion. The median turnaround time for LRP AST was 2 days, significantly faster than that of the BACTEC 460 system (10.5 days). Compared to the BACTEC 460 system, AST with LRPs required less manual labor and monitoring. This is a simple three-step procedure in which cells are initially incubated with antibiotics, followed by phage infection and photon quantification. LRP AST can be even further simplified by adopting an automated plate luminometer that can read many susceptibility tests at once. Although a film-based approach for photographic detection of light has been developed (12), we were unsuccessful in substituting it for the luminometer (data not shown).
In conclusion, we showed that LRP-based assays can be implemented in a reference mycobacterial laboratory in a developing country. We found that if precautions are taken to minimize contamination with other bacteria, the LRPs are comparable in sensitivity, specificity, and speed to the MGIT 960 and BACTEC 460 systems. For AST the LRP-based approach is five times faster than the BACTEC 460 system, making it the fastest AST system available. In selected settings, phage-based assays may greatly increase the timely availability of diagnostic mycobacteriologic results.
ACKNOWLEDGMENTS
N.B. was supported by the Stanford Medical Scholars program. P.F.R. is supported by KO8 AI01628. This work was supported by NIH grants AI35969 and TW01135.
We thank L. B. Heifets for the conventional susceptibility tests, S. H. Siddiqi for providing reagents, E. Desmond for his support, M. Kato-Maeda for purchasing and shipping reagents, and members of the P3 laboratory at INCMNSZ (B. Chavez, A. Bautista, and N. Ortiz) for their assistance and cooperation.
REFERENCES
- 1.Espinal M A, Kim S J, Suarez P G, Kam K M, Khomenko A G, Migliori G B, Baez J, Kochi A, Dye C, Raviglione M C. Standard short-course chemotherapy for drug-resistant tuberculosis: treatment outcomes in 6 countries. JAMA. 2000;283:2537–2545. doi: 10.1001/jama.283.19.2537. [DOI] [PubMed] [Google Scholar]
- 2.Hanna B A, Ebrahimzadeh A, Elliott L B, Morgan M A, Novak S M, Rusch-Gerdes S, Acio M, Dunbar D F, Holmes T M, Rexer C H, Savthyakumar C, Vannier A M. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J Clin Microbiol. 1999;37:748–752. doi: 10.1128/jcm.37.3.748-752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heifets L B, Cangelosi G A. Drug susceptibility testing of Mycobacterium tuberculosis: a neglected problem at the turn of the century. Int J Tuberc Lung Dis. 1999;3:564–581. [PubMed] [Google Scholar]
- 4.Jacobs W R, Jr, Barletta R G, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis G J, Hatfull G F, Bloom B R. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 5.Kanchana M V, Cheke D, Natyshak I, Connor B, Warner A, Martin T. Evaluation of the BACTEC MGIT 960 system for the recovery of mycobacteria. Diagn Microbiol Infect Dis. 2000;37:31–36. doi: 10.1016/s0732-8893(99)00151-0. [DOI] [PubMed] [Google Scholar]
- 6.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 7.Kochi A, Vareldzis B, Styblo K. Multidrug-resistant tuberculosis and its control. Res Microbiol. 1993;144:104–110. doi: 10.1016/0923-2508(93)90023-u. [DOI] [PubMed] [Google Scholar]
- 8.Kubica G P, Dye W E, Cohn M L, Middlebrook G. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodiumhydroxide for culture of mycobacteria. Am Rev Respir Dis. 1963;87:775–779. doi: 10.1164/arrd.1963.87.5.775. [DOI] [PubMed] [Google Scholar]
- 9.Laszlo A, Siddiqi S H. Evaluation of a rapid radiometric differentiation test for the Mycobacterium tuberculosis complex by selective inhibition with p-nitro-α-acetylamino-β-hydroxypropiophenone. J Clin Microbiol. 1984;19:694–698. doi: 10.1128/jcm.19.5.694-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 11.Riska P F, Jacobs W R, Jr, Bloom B R, McKitrick J, Chan J. Specific identification of Mycobacterium tuberculosis with the luciferase reporter mycobacteriophage: use of p-nitro-α-acetylamino-β-hydroxypropiophenone. J Clin Microbiol. 1997;35:3225–3231. doi: 10.1128/jcm.35.12.3225-3231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riska P F, Su Y, Bardarov S, Freundlich L, Sarkis G, Hatfull G, Carriere C, Kumar V, Chan J, Jacobs W R., Jr Rapid film-based determination of antibiotic susceptibilities of Mycobacterium tuberculosis strains by using a luciferase reporter phage and the Bronx Box. J Clin Microbiol. 1999;37:1144–1149. doi: 10.1128/jcm.37.4.1144-1149.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusch-Gerdes S, Domehl C, Nardi G, Gismondo M R, Welscher H M, Pfyffer G E. Multicenter evaluation of the mycobacteria growth indicator tube for testing susceptibility of Mycobacterium tuberculosis to first-line drugs. J Clin Microbiol. 1999;37:45–48. doi: 10.1128/jcm.37.1.45-48.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timme T L, Brennan P J. Induction of bacteriophage from members of the Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum serocomplex. J Gen Microbiol. 1984;130:2059–2066. doi: 10.1099/00221287-130-8-2059. [DOI] [PubMed] [Google Scholar]
- 15.Walters S B, Hanna B A. Testing of susceptibility of Mycobacterium tuberculosis to isoniazid and rifampin by mycobacterium growth indicator tube method. J Clin Microbiol. 1996;34:1565–1567. doi: 10.1128/jcm.34.6.1565-1567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]