Abstract
Autophagy is an intracellular trafficking mechanism by which cytosolic macromolecules and organelles are sequestered into autophagosomes for degradation inside the vacuole. In various eukaryotes including yeast, metazoans, and plants, the precursor of the autophagosome, termed the phagophore, nucleates in the vicinity of the endoplasmic reticulum (ER) with the participation of phosphatidylinositol 3-phosphate (PI3P) and the coat protein complex II (COPII). Here we show that Arabidopsis thaliana FYVE2, a plant-specific PI3P-binding protein, provides a functional link between the COPII machinery and autophagy. FYVE2 interacts with the small GTPase Secretion-associated Ras-related GTPase 1 (SAR1), which is essential for the budding of COPII vesicles. FYVE2 also interacts with ATG18A, another PI3P effector on the phagophore membrane. Fluorescently tagged FYVE2 localized to autophagic membranes near the ER and was delivered to vacuoles. SAR1 fusion proteins were also targeted to the vacuole via FYVE2-dependent autophagy. Either mutations in FYVE2 or the expression of dominant-negative mutant SAR1B proteins resulted in reduced autophagic flux and the accumulation of autophagic organelles. We propose that FYVE2 regulates autophagosome biogenesis through its interaction with ATG18A and the COPII machinery, acting downstream of ATG2.
In Arabidopsis, the proper progression of autophagy requires the effector FYVE2 and its interacting protein SAR1B, a regulator of vesicle budding at the endoplasmic reticulum.
Introduction
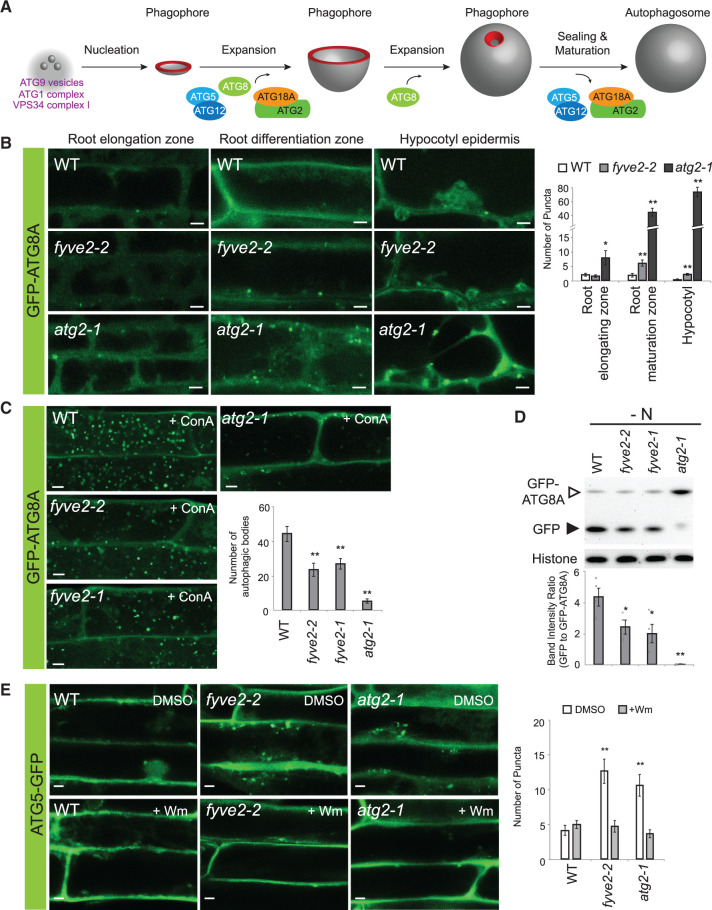
Autophagy is a membrane trafficking pathway for the degradation of cytoplasmic constituents, such as the cytosol, protein complexes and aggregates, organelles, and even invading pathogens (Marshall and Vierstra, 2018). In the best characterized type of autophagy termed macroautophagy (hereafter referred to as autophagy), a cup-shaped precursor called the phagophore expands to sequester a portion of the cytoplasm, finally closing itself to form a double-membrane organelle termed the autophagosome (Figure 1). When the outer membrane of the autophagosome fuses with the limiting membrane of the vacuole (or lysosomes in metazoans), the inner membrane containing autophagic cargo is released as an autophagic body into the vacuolar lumen and is rapidly degraded by the resident hydrolases.
Figure 1.
fyve2 mutants over-accumulate autophagic vesicles and show a reduction in autophagic flux. A, A plant model depicting distinct steps during autophagosome biogenesis. The highly curved rim and neck of the phagophore are highlighted in red. B, Confocal fluorescence images showing the different types of cells in WT (top row), fyve2-2 (middle), and atg2-1 (bottom) seedlings expressing GFP-ATG8A. Seedlings were incubated in N-deficient (−N) liquid medium for 48 h prior to microscopic observation of young cells at the root elongation zone (left column), mature cells at the root differentiation zone (middle), and mature hypocotyl epidermal cells (right). The graph on the right shows the density of GFP-ATG8A puncta per 25,514 µm2. C, Observation of autophagic bodies from WT, fyve2-2, fyve2-1, and atg2-1 root cells expressing the autophagic marker GFP-ATG8A. Seedlings were incubated in −N liquid medium for 48 h and 0.5-µM ConA was added to the liquid medium 16 h prior to confocal microscopy. The graph shows quantification of autophagic bodies per 1,000 µm2 area of the central vacuole. D, Determination of autophagic flux in WT, fyve2-2, fyve2-1, and atg2-1 seedlings by GFP-ATG8A cleavage assay. Seedlings were grown in −N liquid medium for 48 h prior to protein extraction for immunoblot analysis with anti-GFP (upper image) or anti-histone H3 antibodies (lower image for the loading control). The graph shows the quantification of relative band intensities. E, Confocal fluorescence images showing the mature root cells of 9-day-old WT, fyve2-2, and atg2-1 seedlings expressing the phagophore marker ATG5-GFP. Seedlings were incubated in −N liquid medium for 48 h prior to microscopic observation. The graph shows the density of ATG5-GFP puncta per 25,514 µm2. Columns marked with asterisks represent mutants that are significantly different from WT, according to t test. Mean ± se; n = 10–13 images (B), 16–18 images (C), 4 seedling populations (D), 13–15 images (E). *0.01 < P < 0.05; **P < 0.01. Scale bars = 5 µm.
In yeast, autophagy is regulated and executed by a conserved set of core Autophagy-related (Atg) proteins, including the Atg1 kinase complex containing Atg1, Atg13, and either Atg17 or Atg11 as a scaffold protein; the phosphatidylinositol (PI) 3-kinase complex I consisting of Vacuolar Protein Sorting 34 (Vps34), Vps15, Atg6, and Atg14; the transmembrane protein Atg9; the Atg2–Atg18 complex required for phagophore expansion; and the Atg8/12 conjugation system consisting of Atg3, 4, 5, 7, 8, 10, 12, and 16 (Shibutani and Yoshimori, 2014). The Atg8/12 conjugation system attaches the ubiquitin-like Atg8 protein to the membrane lipid phosphatidylethanolamine. Fluorescent tags fused to Atg8 have been used to visualize autophagic organelles, such as phagophores, autophagosomes, and autophagic bodies. Genetic studies of yeast, animals, and plants lacking core Atg genes have revealed essential and conserved roles of autophagy in adaptation to adverse environmental conditions, such as nutrient starvation, pathogen infection, and proteotoxic stress (Avin-Wittenberg et al., 2018; Marshall and Vierstra, 2018). Arabidopsis thaliana atg7 and atg5 mutants are hypersensitive to nutrient limitation but otherwise can grow to maturity and produce progeny. In general, Arabidopsis mutants lacking the functional ATG8/12 conjugation system or ATG2 show more severe defects and a greater reduction in autophagic flux than those lacking either ATG9 or a component of the ATG1 kinase complex (Suttangkakul et al., 2011; Li et al., 2014; Shin et al., 2014; Zhuang et al., 2017; Huang et al., 2019).
How phagophores form is still a matter of active debate. The endoplasmic reticulum (ER) has been proposed as a major site for autophagosome formation in yeast and mammals (Axe et al., 2008; Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009), where autophagosome biogenesis requires the concerted action of PI 3-phosphate (PI3P) and core Atg proteins. The PI3-kinase complex I is responsible for the synthesis of PI3P, which is enriched at the edges of the expanding phagophore (Figure 1A). Atg2 forms a complex with PI3P-binding Atg18 and acts as a tether between phagophores to ER membranes (Chowdhury et al., 2018; Kotani et al., 2018). Yeast and mammalian Atg2 homologs have also been shown to have phospholipid transfer activity (Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019), likely supplying phospholipids from the ER to the phagophore. Atg2 interacts with Atg9, which through its lipid scramblase activity mediates the transport of phospholipids between the luminal and cytoplasmic leaflets of the phagophore membranes (Maeda et al., 2020; Matoba et al., 2020).
Phagophore formation also requires the coat protein complex II (COPII) vesicle budding machinery at the ER exit site (ERES) and the ER–Golgi intermediate compartment in yeast and mammals (Zoppino et al., 2010; Graef et al., 2013; Suzuki et al., 2013; Tan et al., 2013; Ge et al., 2014, 2017; Davis et al., 2016; Shima et al., 2019). Using Arabidopsis as a model for plant autophagy, Zeng et al. (2021) recently showed that phagophore formation requires a subpopulation of COPII vesicles. How COPII vesicles contribute to autophagy remains unclear (Nakatogawa, 2020). It is debatable whether COPII vesicles are a major source for membrane expansion during autophagosome biogenesis. Alternatively, COPII vesicles may supply the phagophore with specific ER proteins or lipids that are essential for autophagosome formation.
Several lines of evidence support the notion that PI3P plays a conserved role in plant autophagy (Chung, 2019). First, treatment with the PI 3-kinase inhibitor wortmannin (Wm) suppresses autophagy in Arabidopsis (Zhuang et al., 2013; Shin et al., 2014). Second, mutations in the components of the PI 3-kinase complexes impair autophagy (Liu et al., 2005; Liu et al., 2020). Third, PI3P-interacting ATG18A and its binding partner ATG2 are essential for plant autophagy (Xiong et al., 2005; Inoue et al., 2006; Kang et al., 2018; Jiang et al., 2020). Last, two putative plant-specific proteins containing a PI3P-binding Fab1p, YOTB, Vac1p, and EEA1 (FYVE) domain, FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING1 (FREE1)/FYVE1 and FYVE2, mediate autophagy in Arabidopsis (Gao et al., 2015; Sutipatanasomboon et al., 2017).
FYVE1/FREE1/VFD1 interacts with VPS23, a component of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery, and is required for normal endosomal and autophagic trafficking, vacuolar dynamics, and plant development (Barberon et al., 2014; Gao et al., 2014, 2015; Kolb et al., 2015; Belda-Palazon et al., 2016). However, why autophagy is impaired in free1/fyve1 mutants remains unclear (Chung, 2019; Zeng et al., 2019). FYVE2, also known as CELL DEATH-RELATED ENDOSOMAL FYVE/SYLF PROTEIN1 (CFS1), also interacts with VPS23A and was proposed to be involved in the fusion of autophagosomes with endosomes (Sutipatanasomboon et al., 2017). However, the dynamics of GFP-ATG8-positive organelles was not analyzed in this study. In addition, fusion between autophagosomes and endosomes has not been demonstrated in plants (Bozhkov, 2018; Chung, 2019). ESCRT components are known to be required for autophagosome formation in both plants and animals by closing the phagophore membrane (Spitzer et al., 2015; Takahashi et al., 2018), providing an alternative interpretation for the role of the FYVE2–VPS23A interaction. Thus, the exact roles of FYVE1 and FYVE2 in autophagosome assembly remain unclear.
Here we report that the plant-specific FYVE2 protein interacts with both core ATG components and the COPII machinery in Arabidopsis. FYVE2 is recruited to autophagic membranes during phagophore expansion and mediates the contribution of PI3P and the COPII machinery to autophagosome formation. Downregulation of either FYVE2 or the COPII machinery culminated in the over-accumulation of autophagic organelles and a reduction in autophagic flux. Our results indicate that FYVE2 and specific COPII components control autophagosome formation in plants.
Results
FYVE2 positively affects autophagic flux downstream of ATG2
To identify PI3P effectors involved in autophagy, we searched for Arabidopsis proteins containing the PI3P-interacting FYVE domain. Out of four proteins with FYVE domains (van Leeuwen et al., 2004), coexpression data (http://atted.jp) indicated that the expression of At3g43230/FYVE2/CFS1 highly correlates with that of several core ATG genes, including ATG11 (Li et al., 2014). The FYVE2/CFS1 protein (van Leeuwen et al., 2004; Sutipatanasomboon et al., 2017) contains an FYVE domain and a C-terminal SYLF (SH3YL1, Ysc84p/Lsb4p, Lsb3p, and plant FYVE) domain (Supplemental Figure S1A), both of which can interact with PI3P independently (Sutipatanasomboon et al., 2017). The SYLF domain, also called the YSC84 actin-binding domain, binds actin filaments and phosphoinositides in yeast (Robertson et al., 2009; Urbanek et al., 2015). To define the functions of FYVE2, we searched for T-DNA insertion mutations in which FYVE2 expression is compromised. We obtained three lines with insertions in FYVE2 (Supplemental Figure S1A) and found that these three fyve2 alleles, designated fyve2-1/cfs1-1, fyve2-2/cfs1-2, and fyve2-3/cfs1-3, lack the full-length FYVE2 transcript (Supplemental Figure S1B).
To determine whether FYVE2 is involved in autophagy, a transgene encoding the autophagy reporter ProUBQ10:GFP-ATG8A (Kim et al., 2013) was introgressed into the fyve2 mutants by genetic crossing. We subjected the GFP-ATG8A transgenic seedlings to nitrogen (N) starvation and monitored starvation-induced autophagy by confocal microscopy. Young fyve2 cells in the root elongation zone showed a density of GFP-ATG8A-labeled autophagic puncta similar to that of wild-type (WT) control cells (Figure 1B). However, fyve2 cells in the root maturation zone and hypocotyls over-accumulated cytoplasmic GFP-ATG8A puncta (Figure 1B). We previously reported that the atg2-1 mutant also accumulated cytoplasmic GFP-ATG8A puncta (Kang et al., 2018) and therefore we used this mutant as a control for impaired autophagy. Compared with atg2, accumulation of GFP-ATG8A-decorated organelles was less pronounced in all types of fyve2 cells examined, including roots, hypocotyls (Figure 1B), and leaves (Supplemental Figure S2A). Time-course analysis of N-starved mature root cells indicated that compared with WT cells, mature epidermal root cells in the fyve2 mutants accumulated 5–20 times more autophagic organelles, whereas in atg2, accumulation of GFP-ATG8A puncta reached 50–100 times more than in WT (Supplemental Figure S2B). Interestingly, in both fyve2 and atg2 seedlings, GFP-ATG8-labeled organelles accumulated even when grown under N-sufficient conditions (Supplemental Figure S2A and S2B).
Homozygous mutants for another fyve2 allele, fyve2-1, also over-accumulated GFP-ATG8A puncta (Supplemental Figure 2C), demonstrating that mutations in FYVE2 are the cause for the over-accumulation of autophagic organelles. The PI 3-kinase inhibitor Wm suppresses the over-accumulation of GFP-ATG8A puncta in atg2 mutants (Kang et al., 2018). Similarly, Wm treatment almost completely blocked the over-accumulation of GFP-ATG8A-positive organelles in fyve2 mutants (Supplemental Figure S2C).
Because the over-accumulation of GFP-ATG8A puncta can result from either the induction of autophagy or impaired autophagosome dynamics (for instance, a delay in membrane expansion, maturation, and/or fusion with vacuoles), we examined how fyve2 mutations affect autophagic flux. WT and mutant seedlings expressing GFP-ATG8A were treated with concanamycin A (ConA), an inhibitor of vacuolar H+-ATPase, to stabilize autophagic bodies inside the vacuoles. The lack of autophagic bodies is expected if autophagic flux is completely blocked, as in atg5 and atg7 mutants (Thompson et al., 2005; Chung et al., 2010). When we treated N-starved seedlings with ConA, we found very few autophagic bodies in atg2, whereas fyve2 seedlings accumulated approximately half the quantity of autophagic bodies inside their vacuoles compared with WT (Figure 1C). This suggests that fewer autophagosomes are delivered to the vacuole in fyve2 than in WT root cells. To confirm this observation, we measured autophagic flux using the GFP-ATG8A cleavage assay (Chung et al., 2010; Suttangkakul et al., 2011; Shin et al., 2014). When GFP-ATG8 proteins are targeted to the vacuole for degradation, the free GFP moiety is released and can be detected by anti-GFP immunoblot analysis (Slavikova et al., 2005; Chung et al., 2010; Marshall et al., 2015). Based on this assay, we found that autophagy was indeed partially inhibited in fyve2 seedlings during N starvation (Figure 1D), whereas autophagic flux was almost completely blocked in atg2, as reported previously (Kang et al., 2018). Thus, the over-accumulation of autophagic puncta in fyve2 root cells is due to impaired progression of autophagosome biogenesis.
The phenotypic consequences of fyve2 mutations were further validated by complementation analysis using transgenic plants coexpressing mCherry-FYVE2 and GFP-ATG8A. As expected, the expression of mCherry-FYVE2 in the fyve2-2 mutant background restored its autophagic flux to a normal level (Supplemental Figure S3A) and suppressed the accumulation of GFP-ATG8A puncta in root cells (Supplemental Figure S3B).
To assess whether FYVE2 affects phagophore initiation, we crossed the atg2 and fyve2 mutants with a transgenic plant expressing the phagophore marker ATG5-GFP (Le Bars et al., 2014) and analyzed their progenies by confocal microscopy. Unlike GFP-ATG8, ATG5-GFP is not targeted to the vacuole (Le Bars et al., 2014) and probably detaches from the autophagic membranes during autophagosome maturation (Figure 1A). We found that both atg2 and fyve2 mutants over-accumulated ATG5-GFP puncta compared with WT (Figure 1E). Consistent with previous data (Le Bars et al., 2014), Wm effectively blocked the formation of ATG5-GFP puncta in both mutants (Figure 1E). These observations suggest that phagophores initiate normally in both atg2 and fyve2 mutants but are arrested as nonfunctional intermediates.
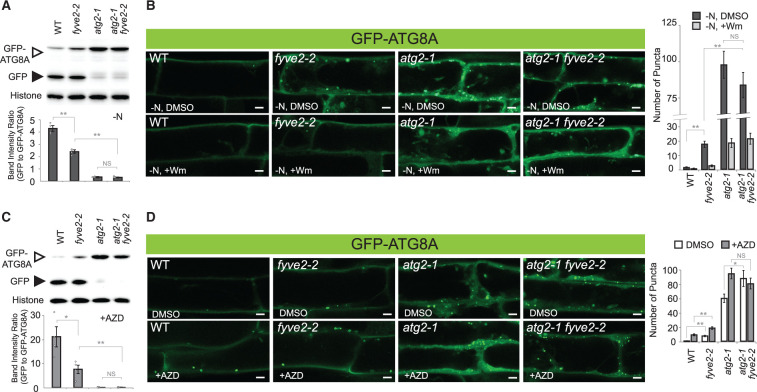
To investigate the genetic interaction between FYVE2 and ATG2, we crossed fyve2-2 with atg2 plants and measured autophagic fluxes in single and double homozygous mutant progeny (Figure 2). The GFP-ATG8A cleavage assay indicated that the autophagic flux in fyve2 atg2 double mutants was comparable to that of atg2 single mutants under N-limiting conditions (Figure 2A). Consistently, imaging analysis showed that both atg2 single and atg2 fyve2 double mutant seedlings over-accumulated GFP-ATG8A puncta at a similar level (Figure 2B). In all cases, this process was dependent on PI3P, as Wm treatment largely reduced the accumulation of GFP-ATG8A puncta (Figure 2B). This epistatic interaction between atg2 and fyve2 suggests that FVYE2 acts downstream of ATG2 to mediate autophagosome formation.
Figure 2.
atg2 is epistatic to fyve2. A and C, GFP-ATG8A cleavage assay to determine the autophagic flux in WT, fyve2, atg2, and fyve2 atg2 seedlings. Immunoblot analysis was performed using anti-GFP (upper image) or anti-histone H3 antibody (lower image for the loading control). Graphs below show the quantification of band intensity ratios (mean ± se; n = 4). B and D, Confocal fluorescence images of mature root cells expressing GFP-ATG8A. Autophagy was induced by exposing 9-day-old seedlings to N starvation for 48 h (A) or 12 h (B) and to 0.5 µM AZD treatment for 12 h (C) or 1 h (D). Prior to microscopic observation in (B), WT, fyve2, atg2, and fyve2 atg2 seedlings were either treated with DMSO or 30 µM Wm for 1 h. The graphs on the right show quantification of GFP-ATG8A puncta. Images (n = 16–18 in (B), 14–16 in (D)) were collected to calculate the density of GFP-ATG8A puncta per 25,514 µm2 (mean ± SE). Columns marked with asterisks represent mutants that are significantly different from WT, according to t test. *0.01 < P < 0.05; **P < 0.01. NS, not significant. Scale bars = 5 µm.
To further analyze the role of FYVE2 under a different autophagy-inducing condition, we treated mutant lines expressing GFP-ATG8A with AZD8055 (AZD), an inhibitor of the Target of Rapamycin (TOR) kinase. AZD has been used as an autophagy inducer in plants (Soto-Burgos and Bassham, 2017; Dauphinee et al., 2019). Our GFP-ATG8A cleavage assay indicated that AZD-induced autophagy was attenuated in fyve2 but completely blocked in atg2 single and atg2 fyve2 double mutant seedlings (Figure 2C).
Confocal microscopy imaging of the transgenic mutant seedlings treated with AZD for 1 h revealed that the density of GFP-ATG8A-positive organelles was significantly increased by AZD in WT, fyve2, and atg2 single mutant seedlings (Figure 2D). When treated with AZD, atg2 single and atg2 fyve2 double mutants accumulated autophagic puncta at a similar level (Figure 2D). Taken together, these data indicate that atg2 is epistatic to fyve2 during both nutrient starvation- and AZD-induced autophagy.
FYVE2 interacts with components of phagophores and COPII vesicles
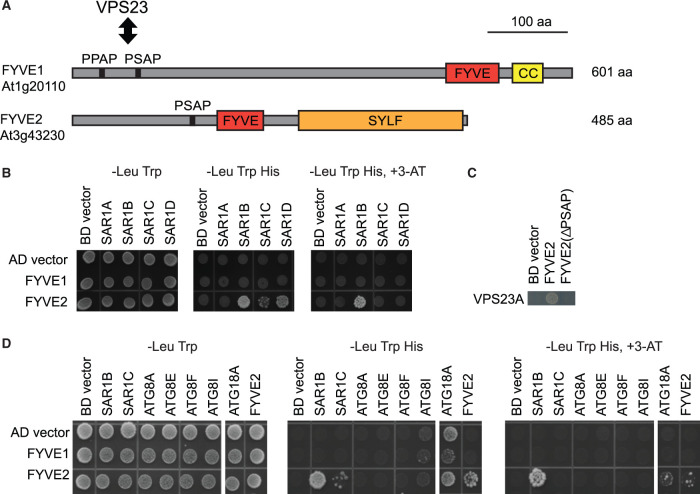
To gain insight into the molecular function of FYVE2, we investigated potential FYVE2-interacting proteins (Figure 3). FYVE2 was shown to interact with the ESCRT protein VPS23A (Sutipatanasomboon et al., 2017). In addition, a study based on yeast two-hybrid (Y2H) assays identified the small GTPase Secretion-associated Ras-related GTPase 1B (SAR1B) as an interactor of FYVE2 (Arabidopsis Interactome Mapping Consortium, 2011). SAR1B is encoded by one of four Arabidopsis SAR1 genes that mediate COPII vesicle formation (Hanton et al., 2008; Zeng et al., 2015). To confirm these interactions, we performed an Y2H assay using SAR1 isoforms as baits and FYVE2 as a prey. We included as a prey FYVE1/FREE1 (van Leeuwen et al., 2004; Gao et al., 2014), which contains a C-terminal FYVE domain (Figure 3A), interacts with VPS23 through its N-terminal P(P/S)AP motif, and has been shown to participate in both autophagy (Gao et al., 2015) and vacuole biogenesis (Gao et al., 2014).
Figure 3.
FYVE2 physically interacts with proteins involved in the formation of autophagosomes and COPII vesicles. A, Diagram of the Arabidopsis FYVE1 and FYVE2 proteins. FYVE and SYLF domains are shown in red and orange boxes, respectively. CC, coiled-coil region. The VPS23A/ELC-interacting PSAP/PPAP motifs in FYVE1 and FYVE2 are indicated by vertical lines. B and D, Y2H interactions of FYVE1 and FYVE2. Mating-based Y2H was used. Gal4BD, Gal4-DNA-binding domain. Gal4AD, Gal4-activation domain. C, Y2H interaction of VPS23A with either WT or ΔPSAP mutant FYVE2 protein. Co-transformation-based Y2H was used.
In our Y2H assay, FYVE2 interacted strongly with SAR1B and weakly with two other isoforms, SAR1C and SAR1D (Figure 3B). In contrast, no interaction was detected between FYVE2 and SAR1A or between FYVE1 and any of the SAR1 isoforms. We also confirmed the interaction between FYVE2 and VPS23A and found that it requires the PSAP motif of FYVE2 (Figure 3C). To identify additional interactors, we tested Y2H interactions with selected ATG proteins. Interestingly, FYVE2 but not FYVE1 interacted with ATG18A and with FYVE2 itself (Figure 3D). Neither FYVE1 (Gao et al., 2015) nor FYVE2 interacted with any of the four ATG8 isoforms (ATG8A/E/F/I) tested in our assay.
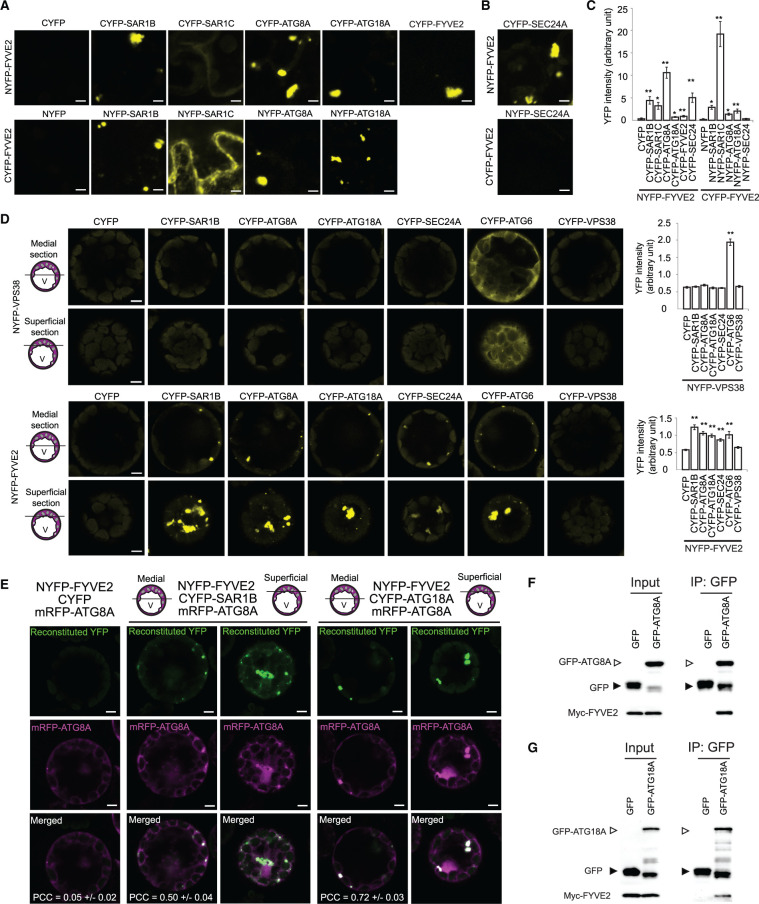
To further test the interactions of FYVE2 with SAR1B, ATG18A, and itself, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaf epidermis, with each pair fused to either the C- or the N-terminal fragments of YFP (NYFP and CYFP; Figure 4A). SAR1C interacted with FYVE2 only when fused to the NYFP. Although FYVE2 did not interact with ATG8A in our Y2H assay (Figure 3D), we were able to detect reconstitution of YFP fluorescence signal when ATG8A and FYVE2 were tested in reciprocal BiFC assays (Figure 4, A and C). It is possible that by binding to ATG18A, FYVE2 is brought in close proximity to ATG8A at the phagophore membrane to allow for YFP reconstitution. We then tested whether FYVE2 interacts with other COPII coat components besides SAR1B and SAR1C. Indeed, we detected an interaction between FYVE2 and SEC24A, a subunit of the COPII inner layer (Faso et al., 2009; Nakano et al., 2009), although only in one combination (NYFP-FYVE2 and CYFP-SEC24A in Figure 4, B and C).
Figure 4.
FYVE2 interacts with SAR1B and ATG18A in planta. A–C, BiFC interactions of FYVE2 using transient expression in N. benthamiana leaves. The indicated proteins fused to the NYFP were co-expressed with the indicated proteins fused to the CYFP. Vector controls (NYFP or CYFP) co-expressed with NYFP-FYVE2 or CYFP-FYVE2 are shown on the left. C, Quantification of reconstituted YFP intensity. D, BiFC interactions of NYFP-VPS38 (control; first and second rows) and NYFP-FYVE2 (third and fourth rows) using transient expression in Arabidopsis leaf protoplasts. Confocal images of medial (first and third rows) and superficial (second and fourth rows) optical sections are shown. V, vacuole. The graphs on the right show quantification of reconstituted YFP intensity. Columns marked with asterisks represent means that are significantly different from the CYFP or NYFP control, according to t test (mean ± se; n = 3–10 images in (A) and (B), n = 39–51 images in (D)). *0.01 < P < 0.05; **P < 0.01. Scale bars = 5 µm. E, Colocalization of mRFP-ATG8A puncta with YFP signal reconstituted from BiFC pairs FYVE2-SAR1B (second and third columns) and FYVE2-ATG18A (fourth and fifth columns). Confocal images of medial (second and fourth columns) and superficial (third and fifth columns) optical sections are shown. The Pearson correlation coefficients (PCCs) were calculated from 21 medial section images (mean ± se). F and G, Co-immunoprecipitation of Myc-FYVE2 with GFP-ATG8A (F) and GFP-ATG18A (G) expressed in Arabidopsis leaf protoplasts. Inputs (immunoblot images on the left) and immunoprecipitates (images on the right) are shown. Protein blots were reacted with anti-GFP (upper blots) and anti-Myc (lower blots) antibodies. Protein samples prepared from protoplasts expressing GFP and Myc-FYVE2 were used as a negative control. A representative set of immunoblots is shown, selected from three independent replicates.
We also tested interactions of FYVE2 with SAR1B, ATG8A, ATG18A, and SEC24A by BiFC assay using transient expression in Arabidopsis leaf protoplasts. Here, we employed VPS38 as a negative control since VPS38, which encodes a subunit of the endosomal PI 3-kinase complex, is critical for endosomal sorting and vacuolar trafficking but dispensable for autophagy (Lee et al., 2018; Liu et al., 2018, 2020). As expected, VPS38 interacted with ATG6 (another subunit of the PI 3-kinase complex) but not with SAR1B, ATG8A, ATG18A, or SEC24A (Figure 4D, upper graph). Consistent with our BiFC assays in N. benthamiana leaves, FYVE2 interacted with SAR1B, ATG8A, ATG18A, and SEC24A (Figure 4D, lower graph). Taken together, these results suggest that FYVE2 interacts with ATG18A, ATG8A, SAR1, and SEC24A.
Interestingly, FYVE2 also interacted with ATG6 but not with VPS38 (Figure 4D, lower graph). Reconstituted YFP signal from FYVE2 interactions was mostly detected in ∼1–2 µm puncta (Figure 4D, third and fourth rows), whereas the fluorescent signal from the VPS38–ATG6 interaction was largely diffuse (Figure 4D, first and second rows). This suggests that FYVE2 interaction mainly occurs at autophagic membranes but not in the cytosol. However, we also detected reconstituted YFP signal in large puncta (∼5–8 µm) located at the peripheral cytoplasm of Arabidopsis protoplasts (Figure 4D, fourth row) and N. benthamiana cells (Figure 4, A and B). These enlarged structures were most likely protein aggregates, since autophagosomes are typically smaller than 2 µm. To examine whether the 1- to 2-µm YFP puncta correspond to autophagosomes, we co-expressed monomeric red fluorescent protein (mRFP)-ATG8A with two BiFC pairs, FYVE2-SAR1B and FYVE2-ATG18A. Reconstituted YFP puncta mostly emitted mRFP-ATG8A fluorescence, indicating that these puncta were indeed autophagic organelles (Figure 4E).
To demonstrate the association of FYVE2 with autophagy proteins in plants by biochemical analysis, we performed a co-immunoprecipitation assay using Arabidopsis protoplasts transiently co-expressing Myc-tagged FYVE2 and either GFP alone, GFP-ATG18A, or GFP-ATG8A. Myc-FYVE2 was identified from co-immunoprecipitates containing GFP-ATG8A (Figure 4F) and GFP-ATG18A (Figure 4G). These BiFC and co-immunoprecipitation data collectively indicate that FYVE2 interacts with ATG8A and ATG18A.
FYVE2 localizes to autophagic membranes near the ER
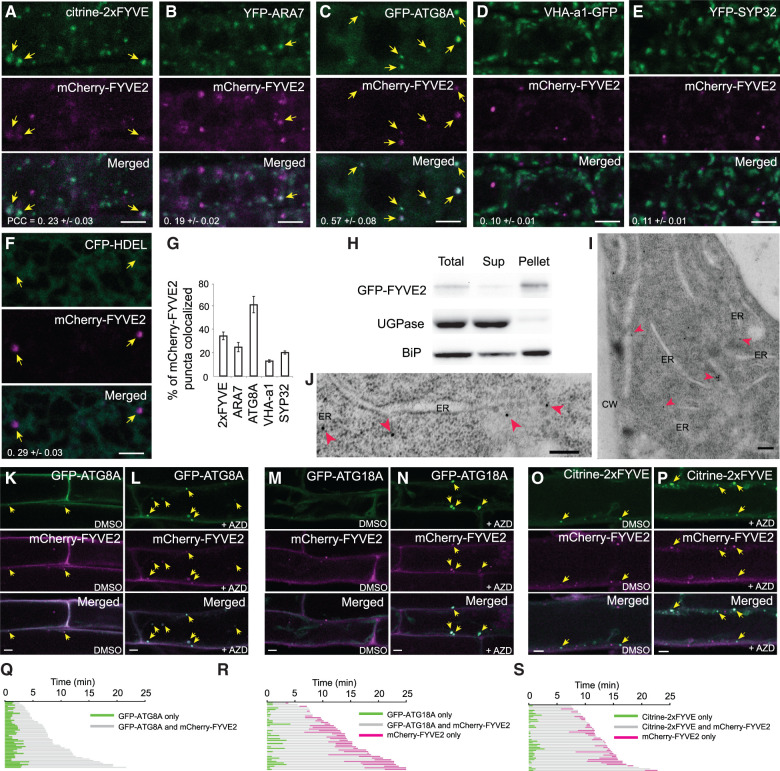
To investigate the subcellular localizations of FYVE2 proteins, we generated Arabidopsis transgenic plants expressing mCherry-FYVE2 together with various organelle markers and analyzed their distribution patterns in young root cells (Figure 5, A–E). Of all the imaged mCherry-FYVE2 puncta, ∼60% were decorated with GFP-ATG8A (Figure 5, C and G). mCherry-FYVE2 showed moderate co-localization with the PI3P biosensor citrine-2xFYVE (Figure 5, A and G). Lower degrees of overlap were detected between mCherry-FYVE2 puncta and the markers for late endosomes (YFP-ARA7), the trans-Golgi network (VHAa1-GFP), and Golgi stacks (YFP-SYP32) (Figure 5, B, D, E, and G). The mCherry-FYVE2 puncta also appeared to associate with the ER network, as labeled by the ER marker cyan fluorescent protein (CFP)-HDEL (Figure 5F). Consistently, membrane fractionation analysis of transgenic plants expressing GFP-tagged FYVE2 indicated that GFP-FYVE2 was associated with membranes (Figure 5H). To validate the association of FYVE2 with endomembranes, root tips of GFP-FYVE2 transgenic plants were high-pressure-frozen/freeze-substituted and immunolabeled with anti-GFP antibodies. Although we were unable to identify double-membrane autophagosomes in our sections, we did detect GFP-FYVE2 labeling associated with the ER membrane and vesicles adjacent to the ER (Figure 5, I and J).
Figure 5.
Fluorescent fusions to FYVE2 are found near the ER and mostly co-localize with autophagic markers. A–F, Localization of mCherry-FYVE2 relative to biosensors and organelle markers. Confocal fluorescence images were acquired from root cells expressing mCherry-FYVE2 (A–F) and either a PI3P biosensor (A), late endosome marker (B), autophagic marker (C), TGN marker (D), Golgi apparatus marker (E), or ER luminal marker (F). Arrows indicate mCherry-FYVE2 puncta overlapping with the respective markers. G, Quantification of mCherry-FYVE2 puncta co-localizing with various markers. Images (n = 10–12) similar to those shown in (A)–(E) were collected to calculate the percentage of mCherry-FYVE2 puncta showing fluorescence at the yellow/green channel. The Pearson correlation coefficients (PCCs) are also provided in (A)–(F) (mean ± se). H, Membrane fractionation analysis of GFP-FYVE2. Anti-GFP antibodies were used to detect GFP-FYVE2. Immunoblots using antibodies reacting with UGPase and ER-resident isoform of heat shock protein 70 (BiP) were used as markers for soluble and microsomal fractions, respectively. I and J, Immunodetection of GFP-FYVE2 on the ER of cells at the root tip. CW, cell wall. Arrowheads indicate gold particles labeled with anti-GFP antibodies. K–P, Confocal fluorescence images of root cells expressing mCherry-FYVE2 and either GFP-ATG8A (K and L), GFP-ATG18A (M and N), or citrine-2xFYVE (O and P). Seedlings were incubated in liquid medium containing DMSO (K, M, and O) or 0.5 µM AZD (L, N, and P) for 1 h prior to microscopy. Arrows indicate mCherry-FYVE2 puncta overlapping with the respective autophagic markers. Q–S, Time-lapse imaging analysis of puncta emitting both mCherry-FYVE2 and either GFP-ATG8A (Q), GFP-ATG18A (R), or citrine-2xFYVE (S). Scale bars = 5 µm (A–E, F, and K–P) or 200 nm (I and J).
To analyze the recruitment dynamics of FYVE2 to autophagic membranes, we induced autophagy by treating seedlings expressing mCherry-FYVE2 and GFP-ATG8A with AZD. Under noninducing conditions, the organelles decorated with both GFP-ATG8A and mCherry-FYVE2 were only occasionally detected (Figure 5K). When autophagy was induced by AZD treatment, mature root cells contained abundant GFP-ATG8A organelles, which were also positive for mCherry-FYVE2 (Figure 5L).
Our finding that FYVE2 interacts with ATG18A (see Figures 3 and 4) prompted us to test whether mCherry-FYVE2 associates with the ATG18A-positive organelles. ATG18A was previously shown to be cytosolic in Arabidopsis but to transiently associate with autophagic membranes upon autophagy induction (Zhuang et al., 2017). We confirmed a diffuse distribution of GFP-ATG18A in control root cells (Figure 5M). Upon autophagy induction by AZD, large puncta emitting both GFP-ATG18A and mCherry-FYVE2 fluorescence were readily detected (Figure 5N). mCherry-FVYE2 showed moderate co-localization with the PI3P biosensor citrine-2xFYVE before and after AZD induction (Figure 5, O and P). Together, these localization analyses indicate that fluorescently tagged FYVE2 strongly associates with autophagic organelles under both basal and autophagy-inducing conditions.
To pinpoint when FYVE2 is recruited to autophagic membrane, we performed time-lapse analysis of root cells co-expressing mCherry-FYVE2 and either GFP-ATG8A or GFP-ATG18A. We focused on tracking relatively static puncta positive for both GFP and mCherry signals. Our time-lapse analysis of autophagic organelles (Movie 1) indicated that mCherry-FYVE2 fluorescence was detected ∼2 min (119.8 ± 7.7 s; n = 48 puncta) after GFP-ATG8A signals first appeared (Figure 5Q), suggesting that mCherry-FYVE2 is recruited to autophagic membranes during phagophore expansion. Similar analysis using transgenic plants co-expressing mCherry-FYVE2 and GFP-ATG18A (Movie 2) indicated that mCherry-FYVE2 fluorescence was detected ∼1 min (66.7 ± 10.6 s; n = 42 puncta) after the appearance of GFP-ATG18A signals (Figure 5R). Unlike GFP-ATG8A, GFP-ATG18A fluorescence disappeared from these foci while mCherry-FYVE2 fluorescence remained. We estimated that GFP-ATG18A disappeared from the mCherry-FYVE2-positive organelles ∼3 min (179.3 ± 22.2 s) before they finally disappeared, likely due to their fusion with the tonoplast. A similar sequential pattern of association and dissociation was observed in roots co-expressing mCherry-FYVE2 and citrine-2xFYVE, with mCherry-FYVE2 being recruited to PI3P-positive membranes at 49.3 ± 6.4 s (n = 43 puncta), followed by the disappearance of mCherry-FYVE2 signals at 87.4 ± 15.8 s before the mCherry-FYVE2 signals finally faded (Figure 5S; Movie 3). Based on these time-lapse imaging data, we conclude that FYVE2 is recruited to the ATG8A-, ATG18A-, and PI3P-positive membranes after phagophore initiation and remains on autophagic membranes after ATG18A dissociation and PI3P turnover.
FYVE2 is recruited to the phagophore and is subsequently targeted to the vacuole through autophagy
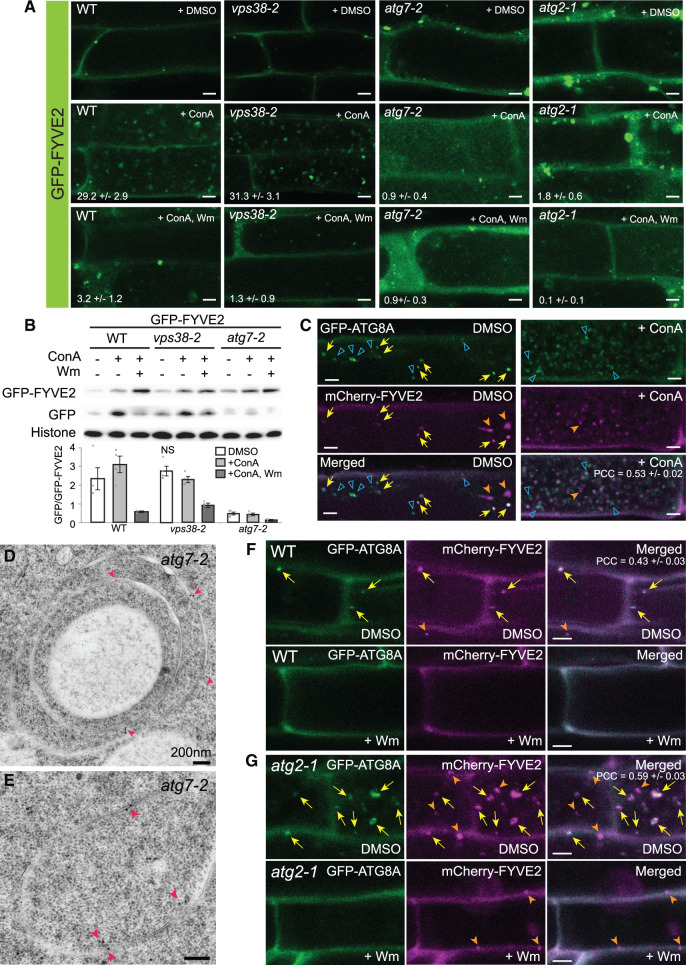
Following its recruitment to the autophagic membrane, FYVE2 could follow two possible fates: detaching from autophagic membranes and returning to the cytosol, like ATG5 and ATG18A; or being sequestered into autophagosomes and degraded in the vacuole as a part of autophagic bodies, similar to ATG8 and ATG1. To test these two possibilities, we investigated whether fluorescently tagged FYVE2 localizes to autophagic bodies. When treated with ConA, WT transgenic seedlings expressing mCherry-FYVE2 (Supplemental Figure S4A) or GFP-FYVE2 (Figure 6A, WT, +ConA) under the control of the Arabidopsis UBQ10 promoter accumulated fluorescent puncta inside the central vacuole, suggesting that FYVE2 is sequestered into autophagosomes and subsequently released into the vacuole as autophagic bodies. Autophagic body-like FYVE2-GFP puncta accumulated in the vacuoles of ConA-treated transgenic plants expressing FVYE2-GFP under the control of the native FYVE2 promoter (Supplemental Figure S4B). Thus, the deposition of GFP-FYVE2 in the vacuole does not seem to be caused by ectopic overexpression.
Figure 6.
GFP-FYVE2 is targeted to the vacuole via the autophagic route. A, Confocal fluorescence images of mature root cells expressing GFP-FYVE2. GFP-FYVE2 seedlings with the indicated genetic background were incubated in N-sufficient liquid medium for 9 days and treated with DMSO, 0.5-µM ConA, or 0.5 µM ConA plus 30 µM Wm for 16 h. Numbers in the second and third rows show the number of autophagic bodies (mean ± se; n = 11–12 images) per 1,000 µm2 area of the central vacuole. B, GFP-FYVE2 cleavage assay. WT, vps38-2, or atg7-2 seedlings expressing GFP-FYVE2 were incubated as described above, and protein extract was prepared for immunoblot analysis using anti-GFP (upper) and anti-histone H3 (lower; for loading control) antibodies. Representative images selected from four repeat experiments are shown. C, Confocal fluorescence images of root cells expressing GFP-ATG8A and mCherry-FYVE2. Seedlings grown in N-rich medium were treated with DMSO or 0.5 µM ConA for 16 h prior to confocal microscopy. The Pearson correlation coefficients (PCCs) were calculated from 12 images (mean ± se). D and E, Immunodetection of GFP-FYVE2 in atg7-2 cells at the root tip. Arrowheads indicate gold particles labeled with anti-GFP antibodies. F and G, Confocal fluorescence images of mature root cells expressing GFP-ATG8A and mCherry-FYVE2 in WT (F) and atg2-1 (G) background. Seedlings were incubated in −N liquid medium for 2 days and treated with DMSO or 30 µM Wm for 1 h prior to microscopic observation. The PCCs were calculated from 11 (F) and 10 (G) images (mean ± se). Arrows (C, F, and G) indicate GFP-ATG8A puncta overlapping with mCherry-FYVE2 signal. Solid and open arrowheads (C, F, and G) indicate nonoverlapping puncta labeled only by mCherry-FYVE2 and GFP-ATG8A, respectively. Scale bars = 5 µm (A, C, F, and G) or 200 nm (D and E).
PI3P is required for autophagosome assembly and the vacuolar delivery of fluorescently tagged ATG8 (Zhuang et al., 2013; Shin et al., 2014). We, therefore, investigated whether PI3P is crucial for vacuolar trafficking of GFP-FYVE2. The accumulation of GFP-FYVE2 puncta in ConA-treated seedlings was prevented by additional treatment with Wm (Figure 6A, WT, +ConA, Wm), indicating that PI3P is important for targeting GFP-FYVE2 to the vacuole. However, PI3P is also important for endosomal function, and Wm is known to disrupt endosomal trafficking. To determine whether endosomal trafficking contributes to the delivery of FYVE2 into vacuoles, we introgressed GFP-FYVE2 into the atg2-1, atg7-2, and vps38-2 mutants. The number of vacuolar GFP-FYVE2 puncta was greatly reduced in ConA-treated atg7-2 and atg2-1, but not in vps38-2 roots (Figure 6A, +ConA), indicating that the vacuolar trafficking of GFP-FYVE2 does not rely on endosomal PI 3-kinase activity but instead requires core ATG components and probably autophagic PI 3-kinase activity.
To further confirm these imaging results with a biochemical assay, we assessed vacuolar cleavage of GFP from GFP-FYVE2 in the atg7-2 and vps38-2 mutants. In immunoblot analysis of control transgenic seedlings expressing GFP-FYVE2, the intensity of free GFP band was increased by ConA treatment but attenuated by additional treatment with Wm (Figure 6B, WT), similar to the GFP cleavage pattern for GFP-ATG8A (Supplemental Figure S4C). The cleavage of GFP from FYVE2-GFP was also seen in the native FYVE2 promoter line (Supplemental Figure S4D). The release of GFP from GFP-FYVE2 in vps38-2 was comparable to that of the control samples, whereas atg7-2 and atg2-1 showed lower levels of free GFP (Figure 6B; Supplemental Figure S4E); these results are consistent with the confocal imaging data (see Figure 6A). The free GFP moiety was stabilized when seedlings were kept in the dark (Supplemental Figure S4E), confirming that the free GFP bands were derived from protein cleavage in the vacuole (Tamura et al., 2003). Notably, some free GFP was detectable in atg7-2 and atg2-1 (Figure 6B; Supplemental Figure S4E), suggesting that canonical autophagy mainly, but not exclusively, mediates the vacuolar trafficking of GFP-FYVE2.
N starvation increases autophagic flux, as measured by the relative levels of free GFP cleaved from GFP-ATG8A (see Figure 2A). Similarly, we detected an increase in free GFP relative to GFP-FYVE2 when control seedlings were subjected to N starvation, whereas no changes were observed in atg7-2 (Supplemental Figure S4F), further supporting the notion that the vacuolar delivery of GFP-FYVE2 largely depends on autophagy.
Next, we examined whether mCherry-FYVE2 and GFP-ATG8A are delivered together to the vacuole for degradation. As expected, mCherry-FYVE2 puncta largely co-localized with GFP-ATG8A in mature WT root cells [indicated by arrows in Figure 6C, dimethyl sulfoxide (DMSO)]. In ConA-treated roots, numerous puncta of mCherry-FYVE2 were detected in the vacuole and mostly overlapped with GFP-ATG8A signal, indicating that they were autophagic bodies (Figure 6C, +ConA). However, 20% of the vacuolar mCherry-FYVE2 puncta lacked GFP-ATG8A signal (indicated by solid arrowheads in Figure 6C, +ConA). These results are consistent with the notion that the vacuolar delivery of GFP-FYVE2 is not solely dependent on ATG7 (see Figure 6B and Supplemental Figure S4E). Likewise, 26% of the vacuolar GFP-ATG8A puncta lacked mCherry-FYVE2 signal (indicated by open arrowheads in Figure 6C, +ConA), indicating that not all autophagosomes delivered to the vacuole contain mCherry-FYVE2.
We further investigated how mutations in core ATG genes affect autophagic trafficking of FYVE2. We analyzed the association of FYVE2 with membranes in the atg7-2 mutants, which fails to lipidate and recruit ATG8 to autophagic membranes. Both control and atg7-2 seedlings accumulated GFP-FYVE2 in membrane fractions (Supplemental Figure S4G), implying that the association of GFP-FYVE2 with membranes does not require ATG8 lipidation. Moreover, we observed a pronounced accumulation of GFP-FYVE2-decorated organelles in the cytoplasm of atg2-1 and to a lesser extent, in atg7-2 root cells compared to the controls (Supplemental Figure S4, H and I; see Figure 6A, + DMSO). This abnormal accumulation of GFP-FYVE2 was partially suppressed by Wm treatment (Supplemental Figure S4, H and I), suggesting that PI3K activity is involved in the formation of GFP-FYVE2-positive puncta. Immunogold labeling with anti-GFP antibodies of atg7-2 root cells expressing GFP-FYVE2 showed the presence of multi-lamellar, double-membrane structures of 1–3 µm decorated with GFP-FYVE2 (Figure 6, D and E). These abnormal membranous structures were not observed in control cells (see Figure 5, I and J).
We subsequently tested whether atg2 affects the recruitment of mCherry-FYVE2 to autophagic vesicles. In atg2 mutants, the phagophore is initiated (see Figure 1E) but most likely fails to expand. We observed puncta with overlapping mCherry-FYVE2 and GFP-ATG8A signals in both WT (Figure 6F) and atg2 (Figure 6G), indicating that ATG2 is not required for the association of mCherry-FYVE2 with autophagic membranes. This result supports the notion that mCherry-FYVE2 is recruited to GFP-ATG8-positive membranes during early phases of phagophore formation (see Figure 5Q).
Treatment with Wm largely eliminated puncta positive for both mCherry-FYVE2 and GFP-ATG8A (Figure 6, F and G), which is consistent with FYVE2 being recruited to autophagic organelles by PI3P. Notably, some mCherry-FYVE2 puncta not labeled with GFP-ATG8A were insensitive to Wm in WT, atg2, and atg7 seedlings (arrowheads in Figure 6, F and G; Supplemental Figure S4, H and I). Taken together, these data suggest that FYVE2 largely localizes to the autophagic organelles in an ATG2-independent but PI3K-dependent manner and that a small fraction of FYVE2 may be recruited to nonautophagic structures via a PI3K-independent pathway.
FYVE2 mediates the function of SAR1B in autophagy
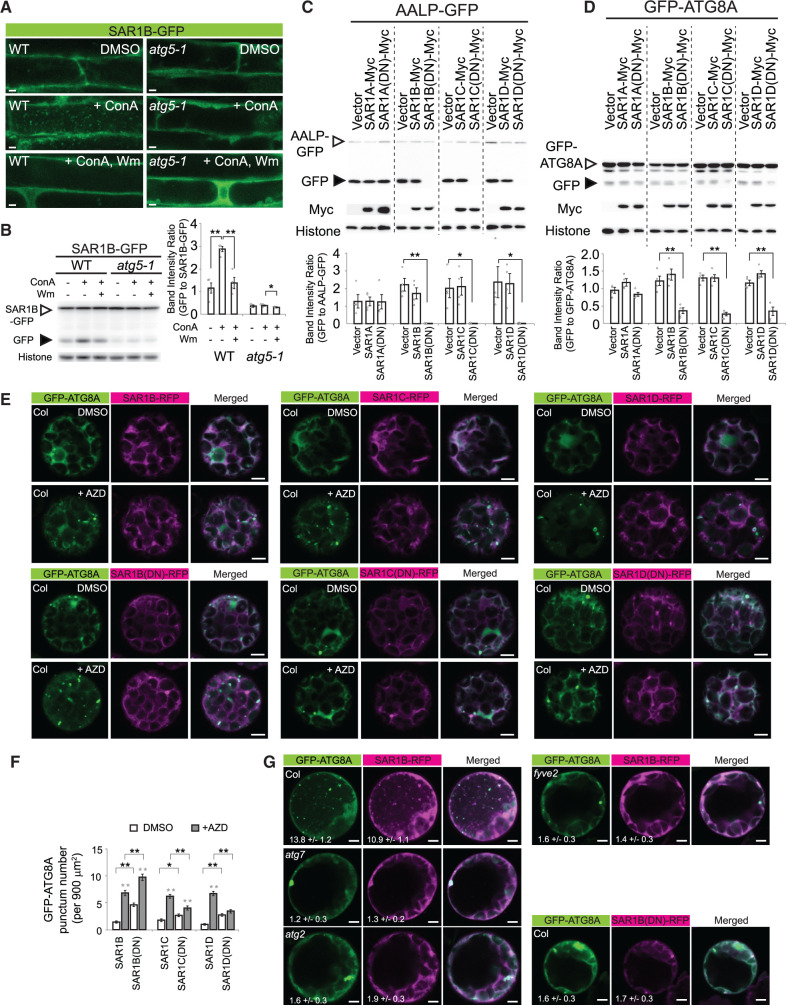
Autophagosome biogenesis in budding yeast and mammals requires the participation of SAR1 and other proteins involved in COPII vesicle formation (Ishihara et al., 2001; Zoppino et al., 2010; Graef et al., 2013; Suzuki et al., 2013; Tan et al., 2013; Ge et al., 2014; Shima et al., 2019). To study the biological relevance of the interaction between FYVE2 and SAR1 isoforms (see Figures 3 and 4) in autophagy, we generated transgenic Arabidopsis lines expressing SAR1B-GFP or SAR1C-GFP. Under control conditions, root cells of seedlings expressing either SAR1 isoform showed a diffuse GFP pattern (Figure 7A; Supplemental Figure S5A; DMSO), which probably represented the cytosolic pool of inactive SAR1 proteins. When these seedlings were treated with ConA, a number of SAR1B/C-GFP puncta were detected inside their vacuoles, but not when they were treated simultaneously with Wm (Figure 7A; Supplemental Figure S5A). These puncta resembling autophagic bodies were not observed in the vacuoles of autophagy-deficient atg5 mutants (Figure 7A), in which ATG8 conjugation is compromised, similar to the case in atg7 (Thompson et al., 2005; Chung et al., 2010). These imaging results were confirmed by anti-GFP immunoblot analysis of SAR1B-GFP and SAR1C-GFP plants (Figure 7B; Supplemental Figure S5B). As expected, WT control seedlings expressing SAR1B-GFP showed a higher ratio of free GFP to SAR1B-GFP than atg5-1 plants, and the ratio in WT seedlings was increased by ConA treatment (Figure 7B). Together, these data indicate that autophagy mediates the degradation of SAR1B-GFP in the vacuole.
Figure 7.
SAR1 function is crucial for autophagy. A, Confocal fluorescent images of WT or atg5-1 root cells expressing SAR1B-GFP. Seedlings grown in N-rich medium for 9 days were treated with DMSO, 0.5-µM ConA, or 0.5-µM ConA plus 30-µM Wm for 16 h prior to microscopic observation. B, Immunoblot analysis of SAR1B-GFP transgenic plants using anti-GFP antibodies. Seedlings were incubated as described above. The graph shows quantification of relative band intensities (mean ± se; n = 4 seedling populations). C and D, Anti-GFP immunoblot analysis of Arabidopsis leaf protoplasts transiently expressing AALP-GFP (C) or GFP-ATG8A (D) for 16–18 h. Immunoblot analysis using anti-GFP (upper), anti-Myc (middle; for the expression control) and anti-histone H3 (lower; for the loading control) antibodies. The graphs below (B–D) show the quantification of relative band intensities (mean ± se; n = 4). Columns marked with asterisks represent means that are significantly different from each other, according to t test. *0.01 < P < 0.05; **P < 0.01. E, Confocal fluorescence images of Col-0 leaf protoplasts transiently expressing GFP-ATG8A and either SAR1B-RFP, SAR1B(DN)-RFP, SAR1C-RFP, SAR1C(DN)-RFP, SAR1D-RFP, or SAR1D(DN)-RFP. Leaf protoplast preparations were incubated in liquid medium containing DMSO or 0.5 µM AZD for 1 h prior to observation. F, Quantification of GFP-ATG8A punctum number per 900 µm2, using images (n = 30–40) similar to those shown in (E). Columns marked with black and gray asterisks represent means that are significantly different from those of paired columns and of the DMSO controls, respectively, according to t test. G, Confocal fluorescence images of Col-0, atg7-2, atg2-1, or fyve2-2 leaf protoplasts transiently expressing GFP-ATG8A and SAR1B-RFP or SAR1B(DN)-RFP. The protoplasts were treated with ConA for 12 h prior to observation. The numbers show the quantification of vacuolar puncta per 400 µm2 area of the vacuole (mean ± se; n = 25–31 images). Scale bars = 5 µm.
To investigate whether SAR1 plays a general role in autophagosome biogenesis in plants, we measured autophagic fluxes in Arabidopsis protoplasts expressing WT or mutated SAR1 isoforms. Specifically, we added a Myc tag to four WT isoforms (SAR1A/B/C/D) and their respective dominant-negative (DN) mutant forms where their conserved His74 residues were replaced by Leu, thus locking SAR1 in the GTP state and blocking protein cargo export from the ER (Takeuchi et al., 2000). As a control, we co-expressed aleurain (AALP)-GFP, a vacuolar cargo whose vacuolar delivery is blocked by overexpression of SAR1C(DN) but not SAR1A(DN) (Zeng et al., 2015). As reported, we confirmed that free GFP released from vacuolar AALP-GFP was blocked by the DN forms of SAR1B/C/D but not by the expression of either their WT forms or SAR1A(DN) (Figure 7C). In protoplasts co-expressing GFP-ATG8A with various SAR1-Myc proteins, autophagic fluxes were specifically inhibited by SAR1B/C/D DN (Figure 7D), indicating that normal SAR1 function, hence COPII trafficking, is critical for general autophagy in Arabidopsis protoplasts. The reduction in autophagic flux was evident as early as 12 h of transient expression (Supplemental Figure S5, C and D), suggesting a direct effect of SAR1 dysfunction. Of note, we could not confirm the effect of SAR1 dysfunction in stable transformants, because the SAR1(DN) mutations cause plant lethality (Takeuchi et al., 2000; Liang et al., 2020).
We further investigated whether the expression of SAR1(DN) isoforms results in the accumulation or reduction of GFP-ATG8A puncta during AZD-induced autophagy. If a specific SAR1 isoform is necessary for phagophore nucleation, its DN mutation will cause a reduction in GFP-ATG8A puncta. If a SAR1 isoform is dispensable for phagophore nucleation but essential for a later step in autophagosome biogenesis, its dysfunction will culminate in the accumulation of GFP-ATG8A puncta, similar to fyve2 and atg2 (see Figure 2D). We isolated protoplasts from Col-0 leaves and transiently co-expressed GFP-ATG8A with either WT or DN mutant proteins of SAR1B/C/D fused to red fluorescent protein (RFP) (Figure 7E). SAR1B/C/D-RFP mostly showed a diffuse distribution, whereas SAR1B/C/D(DN)-RFP often formed large puncta likely representing disrupted ER–Golgi interfaces (Osterrieder et al., 2010). Under both control and autophagy-inducing conditions, GFP-ATG8A puncta over-accumulated in protoplasts expressing SAR1B(DN)-RFP compared with protoplasts expressing SAR1B-RFP (Figure 7, E and F). In contrast, the number of the puncta was reduced in AZD-treated protoplasts expressing SAR1C(DN)-RFP and SAR1D(DN)-RFP compared with those expressing their WT isoforms (Figure 7, E and F). These results are consistent with the possibility that SAR1C and SAR1D play a role in phagophore initiation, whereas SAR1B participates in a later step during autophagosome biogenesis. Because expressing the SAR1B(DN) mutants specifically mimicked fyve2 mutation (see Figure 2D), we focused on the SAR1B isoform.
To further test the functional relevance of the SAR1B–FYVE2 interaction, we transiently expressed both GFP-ATG8A and SAR1B-RFP in Arabidopsis leaf protoplasts prepared from WT, atg7-2, atg2-1, or fyve2-2 plants (Figure 7G). As expected, the central vacuole of WT protoplasts treated with ConA contained autophagic bodies positive both for GFP-ATG8A and SAR1B-RFP (Figure 7G), which were not detected in the protoplasts from atg7 or atg2 leaves. Similarly, few autophagic bodies decorated with GFP-ATG8A and/or SAR1B-RFP were detected in fyve2 protoplasts, indicating that FYVE2 is essential for the autophagy-mediated targeting of SAR1B-RFP to the vacuole. Furthermore, autophagic bodies were rarely detected in WT vacuoles of protoplasts expressing both GFP-ATG8A and SAR1B(DN)-RFP (Figure 7G), which is consistent with the results of the biochemical assay for autophagic flux (Figure 7D). Taken together, these data demonstrate that SAR1 and FYVE2 are crucial for general autophagic flux in Arabidopsis.
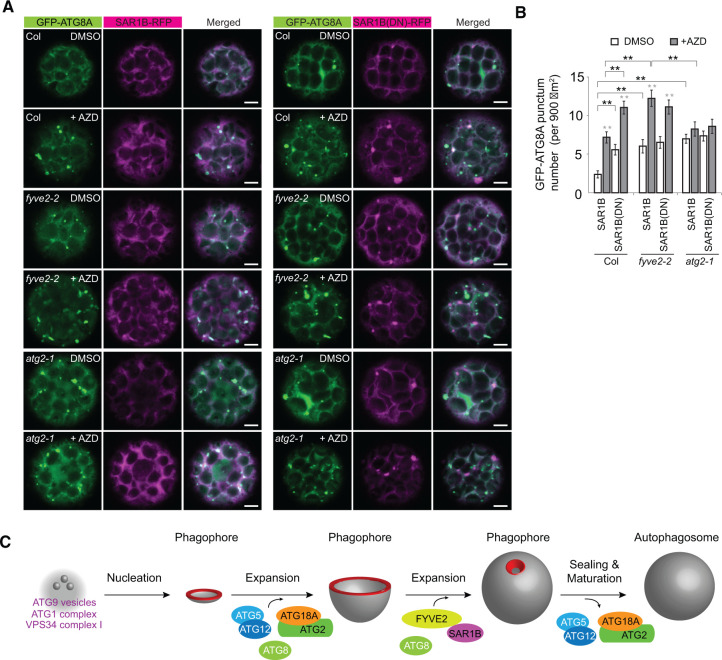
To directly compare the phenotypes of SAR1B dysfunction with the fyve2 and atg2 mutations, we transiently expressed GFP-ATG8A together with either SAR1B-RFP or SAR1B(DN)-RFP in WT, fyve2, and atg2 leaf protoplasts incubated in medium containing AZD or its solvent DMSO (Figure 8A). As expected, compared with the WT controls, fyve2 and atg2 protoplasts over-accumulated GFP-ATG8A puncta in the cytoplasm (Figure 8A and white columns in Figure 8B). AZD treatment significantly increased the accumulation of GFP-ATG8A-positive organelles in WT and fyve2 protoplasts and only slightly in atg2 protoplasts (Figure 8A and gray columns in Figure 8B), compared with the respective DMSO-treated controls (Figure 8A and white columns in Figure 8B). When SAR1B(DN)-RFP was expressed in WT protoplasts, we observed over-accumulation of GFP-ATG8A puncta compared with those expressing SAR1B-RFP (Figure 8, A and B). The increased abundance of GFP-ATG8A puncta in WT protoplasts expressing SAR1B(DN)-RFP was similar to that in fyve2 protoplasts expressing SAR1B-RFP (Figure 8B). Moreover, the expression of SAR1B(DN)-RFP in fyve2 protoplasts did not lead to any further increase in the number of GFP-ATG8A puncta compared to fyve2 protoplasts expressing SAR1B-RFP (Figure 8B). These data indicate that SAR1B dysfunction phenocopies the fyve2 but not the atg2 mutation, which is consistent with the physical interaction of FYVE2 with SAR1B. The common defects caused by the over-expression of SAR1B(DN) and the fyve2 mutation (reduced autophagic flux and increases in the density of GFP-ATG8A-positive organelles) suggest that SAR1B and FYVE2 act during the same step in autophagy progression. In atg2 protoplasts, the expression of SAR1B(DN)-RFP did not cause additional defects in autophagy (Figure 8B), indicating that SAR1B acts downstream of ATG2. We conclude that SAR1B and FYVE2 control phagophore elongation and/or later steps in autophagosome biogenesis (Figure 8C).
Figure 8.
Either fyve2 mutation or expression of the SAR1B DN fluorescent fusion leads to the over-accumulation of autophagic vesicles in the cytoplasm. A, Confocal fluorescence images of WT (first and second rows), fyve2-2 (third and fourth rows), and atg2-1 (fifth and sixth rows) protoplasts expressing GFP-ATG8A and either SAR1B-RFP (left columns) or SAR1B(DN)-RFP (right columns). Leaf protoplast preparations were incubated in liquid medium containing DMSO (first, third, and fifth rows) or 0.5 µM AZD for 1 h prior to observation. Scale bars = 5 µm. B, Quantification of GFP-ATG8A punctum number per 900 µm2, using images (n = 20–26) similar to those shown in (A). Columns marked with black and gray asterisks represent means that are significantly different from those of paired columns and of the DMSO controls, respectively, according to t test. Mean ± se. *0.01 < P < 0.05; **P < 0.01. C, A model for the roles of Arabidopsis FYVE2 and SAR1B in autophagosome biogenesis. The highly curved rim and neck of the phagophore are highlighted in red.
Discussion
The biogenesis of autophagosomes consists of several steps involving core ATG proteins and their associated proteins and membrane lipids. Mammalian and yeast autophagy is initiated by phagophore nucleation, in which the phosphorylation and scaffolding functions of the ATG1 complex and PI3P synthesis by the autophagic PI 3-kinase complex I are particularly important. Recent studies using yeast and mammalian homologs of ATG2 indicated that phagophore expansion is made possible by the lipid transfer activity of ATG2 and its interacting protein ATG18, a PI3P effector for autophagy (Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019). ATG9 not only plays a role in phagophore nucleation but also may distribute phospholipids from a cytosolic to luminal leaflet of the phagophore during its expansion (Maeda et al., 2020; Matoba et al., 2020). Less defined are the precise roles of COPII-coated membrane components and factors involved in later steps, such as phagophore sealing and autophagosome maturation. Furthermore, it is unclear how autophagosomes form in plant cells and whether the functions of known yeast and mammalian ATG proteins and interacting factors are conserved in the biogenesis of plant autophagosomes.
Here, we demonstrated the plant autophagy is mediated by the PI3P-binding protein FYVE2 and its interacting partner SAR1B, a small GTPase important for the formation of COPII vesicles. We further propose that FYVE2 and SAR1B participate in later steps of autophagosome biogenesis, such as phagophore expansion and sealing, and autophagosome maturation (Figure 8C). This conclusion is supported by multiple lines of evidence, including: (1) localization of FYVE2 fluorescent fusions to autophagic vesicles and membranes near the ER (Figure 5); (2) sequential recruitment of tagged FYVE2 proteins to phagophores that are positive for GFP-ATG8A, GFP-ATG18A, and PI3P (Figure 5); (3) interactions of FYVE2 with ATG18A and SAR1 (Figures 3 and 4); (4) vacuolar degradation of FYVE2 and SAR1 via a canonical autophagic route (Figures 6 and 7); (5) epistasis of atg2 to fyve2 (Figure 2); and (6) reduced autophagic flux and accumulation of autophagic organelles in fyve2 and upon the disruption of SAR1 functions (Figures 1, 2, 7, and 8).
We showed that FYVE2 is most likely recruited to the phagophore during its expansion (Movies 1 and 2), but further investigation is needed to explain how this recruitment occurs. mCherry-FYVE2 still co-localized with GFP-ATG8A in the absence of Arabidopsis ATG2 (Figure 6G), which is a putative lipid transfer protein required for phagophore expansion (Kang et al., 2018; Kim et al., 2020). This implies that the recruitment of FYVE2 to autophagic membranes is mediated not only by its interaction with ATG18A (Figures 3, D and 4) but also with PI3P (Sutipatanasomboon et al., 2017) and ATG8 (Figure 4).
During phagophore expansion, FYVE2 may help maintain or remodel phagophore membrane curvature using its interaction with PI3P and actin filaments (Sutipatanasomboon et al., 2017). It is also tempting to speculate that Arabidopsis FYVE2 plays a role in phagophore sealing via interaction with the ESCRT component VPS23A (Sutipatanasomboon et al., 2017). We identified a PSAP motif in FYVE2, which is responsible for the interaction with VPS23A (Figure 3C). The FYVE2–VPS23A interaction may be crucial for recruiting downstream ESCRT components to the membrane neck of the phagophore, because VPS23 commonly acts as an assembly factor essential for a variety of ESCRT-dependent membrane remodeling events (Vietri et al., 2020). The proposed role of FYVE2 in phagophore sealing through interactions with ESCRT proteins is consistent with the finding that fyve2 mutants accumulate phagophores (Figure 1E) and autophagic organelles (Figures 2D and 8), which have also been detected in ESCRT mutants (Spitzer et al., 2015).
A biochemical role of FYVE2 may be inferred from the finding that FYVE1/FREE1 (a plant equivalent to the ESCRT-0 component, Hepatocyte growth factor-regulated tyrosine kinase substrate [HRS]) functions in intralumenal vesicle formation on multivesicular endosomes (Barberon et al., 2014; Gao et al., 2014; 2015; Kolb et al., 2015; Belda-Palazon et al., 2016). Mammalian HRS interacts with PI3P and TSG101/VPS23 via its FYVE domain and the PSAP motif, respectively. HRS also interacts with clathrin coats and ubiquitylated membrane proteins (Raiborg et al., 2002), which is consistent with its role in endosomal sorting. In this regard, it would be interesting to determine whether FYVE2 and the COPII coat complex containing SAR1 and SEC24A recruit other proteins onto the phagophore. One intriguing possibility is that FYVE2 and SAR1 recruit downstream factors for autophagosome maturation and fusion, such as PI3P phosphatases and SNAREs. If this is true, it will provide a mechanism ensuring that autophagosome maturation does not occur prior to phagophore sealing.
In yeast and mammals, COPII vesicles participate in early steps of autophagosome biogenesis, facilitating phagophore expansion and likely, phagophore nucleation as well (Ishihara et al., 2001; Zoppino et al., 2010; Graef et al., 2013; Suzuki et al., 2013; Tan et al., 2013; Ge et al., 2014, 2017; Davis et al., 2016; Shima et al., 2019; Nakatogawa, 2020). During phagophore expansion, vesicles containing a specific set of COPII coat proteins originate from the ERES and supply membrane lipids to the growing phagophore. However, it is questionable whether lipids supplied by COPII vesicles are a major source for phagophore membrane expansion compared to the direct lipid transfer from the ER mediated by ATG2. Notably, autophagosome formation is not completely impaired upon disruption of mammalian SAR1 function (Ge et al., 2013), and not all autophagosomes in budding yeast (Saccharomyces cerevisiae) are decorated with COPII coat proteins (Shima et al., 2019), indicating only a partial requirement of the COPII machinery for autophagy. Consistently, our autophagic flux analysis in Arabidopsis indicated that SAR1 dysfunction led to a partial inhibition of autophagy (Figure 7D). Moreover, fyve2 mutants and SAR1B dysfunction led to over-accumulation of autophagic structures, whereas DN mutants of SAR1C and SAR1D did not (Figures 2, 7, and 8), indicating that the Arabidopsis COPII machinery containing SAR1B may specifically regulate phagophore expansion or a later step in autophagosome biogenesis.
Whether FYVE2 is also involved in phagophore nucleation is less clear. For example, one might envision a scenario where FYVE2 acts as a tether between the ERES and phagophore membranes after phagophore nucleation, facilitating the formation of the ATG2–ATG18A complex at the ERES. Indeed, we observed a small fraction of RFP-FYVE2 puncta in proximity to SAR1B-GFP signals (Supplemental Figure S6A) and foci of the ERES marker GFP-SEC24A (Supplemental Figure S6B), supporting this idea. AZD treatment increased the fraction of GFP-SEC24A foci located ˂1 µm from RFP-FYVE2 puncta (45%–66%; Supplemental Figure S6C), raising the possibility that the inhibition of TOR affects the distribution of the ERES. However, AZD treatment only slightly increased the fraction of RFP-FYVE2 puncta near the GFP-SEC24A (22%–29%; Supplemental Figure S6C). This implies that at a steady state, a majority of FYVE2 is present at the phagophore rather than at the ERES. Furthermore, the effects of fyve2 and SAR1B dysfunction on autophagy differ from that of atg2 (Figures 1, 2, and 8), which is inconsistent with the possible ER-phagophore tethering function of FYVE2-SAR1B.
It is also possible that FYVE2 acts as an autophagic receptor for the selective degradation of COPII proteins by sequestering them into the phagophore through its interaction with SAR1B and ATG8. This scenario is supported by the observation that not all autophagic bodies contain mCherry-FYVE2 (Figure 6C) and that both FYVE2 and SAR1 are delivered to the vacuole through autophagy, as expected for an autophagic receptor and its cargo. However, this scenario is unlikely, as the reduction in the autophagic flux seen in the fyve2 mutant could be entirely explained by the lack of COPII selective autophagy. In addition, plants with a fyve2 mutation and SAR1 dysfunction showed a defect in constitutive autophagy under nutrient-rich conditions (Supplemental Figure S2B; Figure 7D). These data do not support the view that FYVE2 is an autophagy receptor for COPII proteins or vesicles.
Zeng et al. (2021) recently reported a role for Arabidopsis SAR1D in autophagosome biogenesis. SAR1D seems to act as a molecular switch for COPII function in autophagy, since expression of DN SAR1D (but not SAR1A or SAR1C) proteins negatively affects autophagic flux. Upon autophagy induction, WT roots accumulated more autophagic organelles than the sar1d-1 mutant, suggesting that SAR1D-dependent COPII vesicles contribute to early steps in autophagosome biogenesis (Zeng et al., 2021). We confirmed that the expression of DN SAR1D is associated with a reduced number of autophagic organelles during AZD-induced autophagy (Figure 7). In contrast, SAR1B dysfunction specifically led to the accumulation of autophagic organelles. It is possible that SAR1D and SAR1B function with different subpopulations of COPII vesicles in early and late steps of autophagosome biogenesis, respectively. Curiously, we showed that the effect of SAR1C(DN) on AZD-induced autophagy is indistinguishable from that of SAR1D(DN) (Figure 7; Supplemental Figure S5D) and that SAR1C-GFP puncta are also degraded in the vacuole (Supplemental Figure S5, A and B). Further studies will be needed to explore the specialization of different SAR1 isoforms in specific steps of autophagy progression.
FYVE2 is not essential for autophagy, as autophagic flux is only moderately inhibited by fyve2 mutations, relative to atg2 (Figures 1 and 2). Several possible scenarios could explain the apparent dispensability of FYVE2 for autophagy. First, a protein similar to FYVE2 may play redundant functions. The simultaneous deletion of FYVE1 and FYVE2 could result in the absence of GFP-ATG8 puncta and the complete inhibition of autophagic flux, but we were unable to test this hypothesis because fyve1 null alleles cause lethality. FYVE3 (van Leeuwen et al., 2004) also has a FYVE domain and SYLF domain, but no further reduction in autophagic flux was observed in fyve2 fyve3 double mutants (Supplemental Figure S7). Second, the proposed function of FYVE2 in phagophore sealing and/or autophagosome maturation may be compensated by other mechanisms. Lastly, the role of FYVE2 in autophagy may vary according to the subpopulation of autophagosomes or cell types under specific conditions. For example, we noted that the fyve2 mutation has stronger negative effects on autophagy in the root maturation zone, hypocotyls, and leaves, compared with elongating root cells (Figure 1C; Supplemental Figures S2A and S7D).
To summarize, we identified a role for FYVE2 in autophagosome biogenesis in plants. We also defined molecular requirements for vacuolar trafficking of FYVE2 and the COPII-associated GTPase SAR1B. As FYVE2 is a plant-specific protein, our study provides insights into the molecular machinery that mediates autophagy progression in plants. Further studies will be needed to identify the biochemical functions of FYVE2 and SAR1B and to test the putative roles of FYVE2 in recruiting actin filaments, COPII coats, VPS23, and other factors needed for membrane bending, phagophore sealing, autophagosome maturation, and autophagosome fusion.
Materials and methods
Clones and DNA constructs
cDNA and expression clones were obtained from the Arabidopsis Biological Resource Center (ABRC) with the following identifiers: Arabidopsis (A. thaliana) ATG5 (G50285), ATG6 (G19464), FYVE1/FREE1 (G12263), FYVE2 (G18828), SAR1A (GC102951), VPS38 (G82505), SAR1B (G19464), SAR1C (G11541), SAR1D (G19132), SEC24A (U24156), ER-CFP (CD3-954), pEarleyGate103 (CD3-685), pEarleyGate203 (CD3-689), pEarleyGate303 (CD3-694), pIX-Halo-ATG8F (HALO_SFI_23-A01), pIX-Halo-ATG8I (HALO_SFI_71-H06), pMDC43 (CD3-741), pMDC83 (CD3-742), pMDC107 (CD3-748), pSITE-4CA (CD3-1641), pSITE-4NB (CD3-1643), PSAT4-DEST-n(1-174)EYFP-C1 (CD3-1089), PSAT5-DEST-c(175-END)EYFP-C1(B) (CD3-1097), pSITE-nEYFP-C1 (CD3-1648), and pSITE-cEYFP-C1 (CD3-1649). UBC-GFP (Grefen et al., 2010) was obtained from the Plant Science Research Group at University of Glasgow (https://psrg.org.uk/). The stop codons in ATG5, SAR1A, SAR1C, and SAR1D cDNA clones were erased by site-directed mutagenesis using primers listed in Supplemental Table S1. The resulting polymerase chain reaction (PCR) products were treated with DpnI (New England Biolabs, Ipswich, MA, USA) to isolate mutated ATG5(NS), SAR1A(NS), SAR1C(NS), and SAR1D(NS). The same procedure was used to obtain SAR1B(NS) cDNA using the primers listed in Supplemental Table S1 and inserted into the pENTR/TOPO vector (Invitrogen, Waltham, MA, USA). ATG8E, ATG18A, and AALP(NS) cDNA were obtained by PCR using the primers listed in Supplemental Table S1 and cloned into pENTR/TOPO. A genomic DNA fragment containing the FYVE2 promoter and coding regions was amplified by PCR using the primers listed in Supplemental Table S1. The amplified genomic DNA was cloned into pDONR221 via the BP Clonase II reaction (Thermo Fisher Scientific, Waltham, MA, USA) to obtain the entry vector ProFYVE2:FYVE2(NS). SAR1A/B/C/D(DN)(NS) cDNAs encoding DN (His74 to Leu) mutant SAR1 proteins were generated by site-directed mutagenesis using the primers listed in Supplemental Table S1. Expression clones of pIX-Halo-ATG8F and pIX-Halo-ATG8I were recombined with pDONR221 via the BP Clonase II reaction to obtain cDNA entry clones.
Transgenic plants
To generate binary vectors for transgenic plants expressing ProUBQ10:GFP-FYVE2 or ProUBQ10:mCherry-FYVE2, entry clones containing FYVE2 cDNA were recombined with pMDC99-AtUBQ10p-GFP and pMDC99-AtUBQ10p-mCherry (Suttangkakul et al., 2011), respectively, via the LR Clonase II reaction (Thermo Fisher Scientific). Similarly, binary vectors for transgenic plants expressing ProFYVE2:FYVE2-GFP and ProUBQ10:GFP-ATG18A were generated via the LR Clonase II reaction in which the entry clones ProFYVE2:FYVE2(NS) and ATG18A were recombined with pMDC107 (Curtis and Grossniklaus, 2003) and pMDC99-AtUBQ10p-GFP, respectively. To generate a binary vector for transgenic plants expressing ProUBQ10:ATG5-GFP, an entry clone containing ATG5(NS) cDNA was recombined with the UBC-GFP destination vector (Grefen et al., 2010). To prepare binary vectors for the transgenic lines Pro35S:SAR1B-GFP and ProCaMV35S:SAR1C-GFP, entry clones containing SAR1B(NS) and SAR1C(NS) cDNA were recombined with destination vectors pMDC83 (Curtis and Grossniklaus, 2003) and pEarleyGate103 (Earley et al., 2006), respectively.
To generate Arabidopsis stable transformants, the binary vectors mentioned above were introduced into Agrobacterium tumefaciens strain GV3101. The resulting Agrobacterium transformants were used to infect Arabidopsis by the floral dip method (Clough and Bent, 1998).
Plant materials and growth conditions
Arabidopsis T-DNA mutants fyve2-1/cfs1-1 (SALK_018265), fyve2-2/cfs1-2 (SALK_024058C), fyve2-3/cfs1-3 (SALK_068647), fyve3-1(SALK_027752C), atg2-1 (Inoue et al., 2006), and transgenic plants expressing ProUBQ10:YFP-ARA7, ProUBQ10:YFP-SYP32 (Geldner et al., 2009), and ProUBQ10:Citrine-2xFYVE (Simon et al., 2014) were obtained from the ABRC. Arabidopsis T-DNA mutants atg5-1 (Thompson et al., 2005), atg7-2 (Chung et al., 2010), atg11-1 (Li et al., 2014), and vps38-2 (Lee et al., 2018) and transgenic plants expressing ProUBQ10:GFP-ATG8A (Kim et al., 2013) and ProVHA-a1:VHA-a1-GFP (Dettmer et al., 2006) were previously described.
Before germination, seeds were surface-sterilized in 50% (v/v) bleach solution and washed at least three times with sterile water. The seeds were germinated on 1× Murashige and Skoog (MS) solid medium [1× MS salt including vitamin (Duchefa, Haarlem, Netherlands) with 1% (w/v) sucrose, 0.25% (w/v) Phytagel (Sigma, St Louis, MO, USA)]. Alternatively, the seeds were germinated in 1 mL of 1× MS liquid medium (1× MS salts including vitamin with 1% sucrose) with gentle shaking (100 rpm). Plants were grown at 21–23°C under long-day conditions (16-h light/8-h dark; fluorescent lamps; 90 µmol m−2 s−1) for either 9 days (immunoblot and confocal microscopy analysis) or 2–3 weeks (protoplast isolation).
For N starvation treatment, liquid medium containing seedlings was replaced by 1× MS-N liquid medium (1× MS micronutrient solution, 3 mM CaCl2, 1.5 mM MgSO4, 1.25 mM KH2PO4, 5 mM KCl, and 1% sucrose, pH 5.7), and the seedlings were further incubated for various time periods (Chung et al., 2010). If needed, seedlings were treated with 0.5 µM ConA (Cayman, Ann Arbor, MI, USA), 30 µM Wm (Sigma) and 0.5 µM AZD8055 (LC Laboratories, Woburn, MA, USA).
Transient expression in Arabidopsis leaf protoplasts
Transient expression was driven by tandem repeats of the Cauliflower Mosaic Virus (CaMV) 35S promoter (Pro35S). For GFP-ATG8A expression, pUC119-Pro35S:GFP-ATG8A was generated by ligating a PstI-digested pUC119 fragment with a PstI-digested expression cassette Pro35S:GFP-ATG8A (Supplemental Figure S8A). For GFP-ATG18A and GFP-SEC24A expression, pUC119-Pro35S:GFP-ATG18A and pUC119-Pro35S:GFP-SEC24A were obtained by an LR Clonase II reaction in which the ATG18A and SEC24A cDNA clones were recombined with the Gateway destination vector pUC119-Pro35S:GFP-DEST (Supplemental Figure S8A). For RFP-ATG8A and RFP-FYVE2 expression, PSITE-4CA-ATG8A and PSITE-4CA-FYVE2 were obtained by an LR Clonase II reaction in which the ATG8A and FYVE2 cDNA clones were recombined with the Gateway destination vector PSITE-4CA. For AALP-GFP and SAR1B-GFP expression, pUC119-Pro35S:AALP-GFP and pUC119-Pro35S:SAR1B-GFP were obtained by an LR Clonase II reaction in which the AALP(NS) and the SAR1B(NS) cDNA clones were recombined with the Gateway destination vector pUC119-Pro35S:DEST-GFP (Supplemental Figure S8B). For SAR1-RFP and SAR1(DN)-RFP expression, PSITE-4NB-SAR1 and PSITE-4NB-SAR1(DN) were obtained by LR Clonase II reaction in which the SAR1B/C/D(NS) and the SAR1B/C/D(DN)(NS) cDNA clones were recombined with the Gateway destination vector PSITE-4NB. For SAR1-Myc expression, a series of pUC119-Pro35S:SAR1-Myc vectors were obtained by LR Clonase II reactions in which either the SAR1A/B/C/D(NS) or SAR1A/B/C/D(DN)(NS) cDNA clone was recombined with the Gateway destination vector pUC119-Pro35S:DEST-Myc (Supplemental Figure S8C). For Myc-FYVE2 expression, pUC119-Pro35S:Myc-FYVE2 vectors were obtained by LR Clonase II reactions in which the FYVE2 cDNA clone was recombined with the Gateway destination vector pUC119-Pro35S:Myc-DEST (Supplemental Figure S8D). For NYFP-FYVE2 and NYFP-VPS38 expression, PSAT4-n(1-174)EYFP-C1-FYVE2 and PSAT4-n(1-174)EYFP-C1-VPS38 were obtained by LR Clonase II reaction in which the FYVE2 and VPS38 cDNA clones were recombined with the Gateway destination vector PSAT4-DEST-n(1-174)EYFP-C1. For CYFP-SAR1B, CYFP-ATG8A, CYFP-ATG18A, CYFP-SEC24A, CYFP-ATG6, and CYFP-VPS38 expression, a series of PSAT5-c(175-END)EYFP-C1(B)-SAR1B/ATG8A/ATG18A/SEC24A/ATG6/VPS38 were obtained by LR Clonase II reactions in which the SAR1B, ATG8A, ATG18A, SEC24A, ATG6, and VPS38 cDNA clones were recombined with the Gateway destination vector PSAT5-DEST-c(175-END)EYFP-C1(B).
Arabidopsis leaf protoplasts were prepared using a standard procedure (Yoo et al., 2007) with some modifications. Briefly, the leaves of 2- to 3-week-old Arabidopsis plants were used for protoplast isolation. Protoplasts were released in enzyme solution in the dark for 16–19 h. The released protoplasts were placed on 21% (w/v) sucrose solution, and intact protoplasts were obtained by centrifugation at 100g for 6–7 min. After transformation using polyethylene glycol, the protoplasts were resuspended in WI buffer (4 mM MES, 0.5 M Mannitol, 20 mM KCl, pH 5.7). Vectors used for protoplast transformation were as follows: pUC119-Pro35S:GFP-ATG8A, pUC119-Pro35S:AALP-GFP, pUC119-Pro35S:SAR1B-GFP, pUC119-Pro35S:GFP-ATG18A, pUC119-Pro35S:GFP-SEC24A, PSITE-4CA-ATG8A, PSITE-4CA-FYVE2, PSITE-4NB-SAR1B, PSITE-4NB-SAR1B(DN), PSITE-4NB-SAR1C, PSITE-4NB-SAR1C(DN), PSITE-4NB-SAR1D, PSITE-4NB-SAR1D(DN), pUC119-Pro35S:SAR1-Myc, pUC119-Pro35S:Myc-FYVE2, PSAT4-n(1-174)EYFP-C1-FYVE2, PSAT4-n(1-174)EYFP-C1-VPS38, PSAT5-c(175-END)EYFP-C1(B)-SAR1B, PSAT5-c(175-END)EYFP-C1(B)-ATG8A, PSAT5-c(175-END)EYFP-C1(B)-ATG18A, PSAT5-c(175-END)EYFP-C1(B)-SEC24A, PSAT5-c(175-END)EYFP-C1(B)-ATG6, and PSAT5-c(175-END)EYFP-C1(B)-VPS38. After transformation, the protoplasts were incubated at 20–25°C under continuous light conditions (fluorescent lamps; 90 µmol m−2 s−1) for 12–18 h. To observe autophagic bodies in protoplasts, protoplasts were treated with 0.5-µM ConA or an equivalent volume of DMSO for 12 h.
Immunoblot analysis
Seedlings were homogenized in 1× Laemmli buffer and clarified by centrifugation at 16,000g for 10 min at room temperature. Alternatively, seedlings or protoplasts were homogenized in TNPI lysis buffer [50-mM Tris–Cl, 150 mM NaCl, 0.5% (w/v) sodium dodecyl sulfate (SDS), 0.5% (v/v) Triton-X 100, 5% (v/v) glycerol, 2-mM dithiothreitol, 1-mM phenylmethyl sulfonyl fluoride, 10-mM iodoacetamide, pH 8.0] and clarified by centrifugation 16,000g for 10 min at 4°C. The supernatant was diluted with 5× SDS–polyacrylamide gel electerophoresis (PAGE) sample buffer [200 mM Tris–HCl (pH 6.8), 25% (v/v) glycerol, 10% (w/v) SDS, 10% (v/v) 2-mercaptoethanol] (Marshall et al., 2015). After the supernatant was heated at 95°C for 10 min, the resulting total proteins were separated by SDS–PAGE and transferred onto Immobilon-P polyvinylidene fluoride membranes (Millipore, St Louis, MO, USA).
For membrane fractionation, whole seedlings were homogenized in TNPI buffer [50 mM Tris–Cl, 150-mM NaCl, 1 mM phenylmethyl sulfonyl fluoride, 10-mM iodoacetamide, pH 8.0] using a chilled pestle and mortar. After the homogenate was clarified by centrifugation at 500g for 5 min at 4°C, an aliquot of supernatant was set aside as total extract, and remaining supernatant was subsequently separated into soluble and pellet fractions by centrifugation at 20,000g for 15 min at 4°C.
For co-immunoprecipitation, leaf protoplast cells were collected by sequential centrifugation 500g for 5 min and 10,000g for 5 min at room temperature. The cells were homogenized in 550 µL of IP lysis buffer [50 mM Tris–Cl, 150 mM NaCl, 5% (v/v) glycerol, 0.1% (v/v) Triton-X 100, 1-mM phenylmethyl sulfonyl fluoride, 1× plant protease inhibitor cocktail (Sigma)] and clarified by centrifugation 16,000g for 5 min at 4°C. To control for nonspecific interactions, 500 µL of supernatant was incubated with 30 µL of preequilibrated Protein A-agarose (Santa Cruz, Dallas, TX, USA) for 30 min at 4°C. The beads were collected by centrifugation 1,000g for 3 min at 4°C, and 50 µL of supernatant was set aside as an input. Approximately 400 µL of the supernatant was incubated with preequilibrated 30 µL GFP-Trap Agarose (Chromotek) for 3–4 h at 4°C. The beads were collected by centrifugation 1,000g for 3 min at 4°C and washed 3 times with wash buffer [50 mM Tris–Cl, 150 mM NaCl, 5% (v/v) glycerol]. Co-immunoprecipitated samples were extracted from the beads using 50 µL of 2× SDS–PAGE sample buffer [80-mM Tris–HCl (pH 6.8), 10% (v/v) glycerol, 4% (w/v) SDS, 4 mM dithiothreitol, 4% (v/v) 2-mercaptoethanol] followed by heating at 95°C for 10 min.
Immunoblotting was performed as previously described (Kim et al., 2013). Anti-GFP (Sigma-Aldrich, St Louis, MO, USA; 11814460001), Anti-Myc (Cell Signaling Technology 2276S), Anti-H3 (Abcam ab1791), Anti-BiP (Agrisera AS09 481), and Anti-UDP-glucose pyrophosphorylase (UGPase) (Agrisera AS05 086) antibodies were used. Band intensities of the immunoblots were measured by ImageJ (National Institute of Health).
Reverse transcriptase polymerase chain reaction (RT-PCR) analysis
Seedlings grown on MS solid medium for 10 days were homogenized in TRIsure agent (Bioline, London, UK). The RNA extract was treated with DNase I (New England Biolabs), and cDNA was synthesized using RevertAid H Minus reverse transcriptase (Thermo Fisher Scientific) with oligo(dT)20 primers. FYVE2 and FYVE3 cDNAs were amplified by 35 cycles of PCR using a Gene Atlas G02 thermocycler (ASTEC), Solg Taq DNA Polymerase (Solgent, South Korea), and the primers listed in Supplemental Table S1.
Y2H analysis
Entry clones were recombined with the destination vectors pDEST22 (encoding a prey protein containing an activating binding domain), pDEST32 (encoding a bait protein containing the DNA binding domain) via the LR Clonase II reaction. Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA, USA) was used for co-transformation of the MaV203 strain with pDEST22 and pDEST32. Alternatively, single transformation of the AH109 strain and Y187 strain was used for mating-based Y2H analysis.
Y2H using the ProQuest™ system (Invitrogen) was performed according to the manufacturer’s guidelines. To verify protein interactions, several yeast colonies selected on Leu-, Trp-lacking medium were transferred to Leu-, Trp-, His-lacking medium containing 0, 1 (for mating-based Y2H), or 30 (for cotransformation-based Y2H) mM 3-amino-1,2,4-triazole.
BiFC assay
To perform BiFC assays using N. benthamiana leaves, entry clones were recombined into the destination vectors pSITE-nEYFP-C1 and pSITE-cEYFP-C1 (Martin et al., 2009). Sequencing-verified BiFC expression vectors were then introduced into A. tumefaciens strain GV3101. Agrobacterium transformants were infiltrated into 6-week-old N. benthamiana leaves. During agroinfiltration, the p19-containing plasmid pCB301-p1934 was used to prevent gene silencing (Win and Kamoun 2004). Leaf discs were examined 36–48 h after agrobacterium infiltration using confocal microscopy.
For the BiFC assay in Arabidopsis leaf protoplasts, PSAT4-n(1-174)EYFP-C1-FYVE2, PSAT4-n(1-174)EYFP-C1-VPS38, and PSAT5-c(175-END)EYFP-C1(B)-SAR1B/ATG8A/ATG18A/SEC24A/ATG6/VPS38 were used for at least three independent transformation. For BiFC combined with colocalization with RFP-ATG8A, protoplasts were co-transformed with PSITE-4CA-ATG8A.
Immunogold labeling
WT and atg7-2 seedlings expressing GFP-FYVE2 were germinated on 1× MS solid medium. Four-day-old root tips were excised from the seedlings and frozen in a BAL-TEC HPM 010 high-pressure freezer. The root samples were freeze-substituted in 0.2% glutaraldehyde plus 0.2% uranyl acetate in acetone at −90°C for 4 days in an automated freeze-substitution device (Leica Microsystems, Wetzlar, Germany) and embedded in Lowicryl HM20 (Electron Microscopy Sciences Hatfield, PA, USA). Sections of HM20-embedded roots were mounted on formvar-coated nickel grids and blocked with a 5% (w/v) solution of nonfat milk in TBS containing 0.1% Tween-20. The sections were incubated with anti-GFP antibodies (Torrey Pines Antibodies, San Diego, CA, USA) for 1 h, rinsed in TBS containing 0.5% Tween-20, and incubated with the secondary antibody (1:10 anti-rabbit IgG) conjugated to gold particles for 1 h. Samples were imaged in a Philips CM120 transmission electron microscope.
Confocal microscopy, image processing, and quantification
Fluorescence images were obtained under either an LSM 510 or AxioObserver LSM 800 confocal microscope (Carl Zeiss). For LSM 510, 488-nm laser and BP500-530IR emission filter were used to detect GFP signals. To acquire GFP, citrine, and YFP signals using the LSM 800, 488-nm was used for excitation and fluorescence was detected at a 410- to 546-nm range. To detect mRFP and mCherry signals in Arabidopsis roots, the 587-nm laser was used for excitation and fluorescence was detected at a 595- to 700-nm range. In the leaf epidermis and protoplasts, the 587-nm laser was used and fluorescence was detected at a 595- to 617-nm range.
ImageJ (NIH) was used for quantitative analysis of confocal microscope images. The Subtract Background Tool was used to remove background noise from the images and excess cytosolic signal. GFP punctum size and abundance were determined using the Analyze Particles Tool and Multi Measure Tool. Quantification of fluorescent puncta in Arabidopsis tissues was performed using whole frames of fluorescence images, with a frame size of either 25,514 µm2 (159.73 µm × 159.73 µm) or 12,656 µm2 (112.5 µm × 112.5 µm). To measure punctum abundance in leaf protoplasts, a region of interest (ROI) of 900 µm2 (30 µm × 30 µm) was randomly selected from each image of the cytoplasm emitting fluorescence. For fluorescence intensity measurement, ROIs of 1,225 µm2 (35 µm × 35 µm) were randomly selected. For quantification of autophagic bodies in Arabidopsis roots and leaf protoplasts, at least three optical section images at a 1-µm interval were acquired, and ROIs of either 1,000 µm2 (50 µm × 20 µm; for root cell) or 400 µm2 (20 µm × 20 µm; for protoplasts) were randomly selected in the large lumen of the central vacuole.
To quantify co-localization between citrine-2xFYVE, YFP-ARA7, GFP-ATG8A, VHA-a1-GFP, YFP-SYP32, and mCherry-FYVE2, the Comdet plug-in was used [https://imagej.net/Spots_colocalization_(ComDet)]. A whole frame of single-channel fluorescence images (frame size of 25,514 µm2; 159.73 µm × 159.73 µm) was merged using the Merge Channels Tool. To exclude background noise, fluorescence puncta were isolated from cytosolic signals using the Intensity Threshold option of the Comdet plug-in. Punctate signals in the two channels were considered co-localized if the distance between their centroids was ˂300 nm.
For co-localization of GFP-ATG8A-positive autophagic bodies with mCherry-FYVE2 puncta, at least three optical section images at a 1-µm interval were acquired from the root maturation zone, and ROIs of 1,000 µm2 (50 µm × 20 µm) were selected from z-sections including the central vacuole. Due to the rapid Brownian motion of autophagic bodies inside the vacuole, GFP and mCherry punctate signals were considered to overlap if the distance between their centroids was ˂1 µm.
Pearson correlation coefficient (r) was calculated via the Coloc 2 Plug-in (https://imagej.net/Coloc_2) using the processed images described above.
Statistical analysis
The data obtained in this study are presented as means ± standard error (se). The numbers of samples in each experiment are noted in the figure legends. Statistical analysis was performed by two-tailed Student’s t test using SPSS statistics or VassarStats (http://www.vassarstats.net/). Details about statistical analysis are provided in Supplemental Data Set S1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AALP (At5g60360), ATG2 (At3g19190), ATG5 (At5g17290), ATG6 (At3g61710), ATG7 (At5g45900), ATG8A (At4g21980), ATG8E (At2g45170), ATG8F (At4g16520), ATG8I (At3g15580), ATG11 (At4g30790), ATG18A (At3g62770), RABF2b/ARA7 (At4g19640), FYVE1/FREE1 (At1g20110), FYVE2/CFS1 (At3g43230), FYVE3 (At1g29800), SAR1A (At1g09180), SAR1B (At1g56330), SAR1C (At4g02080), SAR1D (At3g62560), SEC24A (At3g07100), SYP32 (At3g24350), UBC9 (At4g27960), VHA-a1 (At2g28520), VPS23A (At3g12400), and VPS38 (At2g32760).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular characterization of Arabidopsis fyve2 mutants.
Supplemental Figure S2. Accumulation of autophagic vesicles in mature roots exposed to N starvation.
Supplemental Figure S3. Expression of the mCherry-FYVE2 transgene complements the fyve2-2 phenotypes.
Supplemental Figure S4. Fluorescent FYVE2 fusion proteins are targeted to the vacuole via the autophagic route.
Supplemental Figure S5. SAR1C-GFP is degraded in the vacuole in a PI3P-dependent manner.
Supplemental Figure S6. RFP-FYVE2 does not localize to the ERES.
Supplemental Figure S7. A mutation in FYVE3 does not inhibit autophagic flux alone or in combination with fyve2.
Supplemental Figure S8. Diagram for the preparation of plant transformation vectors.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplemental Data Set S1. Student’s t test tables.
Supplementary Material
F.L. and T.C. initiated the project and designed the research. J.H.K. performed most experiments. H.N.L. and T.C. initially developed genetic materials. X.H. performed bimolecular fluorescence complementation in N. benthamiana leaves. J.H.K., H.N.L., and H.J. performed autophagic flux analysis. M.S.O. and T.C. performed immunogold labeling experiments. M.S.O., F.L., and T.C. wrote the manuscript with support from J.H.K., H.N.L., X.H., and H.J.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Faqiang Li (fqli@scau.edu.cn) and Taijoon Chung (taijoon@pusan.ac.kr).
Funding
This work was supported by grants from the National Research Foundation of Korea (NRF-2019R1A2C1084203) to T.C., from the Natural Science Foundation of Guangdong Province (grant 2018A0303130348) to X.H., and from US National Science Foundation NSF IOS-1840687 to M.S.O.
Conflict of interest statement. None declared.
References
- Arabidopsis Interactome Mapping Consortium. (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Baluska F, Bozhkov PV, Elander PH, Fernie AR, Galili G, Hassan A, Hofius D, Isono E, Le Bars R, et al. (2018) Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot 69: 1335–1353 [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G (2014) Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA 111: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B, Rodriguez L, Fernandez MA, Castillo MC, Anderson EA, Gao C, Gonzalez-Guzman M, Peirats-Llobet M, Zhao Q, De Winne N, et al. (2016) FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov PV (2018) Plant autophagy: mechanisms and functions. J Exp Bot 69: 1281–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Otomo C, Leitner A, Ohashi K, Aebersold R, Lander GC, Otomo T (2018) Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc Natl Acad Sci USA 115: E9792–E9801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T (2019) How phosphoinositides shape autophagy in plant cells. Plant Sci 281: 146–158 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauphinee AN, Cardoso C, Dalman K, Ohlsson JA, Fick SB, Robert S, Hicks GR, Bozhkov PV, Minina EA (2019) Chemical screening pipeline for identification of specific plant autophagy modulators. Plant Physiol 181: 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Wang J, Zhu M, Stahmer K, Lakshminarayan R, Ghassemian M, Jiang Y, Miller EA, Ferro-Novick S (2016) Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. Elife 5: e21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Faso C, Chen YN, Tamura K, Held M, Zemelis S, Marti L, Saravanan R, Hummel E, Kung L, Miller E, et al. (2009) A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L (2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24: 2556–2563 [DOI] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Cui Y, Fu X, He Y, Zhao Q, Zeng Y, Shen J, Luo M, Jiang L (2015) Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci USA 112: 1886–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R (2013) The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2: e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R (2014) Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 3: e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Kenny SJ, Liu D, Maeda M, Saito K, Mathur A, Xu K, Schekman R (2017) Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep 18:1586–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J (2013) ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell 24: 2918–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hanton SL, Chatre L, Matheson LA, Rossi M, Held MA, Brandizzi F (2008) Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol Biol 67: 283–294 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Huang X, Zheng C, Liu F, Yang C, Zheng P, Lu X, Tian J, Chung T, Otegui MS, Xiao S, et al. (2019) Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31: 2973–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y (2001) Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 12: 3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Zhu L, Wang Q, Hou X (2020) Autophagy-related 2 regulates chlorophyll degradation under abiotic stress conditions in Arabidopsis. Int J Mol Sci 21: 4515 doi: 10.3390/ijms21124515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Shin KD, Kim JH, Chung T (2018) Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep 37: 653–664 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jung H, Chung T (2020) Birth, growth, maturation, and demise of plant autophagic vesicles. J Plant Biol 63: 155–164 [Google Scholar]
- Kolb C, Nagel MK, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E (2015) FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol 167: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H (2018) The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci USA 115: 10363–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW (2014) ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun 5: 4121. [DOI] [PubMed] [Google Scholar]
- Lee HN, Zarza X, Kim JH, Yoon MJ, Kim SH, Lee JH, Paris N, Munnik T, Otegui MS, Chung T (2018) Vacuolar trafficking protein VPS38 is dispensable for autophagy. Plant Physiol 176: 1559–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) AUTOPHAGY-RELATED (ATG)11 plays a critical role in general autophagy and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Li SW, Gong LM, Li S, Zhang Y (2020) COPII components Sar1b and Sar1c play distinct yet interchangeable roles in pollen development. Plant Physiol 183: 974–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hu W, Vierstra RD (2018) The vacuolar protein sorting-38 subunit of the Arabidopsis phosphatidylinositol-3-kinase complex plays critical roles in autophagy, endosome sorting, and gravitropism. Front Plant Sci 9: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hu W, Li F, Marshall RS, Zarza X, Munnik T, Vierstra RD (2020) AUTOPHAGY-RELATED14 and its associated phosphatidylinositol 3-kinase complex promotes autophagy in Arabidopsis. Plant Cell 32: 3939–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Maeda S, Otomo C, Otomo T (2019) The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8: e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T (2020) Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 27: 1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69: 173–208 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al. (2020) Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Nakano RT, Matsushima R, Ueda H, Tamura K, Shimada T, Li L, Hayashi Y, Kondo M, Nishimura M, Hara-Nishimura I (2009) GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21: 3672–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H (2020) Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458 [DOI] [PubMed] [Google Scholar]
- Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, Noda NN (2019) Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26: 281–288 [DOI] [PubMed] [Google Scholar]
- Osterrieder A, Hummel E, Carvalho CM, Hawes C (2010) Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. J Exp Bot 61: 405–422 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Robertson AS, Allwood EG, Smith AP, Gardiner FC, Costa R, Winder SJ, Ayscough KR (2009) The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol Biol Cell 20: 1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, Yoshimori T (2014) A current perspective of autophagosome biogenesis. Cell Res 24: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima T, Kirisako H, Nakatogawa H (2019) COPII vesicles contribute to autophagosomal membranes. J Cell Biol 218: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KD, Lee HN, Chung T (2014) A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 37: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y (2014) A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 77: 322–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G (2005) The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Soto-Burgos J, Bassham DC (2017) SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One 12: e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Li F, Buono R, Roschzttardtz H, Chung T, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS (2015) The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutipatanasomboon A, Herberth S, Alwood EG, Haweker H, Muller B, Shahriari M, Zienert AY, Marin B, Robatzek S, Praefcke GJK, et al. (2017) Disruption of the plant-specific CFS1 gene impairs autophagosome turnover and triggers EDS1-dependent cell death. Sci Rep 7: 8677–017-08577-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y (2013) Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 126: 2534–2544 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, He H, Tang Z, Hattori T, Liu Y, Young MM, Serfass JM, Chen L, Gebru M, Chen C, et al. (2018) An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9: 2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I (2003) Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tan D, Cai Y, Wang J, Zhang J, Menon S, Chou HT, Ferro-Novick S, Reinisch KM, Walz T (2013) The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci USA 110: 19432–19437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek AN, Allwood EG, Smith AP, Booth WI, Ayscough KR (2015) Distinct actin and lipid binding sites in Ysc84 are required during early stages of yeast endocytosis. PLoS One 10: e0136732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ (2019) ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218: 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Okresz L, Bogre L, Munnik T (2004) Learning the lipid language of plant signalling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, Stenmark H (2020) The many functions of ESCRTs. Nat Rev Mol Cell Biol 21: 25–42 [DOI] [PubMed] [Google Scholar]
- Win J, Kamoun S (2004) pCB301-p19: a binary plasmid vector to enhance transient expression of transgenes by agroinfiltration. Kamoun lab: Web supplements. http://www.Kamounlab.net
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocol 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong KB, et al. (2015) Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 14360–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Li B, Lin Y, Jiang L (2019) The interplay between endomembranes and autophagy in plants. Curr Opin Plant Biol 52: 14–22 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Li B, Ji C, Feng L, Niu F, Deng C, Chen S, Lin Y, Cheung KCP, Shen J, et al. (2021) A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 118: e2021293118 doi: 10.1073/pnas.2021293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Chung KP, Cui Y, Lin W, Gao C, Kang BH, Jiang L (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA 114: E426–E435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C., Colombo MI (2010) Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11: 1246–1261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.