Abstract
Outer membrane vesicles (OMVs) are released from the outer membranes of Gram-negative bacteria during infection and modulate host immunity during host–pathogen interactions. The mechanisms by which OMVs are perceived by plants and affect host immunity are unclear. Here, we used the pathogen Xanthomonas campestris pv. campestris to demonstrate that OMV–plant interactions at the Arabidopsis thaliana plasma membrane (PM) modulate various host processes, including endocytosis, innate immune responses, and suppression of pathogenesis by phytobacteria. The lipid phase of OMVs is highly ordered and OMVs directly insert into the Arabidopsis PM, thereby enhancing the plant PM’s lipid order; this also resulted in strengthened plant defenses. Strikingly, the integration of OMVs into the plant PM is host nanodomain- and remorin-dependent. Using coarse-grained simulations of molecular dynamics, we demonstrated that OMV integration into the plant PM depends on the membrane lipid order. Our computational simulations further showed that the saturation level of the OMV lipids could fine-tune the enhancement of host lipid order. Our work unraveled the mechanisms underlying the ability of OMVs produced by a plant pathogen to insert into the host PM, alter host membrane properties, and modulate plant immune responses.
The outer membrane vesicles from bacterial pathogens manipulate plant immunity by entering plants via plant nanodomains and altering the lipid order of the plant plasma membrane.
Introduction
Extracellular vesicle secretion has been described in almost all kingdoms and has recently been recognized as an important channel for interspecies and interkingdom interactions. In Gram-negative bacteria, these extracellular vesicles are often referred to as outer membrane vesicles (OMVs), which are membranous particles that pinch off from the bacterial outer membrane during normal growth (Dorward and Garon, 1990). OMV production occurs during both in vitro growth and host infection. OMVs are an integral part of biofilms in important human pathogens, such as Pseudomonas aeruginosa and Helicobacter pylori, and OMV production also correlates with their fitness and ability to form biofilms (Schooling and Beveridge, 2006; Yonezawa et al., 2009). In addition to their functions in bacterial processes, OMV production is crucial for microbial virulence, as OMVs modulate host immune responses during colonization of the hosts by human or plant pathogens.
The OMV surface and lumen carry a wide variety of lipids and other bacterial products, such as cell wall-degrading enzymes, quorum-sensing signals, hyper-antigenic lipopolysaccharides molecules, peptidoglycan, toxins, and intracellular and outer membrane proteins, together with various components of the periplasmic space (Sidhu et al., 2008; Chowdhury and Jagannadham, 2013; Solé et al., 2015; Feitosa-Junior et al., 2019). Several OMV-triggered host immune responses were reported to occur during bacterial invasion, and they favored either bacterial pathogenesis or activation of host immune responses (Bahar et al., 2016; Chen et al., 2018). The biological significance of bacterial OMVs has been studied extensively for animal-pathogenic bacteria (Cecil et al., 2019; Nagakubo et al., 2020). For instance, in Neisseria gonorrheae, the major porin complex protein PorB is transported into host cells and targeted to the host cell mitochondrial membrane via OMVs, where this complex contributes to macrophage apoptosis in a time-dependent manner to suppress innate immunity (Deo et al., 2018). The stability of OMVs provides bacterial pathogens with unique access to long-distance transport of their virulence factors into host cells without the need for direct contact with targeted tissues (Bomberger et al., 2009).
Much less is known about the mechanistic interactions between OMVs from phytopathogenic bacteria and plant cells. OMVs produced by phytopathogenic bacteria can contain as many components as OMVs from their animal-pathogenic counterparts (Sidhu et al., 2008; Bonnington and Kuehn, 2014; Solé et al., 2015; Feitosa-Junior et al., 2019) and play significant roles in the bacterial life cycle inside host plants (Ionescu et al., 2014). For example, Xanthomonas campestris pv. campestris (Xcc) OMVs can induce defense gene responses in the plant host, and the immune coreceptors BRI1-ASSOCIATED KINASE 1 (BAK1) and SUPPRESSOR OF BIR1-1 (SOBIR1) are involved in this response (Bahar et al., 2016). Recently, two independent works reported the immunotriggering properties of Pseudomonas syringae pv. tomato DC3000 OMVs and OMV-induced protection against the bacterium, as well as against a fungal pathogen (Janda et al., 2021; McMillan et al., 2021). However, it is unclear whether OMVs interact with plant cells beyond the cell wall or plasma membrane (PM) barrier.
OMVs from human pathogenic bacteria can insert into host PMs through lipid rafts/nanodomains, which are specialized regions in the membrane highly enriched in ordered lipids, such as sphingolipids and sterol; the OMVs are internalized via clathrin-mediated endocytosis (CME; Bomberger et al., 2009; O’Donoghue et al., 2017). Plants also have lipid rafts/nanodomains, which contain raft-associated proteins, such as members of the remorin family (Jarsch et al., 2014; Gouguet et al., 2020). This raises the possibility that the integration and delivery of phytopathogen-derived OMVs could occur in a similar manner in plants and animals.
In this study, we focused on the mechanisms by which the OMVs initially integrate into the plant PM and how host integration affects plant immune responses, using the black-rot pathogen Xcc. We used mutants of Arabidopsis thaliana with varying levels of surface nanodomain assembly and performed pharmacological studies on the plant host and coarse-grained molecular dynamics (MD) simulations of the lipid bilayers to dissect the interaction between OMV membranes and plant PMs upon contact and identify the effects of OMVs on various biological activities of the plant PM. Our results supported the hypothesis that Xcc OMV insertion into the plant PM alters the membrane’s physicochemical properties, thereby altering the MD and membrane organization of the plant PM. In addition, microscopy revealed that nanodomain assembly mediates the initial fusion between OMVs and the plant PM, the recruitment of OMVs to nanodomain sites marked by remorins during infection, and the enhanced plant defense responses induced by OMV. Therefore, our work emphasizes not only the functions of remorins and nanodomains but also the various lipid–lipid and lipid–protein interactions occurring between the host and pathogens during infection.
Results
Xcc OMVs directly insert into the Arabidopsis PM but are not internalized.
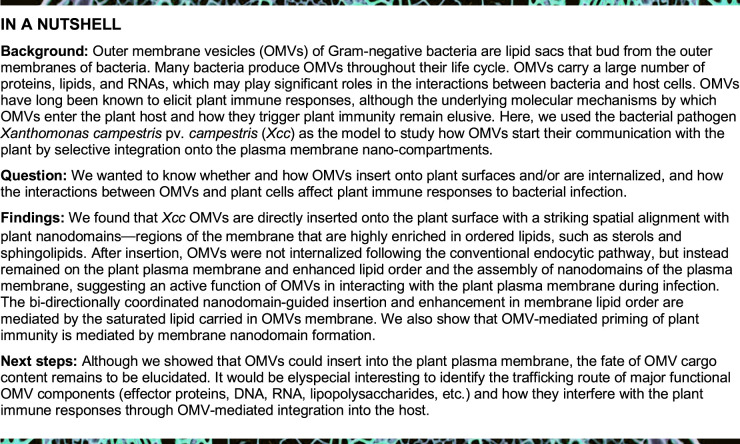
OMVs extracted from Xcc cultures are typical sphere-shaped vesicles, as previously described (Bahar et al., 2016), with an average diameter size of 39.58 ± 0.66 nm (Figure 1A). The hydrodynamic size of Xcc OMVs measured by nanoparticle tracking analysis (NTA) was larger at 112.30 ± 0.93 nm (Figure 1B). OMVs produced by human pathogenic bacteria can readily integrate into host cell PM and deliver their contents into the host cytoplasm (Bomberger et al., 2009; Jäger et al., 2015; O’Donoghue et al., 2017). Therefore, we hypothesized that phytopathogenic bacteria may act similarly to human pathogens and extrude OMVs that integrate into the plant cell PM and deliver their contents inside plant cells.
Figure 1.
Bacterial OMVs’ morphology and direct insertion of OMVs into plant PM. A, TEM micrographs of OMVs extracted from cell-free Xcc culture supernatant from NYG culture (bar, 500 nm in the left and 100 nm in the right). Arrowheads indicate several representative sizes of OMVs observed in the electron micrographs. The frequency distribution of OMV diameter was curve fitted using Gaussian distribution with GraphPad Prism 8.0. B, Diameter of Xcc OMVs measured by NTA. C, Labeling of Xcc OMVs with FM4–64 dye using centrifugal filter (bars, 5 µm and 1 µm) and fusion of FM4–64-labeled Xcc OMV to plant PM. Super-resolution of FM4–64-labeled OMVs was acquired on a Nikon Ti2 system equipped with Live-SR structured illumination microscopy. Xcc OMVs was labeled with FM4–64 dye, then applied onto Arabidopsis YFP-PIP1;4 seedling roots. Seedlings were mounted immediately, and the confocal images of root cells were taken at indicated time-points up to 30 min after incubation. Insets beneath each image are regions marked by white dashed boxes (bar, 5 µm). Relative fluorescence intensity of FM4–64 (OMV) signal on the PM over time was measured by Fiji and normalized to the intensity of the corresponding ROIs in the Green (YFP-PIP1;4) channel. In addition, PBS buffer was spiked with FM4–64 dye, subjected to centrifugation on a centrifugal column and the left-over buffer on the column was used to apply onto YFP-PIP1;4 plants and imaged as control. D, Integration of FM4–64 labeled OMVs into the PM of Arabidopsis Col-0 protoplast. FM4–64-labeled Xcc OMVs were applied on leaf protoplasts for 30 min (bar, 5 µm) prior to imaging.
To explore this hypothesis, we first tested the potential host PM integration of Xcc OMVs by labeling OMVs with the lipophilic dye FM4–64, then incubating the labeled OMVs with roots of Arabidopsis Wave138Y seedlings, which express a fusion of yellow fluorescent protein (YFP) to the PM water channel protein (aquaporin) PLASMA MEMBRANE INTRINSIC PROTEIN 1;4 (YFP-PIP1;4) as a marker of the PM (Geldner et al., 2009). To confirm that our labeling procedure followed by column-based dye removal does not leave trace amounts of free FM4–64 dye, we spiked phosphate-buffered saline (PBS) buffer with FM4–64 and ran the buffer through a centrifugation column. We did not observe any FM4–64 signal with FM4–64-spiked PBS, suggesting that the amount of dye remaining on the column after centrifugation is negligible and that this method can be used to observe OMV uptake (Figure 1C). Next, we used super-resolution microscopy to observe the labeled OMVs, which appeared as individual puncta (Figure 1C). However, because OMV sizes determined by transmission electron microscopy (TEM) and NTA were below the resolution limit of our microscope, it was not possible to discern whether these puncta were individual vesicles or clusters of OMVs.
Intriguingly, after applying FM dye-labeled OMVs onto YFP-PIP1;4 plants, we observed that FM4–64-labeled OMVs integrated into the PM and the FM4–64 signal showed a uniform distribution similar to that of the YFP-PIP1;4 signal as early as 5 min after incubation (Figure 1C). Surprisingly, the OMVs did not appear to be internalized further because the FM4–64 signal from the OMVs remained in the PM up to 30 min after incubation of the Arabidopsis seedling roots and the signal intensity gradually increased over time. In contrast, free FM4–64 could be internalized by endocytosis in the Arabidopsis roots (Supplemental Figure S1A).
To rule out the possibility that OMVs are merely trapped in the plant cell wall, we also treated Arabidopsis protoplasts with labeled OMVs (Figure 1D) and observed similar insertion of FM4–64 OMVs into the protoplast membrane without further internalization of the signal, indicating that OMVs integrate into the PM of plant cells. The above results using relatively immobile FM4–64-labeled OMVs showed that the OMV lipids might be constrained locally on the host cell PM without undergoing free exchange with surrounding plant lipids. Therefore, OMV internalization from insertion sites by plant endocytosis is restricted.
Host-integrated OMVs are not transported via CME but instead accelerate CME
The lack of internalization of OMV that we observed above prompted us to test whether the inability of FM4–64-labeled OMVs to be internalized into plant cells was due to an impairment of overall host cell endocytosis. Plant PM dynamics are tightly linked to various factors, including the underlying cytoskeletal organization, the plant cell wall, and functional endocytic and exocytic machinery (Konopka et al., 2008). As we observed that OMVs could insert effectively into the plant PM, we sought to determine whether OMV treatment inhibited the general endocytic process of plant cells, which might prevent OMV internalization.
To evaluate plant endocytosis, we performed variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) to measure the lifetimes of several endocytic markers (fused to green fluorescent protein, GFP, or red fluorescent protein, RFP) on the PM of seedlings, including the CLATHRIN LIGHT CHAIN 2 marker (CLC2-GFP), the TPLATE marker EH DOMAIN CONTAINING PROTEIN 2 (EH2-GFP), and the dynamin marker DYNAMIN-RELATED PROTEIN 2B (DRP2B)-RFP (Konopka et al., 2008; Fujimoto et al., 2010; Gadeyne et al., 2014). An analysis of kymographs from VA-TIRF time-lapse images showed that compared to PBS-treated controls, OMV-treated seedlings displayed a significant reduction in the lifetime of CLC2, EH2, and DRP2B puncta in a dose-dependent manner (Supplemental Figure S1, B–E), indicating a more efficient CME rate. In PBS-treated plants, the lifetimes of CLC2-GFP, EH2-GFP, and DRP2B-RFP puncta were 15.31 ± 0.34 s, 14.30 ± 0.50 s, and 12.60 ± 0.40 s, respectively. In contrast, in OMV-treated seedlings, the lifetimes of these endocytic markers were reduced significantly (11.12 ± 0.27 s, 6.70 ± 0.19 s, and 6.90 ± 0.27 s, respectively).
OMVs contain immunogenic flagellin molecules that can be recognized by plant surface receptors and trigger receptor-mediated endocytosis (Robatzek et al., 2006; Mbengue et al., 2016; Tran et al., 2020). Therefore, we then determined if such ligands of endocytosed receptors contributed to the unexpected enhancement of CME by OMV. We constructed a bacterial mutant lacking the FliC subunit of the flagellin molecule in Xcc 8004 strain, applied Xcc fliC− OMVs to CLC2-GFP seedlings, and measured the CLC2 lifetime in comparison to seedlings treated with Xcc wild-type (WT) OMVs. Our results showed that the OMV-induced enhancement of endocytosis was independent of flagellin as the lifetime of CLC2-GFP puncta was similarly reduced when plants were treated with Xcc fliC− OMVs (10.86 ± 0.56 s, Supplemental Figure S1B). These results demonstrate that the immobilization of OMVs on the plant PM after insertion is not caused by impairment of plant endocytosis. In fact, Arabidopsis endocytic activity was enhanced in the presence of Xcc OMVs, as indicated by the significantly shortened lifetimes of the endocytic machinery markers CLC2, EH2, and DRP2B. Together, our data demonstrate that Xcc OMVs undergo a unique mode of integration that differs from plant CME but could profoundly affect the host membrane transport pathway by accelerating CME.
Xcc OMVs elevate lipid order, induce clustering of remorin proteins, and enhance nanodomain formation on the plant PM
Based on our observation that Xcc OMVs integrate into the plant PM and accelerate Arabidopsis CME, we next investigated whether the integration of bacterial OMVs influenced plant membrane lipid packing. To address this question, we used the lipid polarity dye di-4-ANEPPDHQ (Owen et al., 2012) to probe for changes in membrane lipid order on the PM of Arabidopsis Col-0 cotyledons treated with PBS or Xcc OMVs. The lipid order is reflected by the spectral shift between the disordered and ordered lipid phases and indicates the degree of membrane lipid packing; therefore, we assessed lipid order based on the generalized polarity (GP) values of the fluorescence signal, with higher GP values corresponding to a more-ordered membrane bilayer phase. After a 30-min incubation with Xcc OMVs, the lipid order of the plant PM increased significantly compared to the PBS-treated control (Figure 2A), suggesting that the packing density of membrane lipids increased after OMV treatment.
Figure 2.
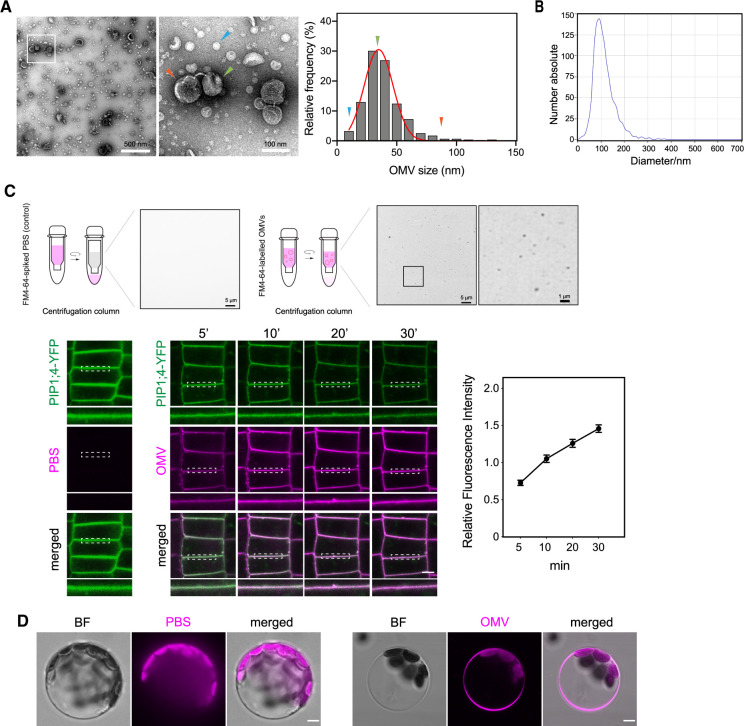
OMVs increase the lipid order of the plant PM in a nanodomain assembly-dependent manner. A, OMVs induced an increase in lipid order of Arabidopsis PM. PM order was monitored in Col-0 and rem1.2 1.3c double mutants treated with PBS or OMVs (30 µg·mL−1, 30 min) using the lipid dye di-4-ANEPPDHQ (N = 3 plants, bar, 5 µm). B, OMV-induced lipid order increase could be reverted by an acute sterol depletion using MβCD. Col-0 seedlings were incubated with PBS, MβCD (N = 3 plants, 2 mM, 30 min), Xcc OMVs (30 µg·mL−1), or MβCD followed by an OMV treatment prior to staining with di-4-ANEPPDHQ dye (bar, 1 µm). C, Density and signal intensity of GFP-REM1.2 foci on Arabidopsis PM. Seven-day-old pREM1.2:GFP-REM1.2 seedlings were treated with PBS, MβCD (2 mM, 30 min), OMV (30 µg·mL−1, 30 min) or a sequential treatment of both MβCD and OMV, then washed with 1/2 MS and imaged using a VA-TIRF microscope. GFP-REM1.2 foci density and signal intensity were quantified using TrackMate in Fiji (N ≥ 3 plants, bar, 1 µm). D, Dynamics of REM1.2 foci on Arabidopsis PM. pREM1.2:GFP-REM1.2 seedlings were tracked and diffusion coefficient was measured with SpatTrack (bar, 1 µm). Representative tracks were shown for 100-frame time-lapse movies, with 100-ms intervals. E, OMVs insertion is dependent on membrane nanodomain. Vertically grown Col-0 and rem1.2 1.3c mutant were inoculated with FM4–64-labeled OMV (100 µg·mL−1) for 30 min before imaging (bar, 5 µm).
Monitoring lipid order enhancement by OMVs stored at 4°C showed that after long storage (4 days after extraction), OMVs lost the ability to induce lipid order changes in Arabidopsis seedlings (Supplemental Figure S1F). Therefore, OMVs were used within three days after extraction for further experiments. We also confirmed that this ability to modulate the host membrane comes from OMVs themselves because only the treatments of Arabidopsis seedlings with OMVs and bacterial culture supernatant containing OMVs enhanced the plant membrane lipid order while the PBS control or ultracentrifuge supernatant, which was depleted of OMVs, did not (Supplemental Figure S1G).
Similar to the OMV-induced enhancement of endocytosis (Supplemental Figure S1B), Xcc fliC− OMVs enhanced the plant PM lipid order (Supplemental Figure S1H), suggesting that the modification of membrane properties is independent of flagellin. Moreover, the dynamics of lipid order changes induced by OMVs versus flg22 peptide (Supplemental Figure S1I) showed that while flg22 elicited only a transient lipid order change in the Arabidopsis PM as previously reported (Sandor et al., 2016), Xcc OMVs induced a slower but much more dramatic increase in plant PM lipid order. This finding indicates that the nature of lipid order enhancement by microbe-associated molecular pattern (MAMP) flg22 and OMVs are distinct. Furthermore, the OMV-mediated change in lipid order was not dependent on the plant cell wall, as evidenced by the higher GP values of cellulose biosynthesis mutants (cev1, prc1-1, je5) or plants pretreated with the cellulose synthase inhibitor isoxaben, once these seedlings were incubated with Xcc OMVs, compared to those of PBS-treated plants (Supplemental Figure S1J).
An increasing number of recent studies suggest that highly ordered plant surface nanodomains play critical roles in plant immune signaling during plant–pathogen communication (Bücherl et al., 2017; Huang et al., 2019). The formation of nanodomains requires the orchestrated assembly of remorin oligomers, phytosterols, and sphingolipids, in a spatially regulated manner, which recruits and compartmentalizes surface biomolecules for different biological activities (Huang et al., 2019; Legrand et al., 2019). Therefore, we sought to determine whether plants with compromised nanodomain assembly would still undergo OMV-induced enhancement of membrane lipid order. We examined the membrane lipid order of the Arabidopsis remorin double mutant rem1.2 1.3c, which lacks the two most transcriptionally active remorins (Huang et al., 2019), using the lipid order dye di-4-ANEPPDHQ. The rem1.2 1.3c mutant did not display any significant change in lipid order after OMV treatment (Figure 2A). This finding suggests that the increase in lipid order caused by OMVs is dependent on the presence of remorin in the plant PM.
To further examine the idea that nanodomain assembly mediates the OMV-induced enhancement of lipid order, we tested whether chemical disruption of plant membrane nanodomains abolished the enhancement of lipid order caused by OMVs. Methyl-β-cyclodextrin (MβCD) is an effective sterol-depleting drug that causes disruption of membrane nanodomains and disassembly of nanodomain-associated proteins (Hao et al., 2014; Huang et al., 2019; Tran et al., 2020). Here, we treated Col-0 seedlings with MβCD and measured the plant PM lipid order in combination with OMV treatment. Lipid order measurements showed that Xcc OMVs were no longer able to enhance the PM lipid order of sterol-depleted plants (Figure 2B).
We next sought to determine whether OMVs enhanced endocytosis in the remorin-deficient mutant. Using CLC2-RFP marker lines in the WT and rem1.2 backgrounds, we measured the CLC2-RFP lifetime on the plant PM using VA-TIRFM. The CLC2-RFP lifetime measured from VA-TIRF kymographs (Supplemental Figure S2A) revealed that compared with the WT, rem1.2 mutant did not show a significant change in the CLC2 lifetime after incubation with OMVs. Consistent with this observation, CLC2-GFP lifetime measurements revealed that sterol depletion similarly abolished the enhancement of endocytosis efficiency induced by OMVs when CLC2-GFP plants were treated with MβCD prior to the OMV treatment (Supplemental Figure S2B). The above evidence suggests that both the OMV insertion into the host PM and OMV-induced enhancement of endocytosis and lipid packing are dependent on REM1.2/REM1.3 assembly.
Next, we asked whether OMVs directly influence the assembly of PM nanodomains after inserting into the plant PM. To address this question, we monitored the signal intensity and density of GFP-REM1.2 foci, which reflect remorin oligomerization during nanodomain assembly and initiation of nanodomain formation. Single-particle analysis of REM1.2 foci demonstrated that OMVs indeed enhanced both the signal intensity and particle density of REM1.2, suggesting the enhancement in the local assembly of nanodomains and their overall formation on the cell surface (Figure 2C). Sterol depletion with MβCD efficiently antagonized this increase in REM1.2 clustering (Figure 2C). Analyzing the dynamics of REM1.2 foci on the PM with TIRF time-lapse imaging showed that OMV reduced the lateral dynamics of REM1.2 puncta, as measured by mean square displacement (MSD; Figure 2D), suggesting the enhanced stability of local molecules along with the macromolecular assembly of both remorin proteins and nanodomain structures. However, in the MβCD-treated plants, the OMV-induced decrease in MSD of REM1.2 particles was much less strong than that observed in untreated plants, thus providing additional evidence for the opposite effects of OMV integration and MβCD treatment on remorin assembly (Figure 2, C and D). Collectively, the nanodomain-dependent lipid order changes, as well as the REM1.2 clustering and dynamics induced by OMVs suggest a clear connection between membrane nanodomain stability and the ability of Xcc OMVs to interact with and cause physicochemical changes in the host PM.
Plant nanodomains mediate progressive membrane integration of OMVs and guide spatial association with nanodomains during bacterial infection
By observing the host nanodomain alterations induced by the integration of bacterial OMVs, we next sought to determine how plant PMs perceive the OMVs. The disruption of nanodomain assembly by MβCD or by genetic mutation reverted the OMV-induced enhancement of REM1.2 assembly and endocytosis, respectively. However, it is unclear whether these effects originated from the impaired insertion of OMVs into the plant PM, or due to the lack of lipid exchange between OMVs and the PM after insertion. By applying FM4–64-labeled OMVs (which enhance endocytosis similarly to unlabeled OMVs; Supplemental Figure S2C) onto rem1.2 1.3c plants and MβCD-treated Col-0 plants, we showed here that the integration of FM4–64-labeled OMVs into plant the PM was less efficient in these plants (Figure 2E). This finding supports the hypothesis that the lack of the OMV-induced increase in lipid order and enhancement of endocytosis in nanodomain-disrupted conditions could be the direct result of the poor integration rate of Xcc OMVs into the plant PM.
Next, we tested whether interfering with the plant surface membrane lipids or cytoskeletal structures would also affect the integration of Xcc OMVs into the plant PM. Similar to the effects of MβCD, disruption of membrane sphingolipid biosynthesis by treatment with myriocin (Miyake et al., 1995) also reduced OMV insertion into the plant PM, whereas the phosphatidylinositol 4-kinase inhibitor phenylarsine oxide (PAO; Simon et al., 2016) had little effect on OMV integration (Supplemental Figure S3A). Surprisingly, the actin polymerization inhibitor Latrunculin B (LatB) did not block OMV insertion. Instead, destabilization of the cortical actin cytoskeleton substantially increased the FM4–64 intracellular signal. In addition, the disruption of plant microtubules by oryzalin drastically increased the FM4–64 dye on internalized vesicles and the PM. As a control, we found that that the tested inhibitors (MβCD, myriocin, PAO, and oryzalin) inhibited FM4–64 endocytosis without an obvious effect on FM4–64 dye integration in the PM (Supplemental Figure S3B). The distinct membrane integration and internalization of FM4–64 and FM4–64-labeled OMVs under different perturbations of the PM and cytoskeleton demonstrated that FM4–64 and OMVs utilized different membrane integration and trafficking mechanisms upon interaction with the plant PM.
In addition to using multiple pharmacological inhibitors, we also examined OMV integration into the PM of Arabidopsis sterol biosynthesis mutants to evaluate the importance of sterol homeostasis for the interaction between Xcc OMVs and plant cells. We showed here that the sterol mutants displayed obvious defects in OMV integration into the plant PM (Supplemental Figure S4, A and B). Altogether, our results demonstrate the roles of the PM–actin cytoskeleton continuum in the insertion and internalization of bacterial OMVs.
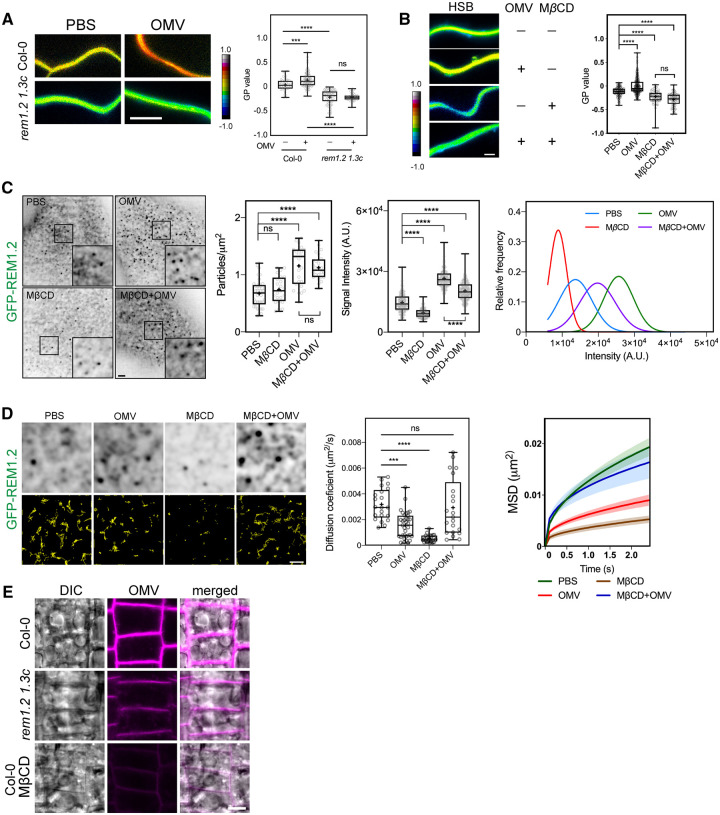
The feedback regulation of nanodomain assembly and OMV integration motivated us to further investigate their relationship during bacterial infection. Since remorin proteins were recently shown to function as host regulators that respond to various biotic and abiotic environmental cues (Perraki et al., 2014; Cheng et al., 2020; Abel et al., 2021), we first investigated whether remorins and nanodomains were modulated by bacterial infection. We performed a flooding assay using Xcc 8004 on Arabidopsis pREM1.2:GFP-REM1.2 plants and monitored the pattern of GFP-REM1.2 foci on the plant PM. TIRFM revealed that the signal intensity of GFP-REM1.2 foci on the PM increased over time after the plants were exposed to pathogenic bacteria, reached a peak at approximately 6 h, and remained high up to 24 h post-inoculation (Figure 3A). Then, we flooded Arabidopsis GFP-REM1.2 plants with Xcc 8004, incubated them for 6 h and then applied Xcc OMVs for another 30 min prior to VA-TIRF imaging. Interestingly, the Xcc-infected plants showed high colocalization of OMVs with REM1.2-marked membrane nanodomains (Figure 3B), suggesting that the enhanced nanodomain assembly triggered by bacterial infection favored the recruitment of OMVs into nanodomain sites.
Figure 3.
Colocalization of remorins with FM4–64-labelled OMVs. A, Time-dependent increment of GFP-REM1.2 intensity on Arabidopsis PM after flooding inoculation with Xcc. Seven-day-old pREM1.2:GFP-REM1.2 seedlings were flooded with 1 × 107 CFU·mL−1 bacterial suspension before TIRFM imaging at the indicated times post-inoculation. Images of GFP-REM1.2 foci were acquired by VA-TIRFM using a Zeiss LSM780 + ELYRA microscope, equipped with a 100× oil objective (NA = 1.46, bar, 5 µm). B, VA-TIRF dual imaging of the colocalization of GFP-REM1.2, YFP-REM1.3, GFP-REM1.2 in the XVE:GFP-REM1.2 seedlings (REM1.2-OE) and under infection condition (flooded with Xcc 8004 and incubated for 6 h) with FM4–64-labeled OMVs. VA-TIRF dual-color images were acquired by a Zeiss LSM780 + ELYRA microscope equipped with a 100× oil-objective lens. XVE:GFP-REM1.2 seedlings were induced with 5-µM β-estradiol for 24 h prior to imaging (bar, 5 µm). Violin plot shows Person’s R values, representing the colocalization between remorins and FM4–64 labeled OMVs. The differences between treatments were determined by one-way ANOVA (ns, not significant, *P≤0.05; **P≤0.01, ***P≤0.001, ****P≤0.0001).
The modulation of the plant PM lipid order and microdomain assembly by Xcc OMVs (Figure 2, A–D) might be the result of enhanced biosynthesis or transport of ordered lipids (such as phytosterols, sphingolipids, ceramides) in the PM, or the direct insertion of OMVs into existing membrane nanodomains, which introduces lateral compression. Although the first scenario is possible, it is less likely to occur within the short timeframe of the OMV treatment (30 min) because changes transcriptional/translational processes generally require more time to alter lipid biosynthesis. Therefore, we investigated whether OMVs could preferentially insert into remorin-occupied nanodomains in the plant PM, such that interruption of these sites may lead to less OMV insertion and abolishment of OMV-induced effects. To this end, FM4–64-labeled Xcc OMVs were applied to seedlings of several remorin marker lines, namely pREM1.2:GFP-REM1.2, pREM1.3:YFP-REM1.3, as well as an estradiol-inducible REM1.2 overexpression line (XVE:REM1.2-GFP, hereby REM1.2-OE). The colocalization of OMVs and remorins was observed using VA-TIRF imaging (Figure 3B; Supplemental Figure S4). TIRF micrographs revealed that labeled Xcc OMVs inserted as distinct and irregular foci on the Arabidopsis PM, and the distribution of OMV insertion sites was not correlated with REM1.2 or REM1.3 foci (Pearson’s R value ∼ 0, Figure 3B). These results suggest that the initial insertion of OMVs does not require physical contact with the outer leaflet of nanodomains, although surface lipid environments maintained by nanodomains are necessary for perceiving OMVs, as shown in the aforementioned results (Figure 2E). However, if we overexpressed remorin proteins to enhance remorin oligomerization and induce nanodomain assembly, OMVs then strongly colocalized with the REM1.2 signal. The enhanced OMV-remorin colocalization occurred in a remorin-clustering and nanodomain formation-dependent manner following progressive β-estradiol-mediated induction over time, or with early infection at 6-h post-inoculation with Xcc (Figure 3B; Supplemental Figure S5). Pearson’s correlation coefficient analysis showed that the colocalization of labeled OMVs with REM1.2 was enhanced significantly with a longer incubation time with β-estradiol starting at 24 h and remained at a similar level up to 48 h after induction.
Taken together, our data demonstrate that OMV insertion into the plant PM requires the native physico-chemical properties of the PM that are maintained by nanodomains. The enhancement in nanodomain assembly is one of the responses following plant–bacterial interactions, as evidenced by the increase in REM1.2-marked nanodomain assembly upon bacterial infection. The observed increase further established that the spatial association between bacterial OMV and host membrane nanodomains caused by OMV insertion and increased lipid order in the plant PM.
Sterol in the plant PM regulates the OMV–PM interaction
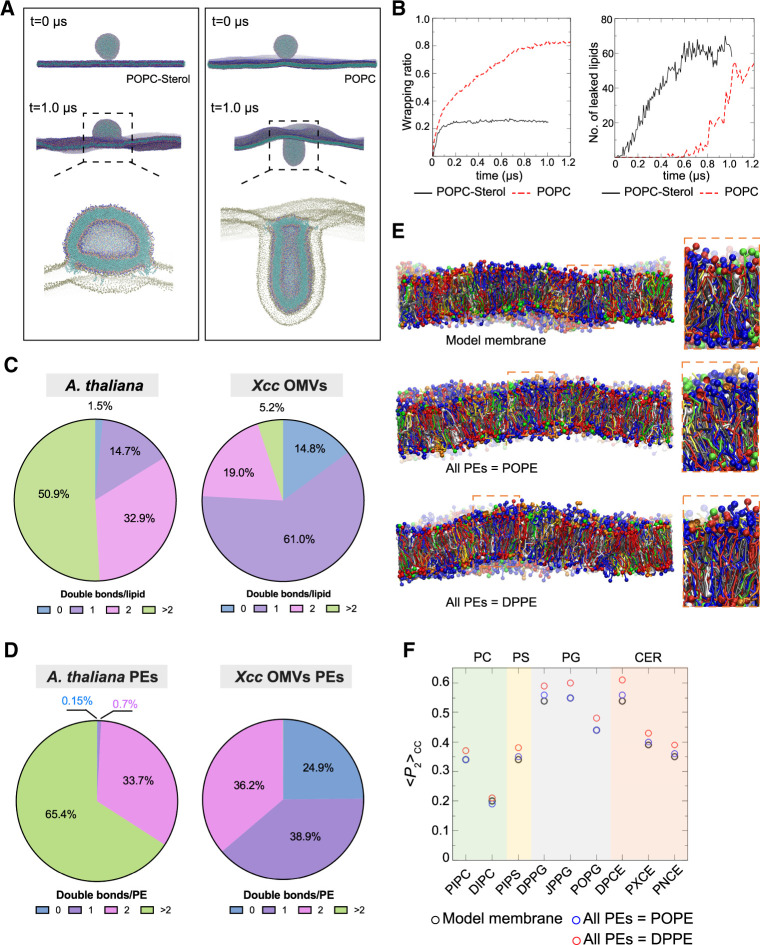
To gain insight into the effect of sterols on OMV–plant PM interactions at the molecular level, we carried out coarse-grained MD simulations. The model phospholipid 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) was chosen as a generic phospholipid for modeling the vesicle because it is one of the major lipid species in Xcc OMVs and, like POPC, more than half of the total lipids in OMVs contain mono-unsaturated tails (Supplemental Data Set S1). In the simulations, a 23-nm POPC (C16:0/C18:1) vesicle was placed atop either a POPC (control) or a POPC:sterol (1:1; mimicking the composition of the plant PM) planar bilayer membrane (Figure 4A). Coarse-grained lipid models in which multiple atoms are grouped into a single particle were used to construct the vesicle and planar membrane (Marrink et al., 2007). Although the POPC vesicle size was smaller than the reported mean diameter of OMVs measured from TEM micrographs (Figure 1A), it was close to the peak of the Gaussian fit to the OMV size distribution data (∼30 nm).
Figure 4.
Simulation of OMV-plant PM interaction and the lipid order modulation in model plant membrane by receiving OMV lipids. A, Molecular simulation snapshots of a 23-nm diameter POPC vesicle placed above a POPC-sterol (1:1) (Left) or POPC (Right) planar membrane at the beginning of the simulation and after 1 μs. Bottom panels show the cross-sectional views. Phospholipid particles are colored as follows: head-group particles in blue, phosphate particles in dark green, glycerol particles in pink, and fatty acid tail particles in cyan. Sterol particles are colored as follows: head-group particles in purple and the rest in cyan. B, The temporal evolution of the wrapping ratio (Left) and number of vesicle lipids leaked into planar membrane (Right) for systems in (A): POPC-sterol membrane (black solid line) and a control system (POPC membrane, red dashed line). C, Saturation levels of lipids in Arabidopsis and Xcc OMVs characterized by lipidomic analyses. Data represent the mean of six biological replicates (N = 6). Results of the Arabidopsis lipidome were reported previously (Tran et al., 2020). D, Saturation levels of PEs in Arabidopsis and Xcc OMVs characterized by lipidomic analyses. Data represent mean of six biological replicates (N = 6). E, Side views of the simplified Arabidopsis PM model (Top) and the ones with all PEs replaced by POPE (Middle) or by DPPE (DP: dipalmitoyl) (Bottom) after 2 μs of simulation. Lipids are in ball-and-stick representation and colored as follows: sterols in gray, PEs in red, PCs in blue, PGs in green, phosphatidylserines (PSs) in yellow, ceramides in white, and galactolipids in orange. Each lipid has one of its head-group particles shown as a sphere (except for sterols). F, Lipid order parameters averaged over fatty acid tail particles from both tails for phospholipids (PC, PS, PG) and ceramides (CER) with PE tails being poly-unsaturated (original: 1-palmitoyl-2-linoleoyl or PI, dilinoleoyl or DI), mono-unsaturated (PO) or saturated (DP). JPPG (C16:1(3t)/16:0 PG) is phosphatidylglycerol lipid with trans-3-hexadecenoic acid tail (sn1) and a palmitoyl (sn2) tail. DPCE, PXCE, and PNCE are ceramides with 18:0, 24:0, and 24:1 fatty acid tail, respectively. See Supplemental Data Set S4 for detailed information on the fatty acid tails.
As shown in Figure 4A, modeling showed that the electrostatic and van der Waals interactions between vesicle lipids and the lipids in the planar membrane drove the planar membrane to wrap around the vesicle as the vesicle binds to the membrane, causing the vesicle to deform. The vesicle turned into a hemisphere when it was bound to the POPC:sterol (1:1) membrane (left panel of Figure 4A; Movie 1). In contrast, the vesicle was wrapped to a much greater extent by the sterol-free membrane, and its shape changed from spherical to rod-like (right panel of Figure 4A; Movie 2). We characterized the extent of wrapping by quantifying the fraction of the vesicle surface in contact with the planar membrane, i.e., the wrapping ratio. The vesicle was continuously wrapped by the sterol-free membrane until ∼80% of its surface was in contact with the membrane (left panel of Figure 4B; Supplemental Movie S1), whereas vesicle wrapping stalled at a wrapping ratio of ∼25% for the POPC:sterol membrane. Such distinct behavior could be due to the stiffness of the planar membrane, as a membrane containing 50% sterol molecules is much stiffer than a membrane with no sterol (Orädd et al., 2009) and therefore, the POPC:sterol membrane cannot easily bend to enclose the vesicle. The stalled wrapping of the vesicle by the POPC:sterol membrane was consistent with our observation of OMV immobilization and localization on the host plant PM (Figure 1B).
Vesicle–planar membrane fusion was observed in both systems by focusing on the early stage of membrane associations, with and without sterol in the membrane, although the onset of fusion was much earlier for the membrane with the sterols (Figure 4A). In the simulation, the vesicle fused with the POPC:sterol membrane soon after binding. Lipid exchange occurred at the fused rim, where vesicle lipids leaked onto the upper leaflet of the planar membrane (Figure 4A). The number of leaked vesicle lipids initially increased linearly with time and reached a plateau after ∼0.6 µs (right panel of Figure 4B). This finding suggests that the lipid contents of OMVs might be transferred to the plant PM via vesicle fusion with the membrane surface without being endocytosed. In contrast, fusion occurred only after almost 80% of the vesicle surface was wrapped by the sterol-free membrane, and a much smaller fused rim was close to the upper membrane compared to the POPC:sterol membrane. Vesicle lipid leakage then accelerated and reached a similar level as that of the POPC:sterol membrane. The results suggest that, in the absence of sterol, significant membrane bending is required before vesicle–membrane fusion can occur. Hence, the delivery of lipids to the PM would be more efficient when OMV binds to membrane patches containing sterol, such as nanodomains.
Saturated phospholipids from OMVs increased membrane lipid order in the plant PM
To better understand the lipid–lipid interaction between the OMV membrane and the plant PM, we characterized the lipid composition of the Xcc OMV membrane. Our targeted lipidomic profiling of Xcc OMVs revealed that Xcc OMVs contained a complex mixture of lipid constituents (Supplemental Data Set S1; Figure 4C), including phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingomyelin, lysophosphatidic acid, and phosphatidylglycerol (PG). Interestingly, we found that of the lipid species detected in our lipidomic analyses, the majority were monounsaturated (61.04%, Supplemental Data Set S1), followed by diunsaturated (19.02%) and saturated lipids (14.84%) and unsaturated lipids with more than two double bonds (5.10%). With regard to the saturation level of individual acyl tails, 52.07% of the acyl tails detected in the lipidomic profile of Xcc OMVs were saturated, followed by 40.31% with monounsaturated tails, 4.77% diunsaturated, and 2.85% with unsaturated tails and more than two double bonds (Supplemental Data Set S1). Therefore, the saturation level of OMV lipids is much higher than that of the A. thaliana lipidome (Tran et al., 2020), which has only 1.51% saturated, 14.72% monounsaturated, 32.89% diunsaturated, and 50.90% unsaturated lipids with more than two double bonds (Figure 4C; Supplemental Data Set S1).
Our experiments found that >14% of the total lipids in OMVs contain saturated tails (Figure 4C; Supplemental Data Set S1), and incubation of OMVs with plant cells increased the plant PM lipid order (Figure 2A). Within the PE group, Xcc OMVs contained more saturated PEs (24.9%) compared with Arabidopsis PEs (0.15%, Figure 4D). We hypothesized that the introduction of phospholipids with saturated fatty acid tails from OMVs into the plant PM might account for the observed increase in plant PM lipid order. We tested this hypothesis by constructing a model membrane using coarse-grained lipid models based on the membrane lipid composition determined by Uemura et al. (1995). In our model, sterol accounted for 46% of the total lipids, and phospholipids accounted for 47%, with PE and PC representing the dominant phospholipids (Supplemental Data Set S2). The various lipids were uniformly distributed in the model. The side-views of the model after equilibration are shown in the top panel of Figure 4E. The magnified view of the membrane model with all PE lipids as dipalmitoyl phosphoethanolamine (DPPE) showed that all lipids appeared more straightened and aligned as compared to the original model, thus leading to a slightly larger average membrane thickness (based on the locations of phosphate particles) of 4.23 ± 0.014 nm versus 4.11 ± 0.026 nm in the original model. The model with all PE lipids as POPE showed an intermediate alignment of lipids (middle panel of Figure 4E).
We further quantitatively characterized the packing of each type of lipid by computing the value, which represents the lipid order through the alignment of individual molecular bonds to the axis perpendicular to the membrane. As shown in Figure 4F, the values were generally higher for tails with a higher degree of saturation, i.e. fewer carbon–carbon double bonds. For example, the averaged lipid order parameter for 1-palmitoyl-2-linoleoyl PC (PIPC) was higher than that for dilinoleoyl PC (DIPC) because one of the tails of PIPC is fully saturated while both of the tails of DIPC are unsaturated. 1,2-Dipalmitoyl-sn-glycero-3-phosphoglycerol and C(d18:1/18:0) Ceramide (DPCE) had some of the highest lipid orders because both of their tails are fully saturated.
We focused on the PE group because PEs are conical in shape and have smaller headgroups, thus increasing their likelihood of migrating into the host PM upon fusion (Jouhet, 2013). After we replaced all PE lipids with POPE (one saturated tail and one monounsaturated tail), the lipid order of all other lipids remained the same or increased slightly (blue symbols in Figure 4F). However, when we replaced all PE lipids with DPPE (two saturated tails), the lipid order of all other lipids increased (red symbols in Figure 4F). The increased lipid order is consistent with the enhanced lipid tail alignment shown in Figure 4E. This result strongly suggests that the introduction of lipids with saturated tails would increase the overall lipid order, supporting our hypothesis that the transfer of saturated phospholipids into the plant PM via fusion with OMVs might account for the observed increase in PM lipid order.
Furthermore, we estimated the elastic modulus of the three simplified membranes from our simulations and found that both the original model membrane and membrane with all PEs as POPE had elastic moduli of ∼160 mN·m−1, while the membrane with all PEs as DPPE had a much higher elastic modulus of 390 mN·m−1. The range of the predicted elastic modulus values is consistent with previous studies (Doktorova et al., 2019). This enhancement of the membrane elastic modulus results from the enhanced membrane lipid order.
OMVs enhance plant immunity in a remorin- and sterol-dependent manner
Recent studies have demonstrated that OMVs produced by a number of plant-interacting bacteria, such as the phytopathogen Pseudomonas syringae pv. tomato and the biocontrol bacterium Pseudomonas fluorescens (Janda et al., 2021; McMillan et al., 2021) elicit plant innate immunity and protective effects. However, there is little evidence connecting the interaction between OMVs and plant cells with the outcome of the pathogen–plant interaction (Bahar et al., 2016). Moreover, various studies highlighted membrane nanodomain assembly and endocytosis as major contributors to the outcome of diverse plant–microbe interactions (Mbengue et al., 2016; Sandor et al., 2016). As OMVs exert various effects on Arabidopsis cells and these effects are nanodomain-dependent, we were particularly motivated to determine (1) whether Xcc OMVs could contribute to the outcome of the plant–bacteria interaction and (2) whether Xcc OMV-mediated protection is also dependent on membrane nanodomain assembly.
Reactive oxygen species (ROS) bursts are a hallmark of the plant immune response. Therefore, to examine the effect of Xcc OMVs on plant innate immune responses, we first measured ROS bursts in Arabidopsis Col-0 leaf strips pre-treated with bacterial OMVs and those treated with the defense elicitor flg22 or PBS/water control. Our results showed that Xcc OMV treatment triggered a small ROS burst, although to a much lower extent than that triggered by the flg22 peptide, in agreement with a previous study (Bahar et al., 2016; Supplemental Figure S6A).
We next investigated whether the induction of ROS caused by OMVs could also lead to inhibition of other receptor-mediated immune responses. In the plant defense response, callose deposition reinforces the cell wall and limits the spread of pathogens upon infection (Gómez-Gómez and Boller, 2000). Xcc OMVs triggered significantly less callose deposition in Arabidopsis seedling leaves compared to flg22 elicitation (Supplemental Figure S6B). Consistent with the induction of ROS and callose deposition by Xcc OMVs, we found that MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) phosphorylation in Arabidopsis leaves was also activated shortly after leaves were treated with OMVs (5 min), and the pattern was similar to that of p-MAPK activation by flg22 but at a lower level (Supplemental Figure S6C). MAPK activation by Xcc OMVs is in agreement with the enhanced FLG22-INDUCED RECEPTOR-LIKE KINASE 1 (FRK1) expression (downstream of MAPK phosphorylation) in a previous report (Bahar et al., 2016).
To further dissect the contribution of MAMPs, especially the potential flagellin carried in OMVs, to the ROS burst induced in host tissue, we monitored ROS responses in Arabidopsis plants lacking the flagellin sensor receptor kinase FLAGELLIN-SENSITIVE 2 (FLS2). To this end, we used Col-0 and fls2 leaf strips elicited with Xcc WT or fliC− OMVs. We found that the ROS response triggered by WT OMVs on the fls2 mutant was lower, but not significantly different from that of WT OMVs on Col-0 plants (Supplemental Figure S6D). A similar trend was observed for fliC− OMVs on Col-0 plants, suggesting that flagellin, even when present in the OMV preparation, is not a major source of the ROS burst induced by Xcc OMVs.
Flooding inoculation of Arabidopsis Col-0 seedlings pretreated with Xcc WT and fliC− OMVs showed that both types of OMVs could induce significant protection against Xcc infection, as indicated by the lower bacterial population in OMV-treated plants (Supplemental Figure S6E). However, the remaining MAMP flg22 in OMVs might still contribute slightly to OMV-mediated protection because fliC- OMV-mediated protection was slightly reduced (Supplemental Figure S6E). Similarly, without flg22-mediated protection, the fls2 seedlings displayed a reduction in Xcc populations after being treated with WT OMVs (Supplemental Figure S6E). Our results suggest the MAMP flg22-independent protection against bacterial pathogens occurs with OMVs.
A previous study showed that Xcc OMVs could simultaneously trigger defense gene activation through multiple pattern-recognition receptors (PRRs). BAK1, the co-receptor of several PRRs involved in innate immunity signaling (Xue et al., 2020), is critical for OMV-mediated defense gene activation (Bahar et al., 2016). Here, using a flooding assay on Arabidopsis seedlings, we showed that OMV treatment of fls2 and efr (EF-Tu RECEPTOR) seedlings still provided protection against Xcc infection (Supplemental Figure S6F). Unexpectedly, OMV pretreatment also protected the bak1-4 seedlings against bacterial infection (Supplemental Figure S6F). Together, our data suggest that the OMV-induced protection against bacterial infection is complex and not mediated by a single PRR or co-receptor.
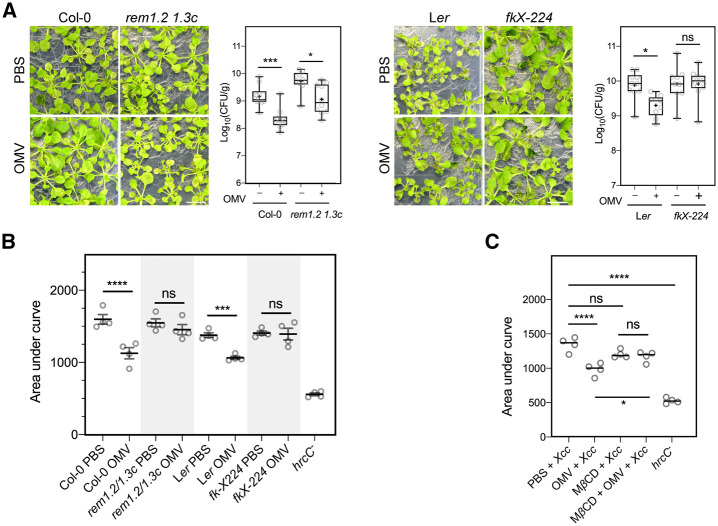
We next evaluated whether PM biophysical properties, such as membrane compartmentalization, contribute to the effects of OMVs. Thus, we evaluated OMV-mediated protection against Xcc in Arabidopsis mutants defective in nanodomain assembly (rem1.2 1.3c, Col-0 background) and sterol biosynthesis (fk-X224, Ler background). The fk-X224 mutant was chosen for this assay because of its relatively normal growth compared to that of other sterol mutants. Three days post-inoculation (DPI), we observed water-soaking and chlorosis symptoms on infected plants. Quantification of bacterial populations at 3 DPI showed that compared with PBS-treated plants, OMV-treated Arabidopsis Col-0 and Ler plants harbored significantly fewer bacteria and the plant tissues were not as damaged with water-soaked lesions (Figure 5, A and B), thus demonstrating the protective effect of Xcc OMVs on against Xcc infection. In addition, we observed this protection to a lesser extent in rem1.2 1.3c and fk-X224 mutants, indicating that remorin assembly and phytosterols, which are key components of nanodomains, are important for OMV-mediated plant resistance against bacterial infection, possibly due to the reduced OMV integration into the PM of these mutants (Supplemental Figure S4, A and B).
Figure 5.
Protective effect of Xcc OMVs against bacterial infection. A, OMVs protect Arabidopsis seedlings from Xcc infection. OMVs from Xcc 8004 were extracted as indicated above. Arabidopsis Col-0, rem1.2 1.3c, Ler, and fk-X224 seedlings were incubated with 10 µg·mL−1 Xcc OMVs or PBS (control) for 24 h prior to flooding inoculation with 20 mL of 107 CFU·mL−1 Xcc 8004 bacterial suspension. Seedlings were sampled at 3 DPI and the bacterial population was quantified by serial dilution plating on NYG plates (three plants were pooled as a single technical replicate, N ≥ 9 replicates/treatment). Statistical differences between treatments were determined by one-way ANOVA using GraphPad Prism 8.0 (ns, not significant, *P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001). OMV-induced protection of cell damage from pathogen infection is dependent on Remorin and sterol balance (bars, 1 cm). B, Arabidopsis leaf discs of WT ecotypes (Col-0, Ler), remorin double mutant (rem1.2 1.3c), sterol biosynthesis mutant (fk-X224 – Ler background) were incubated with Xcc OMV (30 µg·mL−1) for 24 h, then infiltrated with Xcc 8004. Xcc hrcC− was infiltrated into Col-0 leaf discs as the negative control. AUC of conductivity increase was calculated by GraphPad Prism 8.0 and statistical differences between treatments were determined by one-way ANOVA (ns, not significant, *P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). C, Ion leakage measurement from Arabidopsis Col-0 leaf discs incubated with OMVs (30 µg·mL−1), MβCD (2 mM), or a combination of both (MβCD +OMV) prior to vacuum infiltration with Xcc 8004 (WT). The Xcc hrcC- mutant was used as the negative control. Statistical differences between treatments were determined by one-way ANOVA (ns, not significant, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P < 0.0001).
Electrolyte leakage during bacterial infection is one of the hallmarks of plant cell death, measured as an increase in the electrolyte concentration in an aqueous bathing solution containing the plant tissues (Johansson et al., 2015). Here, we used leaf infiltration to introduce Xcc bacteria into Arabidopsis leaf discs and measured the conductivity increase in the bathing solution resulting from electrolyte leakage. First, we found that OMV could significantly lower the amount of electrolyte leakage from leaf discs after infiltration with Xcc, suggesting that OMV treatment could prevent membrane damage during infection (Figure 5B; Supplemental Figure S6G). Consistent with disease assay results, rem1.2 1.3c and fk-X224 leaf discs treated with OMVs showed a similar degree of ion leakage in OMV treatment compared to their corresponding PBS controls, indicating that proper phytosterol content and nanodomain assembly are critical for OMV-induced plant immunity against the bacterial pathogen. This finding was further confirmed by measuring the electrolyte leakage in leaf discs treated with OMVs in combination with MβCD. MβCD, which acutely depletes sterols from the plant PM, could similarly abolish the OMV-induced reduction of ion leakage during infection, thereby providing additional evidence showing that the protective effect of bacterial OMVs on plant immune defenses is dependent on the membrane nanodomain assembly (Figure 5C; Supplemental Figure S6H).
Discussion
The bacterial OMVs carry a wide variety of immunogenic molecules, such as lipopolysaccharides, peptidoglycan, toxins, and effectors (Sidhu et al., 2008; Chowdhury and Jagannadham, 2013; Solé et al., 2015), which could all induce a wide range of host responses either favoring bacterial pathogenesis or activating host immune responses (Bahar et al., 2016; Chen et al., 2018). Here, we have revealed a mechanism by which OMVs interact with host cells and directly modulate the plant PM physicochemical properties, which subsequently enhances host immunity against bacterial infection.
Immunogenic nature of Xcc OMVs
In this study, we demonstrated that X. campestris pv. campestris OMVs induced and enhanced defense responses in Arabidopsis plants by integrating and remaining in the host PM, thereby modulating plant PM compartmentalization. Plant PM lipids and remorin proteins in the host nanodomains orchestrate the integration of OMVs and influence host lipid packing and membrane compartmentalization. Recent studies reported the elicitation of an ROS burst in Arabidopsis and activation of MAMP-triggered immunity (MTI)-related defense genes, including the activation of FRK1, as well as protection against bacterial and fungal infection in Pto OMV-pretreated plants (Bahar et al., 2016; Janda et al., 2021; McMillan et al., 2021). Similarly, Bahar et al. (2016) showed that the OMV-induced defense responses depend on multiple factors carried by OMVs, including EF-Tu, flagellin, and eMAX because single, double, and triple mutants of receptors for these MAMPs still react to Xcc OMVs.
Here, our studies suggest a mechanism that plays an essential role in microbe–host interactions independent of MAMPs. Due to the inevitable impurity of OMV preparations, which include a trace amount of flagellin molecules, we observed a small ROS burst, although not strictly dependent on bacterial flagellin (Supplemental Figure S6D). Interestingly, Xcc fliC− OMV is still able to protect plants from the bacterial pathogen. This result shows that flagellin is apparently not the only contributor to the protection against Xcc during infection (Supplemental Figure S6, D and E), which strongly supports the MAMP-independent enhancement of host immunity by OMV against bacterial pathogens (Supplemental Figure S6E). Future studies are needed to dissect the detailed immune signaling pathways of plants that are modulated by bacterial OMVs and result in enhanced defense against bacterial infection.
Based on the limited reports that are currently available, one of the possible mechanisms underlying the OMV-mediated protection against bacterial infection is the changes in plant PM’s physico-chemical properties that prevented cell death, a process that may benefit pathogens from the release of nutrients (Kraepiel and Barny, 2016). Cross-protection through priming of shared co-receptors (Gong et al., 2019) or activation of plant immunity by pathogen’s effectors could also underlie the protection of OMVs during infection (Cheng et al., 2015). More biophysical, cell biology, and pathology experiments will hopefully validate the above-mentioned speculations.
The observed OMV-mediated protection through nanodomain assembly creates new plant–pathogen communication machinery that supports the emerging mechanisms underlying plant immune responses by fine-tuning plant PM compartmentalization and organization (Bücherl et al., 2017; Ma et al., 2021). Here, our results showed several striking aspects of the host nanodomains and bacterial OMVs, in a spatiotemporally regulated manner during bacterial infection. In general, OMVs could integrate into the plant PM directly, and this process requires compartmentalized plant PM with appropriate nanodomain compositions, such as phytosterol and remorin proteins. During early bacterial infection, the nanodomain is rapidly modulated to increase assembly, which further facilitates OMV insertion onto the plant surface. Subsequently, progressive OMV integration over time during infection enhances nanodomain assembly, thereby establishing a feedback loop to allow spatial coordination in recruiting OMVs to the nanodomain sites and eventually increasing capabilities defense responses. Membrane compartmentalization and lipid order enhancement probably played an essential role in fine-tuning signaling scaffolds on the plant PM in response to abiotic and biotic environmental cues, as shown in recent studies (Lefebvre et al., 2010; Bücherl et al., 2017; Huang et al., 2019; Tran et al., 2020; Ke et al., 2021; Ma et al., 2021). Therefore, whether and how such OMV-enhanced nanodomain assembly further facilitates the surface receptor-mediated MTI signaling is worthy of further study to gain a more comprehensive understanding of the complex communication between bacterial OMVs and host immune signal transduction.
What enables OMV insertion into plant PM?
Although the ability of OMVs produced by human pathogenic bacteria to insert into the mammalian cell PM and serve as a gateway for virulence factor delivery was established more than a decade ago (Bomberger et al., 2009; Kaparakis et al., 2010), whether phytobacterial OMVs integrate into the plant cell PM and enter cells in a similar manner remained unknown (Bahar, 2020). Here, we reported that OMVs secreted by a bacterial plant pathogen could insert directly into the PM of host cells (Figure 1, C and D). This was rather surprising because the small 50 Å pore size of the plant cell wall mesh (Carpita et al., 1979) was commonly believed to hinder the passage of OMVs and any interaction with the plant PM (Katsir and Bahar, 2017; Bahar, 2020). However, OMVs from several plant pathogenic bacteria also package cell wall-degrading enzymes, such as cellulase, xylanase (Sidhu et al., 2008), pectin lyase, and lipases/esterases (Feitosa-Junior et al., 2019). The release of these components could prime the cell wall for further enzymatic attack (Adani et al., 2011), facilitating cell wall loosening and the passing of OMVs (especially smaller vesicles) through the cell wall mesh. It remains to be determined if the translocation rates of OMVs through the plant cell wall would be similar for OMVs of different sizes, and whether their lipid components would differ significantly.
We found that OMV insertion occurred quickly in minutes, and it appeared that the lipid membrane of Xcc OMVs integrated but did not homogenize with the plant membrane phospholipid bilayer, as suggested by the distinct OMV foci on the plant PM and the lack of intracellular signal from the FM4–64-labeled OMVs. The initial insertion of OMVs into the plant PM was explained by our computational simulation, which suggested that lipid exchange resulting from membrane fusion between OMV and the PM may occur in a sterol-sensitive manner. Lipid exchange was also observed when a model POPE–POPG vesicle binds and fuses with a multicomponent PM model (containing 25% POPC, 25% POPE, and 25% sterol) in another simulation (Jefferies and Khalid, 2020). Without sterol molecules, the lipid bilayers showed less fusion and lipid exchange with OMV, although the model membrane’s lipid bilayer can be more flexible and bent when coming into contact with the model OMV. However, we would imagine that such PM bending would be restricted by the plant cell wall in vivo.
In addition, our lipidomic analysis also showed that the Xcc OMV membrane is highly enriched in a more saturated acyl tail moiety (Supplemental Data Set S1), which is similar to what was observed on P. aeruginosa OMVs, in which saturated fatty acid accounted for >71% of phospholipids in both early and late stationary stages (Tashiro et al., 2011). In contrast, our previous study (Tran et al., 2020), showed that the Arabidopsis lipidome possessed mainly unsaturated lipids (98.5%, Supplemental Data Set S1). Saturation and symmetry levels of phospholipids contribute to the order and fluidity of lipid bilayers (Smith et al., 2020). Therefore, the insertion of saturated lipids from bacterial OMVs may introduce a general increase in the lipid order in the plant PM. Consistent with this notion, our molecular simulation revealed that by changing only the saturation level of a single minor lipid constituent (PE), the order of all other species could be strongly affected, indicating that membrane lipid order manipulation is a mechanism of interkingdom communication between plants and microbial pathogens.
Previous studies demonstrated that OMVs secreted by human pathogenic Escherichia coli, P. aeruginosa, H. pylori, and N. gonorrhea integrate into the host PM and then internalize (Bomberger et al., 2009; Kaparakis et al., 2010; O’Donoghue et al., 2017). Lipopolysaccharide structures on the OMV surface could immensely influence the entry kinetics of the vesicles (O’Donoghue et al., 2017; Jefferies and Khalid, 2020), suggesting that the inability of Xcc OMVs to pass through the plant PM might stem from the differences in the structures of lipopolysaccharides on the Xcc OMV surface compared to that of other bacteria or the different nanodomain organization of plant and mammalian cells.
Using multiple pharmacological inhibitors that target different cellular components, we identified the cellular components that influence the rate of insertion and internalization of Xcc OMVs. MβCD and myriocin deplete sterols from the PM and inhibit sphingolipid biosynthesis, respectively, which could effectively block OMV insertion. This is consistent with several previous studies that showed that pharmacological disruption of membrane lipid rafts by depleting sterols or sphingolipids could effectively block the entry and NUCLEAR FACTOR KAPPA B (NF-κB) inducing activity of OMVs (Bomberger et al., 2009; Kaparakis et al., 2010). Surprisingly, we also found that treatment with the actin polymerization inhibitor LatB could induce the internalization of FM4–64-labeled OMVs (Supplemental Figure S3). Contrary to a previous study, which reported that the entry of OMVs into airway epithelial cells depends on the actin cytoskeleton (Bomberger et al., 2009), we found that disrupting the actin cytoskeleton and microtubule networks indeed enhanced the internalization of OMV into plant cells. We speculate that PM tethering by the underlying actin cytoskeleton as an interconnected continuum may create more sophisticated biomechanical regulations during immune response signaling (Ma et al., 2021), which also regulates the internalization of OMVs from the plant PM. Interestingly, previous work also showed that actin filaments are often associated with surface nanodomain sites in the plant (Bücherl et al., 2017). This complex regulation of OMV integration into the plant PM and trafficking shared some similarities with the mammalian system, but showed some distinct mechanisms due to the unique mechanical scaffolding structures at plant cell surface and the regulation of trafficking pathways for OMV uptake by the continuum across the cell wall–plant PM–actin cytoskeleton.
Macromolecular remorin assembly and structural assembly of nanodomains mediate OMV interactions with plant cells
We observed that REM1.2 density and intensity was enhanced in OMV-treated plants. In contrast, MβCD significantly reduced REM1.2 signal intensity but only induced a slight but not significant increase in REM1.2 density. Although the increase in REM1.2 density by MβCD was not significant as observed in a previous study (Huang et al., 2019), the difference could be due to the tissue type and imaging technique used.
Both remorin and sterol molecules are actively involved in mediating the integration of OMVs into the plant PM, which is supported by the greatly reduced FM4–64-labeled OMV signal on the plant PM in MβCD-treated seedlings and in rem1.2 1.3c plants. This is in agreement with an earlier work on pathogenic E. coli OMVs demonstrating that MβCD and filipin, which disrupt lipid rafts, could significantly reduce the internalization of OMVs into host cells (O’Donoghue et al., 2017). It was not clear how the lipid raft structure or its assembly contributed to OMV insertion into the host PM at the earlier stage of the OMV–PM interaction.
Here, we showed that macromolecular assembly of remorins and phytosterols, the factors directly controlling nanodomain assembly, are critical for the integration of Xcc OMVs into the plant PM, and that Xcc OMVs are specifically recruited to nanodomain sites occupied by AtREM1.2 when AtREM1.2 clustering is enhanced by overexpression, or during bacterial infection. Remorins have gained much attention in the last several years as accumulating evidence points to the functionality of this protein family in the macromolecular assembly of membrane nanodomains (Gouguet et al., 2020). The critical roles of this protein family and nanodomains in plant development and responses to environmental cues have also been studied extensively, including hormonal responses (Huang et al., 2019; Ke et al., 2021), defense response signaling (Perraki et al., 2017; Tran et al., 2020), symbiosis (Lefebvre et al., 2010; Liang et al., 2018; Su et al., 2020), and interactions with pathogens (Raffaele et al., 2009; Perraki et al., 2014, 2017; Fu et al., 2018; Albers et al., 2019; Khoza et al., 2019). Our results demonstrate that the proper nanodomain organization on the PM is essential for initial OMV insertion, although OMVs are not necessarily recruited to the nanodomain sites. Additional evidence is needed to shed light on the connection between OMV-induced enhancement of endocytosis, lipid packing, and nanodomain assembly because it is not solely limited to AtREM1.2/AtREM1.3-occupied nanodomains.
OMVs increase nanodomain assembly and result in positioning OMVs at the nanodomain sites based on feedback regulation. We found that plant responses to bacterial infection amplified the entrance of bacterial OMVs based on the spatial recruitment of OMVs at the site of the host nanodomains at the early stages of infection, which is when MTI is the primary defense response in progress (Smith and Heese, 2014; Ngou et al., 2021). Such enhanced contact between nanodomain and OMVs by a positive feedback regulation might stimulate better immune responses because several immune receptors and their interacting partners were shown to preferentially occupy nanodomains once receptor-signaling was mounted (Lherminier et al., 2009; Keinath et al., 2010; Qi and Katagiri, 2012). Future studies are needed to identify possible MTI enhancement by OMV integration by studying PRR dynamics and membrane-bound immune receptor signaling capacity.
How OMV insertion changes membrane dynamics within and outside of nanodomains, such as the observed acceleration of endocytosis in plants, has not been clarified. Our computational simulation suggested constrained diffusion of the OMV lipids into the simulated plant PM lipid bilayer. This stage likely had not reached its equilibrium due to the short simulation time and computational power available for our simulation. One plausible explanation for the changes in endocytosis is the multivalent nature of OMV antigens. It is well established that CME mediates ligand-induced endocytosis of immune receptors from the surface of plant cells once they are bound to corresponding ligands (Robatzek et al., 2006; Mbengue et al., 2016; Erwig et al., 2017). Thus, the contact of OMVs, carrying receptor ligands at the surface, with plant cells might accelerate the general endocytosis pathway through the simultaneous introduction of multiple MAMPs (Robatzek et al., 2006; Mbengue et al., 2016). In addition, it is possible that OMV insertion introduced nonequilibrium and nonbalanced flow/dynamics of membrane lipids via the enhanced membrane packing within the nano-compartments, as well as the OMV-exclusive membrane fractions (Cohen and Shi, 2020). Although it remains to be experimentally and systematically validated, one possible explanation is the overall increase in dynamics of the membrane in non-OMV membrane compartments, thereby enhancing the endocytosis rate on the Arabidopsis PM.
In summary, mounting evidence has shown that the interactions between bacterial OMVs and hosts are complex and possibly conveyed through many layers, namely, bacterial physiology and intra- and interspecies interactions (Ionescu et al., 2014; Cooke et al., 2020; Knoke et al., 2020), lipid–lipid interaction (O’Donoghue et al., 2017; Jefferies and Khalid, 2020), MAMP–PRR interaction (Shen et al., 2012; Bahar et al., 2016; Janda et al., 2021), effectors–R proteins interactions (Chowdhury and Jagannadham, 2013). OMVs, therefore, are an essential part of bacterial pathogenesis as well as the basis of host immunity. Here, we reported that Xcc OMVs could directly interact with the Arabidopsis PM, increased the cellular endocytosis rate and PM lipid order, and protect plants from electrolyte leakage and bacterial infection. These phenotypes are dependent on the plant membrane nanodomain assembly mediated by remorins, thus highlighting the important role of remorin-mediated nanodomains as the signaling hubs in regulating plant immunity during plant–microbe interactions. Further investigation is needed to shed light on the complex mechanisms by which diverse pathogen signals, such as OMVs, MAMPs, quorum-sensing molecules, and effectors, orchestrate the communication with plant cells by spatiotemporally fine tuning the macromolecular assembly of remorin complexes and nanodomains. Similar to OMVs produced by human pathogens, the stability of phytopathogen-produced OMVs and their ability to trigger multiple defense response pathways will open new opportunities to engineer OMVs as an underexplored gateway to boost plant immunity and combat plant diseases (Lee et al., 2011; Shen et al., 2012).
Materials and methods
Plant and bacterial culture conditions
Arabidopsis (A. thaliana [L.] Heynh.) plants were maintained regularly on 1/2-strength Murashige–Skoog agar (1/2 MS) at 22°C, under long-day photoperiod (16-h light/8-h dark, 150 μmol m−2 s−1 white light) in a growth chamber (Percival AR95-L2). Plants used for ROS assays, conductivity assays, and immunoblot experiments were grown on soil under short-day photoperiod (8-h light/16-h dark, 120 μmol m−2 s−1 white light) in a growth chamber (Panasonic MLR-352H-PE) to facilitate vegetative growth. Col-0, fls2, efr, and bak1-4 Wave 138Y (YFP-PIP1;4; Geldner et al., 2009) seeds were obtained from the Arabidopsis Resource Center stock center (Ohio, USA). Arabidopsis pREM1.3:YFP-REM1.3, pREM1.2:GFP-REM1.2, XVE:REM1.2-GFP, CLC2-GFP, rem1.2 1.3c, DRP2B-RFP, CLC2-RFP, rem1.2 CLC2-RFP marker lines; and sterol mutants were described previously (Carland et al., 1999; Diener et al., 2000; Schrick et al., 2000; Konopka et al., 2008; Men et al., 2008; Fujimoto et al., 2010; Gadeyne et al., 2014; Jarsch et al., 2014; Huang et al., 2019; Ke et al., 2021).
For cell biology imaging, the seeds were surface-sterilized with 30% bleach and ethanol, then washed with sterile water. Sterilized seeds were vernalized in water at 4°C for 2 days, then sown on 1/2 MS agar plates and grown at 22°C for 6–7 days (16-h light/8-h dark) prior to imaging. For visualization of OMV insertion in Arabidopsis roots, plants were germinated vertically on 1/2 MS agar and imaged at 4 days after sowing. For visualization of OMV insertion in mesophyll protoplasts, the protoplasts were extracted from 4- to 5-week-old plants as described previously (Yoo et al., 2007). Protoplasts or intact seedlings were incubated with labeled OMVs (100 µg·mL−1) for 30 min before being mounted for imaging.
Xcc 8004 was maintained regularly on NYG medium at 28°C. A list of plant, plasmids, and primers used in this study is provided in Supplemental Data Set S3.
Bacterial mutagenesis
The unmarked Xcc deletion mutants Xcc fliC− and hrcC− were constructed by homologous recombination. Briefly, upstream and downstream regions of the Xcc fliC gene were amplified by PCR from Xcc 8004 genomic DNA, then cloned into pK18mobsacB by using the ClonExpress Ultra OneStep Cloning Kit (Vazyme, China). Transformants were selected on LB + Kanamycin plates. Positive clones were then counter-selected on sucrose-supplemented NYG medium for double recombination events. Colonies that grew on sucrose were subsequently verified by Kanamycin susceptibility, as well as PCR and sequencing.
OMV extraction and quantification
Bacterial cultures were grown in NYG medium at 28°C with agitation (200 rpm) overnight. The bacteria were then sub-cultured into 1 L of fresh NYG with a 1:100 dilution from the overnight culture and grown for 24 h with agitation (200 rpm). The bacterial suspension was pelleted at 9,000g for 15 min, and the supernatant was collected and filtered through a 0.22-µm membrane to remove the remaining bacterial cells. The cell-free supernatant was then centrifuged at 120,000g for 1.5 h to pellet the OMVs, using a Beckmann Ti-45 rotor and a Beckmann Ultracentrifuge OPTIMA L-100XP. The OMV pellets were resuspended in 1 mL of PBS buffer and passed through a syringe-mounted 0.22-µm filter. Purified OMVs were stored at 4°C and used within 3 days after the extraction. OMV concentrations represented by total protein content were determined by the Pierce BCA Protein assay kit (Thermo Scientific) using a microtiter plate reader (Biotek, VT).
NTA
To measure the OMV particle size, Xcc OMVs were first diluted in PBS buffer to a final concentration of 1×108 particles·mL−1. One mL of OMVs was then injected into a Nanoparticle Tracking Video microscope (ZetaView PMX-120, Particle Metrix). OMV particle diffusion and size distribution were recorded over 11 positions and analyzed by ZetaView Particle Metrix software.
Labeling of OMVs with FM4–64
Purified OMVs from the cell-free culture supernatant of Xcc 8004 from NYG culture were labeled with 5-mM FM4–64 dye (Life Technologies) for 10 min in the dark at 4°C, then transferred to a 5K Vivaspin column and centrifuged for 30 min at 4°C to remove the free FM4–64 dye. Labeled OMVs were collected from the top of the centrifugation column. The concentrations of OMVs were determined using the Pierce BCA protein assay kit (Thermo Fisher). Labeled OMVs were prepared right after extraction, stored in the dark at 4°C and used within three days after the extraction. As a control, FM4–64 was added to 1× PBS and subjected to centrifugal filtration, then the remaining PBS in the column was used to apply onto Arabidopsis seedlings and imaged with similar settings.
Microscopy
Confocal microscopy
Confocal microscopy was conducted on a Zeiss ELYRA PS.1 + LSM 780 system with a 63X Plan-Apochromat oil-objective (NA = 1.4). The fluorophores GFP/YFP and FM4–64 were excited with 488-nm and 514-nm laser, and fluorescence signals were collected at 493–594 nm and 612–758 nm, respectively.
2D structure-illumination microscopy
Superresolution imaging of FM4–64-labeled OMVs was performed on a spinning disk confocal (SDC) Nikon Ti2 microscope with Live-SR structured illumination microscopy (SDC-SIM, GATACA). OMVs were mounted on a glass slide with acid-cleaned coverslip before imaging. Multifocal structured illumination microscopy (York et al., 2012) achieved with this system allows us to combine the doubled resolution together with the physical optical sectioning of confocal microscopy. The system is equipped with a CSU-W1 confocal spinning head, a 100X Plan-Apo oil-objective (NA = 1.49). The maximum resolution of the system is 128 nm with a pixel size of 64 nm in super resolution mode. Excitation light was provided by 488 nm/150 mW (Vortran) and emissions were collected at 672–812 nm. Image acquisition was from a back-illuminated sCMOS camera (Prime95B, Teledyne Photometrics) and image processing was controlled by MetaMorph 7.10 (Molecular Device) software.
Total internal reflection microscopy
VA-TIRFM imaging of PM markers: CLC2-GFP, EH2-GFP, DRP2B-RFP, CLC2-RFP, rem1.2 1.3c CLC2-RFP, was performed on a Zeiss ELYRA PS.1 + LSM 780 system equipped with a 100X Plan-Apochromat oil-objective (NA = 1.46). To monitor the lifetime of PM markers, time-lapse movies of CLC-GFP, EH2-GFP, and DRP2B-RFP were acquired for 60 frames with 1,000-ms exposure at 1% laser power. TIRF images were deconvolved using Huygens Essential (Scientific Volume Imaging) using theoretical point spread functions. The lifetime of the endocytic markers was measured from kymographs derived from time-lapse TIRF movies in Fiji (Schindelin et al., 2012) with at least 200 individual endocytic events.
TIRF imaging of FM4–64-labeled OMVs with Remorin marker lines: pREM1.2:GFP-REM1.2, pREM1.3:YFP-REM1.3, REM1.2-GFP overexpression line (XVE:REM1.2-GFP) were performed on a Zeiss ELYRA PS.1 + LSM 780 ELYRA system equipped with a 100X Plan-Apochromat oil-objective (NA = 1.46). TIRF images were deconvolved using Huygens Essential (Scientific Volume Imaging) and colocalization between remorins and OMVs was performed using Coloc2 plug-in in Fiji.
Transmission electron microscopy (TEM)
Negative staining of OMVs was performed by directly applying 5 μL of OMVs, at a concentration of 70 μg·mL−1, to the glow discharged carbon-coated copper grid (200 mess, Electron Microscopy Sciences Inc. USA), followed by a 30 s staining with 1% uranyl acetate in water. Air-dried samples were examined at 120 kV with an FEI Tecnai 12 TEM, and images were recorded using an Ultrascan 1000 CCD camera (Gatan, Inc.). OMV sizes were measured from TEM micrographs using Fiji software.
Remorin single-particle analyses
To quantify GFP-REM1.2 foci intensity and dynamics, we captured time-lapse images of REM1.2 seedlings using VA-TIRFM. 5×5 µm2 regions-of-interest (ROIs) in evenly illuminated areas were then cropped from the original images and analyzed in TrackMate (Tinevez et al., 2017) and SpatTrack (Lund et al., 2014) to quantify the signal intensity and MSD of the GFP-REM1.2 foci, respectively. Diffusion coefficient D (µm2·s-1) was defined by the formula: (Δt) = 4D . Δtα in which α represents the diffusion exponent.
Quantification of plant PM lipid order
The lipid order dye di-4-ANEPPDHQ (Thermo Fisher, cat. D36802) was used for imaging and quantification of membrane lipid order. Seven-day old Arabidopsis seedlings were incubated with PBS or 30 μg·mL−1 OMV for 30 min, then washed with 1/2 MS and stained with 5-μM di-4-ANEPPDHQ for 5 min diluted in 1/2 MS. After staining, the seedlings were washed three times with fresh 1/2 MS, mounted and imaged right away using a Zeiss ELYRA PS.1 + LSM 780 system with a 63X Plan-Apochromat oil-objective (NA = 1.4). The dye was excited with single-photon illumination at 488 nm and emission was collected at 500–580 nm (ordered lipid) and 620–750 nm (disordered lipid). Image analysis was performed in Fiji software (Fiji is just ImageJ) as described previously (Owen et al., 2012). For imaging with di-4-ANEPPDHQ dye, three individual seedlings were imaged, with ≥8 images taken for each plant.
Immunoblot of p-MAPK
Leaf discs from 5- to 6-week-old Arabidopsis Col-0 plants were cut into 1-mm strips and floated on the water under constant light for 16 h to reduce stress from wounding. The leaf tissues were elicited with either 10-μM flg22 or 30-μg·mL−1 Xcc OMVs. The leaf strips were taken out at the indicated time-points, washed with deionized water, blotted dry, and flash-frozen in liquid nitrogen. Total protein extraction and immunoblotting using anti-p44/42-MAPK antibody (1:3,000; Cell Signaling Technologies) was carried out as described previously (Tran et al., 2020). Membranes were stained with Ponceau S as the loading control. The experiment was repeated three times with a similar result.
Measurement of ROS
We measured the production of Reactive Oxygen Species in Arabidopsis leaf according to a previously described protocol (Smith et al., 2014). Leaf discs of 0.7 cm were punched from 5- to 6-week-old Arabidopsis Col-0 plants and cut into five strips, then floated on sterile water for 24 h in a 96-well solid-white plate, under constant light to reduce wounding stress. Flg22 (1 μM) or Xcc OMVs (30 μg·mL−1) were mixed into the elicitation solution containing 20 μg·mL−1 horseradish peroxidase and 34 μg mL−1 Luminol and added to leaf strips. The luminescence was monitored immediately using a plate reader (Biotek Cytation 5). The ROS measurement was performed in duplicate, with each treatment performed on N ≥ 4 cut leaf discs.
Quantification of callose deposition in Arabidopsis leaves
Callose deposition in response to OMVs or flg22 was performed on 2-week-old Col-0 plants. The seedlings were placed in six-well plates with 4 mL of ½ MS, and then treated with either 34 μg·mL−1 Xcc OMVs, 1-μM flg22, or PBS (control) for 24 h. The plants were then destained with a solution of acetic acid: ethanol (1:3) overnight, then washed with 150-mM K2HPO4 for 30 min. The leaves were stained with 0.01% aniline blue for 2 h and mounted on glass microscope slides in 50% glycerol. Callose were imaged on a Zeiss ELYRA PS.1 + LSM780 system equipped with a 10× objective (excitation:emission, 370:509 nm). The experiments were repeated twice on eight plants per treatment (N ≥ 20 images). Callose puncta were quantified using Fiji software.
Flooding assay on Arabidopsis
Two-week-old Arabidopsis plants grown on ½ MS agar (22°C, 16-h light/8-h dark) were used for flood-inoculation with Xcc 8004 as described previously (Ishiga et al., 2011). Briefly, plants were flooded with 40 mL of 10-mM MgCl2 solution containing 30 µg·mL−1 OMV or PBS (control). After 1 day, the liquid was decanted, and the seedlings were flooded with a bacterial suspension of 1 × 107 CFU·mL−1 Xcc 8004 prepared in 10-mM MgCl2 for 3 min. Afterward, the bacterial inoculum was removed from the plates and the Arabidopsis seedlings were returned to the growth chamber. Bacterial population was determined at 3 DPI by dilution plating on NYG medium.
Measurement of electrolyte leakage in Arabidopsis leaf discs
Leaf discs from 5- to 6-week-old Arabidopsis seedlings were collected in 50-mL conical tubes filled with 20 mL of deionized water (eight discs/tube). For treatments with OMVs, the leaf discs were incubated with 30 µg·mL−1 OMVs for 24 h prior to bacterial infiltration. For treatment with MβCD, leaf discs were treated for an additional hour prior to the addition of OMVs, or water (as a control for MβCD treatment). After OMV or MβCD treatments, the leaf discs were vacuum infiltrated with Xcc 8004 or the T3SS hrcC− mutant (as negative control). Fully infiltrated leaf discs (discs that sank to the bottom of the tubes after infiltration) were washed with deionized water to remove leaked electrolytes caused by wounding, then distributed into new conical tubes filled with 20 mL of deionized water. Electrolyte leakage was measured by a conductivity meter (EC210, Horiba) for 30 h. Area under curves of the conductivity increase were calculated by GraphPad Prism 8.0. Differences between treatments were analyzed by one-way analysis of variance (ANOVA) (GraphPad Prism). The experiments were repeated twice with similar results.
Lipidomic analysis of Xcc OMVs
Xcc OMVs were extracted from liquid NYG cultures as described above. Supernatant from 800 mL of NYG culture was centrifuged and pooled into a single OMV pellet as a biological replicate. Vesicle pellet samples (N = 6) was treated with 1.5 mL of methanol to extract the compounds and followed by sonication for 10 min in ice. After centrifugation at 21.1g for 10 min at 4°C, the supernatant was transferred to a new tube and samples were dried using Speed Vac. Dried extracted samples were stored at −20°C and, then 160 µL of methanol and 40 µL of the internal standard were added to the samples before analysis.
Phospholipids were identified using ACQUITY UPLC System (Waters) coupled to Xevo TQ-S triple quadrupole mass spectrometer (Waters). Chromatographic separation was performed on an ACQUITY UPLC BEH C18 (2.1 mm × 100 mm, 1.7 µm) column (Waters) using isocratic elution of solvent A (methanol/water 98/2, vol/vol, 5-mM ammonium acetate, 0.01% formic acid) for positive MS mode and solvent B (methanol/water 98/2, vol/vol, 10-mM ammonium acetate, 0.01% formic acid) for negative MS mode. The flow rate was 0.4 mL·min−1, column temperature was 45°C, and the injection volume was 3 µL. The ion source for both positive and negative modes was operated at the following conditions: capillary voltage, 3.5 kV; desolvation temperature, 350°C; cone gas flow, 150 L·h−1; desolvation gas flow, 650 L·h−1; source temperature, 150°C. The injection order of six samples was performed together with injections of pooled quality control and extracted blank samples to monitor system stability and carryover.
Molecular simulation of the interaction of lipid vesicles with a planar membrane
To study how OMV interacts with the plant host PM, we constructed molecular models of simplified OMV-mimic and plant PM mimic. For the OMV-mimic, we constructed a 23-nm-diameter vesicle composed of POPC phospholipids based on the coarse-grained MARTINI model (Marrink et al., 2004, 2007), following the same protocol published elsewhere (Chng et al., 2021). The solvent-free Dry MARTINI version 2.1 (Arnarez et al., 2015) was used in order to reduce computational cost. The initial configuration and simulation set-up files were generated using the Martini Bilayer Maker (Qi et al., 2015) within the CHARMM-GUI web-based platform (Jo et al., 2008). GROMACS version 4.6.7 MD software was used for the simulations (Abraham et al., 2015). Six 2-nm-diameter pores were symmetrically placed in the vesicle to allow the exchange of inner and outer water as well as lipid flip-flops between the inner and outer leaflets in the initial configuration. After energy minimization steps to remove any steric clashes present, MD simulations were carried out with progressively reduced restraints on lipid tail positions to progressively close the pores. Unrestrained simulations were subsequently carried out for 200 ns for equilibration of the vesicle.
For the plant PM mimic, we constructed a 160 × 160 nm patch of planar bilayer membrane composed of 50% POPC and 50% sterol using the INSANE Python script (Wassenaar et al., 2015). We used cholesterol to replace sitosterol as the latter is not available in the MARTINI model and their molecular structures are similar to each other. As a control, we also constructed a 160 × 160 nm patch of planar bilayer membrane composed only of POPC. Same as the vesicle, the Dry MARTINI model was used to simulate the planar membranes. After energy minimization steps, MD simulations were carried out with progressively reduced restraints on lipid head-group positions. Unrestrained simulations were subsequently carried out for 250 ns for each membrane for further equilibration. After equilibration, the POPC:sterol membrane size was reduced to 130 nm × 130 nm, while the control was increased slightly to 164 nm × 164 nm.
The vesicle was then placed atop either the POPC or POPC:sterol membrane, and three short MD simulation runs of 2 ns each with progressively reduced restraints on lipid head-group positions were performed to ease the vesicle onto the membrane surface. Unrestrained simulations were subsequently carried out for 1,000 ns for the POPC:sterol membrane and 1,200 ns for the POPC membrane.
For both simulations, electrostatic interactions were computed using the Reaction-Field method with the dielectric constant of 15 and cut-off distance of 1.1 nm. Van der Waals interactions were computed using cut-off method with the same distance of 1.1 nm. System temperature was maintained at 310 K with the velocity-rescale thermostat. The planar membrane was maintained at a constant tension at a physiologically relevant level of σ ≈ 0.1 mN·m-1 (Gauthier et al., 2012) by applying an in-plane pressure of −0.2 bar. The pressure was maintained using the Berendsen barostat with semi-isotropic coupling (X–Y plane coupled separately from the Z or bilayer normal direction), with a time-constant of 4 ps and compressibility of 3.4 × 10−4 bar−1 (Arnarez et al., 2015). The simulation time-step was 40 fs for simulation with POPC membrane and 20 fs for simulation with POPC:sterol membrane.
Molecular simulation of model A. thaliana PM
We constructed a simplified model of the Arabidopsis PM based on the lipid composition determined by Uemura et al. (1995). Detailed lipid compositions are listed in Supplemental Data Set S2. Martini Bilayer Maker within the CHARMM-GUI web-based platform was used to construct the model bilayer membrane using the standard MARTINI version 2.2 model with the various lipids uniformly distributed within each monolayer. Cholesterol was used as a replacement for sitosterol, which is not available in the standard MARTINI model. Energy minimization steps and restrained MD simulations were carried out following the same protocol as introduced above for the plant PM mimic. Unrestrained simulations were subsequently carried out for 2,000 ns for each membrane for further equilibration. As the simulation domain contained water particles (and neutralizing ions) above and below the membrane, the system pressure was maintained at 1 bar with the same Berendsen barostat with semi-isotropic coupling (X–Y plane coupled separately from the Z or bilayer normal direction), with time-constant of 5 ps and compressibility of 3.4 × 10−4 bar−1. The system temperature was maintained at 295 K with the velocity-rescale thermostat. The simulation time-step was 10 fs.
Calculation of lipid order parameter and membrane elastic modulus of a planar membrane
The order parameter was calculated as a measure of lipid order in the membrane, where is the angle between a selected lipid bond and the axis perpendicular to the bilayer membrane. To calculate the value for a specific bond within a specific type of lipids, this angle was averaged over all lipids of the same type in the membrane over a simulation time interval of 1,900–2,000 ns. The order parameter takes a maximum value of 1 if the bond is perfectly aligned along the membrane normal, and a minimum value of −1/2 if the bond is perpendicular to the membrane normal. The overall lipid order () of a specific lipid type was calculated by averaging the values for all the bonds along with the lipid tails of the same type of lipids. The order parameter was calculated using an in-house TCL script running within the Visual Molecular Dynamics software environment, which was also used to generate the molecular figures in Figure 4 (Humphrey et al., 1996). The abbreviations of lipids included in the calculation of lipid order are listed in Supplemental Data S4 and Figure 4 legend.
As a mechanical property of the membrane used to characterize its resistance to in-plane stretching, the elastic modulus (also known as area compression modulus) of our model A. thaliana PM and its PE variants was estimated based on the fluctuations in the projected membrane area () as (Marrink et al., 2004), where denotes averaging the area over simulation time interval (1,900–2,000 ns), is the Boltzmann constant and is the system temperature in Kelvin.
Quantification and statistical analysis
Statistical analyses and data visualization were conducted in GraphPad Prism 8.0 (GraphPad, CA, USA). Mean comparisons were performed using one-way ANOVA (ns, not significant, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P < 0.000). Unless specified, data were presented as box-and-whiskers plots with bars indicating the median, 25% and 75% quartiles. Means are indicated by “+” symbol in box-and-whiskers plots. Raw data and tables for statistical analysis are provided in Supplemental Data Set S4.
Accession numbers
Major genes described in this work can be found in the Genbank/TAIR database under the following accession numbers: FLS2 (AT5G46330), EFR (AT5G20480), BAK1 (AT4G33430), REM1.2 (AT3G61260), REM1.3 (AT2G45820), FACKEL (AT3G52940), SMT1 (AT5G13710), CPI1 (AT5G50375), CVP1 (AT1G20330), PIP1;4 (AT4G00430), EH2 (AT1G21630), CLC2 (AT2G40060), DRP2B (AT1G59610).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Xcc OMVs enhanced endocytosis in A. thaliana.
Supplemental Figure S2. Nanodomain-dependent acceleration of plant CME by Xcc OMVs.
Supplemental Figure S3. The effect of pharmacological inhibitors and mutations in the sterol biosynthesis pathways on OMVs uptake by Arabidopsis seedlings.
Supplemental Figure S4. Sterols regulate membrane integration of Xcc OMVs.
Supplemental Figure S5. Colocalization of REM1.2 in XVE:GFP-REM1.2 seedlings (REM1.2-OE) with FM4–64-labelled OMVs after β-estradiol induction.
Supplemental Figure S6. Xcc OMVs trigger various plant defense responses.
Supplemental Data Set S1. Composition and saturation level of lipids from Xcc OMVs and Arabidopsis. Data of the Arabidopsis lipidomic profile was reported in our previous study (Tran et al., 2020).
Supplemental Data Set S2. Lipid species in the simplified plant membrane model for simulation.
Supplemental Data Set S3. Primers, plasmids, and plant materials.
Supplemental Data Set S4. Raw data and statistical analyses.
Supplementary Material
Acknowledgments
The authors thank Tingyan Dong for help with constructing the hrcC- and fliC− deletion mutants. We thank Dr Thomas Ott (University of Freiburg) for sharing the Arabidopsis pREM1.3:YFP-REM1.3 line, Dr Xu Chen (Fujian Agriculture and Forestry University) for sharing the Arabidopsis rem1.2 1.3c, XVE:REM1.2-GFP, pREM1.2:GFP-REM1.2, CLC2-RFP, rem1.2 1.3c CLC2-RFP seeds; Dr Kathrin Schrick (Kansas State University) and Dr Jyan-Chun Jang (Ohio State University) for sharing the Arabidopsis sterol biosynthesis mutants, Dr Han Wei Hou (Nanyang Technological University) for helping with the NTA assay. We also deeply appreciate Dr Jian-Feng Li (Sun Yat-sen University) for sharing the efr and bak1-4 lines in time for the revision of this paper. We thank Dr Yulan Wang, Dr Liang Xu, and Yeong Cheng Cheah (Singapore Phenome Centre) for assistance with the OMV lipidomics analysis. We thank the Advance Biofilm Imaging Facility at Singapore Centre for Environmental Life Sciences Engineering for providing the platform and technical assistance for the cell biology imaging in this work. We also thank Dr Eunyoung Chae (National University of Singapore) for critical feedback on the manuscript.
Author contributions
TMT, XP, YM designed and conceived the experiments. TMT, XP, ZM, XL, XH, performed the experiments. CC and CH performed the computational simulations. TMT, CC, CH and YM wrote the paper with feedback from other authors.
Funding
This study was supported by Nanyang Technological University startup grant (M4081533), Ministry of Education (MOE) Tier 1 (RG32/20; RT11/20), and Tier 3 (MOE2019-T3-1-012) grants to YM; Nanyang Technological University startup grant (M4082352), MOE Tier 1 (RG92/19), and Accelerating Creativity and Excellence grant (NTU–ACE2020-07) to CH. The computational work in this study was fully performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg).
Conflict of interest statement. The authors declared no conflict of interest.
T.M.T., X.P., and Y.M. designed and conceived the experiments. T.M.T., X.P., Z.M., X.L., and X.H. performed the experiments. C.C. and C.H. performed the computational simulations. T.M.T., C.C., C.H., and Y.M. wrote the paper with feedback from other authors.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Yansong Miao (yansongm@ntu.edu.sg) and Changjin Huang (cjhuang@ntu.edu.sg)
References
- Abel NB, Buschle CA, Hernandez-Ryes C, Burkart SS, Deroubaix A-F, Mergner J, Gronnier J, Jarsch IK, Folgmann J, Braun KH (2021) A hetero-oligomeric remorin-receptor complex regulates plant development. bioRxiv, 10.1101/2021.01.28.428596 (January 29, 2021) [DOI]
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1: 19–25 [Google Scholar]
- Adani F, Papa G, Schievano A, Cardinale G, D’Imporzano G, Tambone F (2011) Nanoscale structure of the cell wall protecting cellulose from enzyme attack. Environ Sci Technol 45: 1107–1113 [DOI] [PubMed] [Google Scholar]
- Albers P, Üstün S, Witzel K, Kraner M, Börnke F (2019) A remorin from Nicotiana benthamiana interacts with the Pseudomonas type-III effector protein HopZ1a and is phosphorylated by the immune-related kinase PBS1. Mol Plant-Microbe Interact 32: 1229–1242 [DOI] [PubMed] [Google Scholar]
- Arnarez C, Uusitalo JJ, Masman MF, Ingólfsson HI, De Jong DH, Melo MN, Periole X, De Vries AH, Marrink SJ (2015) Dry Martini, a coarse-grained force field for lipid membrane simulations with implicit solvent. J Chem Theory Comput 11: 260–275 [DOI] [PubMed] [Google Scholar]
- Bahar O (2020) Membrane vesicles from plant pathogenic bacteria and their roles during plant–pathogen interactions. In M Kaparakis-Liaskos, TA Kufer, eds, Bacterial Membrane Vesicles. Springer, New York, pp 119–129 [Google Scholar]
- Bahar O, Mordukhovich G, Luu DD, Schwessinger B, Daudi A, Jehle AK, Felix G, Ronald PC (2016) Bacterial outer membrane vesicles induce plant immune responses. Mol Plant-Microbe Interact 29: 374–384 [DOI] [PubMed] [Google Scholar]
- Bomberger JM, MacEachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA (2009) Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathogens 5: e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington K, Kuehn M (2014) Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 1843: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland FM, Berg BL, FitzGerald JN, Jinamornphongs S, Nelson T, Keith B (1999) Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 11: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N, Sabularse D, Montezinos D, Delmer DP (1979) Determination of the pore size of cell walls of living plant cells. Science 205: 1144–1147 [DOI] [PubMed] [Google Scholar]
- Cecil JD, Sirisaengtaksin N, O'Brien-Simpson NM, Krachler AM (2019) Outer membrane vesicle‐host cell interactions. Microbiol Spectr 7: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yang D, Wen Y, Jiang Z, Zhang L, Jiang J, Chen Y, Hu T, Wang Q, Zhang Y (2018) Dysregulated hemolysin liberates bacterial outer membrane vesicles for cytosolic lipopolysaccharide sensing. PLoS Pathogens 14: e1007240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Yang Z, Zhang H, Zhang J, Xu J (2020) Remorin interacting with PCaP1 impairs Turnip mosaic virus intercellular movement but is antagonised by VPg. New Phytol 225: 2122–2139 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Li J-F, Niu Y, Zhang X-C, Woody OZ, Xiong Y, Djonović S, Millet Y, Bush J, McConkey BJ (2015) Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng C-P, Sadovsky Y, Hsia KJ, Huang C (2021) Curvature-regulated lipid membrane softening of nano-vesicles. Extreme Mech Lett 43: 101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Jagannadham MV (2013) Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta 1834: 231–239 [DOI] [PubMed] [Google Scholar]
- Cohen AE, Shi Z (2020) Do cell membranes flow like honey or jiggle like jello? BioEssays 42: 1900142. [DOI] [PubMed] [Google Scholar]
- Cooke AC, Florez C, Dunshee EB, Lieber AD, Terry ML, Light CJ, Schertzer JW (2020) Pseudomonas quinolone signal-induced outer membrane vesicles enhance biofilm dispersion in Pseudomonas aeruginosa. Msphere 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo P, Chow SH, Hay ID, Kleifeld O, Costin A, Elgass KD, Jiang J-H, Ramm G, Gabriel K, Dougan G (2018) Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathogens 14: e1006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W-x, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktorova M, LeVine MV, Khelashvili G, Weinstein H (2019) A new computational method for membrane compressibility: Bilayer mechanical thickness revisited. Biophys J 116: 487–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward DW, Garon CF (1990) DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl Environ Microbiol 56: 1960–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig J, Ghareeb H, Kopischke M, Hacke R, Matei A, Petutschnig E, Lipka V (2017) Chitin‐induced and Chitin Elicitor Receptor Kinase1 (CERK1) phosphorylation‐dependent endocytosis of Arabidopsis thaliana Lysin Motif‐Containing Receptor‐Like Kinase5 (LYK5). New Phytol 215: 382–396 [DOI] [PubMed] [Google Scholar]
- Feitosa-Junior OR, Stefanello E, Zaini P, Nascimento R, Pierry PM, Dandekar A, Lindow S, da Silva AM (2019) Proteomic and metabolomic analyses of Xylella fastidiosa OMV-enriched fractions reveal association with virulence factors and signaling molecules of the DSF family. Phytopathology 109: 1344–1353 [DOI] [PubMed] [Google Scholar]
- Fu S, Xu Y, Li C, Li Y, Wu J, Zhou X (2018) Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol Plant 11: 269–287 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S-i, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N (2010) Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704 [DOI] [PubMed] [Google Scholar]
- Gauthier NC, Masters TA, Sheetz MP (2012) Mechanical feedback between membrane tension and dynamics. Trends Cell Biol 22: 527–535 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud‐Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gong B-Q, Guo J, Zhang N, Yao X, Wang H-B, Li J-F (2019) Cross-microbial protection via priming a conserved immune co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe 26: 810–822.e7 [DOI] [PubMed] [Google Scholar]
- Gouguet P, Gronnier J, Legrand A, Perraki A, Jolivet M-D, Deroubaix A-F, German-Retana S, Boudsocq M, Habenstein B, Mongrand S (2021) Connecting the dots: from nanodomains to physiological functions of REMORINs. Plant Physiol 185: 632–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Fan L, Chen T, Li R, Li X, He Q, Botella MA, Lin J (2014) Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26: 1729–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Sun Y, Ma Z, Ke M, Cui Y, Chen Z, Chen C, Ji C, Tran TM, Yang L (2019) Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc Natl Acad Sci 116: 21274–21284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graphics 14: 33–38 [DOI] [PubMed] [Google Scholar]
- Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE (2014) Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci USA 111: E3910–E3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS (2011) Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger J, Keese S, Roessle M, Steinert M, Schromm AB (2015) Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell Microbiol 17: 607–620 [DOI] [PubMed] [Google Scholar]
- Janda M, Ludwig C, Rybak K, Meng C, Stigliano E, Botzenhardt L, Szulc B, Sklenar J, Menke FL, Malone JG (2021). Biophysical and proteomic analyses suggest functions of Pseudomonas syringae pv tomato DC3000 extracellular vesicles in bacterial growth during plant infection. bioRxiv. [DOI] [PMC free article] [PubMed]
- Jarsch IK, Konrad SSA, Stratil TF, Urbanus SL, Szymanski W, Braun P, Braun K-H, Ott T (2014) Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26: 1698–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies D, Khalid S (2020) To infect or not to infect: molecular determinants of bacterial outer membrane vesicle internalization by host membranes. J Mol Biol 432: 1251–1264 [DOI] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W (2008) CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J Comput Chem 29: 1859–1865 [DOI] [PubMed] [Google Scholar]
- Johansson ON, Nilsson AK, Gustavsson MB, Backhaus T, Andersson MX, Ellerström M (2015) A quick and robust method for quantification of the hypersensitive response in plants. PeerJ 3: e1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J (2013) Importance of the hexagonal lipid phase in biological membrane organization. Front Plant Sci 4: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J (2010) Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol 12: 372–385 [DOI] [PubMed] [Google Scholar]
- Katsir L, Bahar O (2017) Bacterial outer membrane vesicles at the plant–pathogen interface. PLoS Pathogens 13: e1006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, Ma Z, Wang D, Sun Y, Wen C, Huang D, Chen Z, Yang L, Tan S, Li R (2021) Salicylic acid regulates PIN2 auxin transporter hyperclustering and root gravitropic growth via Remorin‐dependent lipid nanodomain organisation in Arabidopsis thaliana. New Phytol 229: 963–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoza TG, Dubery IA, Piater LA (2019) Identification of candidate ergosterol-responsive proteins associated with the plasma membrane of Arabidopsis thaliana. Int J Mol Sci 20: 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoke LR, Abad Herrera S, Götz K, Justesen BH, Günther Pomorski T, Fritz C, Schäkermann S, Bandow JE, Aktas M (2020) Agrobacterium tumefaciens small lipoprotein atu8019 is involved in selective outer membrane vesicle (OMV) docking to bacterial cells. Front Microbiol 11: 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Backues SK, Bednarek SY (2008) Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel Y, Barny MA (2016) Gram‐negative phytopathogenic bacteria, all hemibiotrophs after all? Mol Plant Pathol 17: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Kim S-H, Kang W, Choi YS, Lee S-H, Lee S-R, You S, Lee HK, Chang K-T, Shin E-C (2011) Adjuvant effect of bacterial outer membrane vesicles with penta-acylated lipopolysaccharide on antigen-specific T cell priming. Vaccine 29: 8293–8301 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand A, Martinez D, Grélard A, Berbon M, Morvan E, Tawani A, Loquet A, Mongrand S, Habenstein B (2019) Nanodomain clustering of the plant protein remorin by Solid-State NMR. Front Mol Biosci 6: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lherminier J, Elmayan T, Fromentin J, Elaraqui KT, Vesa S, Morel J, Verrier J-L, Cailleteau B, Blein J-P, Simon-Plas F (2009) NADPH oxidase-mediated reactive oxygen species production: subcellular localization and reassessment of its role in plant defense. Mol Plant-Microbe Interact 22: 868–881 [DOI] [PubMed] [Google Scholar]
- Liang P, Stratil TF, Popp C, Marín M, Folgmann J, Mysore KS, Wen J, Ott T (2018) Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc Natl Acad Sci USA 115: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund FW, Jensen MLV, Christensen T, Nielsen GK, Heegaard CW, Wüstner D (2014) SpatTrack: an imaging toolbox for analysis of vesicle motility and distribution in living cells. Traffic 15: 1406–1429 [DOI] [PubMed] [Google Scholar]
- Ma Z, Liu X, Nath S, Sun H, Tran TM, Yang L, Mayor S, Miao Y (2021) Formin nano-clustering mediated actin assembly during plant flagellin and DSF- signalings. Cell Rep 34: 108884. [DOI] [PubMed] [Google Scholar]
- Marrink SJ, De Vries AH, Mark AE (2004) Coarse grained model for semiquantitative lipid simulations. J Phys Chem B 108: 750–760 [Google Scholar]
- Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, De Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111: 7812–7824 [DOI] [PubMed] [Google Scholar]
- Mbengue M, Bourdais G, Gervasi F, Beck M, Zhou J, Spallek T, Bartels S, Boller T, Ueda T, Kuhn H (2016) Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci USA 113: 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan HM, Zebell SG, Ristaino JB, Dong X, Kuehn MJ (2021) Protective plant immune responses are elicited by bacterial outer membrane vesicles. Cell Rep 34: 108645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof Y-D, Hartmann M-A, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T (1995) Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun 211: 396–403 [DOI] [PubMed] [Google Scholar]
- Nagakubo T, Nomura N, Toyofuku M (2020) Cracking open bacterial membrane vesicles. Front Microbiol 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn H-K, Ding P, Jones JD (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592: 110–115 [DOI] [PubMed] [Google Scholar]
- O’Donoghue EJ, Sirisaengtaksin N, Browning DF, Bielska E, Hadis M, Fernandez-Trillo F, Alderwick L, Jabbari S, Krachler AM (2017) Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathogens 13: e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orädd G, Shahedi V, Lindblom G (2009) Effect of sterol structure on the bending rigidity of lipid membranes: a 2H NMR transverse relaxation study. Biochim Biophys Acta 1788: 1762–1771 [DOI] [PubMed] [Google Scholar]
- Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, Gaus K (2012) Quantitative imaging of membrane lipid order in cells and organisms. Nat Protoc 7: 24. [DOI] [PubMed] [Google Scholar]
- Perraki A, Binaghi M, Mecchia MA, Gronnier J, German-Retana S, Mongrand S, Bayer E, Zelada AM, Germain V (2014) StRemorin1. 3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett 588: 1699–1705 [DOI] [PubMed] [Google Scholar]
- Perraki A, Gronnier J, Gouguet P, Boudsocq M, Deroubaix A-F, Simon V, German-Retana S, Zipfel C, Bayer E, Mongrand S (2017) The plant calcium-dependent protein kinase CPK3 phosphorylates REM1. 3 to restrict viral infection. bioRxiv, 205765.
- Qi Y, Katagiri F (2012) Membrane microdomain may be a platform for immune signaling. Plant Signal Behav 7: 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Ingólfsson HI, Cheng X, Lee J, Marrink SJ, Im W (2015) CHARMM-GUI Martini maker for coarse-grained simulations with the Martini force field. J Chem Theory Comput 11: 4486–4494 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, Retana SG, Boubekeur T, Leborgne-Castel N, Carde J-P, Lherminier J, Noirot E (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor R, Der C, Grosjean K, Anca I, Noirot E, Leborgne-Castel N, Lochman J, Simon-Plas F, Gerbeau-Pissot P (2016) Plasma membrane order and fluidity are diversely triggered by elicitors of plant defence. J Exp Bot 67: 5173–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188: 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK (2012) Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu VK, Vorhölter F-J, Niehaus K, Watt SA (2008) Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MLA, Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud M-C, Jaillais Y (2016) A PtdIns (4) P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Heese A (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A (2014) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol 164: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Owen DM, Lorenz CD, Makarova M (2020) Asymmetric phospholipids impart novel biophysical properties to lipid bilayers allowing environmental adaptation. bioRxiv.
- Solé M, Scheibner F, Hoffmeister A-K, Hartmann N, Hause G, Rother A, Jordan M, Lautier M, Arlat M, Büttner D (2015) Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps-type II secretion system and outer membrane vesicles. J Bacteriol 197: 2879–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Klein M-L, Hernández-Reyes C, Batzenschlager M, Ditengou FA, Lace B, Keller J, Delaux P-M, Ott T (2020) The Medicago truncatula DREPP protein triggers microtubule fragmentation in membrane nanodomains during symbiotic infections. Plant Cell 32: 1689–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima-Kambe T, Uchiyama H, Nomura N (2011) Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa. Biosci Biotechnol Biochem 75: 605–607 [DOI] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017) TrackMate: an open and extensible platform for single-particle tracking. Methods 115: 80–90 [DOI] [PubMed] [Google Scholar]
- Tran TM, Ma Z, Triebl A, Nath S, Cheng Y, Gong B-Q, Han X, Wang J, Li J-F, Wenk MR, et al. (2020) The bacterial quorum sensing signal DSF hijacks Arabidopsis thaliana sterol biosynthesis to suppress plant innate immunity. Life Sci Alliance 3: e202000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL (1995) Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol 109: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP, Marrink SJ (2015) Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11: 2144–2155 [DOI] [PubMed] [Google Scholar]
- Xue J, Gong BQ, Yao X, Huang X, Li JF (2020) BAK1‐mediated phosphorylation of canonical G protein alpha during flagellin signaling in Arabidopsis. J Integr Plant Biol 62: 690–701 [DOI] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S (2009) Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol 9: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York AG, Parekh SH, Dalle Nogare D, Fischer RS, Temprine K, Mione M, Chitnis AB, Combs CA, Shroff H (2012) Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nature Methods 9: 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.