Abstract
The basidiomycetous yeast Cryptococcus neoformans is an important human fungal pathogen. Two varieties, C. neoformans var. neoformans and C. neoformans var. gattii, have been identified. Both are heterothallic with two mating types, MATa and MATα. Some rare isolates are self-fertile and are considered occasional diploid or aneuploid strains. In the present study, 133 isolates, mostly from Italian patients, were investigated to detect the presence of diploid strains in the Igiene Università Milano culture collection. All of the diploid isolates were further investigated by different methods to elucidate their origins. Forty-nine diploid strains were identified by flow cytometry. PCR fingerprinting using the (GACA)4 primer showed that the diploid state was associated with two specific genotypes identified as VN3 and VN4. Determination of mating type on V8 juice medium confirmed that the majority of the strains were sterile. PCR and dot blotting using the two pheromone genes (MFa and MFα) as probes identified 36 of the 49 diploid isolates as MATa/α. The results of pheromone gene sequencing showed that two allelic MFα genes exist and are distinct for serotypes A and D. In contrast, the MFa gene sequence was conserved in both serotype alleles. Amplification of serotype-specific STE20 alleles demonstrated that the diploid strains contained one mating locus inherited from a serotype A parent and one inherited from a serotype D parent. The present results suggest that diploid isolates may be common among the C. neoformans population and that in Italy and other European countries serotype A and D populations are not genetically isolated but are able to recombine by sexual reproduction.
The basidiomycetous yeast Cryptococcus neoformans is an important pathogen that causes meningitis in both immunocompromised and immunocompetent patients. At present two varieties have been recognized: C. neoformans var. neoformans (serotypes A, D, and AD) and C. neoformans var. gattii (serotypes B and C). Serotype A has recently been proposed to be separated from C. neoformans var. neoformans into a new distinct variety named C. neoformans var. grubii (3). This classification, however, has not been completely agreed upon among mycologists and needs further investigation.
Both varieties of C. neoformans are heterothallic with two mating types, MATa and MATα. Mating type α was found to be more frequent than MATa among clinical as well as environmental isolates (6). In addition, some rare isolates were identified as MATa/α and were considered occasional diploid or aneuploid strains (7, 8). Later, a self-fertile progeny obtained by crossing two haploid strains was demonstrated to be diploid by fluorescence microscopy (18). More recently, a serotype A strain and a serotype D strain were crossed and serotype AD progeny were recovered (13). All of the serotype AD strains isolated were shown to be diploid by flow cytometry analysis, and some of them were self-fertile. Previously, the same group reported a high incidence of diploidy in Japanese natural isolates (12). In another study, many clinical isolates, identified as having VN3 and VN4 genotypes, were found to be diploid (2). These genotypes presented PCR fingerprinting banding patterns intermediate between those of VN1 and VN6, which were always associated with serotypes D and A, respectively. Finally, a 33% frequency of genotypes VN3 and VN4 was reported in Europe from a recent epidemiological survey of cryptococcosis (16). These last results suggest that diploid isolates may be common among the C. neoformans population.
In the present study, a large number of clinical isolates, mostly from Italian patients, were investigated in order to detect the presence of diploid strains in a representative sample of the Igiene Università Milano (IUM) culture collection. In addition, the origins of the strains identified as diploid were investigated, and the mating types were determined by crossing on V8 juice medium, PCR of MF and STE20 genes, dot blot hybridization, and sequencing of MFa and MFα genes.
MATERIALS AND METHODS
Isolates.
One hundred twenty-two isolates of C. neoformans var. neoformans from the IUM culture collection (Istituto di Igiene e Medicina Preventiva, Università degli Studi di Milano, Milan, Italy) were studied. One hundred twenty strains were isolated from Italian patients before 1997 and two were isolated from the environment (Table 1). Among these strains, 13 were included because they were serotype AD. The remaining isolates were chosen randomly among the strains serotyped as A (43 isolates) and D (68 isolates). Six additional AD strains were kindly supplied by Danielle Swinne (Institut de Médicine Tropical, Antwerp, Belgium) and included in the present study. Five strains were used as reference serotypes: NIH B4500 and NIH B4476 as references for serotype D MATα and serotype D MATa, respectively; H99 and CDC B551 as references for serotype A; and CBS 132 as a reference for serotype AD. Saccharomyces cerevisiae DKY1 and Candida albicans SC5314 were used as negative controls. Serotyping, genotyping, determination of DNA content, detection of pheromone genes, and mating on V8 juice medium were performed for all of the isolates. Dot blot hybridization, sequencing, and PCR of STE20 genes were performed only for diploid strains and some controls.
TABLE 1.
C. neoformans var. neoformans strains studieda
| Geographical origin | No. of strains
|
|||
|---|---|---|---|---|
| Total | Clinical
|
Environmental | ||
| HIVb positive | HIV negative | |||
| Italyc | 122 | 107 | 13 | 2 |
| Belgiumd | 4 | 2 | 2 | |
| Africad | 2 | 2 | ||
The reference strains (NIH B4476, NIH B4500, CDC B551, H99, and CBS 132) are not included
HIV, human immunodeficiency virus.
Istituto di Igiene e Medicina Preventiva, Università degli Studi di Milano, Milan Italy (IUM collection; strains isolated before 1997).
Raymond Vanbreuseghem, Institut de Médicine Tropical, Anvers, Belgium (RV collection).
Serotyping.
Serotyping was performed by the slide agglutination test using the Crypto Check Kit (Iatron Labs, Tokyo, Japan). Before each assay, the strains were streaked for isolation, and then a single colony was selected for analysis.
Genotyping by PCR fingerprinting.
Genomic DNA was extracted as previously described (17), and PCR conditions were as follows. The reaction mixture consisted of 100 pmol of the repetitive oligonucleotide (GACA)4 as a single primer; 400 μM (each) dATP, dCTP, dGTP, and dTTP (Boehringer Mannheim GmbH, Mannheim, Germany); 3 mM MgCl2 (Applied Biosystems, Monza, Italy); 10× reaction buffer (500 mM KCl, 100 mM Tris-HCl, pH 8.3) (Applied Biosystems); 2.5 U of AmpliTaq DNA (Applied Biosystems); and 400 ng of DNA sample. PCR was performed in a GeneAmp PCR system 2400 thermal cycler (Applied Biosystems) with an initial cycle of 5 min at 94°C; 38 cycles of 30 s at 94°C, 30 s at 47°C, and 1 min at 72°C; and a final cycle of 5 min at 72°C. The amplification products were visualized by electrophoresis on 1.4% agarose gels in 1× TBE (0.089 M Trizma base, 0.089 M boric acid, 0.002 M EDTA, pH 8.4) (Sigma-Aldrich, Milan, Italy) at 40 V for 17 h and stained with ethidium bromide (Sigma-Aldrich).
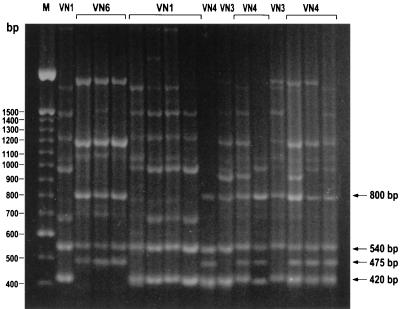
The PCR fingerprinting genotypes, VN1, VN3, VN4, and VN6, were identified according to the different combinations of four major bands (800, 540, 475, and 420 bp) as described previously (2, 17).
DNA content determination.
DNA content was determined by flow cytometry according to the procedure described by Tanaka et al. (14). NIH B4476 and NIH B4500 were used as haploid reference strains. Strains from a 2-day culture at 35°C on YPG (yeast extract, 1%; Bacto peptone, 2%; and glucose, 2%) agar were inoculated into 20 ml of YPG broth. Cells (107/ml) were harvested, washed in distilled water, and fixed in 70% ethanol overnight at 4°C. The cells were then washed, stained with propidium iodide (10 μg/ml) (Sigma-Aldrich) dissolved in NS buffer (10 mM Tris-HCl [pH 7.2] 0.25 M sucrose, 1 mM EDTA, 1 mM MgCl2, 0.1 mM CaCl2, 0.1 mM ZnCl2, 0.4 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol) containing RNase (1 mg/ml) (Sigma-Aldrich), and incubated at 37°C for 90 min. Stained cells were then checked with an Axioskop fluorescence microscope (Zeiss, Oberkochen, Germany). Before cytofluorimetric analysis, cells were sonicated with a homogenizer (Vibracell Sonics & Materials, Danbury, Conn.) for 10 s. Flow cytometry was performed by counting 10,000 cells using a cytofluorimeter (Becton Dickinson, Meylan, France) at 488 nm.
PCR to determine mating type.
The primer sequences for the MFα1 fragment (accession no. S56460) were 5′-ATGGACGCCTTCACTGCCATC-3′ (forward) and 5′-GGCGATGACACAAAGGGTCATG-3′ (reverse). This pair amplifies 113 bp of the MFα coding region. PCRs were performed using Taq polymerase (Gibco/BRL, Gaithersburg, Md.) in a PTC-100 thermocycler (MJ Research, Inc., Watertown, Mass.) with an initial denaturation step of 94°C for 1 min and then 30 cycles of denaturation at 94°C for 40 s, annealing at 64°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 5 min. A 128-bp MFa1 PCR product (accession no. AF305931) was obtained using 5′-ATGGACGCCTTCACTGCTATC-3′ as the forward primer and 5′-TTAAGCAATAACGCAAGAGTAAGT-3′ as the reverse primer. PCR conditions were the same as for MFα except that the annealing temperature was 62°C. STE20a (serotype A and D alleles) and STE20α (serotype A and D alleles) genes were amplified as described elsewhere (10). PCR products were separated on a 1.5% agarose gel.
Sequencing of MFα and MFa genes.
The MFα and MFa genes in 42 diploid and 24 haploid strains were sequenced (see Table 4). Genomic DNA was first amplified as described above, and then the PCR products were purified and sequenced with an ABI Prism 310 automated sequencer (Applied Biosystems) using Big Dye terminators (Applied Biosystems). The following primers were used: 5′-CGCCTTCACTGCTACCTTCT-3′ and 5′-AACGCAAGAGTAAGTCGGGC-3′ to sequence the forward and reverse strands of the MFa gene and 5′-ATGGACGCCTTCACTGCCATC-3′ and 5′-GGCGATGACACAAAGGGTCATG-3′ to sequence the forward and reverse strands of the MFα gene.
TABLE 4.
PCR, sequencing, and dot blotting results
| Straina | Serotypeb | Genotypec | Ploidyd | MFa genee | MFα genee | STE20af | STE20αf | MFα sequenceg | Dot bloth |
|---|---|---|---|---|---|---|---|---|---|
| CDC B551 | A | VN6 | Haploid | − | + | NDi | ND | 1 | ND |
| IUM 91-4437 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 94-3443 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 91-5650 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 89-6538 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 87-3588 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 94-5982 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 86-0912 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 90-2798 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 92-4755 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 91-6422 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 95-2499 | A | VN6 | Haploid | − | + | ND | ND | 1 | ND |
| IUM 94-5984 | A | VN6 | Diploid | − | + | n | A | 1 | C6 |
| IUM 95-0552 | A | VN6 | Diploid | − | + | n | A | 1 | C7 |
| IUM 89-6639 | A | VN6 | Diploid | − | +j | n | A | 1 | E2 |
| IUM 88-1358 | AD | VN4 | Diploid | +k | + | D | A | 1 | A4 |
| IUM 89-4797 | A | VN4 | Diploid | +k | + | D | A | 1 | A7 |
| IUM 90-7031 | AD | VN4 | Diploid | +k | + | D | A | 1 | A11 |
| IUM 91-1871 | AD | VN4 | Diploid | +k | + | D | A | 1 | B2 |
| IUM 91-5480 | AD | VN4 | Diploid | +k | + | D | A | 1 | B3 |
| IUM 92-2562 | D | VN4 | Diploid | +k | + | D | A | 1 | B5 |
| IUM 92-4734 | A | VN4 | Diploid | +k | + | D | A | 1 | B8 |
| IUM 93-1666 | D | VN4 | Diploid | +k | + | D | A | 1 | B10 |
| IUM 93-1667 | AD | VN4 | Diploid | + | + | D | A | 1 | B11 |
| RV 53164 | AD | VN4 | Diploid | +k | + | D | A | 1 | C1 |
| IUM 94-3009 | D | VN4 | Diploid | +k | + | D | A | 1 | C3 |
| IUM 94-5754 | AD | VN4 | Diploid | +k | + | D | A | 1 | C4 |
| RV 53565 | AD | VN4 | Diploid | +k | + | D | A | 1 | C5 |
| IUM 95-1584 | AD | VN4 | Diploid | − | + | n | A | 1 | C8 |
| IUM 98-3350 | AD | VN4 | Diploid | +k | + | D | A | 1 | D1 |
| IUM 98-3357 | D | VN4 | Diploid | +k | + | D | A | 1 | D3 |
| RV 29192 | AD | VN4 | Diploid | − | + | n | A | 1 | D7 |
| IUM 98-3351 | AD | VN3 | Diploid | +k | + | D | A | 1 | D2 |
| IUM 98-3359 | D | VN3 | Diploid | +k | + | D | A | 1 | D4 |
| IUM 98-3362 | A | VN3 | Diploid | +k | + | D | A | 1 | D5 |
| RV 52755 | AD | VN3 | Diploid | − | + | n | A | 1 | D9 |
| CBS 132 | AD | VN3 | Diploid | +jk | + | D | A | 1 | E1 |
| IUM 87-2003 | U | VN4 | Diploid | +k | + | A | D | 2 | A3 |
| IUM 89-0130 | D | VN4 | Diploid | +k | + | A | D | 2 | A5 |
| IUM 89-0384 | D | VN4 | Diploid | − | + | n | D | 2 | A6 |
| IUM 88-6669 | U | VN4 | Diploid | +k | + | A | D | 2 | A8 |
| IUM 92-2736 | D | VN4 | Diploid | +k | + | A | D | 2 | B6 |
| RV 28949 | AD | VN4 | Diploid | − | + | n | D | 2 | D6 |
| IUM 90-0243 | D | VN3 | Diploid | + | + | A | D | 2 | A9 |
| IUM 90-5644 | D | VN3 | Diploid | + | + | A | D | 2 | A10 |
| IUM 91-0804 | AD | VN3 | Diploid | + | + | A | D | 2 | A12 |
| IUM 92-1077 | D | VN3 | Diploid | + | + | A | D | 2 | B4 |
| IUM 93-1920 | D | VN3 | Diploid | +k | + | A | D | 2 | B12 |
| IUM 95-4426 | A | VN3 | Diploid | +k | + | A | D | 2 | C9 |
| IUM 95-5227 | AD | VN3 | Diploid | + | + | A | D | 2 | C10 |
| IUM 95-6038 | D | VN3 | Diploid | + | + | A | D | 2 | C11 |
| IUM 96-4769 | D | VN3 | Diploid | − | + | n | D | 2 | C12 |
| IUM 84-3040 | AD | VN4 | Diploid | +k | − | D | n | ND | A1 |
| RV 33370 | AD | VN4 | Diploid | +k | + | D | A | ND | D8 |
| IUM 91-0835 | D | VN3 | Diploid | + | + | A | D | ND | B1 |
| IUM 92-4957 | D | VN3 | Diploid | + | + | A | D | ND | ND |
| IUM 94-1912 | AD | VN3 | Diploid | + | − | A | n | ND | C2 |
| IUM 85-2574 | D | VN4 | Diploid | − | − | ND | ND | ND | ND |
| IUM 86-3543/2 | D | VN1 | Diploid | +k | − | D | n | ND | A2 |
| IUM 92-4686 | D | VN1 | Diploid | +k | + | A | D | 2 | B7 |
| IUM 92-6198 | D | VN1 | Diploid | + | + | A | D | 2 | B9 |
| NIH B4476 | D | VN1 | Haploid | + | − | D | n | 2 | E3 |
| NIH B4500 | D | VN1 | Haploid | − | + | n | D | 2 | E4 |
| IUM 93-1545 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 89-4478 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 93-3231 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 92-6093 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 88-3921 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 89-0469 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 90-2877 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 91-6367 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 93-0323 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| IUM 94-0047 | D | VN1 | Haploid | − | + | ND | ND | 2 | ND |
| C. albicans SC5314 | − | − | ND | ND | ND | E5 | |||
| S. cerevisiae DKY1 | − | − | ND | ND | ND | E6 |
IUM, Istituto di Igiene e Medicina Preventiva, Università degli Studi di Milano, Milan, Italy; CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; NIH, National Institutes of Health, Bethesda, Md.; RV, Raymond Vanbreuseghem, Institut de Médicine Tropical, Anvers, Belgium; CDC Centers for Disease Control and Prevention, Atlanta, Ga.
Determined by the Crypto Check slide agglutination test. U, untypeable.
Determined by PCR fingeprinting using the (GACA)4 primer.
Determined by flow cytometry.
+, gene amplified by PCR and hybridized by dot blotting; −, gene not amplified by PCR and not hybridized by dot blotting.
A, amplification of serotype A allele; D, amplification of serotype D allele; n, not amplified.
1, sequence associated with serotype A, genotype VN6; 2, sequence associated with serotype D, genotype VN1.
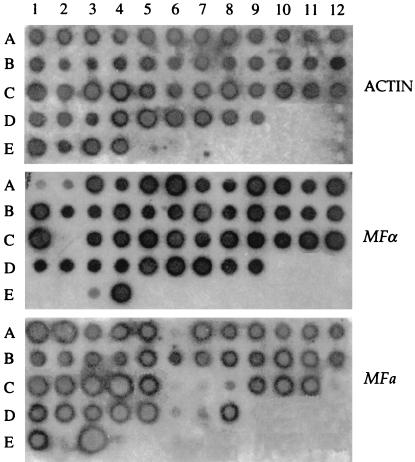
Coordinates of the strains tested by dot blotting and reported in Fig. 3.
ND, not done.
Gene amplified by PCR but not detected by dot blotting. Repeat hybridizations confirmed that the classifications by PCR were correct.
Strain in which the MFa gene was sequenced.
Genomic DNA minipreparations.
DNA preparations for dot blots were made using the bead beating method. Cells were grown on a YPD plate for 1 to 3 days and then streaked onto a new plate. The plate was incubated for 16 h at 30°C. Cells were scraped from the plates and placed in Eppendorf tubes, on ice, containing 1.0 ml of distilled water at one-half plate per tube. After being washed twice, the cells were resuspended in 0.4 ml of cold breaking buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]). The suspension was transferred to a 2.0-ml screw cap tube (Fisher Scientific, Pittsburgh, Pa.), on ice, containing 0.4 ml (wt/vol) of 0.5-mm-diameter glass beads (Biospec Products, Bartlesville, Okla.) and 0.4 ml of phenol-chloroform-isoamyl alcohol. Tubes were vortexed on a standard hand vortexer (Vortex Genie; Fisher Scientific) at the highest setting for 2 min.
The tubes were then centrifuged for 5 min. The supernatant was removed to a new tube and precipitated with 2 volumes of room temperature 100% ethanol. The DNA was pelleted, washed twice in cold 80% ethanol, and dried lightly. The pellets were dissolved in 100 μg of RNase per ml and incubated at 65°C for 2 h. Following this incubation period, samples were extracted one time with chloroform-isoamyl alcohol, made 0.1 M in NaCl, and precipitated with 2 volumes of 100% ethanol by incubation at −20°C for 0.5 to 1 h. Pellets were washed as before, dried lightly, and then resuspended in 200 μl of Tris-EDTA. DNA was quantitated by UV absorbance and confirmed to be high molecular weight by gel electrophoresis.
Dot blot hybridizations.
Dot blotting was performed in a Bio-Dot apparatus (Bio-Rad, Richmond, Calif.) on nylon membranes using 2.5 μg of each sample per dot according to the instructions of the manufacturer. Southern hybridizations were performed according to the instructions of the manufacturer (Schleicher and Schuell, Keene, N.H.). Probes were prepared from PCR products derived from the actin gene (accession no. U10867), which was used as a control, and the MFa1 and MFα1 genes. The 1.3-kb actin gene fragment was amplified as described above using an annealing temperature of 56°C and 5′-ATGGAAGAAGAAGGTACGTTC-3′ (forward) and 5′-TTAGAAACACTTTCGGTGGAC-3′ (reverse) oligonucleotides as primers. Probes were prepared from PCR products recovered from gels by glass milk purification (Elu-Quik; Schleicher and Schuell). Fragments were labeled by random priming (High Prime; Boehringer Mannheim, Indianapolis, Ind.) with [α-32P]dCTP (NEN Life Science Products, Boston, Mass.) according to the manufacturer's instructions.
Mating on V8 juice medium.
Each isolate was suspended in distilled water (about 108.cells/ml) and streaked on the agar surface of a plate containing V8 juice medium (9) by three patches. The first patch was streaked with a loopful of the test strain and a loopful of MATa reference strain, the second patch was streaked with a loopful of the test strain and a loopful of MATα reference strain, and the third patch was streaked with a loopful of the test strain as a control. The plates were then incubated at 24°C for 3 to 4 weeks and observed macro- and microscopically weekly. Mating types were identified by development of hyphae with clamp connections and basidiospores.
Nucleotide sequence accession numbers.
The sequences of MFα1A, MFα1D, and MFa have been deposited in GenBank with accession numbers AF376990 to AF377026, AF376963 to AF376989, and AF377027 to AF377054, respectively.
RESULTS
PCR fingerprinting.
The serotypes and genotypes of the 133 C. neoformans strains studied are reported in Table 2 and Fig. 1. Thirty-nine of the 43 serotype A isolates were identified as genotype VN6, while four had the intermediate genotype VN3 or VN4. Similarly, most of the serotype D isolates (51 strains) were identified as genotype VN1; however, 17 had the VN3 or VN4 genotype. All of the serotype AD strains and the two untypeable strains were genotyped as VN3 or VN4.
TABLE 2.
Distribution among genotypes of the 133 C. neoformans var. neoformans strains selected for the study, including the reference strains
| Serotype | No. of strains | No. with genotype:
|
|||
|---|---|---|---|---|---|
| VN1 | VN3 | VN4 | VN6 | ||
| A | 43 | 2 | 2 | 39 | |
| AD | 20 | 6 | 14 | ||
| D | 68 | 51 | 9 | 8 | |
| Ua | 2 | 2 | |||
U, untypeable.
FIG. 1.
PCR fingerprints of 16 representative C. neoformans var. neoformans strains. The four major bands are indicated by the arrows, and their different combination recognize the four genotypes: VN1 (540 and 420 bp), VN3 (800, 540, and 420 bp), VN4 (800, 540, 475, and 420 bp), and VN6 (800, 540, and 475 bp). Lane M, 100-bp marker (GIBCO Life Technologies, Milan, Italy).
Flow cytometry analysis.
The cytofluorimetric determination of the DNA contents of the haploid reference strains (NIH B4476 and NIH B4500) yielded a graphic representation (profile R [white] in Fig. 2) with two peaks, one centered around 200 (G1 phase) and one centered around 400 (G2 phase). The graphic of the 131 isolates studied was compared with those of the haploid reference strains. Eighty-two strains had the same DNA amount and the same profile as the reference strains, whereas 49 strains had twice as much DNA and had the G1 peak overlapping the G2 peak of the reference strains (profile 2R [black] in Fig. 2). All 43 isolates genotyped as VN3 and VN4, including CBS 132, showed the 2R profile, as did 3 strains each among VN1 and VN6 isolates (Table 3).
FIG. 2.
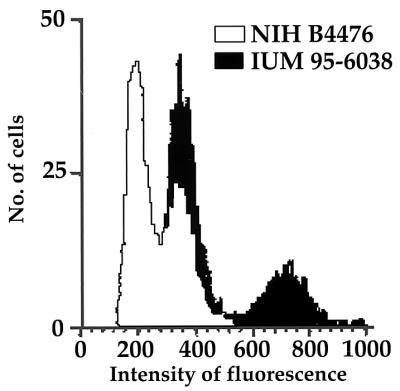
Flow cytometric graphics of two representative isolates showing profile R (white) and profile 2R (black).
TABLE 3.
Comparison of genotypes and ploidies of the 133 C. neoformans var. neoformans strains studied, including reference strains
| Genotype | No. of strains | No. with ploidy:
|
|
|---|---|---|---|
| Haploid | Diploid | ||
| VN1 | 51 | 48 | 3 |
| VN3 | 17 | 17 | |
| VN4 | 26 | 26 | |
| VN6 | 39 | 36 | 3 |
PCR and dot blot results.
Both PCR and dot blot hybridization of the two pheromone genes yielded the same results reported in Tables 4 and 5 and Fig. 3. Thirty-six isolates previously identified as diploid by flow cytometry were confirmed to be MATa/α. In contrast, only one of the two pheromone genes was detected in 12 diploid strains; 9 showed only the α pheromone, and 3 showed only the a pheromone.
TABLE 5.
Mating types of the 133 C. neoformans var. neoformans isolates, including reference strains, determined by crossing on V8 juice medium and PCR of the pheromone genes
| Genotype | No. of strains | No. with the indicated mating typea determined by:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Crossing on V8 juice medium
|
PCR
|
|||||||
| a | α | nM | a | α | a/α | N | ||
| VN1 | 51 | 2 | 5 | 44 | 2 | 47 | 2 | |
| VN3 | 17 | 2 | 15 | 1 | 2 | 14 | ||
| VN4 | 26 | 4 | 22 | 1 | 4 | 20 | 1 | |
| VN6 | 39 | 14 | 25 | 39 | ||||
nM, nonmater; N, no amplification.
FIG. 3.
Dot blot hybridization of 47 C. neoformans diploid isolates, two haploid reference strains, and two negative controls, C. albicans and S. cerevisiae. The coordinates of the isolates tested are reported in Table 4. Strains in the E1 and E2 wells hybridized to both MFa and MFα in a repeated test, confirming results obtained by PCR.
Table 4 lists the results of STE20a and STE20α amplifications. Thirty-four VN3 and VN4 diploid strains, including reference strain CBS 132, were heterozygous for STE20a and STE20α. Twenty strains were serotype D STE20a/serotype A STE20α, while 14 isolates were serotype A STE20a/serotype D STE20α. Eight strains lacked the STE20α or STE20a gene.
Only the serotype A STE20α sequence was amplified in the VN6 diploid isolates, whereas one of the three VN1 diploid strains was positive only for the serotype D STE20a sequence. The two remaining VN1 diploid strains were serotype A STE20a/serotype D STE20α heterozygotes.
Pheromone gene sequence analysis.
The sequence analysis of the MFα gene showed two sequences, one associated with serotype A, genotype VN6 isolates (MFα1A) and one associated with serotype D, genotype VN1 isolates (MFα1D). The two sequences differed in eight nucleotides. When the MFα genes of VN3 and VN4 strains were sequenced, 22 isolates were found to contain the MFα1A sequence and 15 strains were found to contain the MFα1D sequence (Table 4). The diploid isolates that had the MFα1A sequence were always linked to serotype A STE20α, whereas those that contained the MFα1D sequence were always linked to serotype D STE20α. However, the MFa sequence determined for 28 isolates (Table 4) was conserved regardless of the serotype from which the MATa locus was derived.
Mating type on V8 juice medium.
The results obtained from mating on V8 juice medium are reported in Table 5. One hundred six isolates did not mate, 25 isolates were identified as mating type α, and only 2 strains were mating type a (both groups included the reference strains). None of the strains mated as both a and α, nor were any self-fertile on V8 juice agar.
DISCUSSION
The present study further contributes to the characterization of C. neoformans var. neoformans haploid strains and to the understanding of the origin of C. neoformans var. neoformans diploid isolates. Genotype VN1 was always associated with serotype D, whereas genotype VN6 was associated with serotype A. Genotypes VN3 and VN4, which display a PCR fingerprinting banding pattern intermediate between those of genotypes VN1 and VN6, were always associated with the diploid state. All serotype AD isolates were included in this last group. Additionally, 23 strains serotyped as A or D or untypeable were genotyped as VN3 or VN4. As previously shown (17), genotyping is more reproducible and reliable than serotyping in identifying the intermediate strains. An example is provided by the genotype reproducibility for the CBS 132 isolate, which was always genotyped as VN3 (references 2 and 17 and the present study), while serotyping results differed in various laboratories (3, 4, 10).
The diploid state of 36 isolates was also confirmed by PCR and dot blot hybridization, which detected both pheromone genes (MFa and MFα). In contrast, only one of the two pheromone genes was detected in the remaining 12 isolates shown to be diploid by flow cytometry. These results support the hypothesis that the diploid isolates identified as MATa/α originated by a sexual event. In addition, the results of STE20a and STE20α gene amplifications demonstrated that VN3 and VN4 diploid isolates originated from the crossing of serotype A, genotype VN6 and serotype D, genotype VN1 haploid strains and confirmed that the STE20a and STE20α genes are linked to the MFa and MFα genes, respectively (5). The same conclusion was reported by other authors (10), who tested 10 serotype AD isolates. They amplified several genes which showed different alleles for serotypes A and D and found that all of the strains were heterozygous for most of the genes studied.
The MFα and MFa gene sequences determined in the present study are in agreement with those reported elsewhere (1), confirming the polymorphism of MFα. The linkage of MFα1A to serotype A STE20α and of MFα1D to serotype D STE20α suggests that the two MFα alleles are really associated with serotypes A and D, respectively. In accordance with previous studies (8), self-fertile diploid strains were shown to be able to produce MATa/α as well as MATα diploid progeny (13). The 12 diploid isolates in the present study which show only one pheromone gene and one STE20 gene (Table 4) might represent the progeny of self-fertile isolates. Alternatively, it could be that the primers and probes used in this study were not able to amplify or hybridize to the diverged mating type sequences, or it simply could be that one of the two mating loci was lost during the meiotic process. A further possible explanation is that these isolates are homozygous for one mating locus (a/a or α/α).
The two diploid VN1 isolates (IUM 92-4686 and IUM 92-6198), identified as hybrids of serotypes A and D, were not recognized as either VN3 or VN4 genotypes. However, a detailed analysis that considered all of the PCR fingerprinting bands (data not shown) revealed that these isolates are more related to the hybrid group (VN3 and VN4) than to the VN1 group.
The fertility of all of the diploid isolates was tested on V8 juice medium, but the majority of the strains (106 of 133) did not mate. The frequent subculturing of these strains may be the cause of sterility, as previously suggested (8). Alternatively, the presence of both mating type loci in the same cell may inhibit mating, or the tester strain may simply not be compatible. However, 18 of the 25 self-sterile isolates were MATα and haploid, whereas 7 strains were diploid. Four of these last strains contained both pheromone genes as determined by PCR and dot blotting, and three strains, despite being diploid, were shown to contain only the MFα gene.
The present study identified the presence of the serotype A MATa locus in many diploid strains, supporting the hypothesis that serotype A MATa haploid strains might not have been extinct. This hypothesis was recently confirmed by two reports concerning the isolation of C. neoformans serotype A MATa strains (11, 15).
Finally, the high percentage of VN3 and VN4 hybrid strains reported by the European Confederation of Medical Mycology epidemiological survey of cryptococcosis (16) suggests that in Italy and other European countries serotype A and D populations are not genetically isolated but are able to recombine by sexual reproduction. This is in contrast to the hypothesis that serotype A and D populations are diverging towards a clonal evolution (3) and suggests that they are members of a unique variety, C. neoformans var. neoformans.
ACKNOWLEDGMENTS
This work was supported in part by a grant from IRCCS Ospedale Maggiore di Milano (grant RC 1997, 500/02) and by MURST 60%, 2000. Brian L. Wickes is a Burroughs-Wellcome New Investigator in Molecular Pathogenic Mycology and is supported by Public Health Service grant R29AI43522 from the National Institutes of Health.
We thank Danielle Swinne for the C. neoformans strains from the RV collection, John Perfect for supplying the C. neoformans H99 strain, Dave Kolodrubetz for the S. cerevisiae DKY1 strain, and Bill Fonzi for the C. albicans SC5314 strain.
REFERENCES
- 1.Chaturvedi S, Rodeghier B, Fan J, McClelland C M, Wickes B L, Chaturvedi V. Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000;38:2007–2009. doi: 10.1128/jcm.38.5.2007-2009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cogliati M, Allaria M, Liberi G, Tortorano A M, Viviani M A. Sequence analysis and ploidy determination of Cryptococcus neoformans var. neoformans. J Mycol Med. 2000;10:171–176. [Google Scholar]
- 3.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal state for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J Clin Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karos M, Chang Y C, McClelland C M, Clarke D L, Fu J, Wickes B L, Kwon-Chung K J. Mapping of the Cryptococcus neoformans MATα locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs. J Bacteriol. 2000;182:6222–6227. doi: 10.1128/jb.182.21.6222-6227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon-Chung K J, Bennett J E. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 7.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea & Febiger; 1992. pp. 430–431. [Google Scholar]
- 8.Kwon-Chung K J. Proceedings of the Fourth International Conference on Mycoses. Pan American Health Organization Scientific Publication no. 356. Washington, D.C.: Pan American Health Organization; 1978. Heterothallism vs. self-fertile isolates of Filobasidiella neoformans (Cryptococcus neoformans) pp. 204–213. [Google Scholar]
- 9.Kwon-Chung K J. Mating procedures in Cryptococcus neoformans (Filobasidiella neoformans) In: Maresca B, Kobayashi G S, editors. Molecular biology of pathogenic fungi. New York, N.Y: Telos Press; 1994. pp. 341–344. [Google Scholar]
- 10.Lengeler K B, Cox G M, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun. 2001;69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengeler K B, Ping W, Cox G M, Perfect J R, Heitman J. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc Natl Acad Sci USA. 2000;97:14455–14460. doi: 10.1073/pnas.97.26.14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeo K, Tanaka R, Taguchi H, Nishimura K. Analysis of ploidy and sexual characteristics of natural isolates of Cryptococcus neoformans. Can J Microbiol. 1993;39:958–963. doi: 10.1139/m93-144. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka R, Nishimura K, Miyaji M. Ploidy of serotype AD strains of Cryptococcus neoformans. Jpn J Med Mycol. 1999;40:31–34. doi: 10.3314/jjmm.40.31. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka R, Taguchi H, Takeo K, Miyaji M, Nishimura K. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol. 1996;34:299–301. [PubMed] [Google Scholar]
- 15.Viviani M A, Esposto M C, Cogliati M, Montagna M T, Wickes B L. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Med Mycol. 2001;39:383–386. doi: 10.1080/mmy.39.5.383.386. [DOI] [PubMed] [Google Scholar]
- 16.Viviani M A, Swinne D A, et al. Survey of cryptococcosis in Europe. The ECMM working group report. Rev Iberoam Micol. 2000;17:S115. [Google Scholar]
- 17.Viviani M A, Wen H, Roverselli A, Caldarelli-Stefano R, Cogliati M, Ferrante P, Tortorano A M. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J Med Vet Mycol. 1997;35:355–360. [PubMed] [Google Scholar]
- 18.White C W, Jacobson E S. Occurrence of diploid strains of Cryptococcus neoformans. J Bacteriol. 1985;161:1231–1232. doi: 10.1128/jb.161.3.1231-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]