ABSTRACT
Acinetobacter baumannii is a common nosocomial pathogen that utilizes numerous mechanisms to aid its survival in both the environment and the host. Coordination of such mechanisms requires an intricate regulatory network. We report here that A. baumannii can directly regulate several stress-related pathways via the two-component regulatory system BfmRS. Similar to previous studies, results from transcriptomic analysis showed that mutation of the BfmR response regulator causes dysregulation of genes required for the oxidative stress response, the osmotic stress response, the misfolded protein/heat shock response, Csu pilus/fimbria production, and capsular polysaccharide biosynthesis. We also found that the BfmRS system is involved in controlling siderophore biosynthesis and transport, and type IV pili production. We provide evidence that BfmR binds to various stress-related promoter regions and show that BfmR alone can directly activate transcription of some stress-related genes. Additionally, we show that the BfmS sensor kinase acts as a BfmR phosphatase to negatively regulate BfmR activity. This work highlights the importance of the BfmRS system in promoting survival of A. baumannii.
IMPORTANCE Acinetobacter baumannii is a nosocomial pathogen that has extremely high rates of multidrug resistance. This organism’s ability to endure stressful conditions is a key part of its ability to spread in the hospital environment and cause infections. Unlike other members of the gammaproteobacteria, A. baumannii does not encode a homolog of the RpoS sigma factor to coordinate its stress response. Here, we demonstrate that the BfmRS two-component system directly controls the expression of multiple stress resistance genes. Our findings suggest that BfmRS is central to a unique scheme of general stress response regulation by A. baumannii.
KEYWORDS: Acinetobacter baumannii, BfmRS, two-component system, stress response
INTRODUCTION
Acinetobacter baumannii is a Gram-negative opportunistic bacterium that is considered one of the most serious nosocomial pathogens worldwide (1, 2). This pathogen is predominantly responsible for ventilator-associated pneumonia and also causes bloodstream, urinary tract, and skin/soft tissue infections (3, 4). The majority of these infections occur in critically ill individuals, with patients in intensive or long-term care being at greatest risk (4, 5). Unfortunately, high rates of antibiotic resistance have greatly limited therapeutic options for these infections (6–9). It is estimated that 80% of A. baumannii pneumonia infections are caused by multidrug-resistant strains (10). Thus, the World Health Organization has labeled A. baumannii a critical priority pathogen due to the urgent need for the development of novel antimicrobial agents (7). In addition to its ability to develop antibiotic resistance, A. baumannii’s ability to persist in the hospital environment is key to its success as a nosocomial pathogen. This organism is capable of prolonged survival in unfavorable environments, including on inanimate objects such as hospital beds, doorknobs, and medical equipment, allowing indirect patient-to-patient transmission (11–13). One system in A. baumannii that controls both antibiotic resistance and survival during stressful conditions is the BfmRS two-component regulatory system.
Found in both prokaryotes and plants, two-component regulatory systems are used to sense and respond to specific signals, allowing adaptation to changing environmental conditions (14). These systems are usually composed of a membrane-associated sensor kinase and a corresponding response regulator. In a typical two-component regulatory system, the sensor kinase autophosphorylates at a conserved histidine residue in response to a signal. A subsequent phosphotransfer from the sensor kinase to a conserved aspartate residue on the response regulator triggers a conformational change that promotes activation of the response regulator. For OmpR/PhoB family response regulators such as BfmR, phosphorylation typically alters the ability of these proteins to interact with DNA, stimulating induction or repression of transcription (15–18). However, many details of the BfmRS regulatory mechanism, including the regulation of the system itself, remain unclear. Experiments performed using genetic approaches suggest that the BfmRS system may not function using the typical scheme described above. These studies show that the sensor kinase BfmS exerts a negative effect upon BfmR-regulated gene expression (19–21), but the details of the phosphotransfer signals have not been verified. Furthermore, electrophoretic mobility shift assay (EMSA) analysis showed that BfmR treated with the phosphomimic beryllium fluoride (BeF3−) binds to the bfmRS promoter with a lower affinity than untreated BfmR (22). This was unexpected, since activation of response regulators usually increases DNA-binding ability (16), although it is still unclear whether phosphorylated BfmR (BfmR∼P) or unphosphorylated BfmR is the active form.
The BfmRS system is known to control multiple phenotypes in A. baumannii. Originally, BfmR was identified due to its ability to promote biofilm formation and regulate the expression of the Csu pili, which are involved in attachment to abiotic surfaces (23). Surface attachment and biofilm formation are important for sustained colonization and have been implicated in the spread of A. baumannii within nosocomial environments (24). Furthermore, multiple studies have found that the BfmRS system confers resistance to host-mediated defenses. It was shown that a BfmR transposon mutant had reduced survival in human serum and human ascites fluid (25). Disruption of BfmR by transposon mutagenesis also reduced in vivo survival of A. baumannii in a rat subcutaneous abscess model (26) and in a neutropenic murine bacteremia model (27). Similarly, both BfmR and BfmS mutations reduced A. baumannii’s survival in a murine pneumonia model (28). BfmR and BfmS have also been implicated in controlling resistance to a variety of antibiotics, including penicillins, carbapenems, fluoroquinolones, aminoglycosides, and macrolides (20, 25, 29, 30). Finally, we previously found that BfmR is required for A. baumannii to endure a variety of stresses, including desiccation, osmotic challenge, exposure to hydrogen peroxide, and starvation (31). Together, these studies indicate that the BfmRS system controls phenotypes that are critical for the survival of A. baumannii under stressful conditions that occur either in the environment or in the host during infection.
In bacteria, stressful conditions can trigger specific responses to combat a particular stressor or can stimulate a broader protective response known as the general stress response or the stationary-phase response. Under laboratory growth conditions, this response is observed in batch cultures after extended growth, when the available nutrient sources are depleted. However, it can also be induced by suddenly exposing growing cells to stressful conditions that can affect growth, such as osmotic shock or heat shock. In most gammaproteobacteria, this response is coordinated by the sigma factor RpoS, which directs the transcription of numerous stress-related genes (32). Notably, A. baumannii does not encode an RpoS homolog (33). This implies that the general stress responses are controlled by an alternative mechanism in this organism. Most interestingly, many of the functions that are controlled by RpoS in other species appear to be regulated by the BfmRS system in A. baumannii. Transcriptome analysis in A. baumannii strain ATCC 17978 showed that the BfmRS system controls the expression of many genes associated with the transition to stationary phase (20). Similarly, we found that BfmR regulates the expression of stationary-phase-induced genes in strain ATCC 17961 (31). We also found that BfmR is necessary for starvation-induced cross-protection against drying (31), which is also seen with RpoS (34, 35). These findings suggest that the BfmRS system plays a prominent role in controlling the general stress response in A. baumannii. However, mutations affecting other factors and regulatory systems have also been shown to broadly affect A. baumannii’s stress resistance (36–42), making the exact role of the BfmRS system in controlling these responses unclear. In this study, we demonstrate that BfmR∼P can regulate the A. baumannii stress response by directly binding to promoters to activate the transcription of genes in multiple stress response pathways. Additionally, we provide evidence that BfmS is a BfmR phosphatase that negatively affects BfmR-regulated responses.
RESULTS
Mutation of bfmR alters the expression of multiple stress-related pathways.
Our previous findings showed that deletion of bfmR caused A. baumannii to become much less resistant to multiple stresses. To gain a better understanding of how BfmR controls protective responses in A. baumannii, we first compared the transcriptomes of strains ATCC 17961 and 17961-ΔbfmR using transcriptome sequencing (RNA-seq) analysis. For this analysis, we collected RNA from samples of cultures at the onset of stationary growth phase (6 h of growth in LB medium at 37°C with constant aeration). These results showed that the expression of 1,259 genes was significantly altered >2-fold (P < 0.03; Q < 0.05) in the ΔbfmR mutant strain, compared to the wild-type strain. Of these 1,259 genes, 631 genes had reduced expression in the ΔbfmR mutant strain, and 628 genes had increased expression in the mutant (see Table S1 in the supplemental material). We examined the set of differentially regulated genes and performed pathway enrichment analysis on this data set. Table 1 summarizes the key groups of genes that we found to be significantly dysregulated in the ΔbfmR mutant. We observed that numerous genes related to stress responses had decreased expression in the ΔbfmR mutant strain. These included genes involved in protection against osmotic stress and oxidative stress and genes involved in the misfolded protein response (Table 1). Consistent with other studies, we also found that the csu genes for pilus assembly and genes involved in capsular polysaccharide production had reduced expression in the ΔbfmR mutant strain (20, 23). The mutant also had increased expression of genes involved in the biosynthesis and transport of the siderophore acinetobactin, which is a virulence factor that is needed for A. baumannii to grow within the host (43, 44). In addition, we observed that the ΔbfmR mutant strain had increased expression of genes related to type IV pilus production, which has been linked to motility in the closely related species Acinetobacter nosocomialis (45).
TABLE 1.
Differentially regulated genes in the ΔbfmR mutant strain versus the wild-type strain ATCC 17961 in key functional groupsa
| Category and locus tag | Gene | Log2 fold change | P value | Predicted function |
|---|---|---|---|---|
| Stress response—osmotic stress | ||||
| I5593_15090 | otsB | −8.05 | 1.17E−106 | Trehalose-6-phosphate phosphatase |
| I5593_15095 | otsA | −7.48 | 1.04E−136 | Trehalose-6-phosphate synthase |
| I5593_15260 | mscS | −5.24 | 2.03E−95 | Small-conductance mechanosensitive channel |
| I5593_07035 | mscM | −4.77 | 1.22E−70 | Mechanosensitive ion channel |
| I5593_06310 | proP | −3.20 | 3.54E−17 | Glycine betaine/l-proline transporter |
| Stress response—oxidative stress | ||||
| I5593_02915 | −6.13 | 6.49E−112 | Alkyl hydroperoxide reductase subunit C-like protein | |
| I5593_16290 | acnA | −4.89 | 1.09E−71 | Aconitate hydratase |
| I5593_01690 | sodC | −4.64 | 2.42E−64 | Superoxide dismutase |
| I5593_11145 | katE | −4.61 | 1.83E−74 | Catalase HPII |
| Stress response—misfolded protein response | ||||
| I5593_19145 | grpE | −3.72 | 5.59E−24 | Nucleotide exchange factor |
| I5593_19150 | dnaK | −3.60 | 7.06E−23 | Molecular chaperone |
| I5593_03940 | groS | −3.35 | 3.34E−24 | Cochaperone |
| I5593_03945 | groL | −3.29 | 4.62E−23 | Chaperonin |
| I5593_17515 | htpG | −3.22 | 2.08E−41 | Molecular chaperone |
| I5593_00180 | dnaJ | −2.80 | 4.55E−21 | Molecular chaperone |
| I5593_13790 | lon | −2.65 | 1.35E−18 | Endopeptidase La |
| I5593_08315 | clpS | −2.32 | 9.07E−18 | ATP-dependent Clp protease adapter |
| I5593_08320 | clpA | −2.23 | 4.50E−14 | ATP-dependent Clp protease ATP-binding subunit |
| I5593_17185 | −2.28 | 3.89E−24 | DnaJ domain-containing protein | |
| I5593_08020 | −2.17 | 1.48E−12 | Similar to ribosome-associated heat shock protein 15 | |
| Stress response—miscellaneous | ||||
| I5593_11130 | absA | −11.20 | 2.9E−201 | A. baumannii stress-related protein A |
| I5593_11155 | cinA 1 | −5.49 | 7.64E−78 | Damage-inducible protein |
| Csu pili/fimbriae | ||||
| I5593_06275 | csuAB | −4.13 | 4.2E−54 | Csu fimbrial major subunit |
| I5593_06280 | csuA | −3.59 | 4.00E−06 | Csu fimbrial biogenesis protein |
| I5593_06290 | csuC | −1.29 | 0.006209 | Csu fimbrial biogenesis chaperone |
| Capsule/polysaccharide biosynthesis | ||||
| I5593_18825 | pgm | −2.37 | 3.62E−18 | Phosphomannomutase/phosphoglucomutase |
| I5593_18835 | gpi | −2.20 | 4.49E−24 | Glucose-6-phosphate isomerase |
| I5593_18840 | ugd | −2.77 | 8.43E−34 | UDP-glucose 6-dehydrogenase |
| I5593_18910 | wza | −1.31 | 1.04E−06 | Polysaccharide biosynthesis/export family protein |
| I5593_18915 | wzb | −1.28 | 5.12E−05 | Low-mol-wt phosphotyrosine protein phosphatase |
| Siderophore biosynthesis and transport | ||||
| I5593_05095 | bauB | 5.81 | 2.70E−28 | Siderophore-binding periplasmic lipoprotein |
| I5593_05085 | bauC | 5.70 | 1.68E−19 | Ferric acinetobactin ABC transporter permease subunit |
| I5593_05080 | bauD | 5.50 | 1.44E−26 | Ferric acinetobactin ABC transporter permease subunit |
| I5593_05090 | bauE | 5.03 | 5.55E−19 | Ferric acinetobactin ABC transporter ATP-binding protein |
| I5593_05075 | basB | 4.81 | 7.81E−57 | Acinetobactin nonribosomal peptide synthetase subunit |
| I5593_05120 | basF | 4.44 | 1.13E−41 | Acinetobactin biosynthesis bifunctional isochorismatase/aryl carrier protein |
| I5593_05115 | basE | 4.14 | 1.27E−52 | (2,3-Dihydroxybenzoyl)adenylate synthase |
| I5593_05125 | basG | 3.67 | 1.08E−23 | Acinetobactin biosynthesis histidine decarboxylase |
| I5593_05110 | basD | 3.26 | 1.83E−29 | Acinetobactin nonribosomal peptide synthetase subunit |
| I5593_05100 | bauA | 3.02 | 1.12E−24 | TonB-dependent ferric acinetobactin receptor |
| I5593_05065 | bauF | 2.66 | 1.14E−26 | Acinetobactin utilization protein |
| I5593_14045 | 2.65 | 8.72E−32 | TonB-dependent siderophore receptor | |
| I5593_05130 | barA | 2.34 | 1.13E−10 | Acinetobactin export ABC transporter permease/ATP-binding subunit |
| I5593_05070 | basA | 2.23 | 9.64E−12 | Acinetobactin nonribosomal peptide synthetase subunit |
| I5593_05135 | barB | 2.15 | 1.22E−09 | Acinetobactin export ABC transporter permease/ATP-binding subunit |
| Type IV pili | ||||
| I5593_01560 | fimT | 8.76 | 2.74E−58 | GspH/FimT family pseudopilin |
| I5593_01565 | pilV | 6.57 | 2.05E−46 | Type IV pilus modification protein |
| I5593_01440 | pilN | 5.61 | 2.59E−34 | PilN domain-containing protein |
| I5593_01445 | pilO | 5.28 | 3.75E−33 | Type 4a pilus biogenesis protein |
| I5593_01570 | pilW | 4.22 | 7.06E−30 | PilW family protein |
| I5593_01450 | pilP | 4.19 | 9.38E−23 | Pilus assembly protein |
| I5593_01575 | pilX | 3.92 | 2.26E−31 | Pilus assembly protein |
| I5593_01435 | pilM | 3.45 | 3.73E−42 | Pilus assembly protein |
| I5593_01585 | 2.76 | 6.15E−17 | Prepilin-type N-terminal cleavage/methylation domain-containing protein | |
| I5593_01580 | pilY | 2.67 | 6.01E−25 | VWA domain-containing protein |
| I5593_17355 | pilB | 2.56 | 1.04E−20 | Type IV-A pilus assembly ATPase |
| I5593_14635 | 2.02 | 8.34E−09 | Type IV pilus twitching motility protein | |
The complete set of differentially regulated genes is available in Table S1.
We compared our RNA-seq results to those of a previously published transcriptome study, performed by Geisinger et al. (20), that analyzed a ΔbfmR mutant in A. baumannii strain ATCC 17978 during logarithmic growth phase. That study, performed at an earlier time point than our study, found that the expression of 1,774 genes was altered in their ΔbfmR mutant strain. Similar to our data set, they found decreased expression of genes related to osmotic and oxidative stress responses in the ΔbfmR mutant strain, although generally to a lesser degree than what we observed. Deletion of bfmR also caused decreased expression of genes encoding β-lactamases (adc and oxa51) in strain ATCC 17961 (Table S1) and strain ATCC 17978 (20). Additionally, genes related to siderophore biosynthesis and type IV pili had significantly increased expression in the 17978-ΔbfmR mutant strain (20), which is similar to what we found in strain ATCC 17961 (Table 1). One main difference between these strains was in a broad set of genes related to cell division and peptidoglycan metabolism that had reduced expression in the 17978-ΔbfmR mutant strain (20). We found that some of these genes had decreased expression in the 17961-ΔbfmR mutant, but we did not find any significant enrichment of these pathways in our analysis. This could be due to the fact that we performed our transcriptome analysis using samples from cultures that were entering stationary phase (optical density at 600 nm [OD600], 4.5 to 5.0), where a majority of the cells had likely stopped dividing (as opposed to logarithmic phase [OD600, 0.5], as in the study by Geisinger et al. [20]). Overall, these analyses showed that BfmR appears to have consistent roles in regulating genes related to stress responses, antibiotic resistance, capsular polysaccharide production, biofilm formation, and iron acquisition in multiple strains of A. baumannii.
BfmR and BfmS coordinately regulate gene expression.
Since the data presented above identified numerous BfmR-controlled stress-related genes, we wanted to gain a better understanding of how the BfmRS system coordinates the regulation of these genes. Focusing on genes that had the greatest degree of dysregulation in the ΔbfmR mutant strain based on the RNA-seq results, we constructed plasmids carrying transcriptional (gfp) reporter fusions using the predicted promoter regions for representative genes from several different functional gene groups (Table 1). We then assayed the activity of these fusions during stationary phase in A. baumannii strains with different mutations affecting the BfmRS system.
To confirm the RNA-seq results, we tested the activity of each fusion in the ΔbfmR mutant strain. However, in some cases, deletion of a response regulator can allow increased interactions between its cognate sensor kinase and other noncognate response regulators. This amplification of cross talk between systems can cause indirect regulatory effects that are not controlled by the deleted response regulator (46). To account for this, we also tested the activity of the reporter fusions in a ΔbfmRS mutant strain that lacks both the cognate sensor kinase and response regulator. Regulatory effects that result from the coordinated activity of BfmS and BfmR would be expected to be similar in the ΔbfmR and ΔbfmRS mutant strains because both lack BfmR. In contrast, effects in the ΔbfmR mutant that are due to amplified cross talk between BfmS and other regulators should not be observed in the ΔbfmRS mutant that lacks BfmS.
Additionally, it was previously noted that deletion of bfmS tended to cause regulatory effects that were opposite of that seen when bfmR was deleted, implying that BfmS functions to inactivate BfmR (19–21). Therefore, we also assayed the activity of the reporter fusions in a ΔbfmS mutant strain. Finally, we assayed the reporter fusions in a strain where the coding region of bfmR was altered to carry a point mutation that changed the aspartate residue 58 of BfmR to an alanine (referred to as bfmR D58A). Aspartate 58 is the conserved phosphorylation site in the BfmR receiver domain (25). Thus, mutation of this residue in BfmR should prevent it from becoming activated by phosphorylation (22, 47, 48).
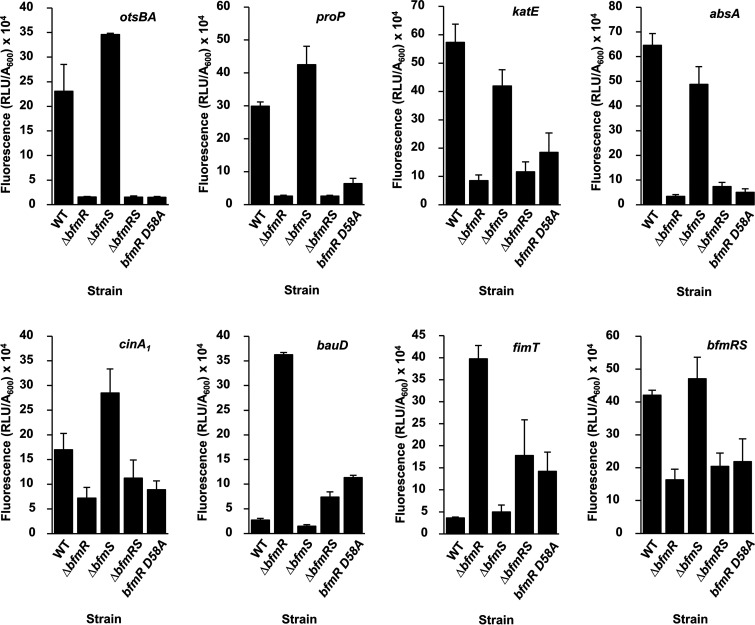
Green fluorescent protein (GFP) reporter plasmids were transformed into the ATCC 17961 wild-type, ΔbfmR, ΔbfmS, ΔbfmRS, and bfmR D58A strains, and each was assayed for gene expression as described in Materials and Methods. Compared to the wild-type strain, the ΔbfmR mutant strain had significantly reduced reporter activity from promoters of the osmotic stress genes otsB/otsA and proP, the oxidative stress catalase gene katE, the general stress-related gene absA, and the damage-inducible gene cinA1 (Fig. 1). These results agree with our RNA-seq data, confirming that BfmR is involved in activating the expression of a variety of stress pathways. Next, we examined the effects of deleting bfmS. Expression of the otsBA, proP, and cinA1 reporter fusions was significantly higher in the ΔbfmS mutant strain than the wild-type strain, but for the katE and absA reporter fusions, expression was slightly decreased (Fig. 1). These data indicate that BfmS is not required for BfmR to activate these genes and suggest that BfmS acts to limit the expression of otsBA, proP, and cinA1. Additionally, we observed that compared to the wild-type strain, expression of otsBA, proP, katE, and absA reporters was significantly decreased in the double ΔbfmRS mutant strain. These results were similar to the reporter activity in the ΔbfmR mutant strain, indicating that this regulatory effect is due to coordinated activity of BfmR and BfmS and is not due to cross talk between BfmS and other regulators.
FIG 1.
BfmR regulates stress-related genes in A. baumannii. ATCC 17961 wild type (WT) and the ΔbfmR, ΔbfmS, ΔbfmRS, and bfmR D58A mutants carrying the indicated gfp transcriptional fusion plasmids were grown to stationary phase as described in Materials and Methods. Fluorescence was measured and reported values were calculated as described in Materials and Methods. Data are means and SD of results from at least three independent experiments.
Next, we examined gene expression in the nonphosphorylatable bfmR D58A strain. Reporter activity for these six transcripts in the bfmR D58A strain was similar to the ΔbfmR mutant data (Fig. 1). This suggests that phosphorylated BfmR (BfmR∼P) is the active form responsible for inducing gene expression of these transcripts. However, these assays do not clarify whether BfmR∼P activates these genes directly.
We also examined expression from reporter fusions for genes that had evidence of negative control by BfmR. These included GFP fusions to the predicted promoter regions of the acinetobactin transporter gene bauD and the fimbrial gene fimT. Similar to the RNA-seq data, we found that expression from the bauD and fimT reporters in the ΔbfmR mutant strain was significantly increased over 10-fold compared to the wild-type strain (Fig. 1). These results suggested that BfmR is also involved in controlling virulence traits such as iron acquisition. However, we observed a lower degree of activation in the ΔbfmRS strain (approximately 2.5-fold for the bauD reporter and 5-fold for the fimT reporter), showing that BfmS was required for the high level of activation of these genes seen in the absence of bfmR alone. This could be an example of cross talk between BfmS and a noncognate response regulator when bfmR is deleted. When we examined expression in the ΔbfmS mutant strain, we observed a significant decrease in bauD promoter activity and no change in fimT promoter activity. Interestingly, we found that bauD- and fimT-GFP expression was significantly increased in the bfmR D58A mutant strain compared to that in the wild-type strain, at a level similar to that seen in the ΔbfmRS mutant strain, but not to the level seen in the ΔbfmR mutant strain. These results indicate that unphosphorylated BfmR is unable to fully repress expression from the bauD and fimT promoters. This could be due to either changes in the DNA-binding ability of BfmR D58A or a decreased ability of BfmR D58A to interact with BfmS, allowing some degree of cross talk to occur.
Finally, since response regulators often possess autoregulatory activity (49), and our previous data showed that BfmR can bind to the bfmRS promoter region (22), we assayed the GFP reporter activity of the bfmRS promoter. Figure 1 shows that the ΔbfmR mutant strain has significantly reduced expression of bfmRS-GFP compared to the wild-type strain, demonstrating that BfmR can indeed activate its own expression. Expression in the ΔbfmS mutant strain is similar to that of the wild-type strain, indicating that BfmS is not required to activate expression of the bfmRS promoter. We also observed that the ΔbfmRS mutant strain has significantly decreased expression of the bfmRS reporter fusion compared to the wild-type strain. This is similar to results obtained for the ΔbfmR mutant strain, indicating that this regulatory activity is a direct result of mutating bfmR. As expected, the bfmR D58A strain also showed reduced expression, confirming that phosphorylation is required for autoinduction.
BfmR directly activates numerous stress-related transcripts, and activation is suppressed by BfmS.
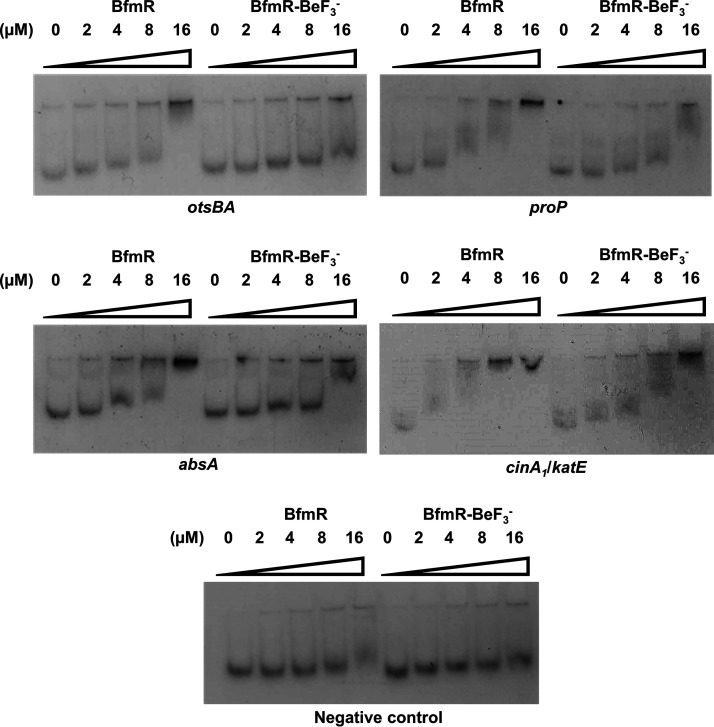
Thus far, the data have demonstrated that BfmR can activate numerous stress response pathways. Usually, OmpR/PhoB family response regulators such as BfmR activate transcription by directly binding to DNA promoter regions (18). In order to determine whether regulation by BfmR is via a direct or an indirect mechanism, we performed electrophoretic mobility shift assays (EMSAs) using the promoter regions of stress-related genes whose assay results are shown in Fig. 1. DNA-binding ability was tested using both untreated BfmR protein and BfmR treated with the phosphomimic beryllium trifluoride (BeF3−). BeF3− is frequently used to promote dimerization and activation of response regulators to enhance DNA binding (22, 50, 51). Most interestingly, we saw that both BfmR and BfmR-BeF3− can bind to the promoter regions of otsBA, proP, absA, and cinA1/katE (Fig. 2; note that cinA1 and katE are divergently transcribed and their regulatory regions likely overlap). We also observed that a mobility shift occurred at approximately 2- to 4-fold-lower concentrations of BfmR than BfmR-BeF3−. This suggests that untreated BfmR binds with a higher affinity than BfmR-BeF3−. Taken together, these data showed that BfmR can directly bind to stress-related promoter regions.
FIG 2.
BfmR directly interacts with promoter regions of several stress-related genes. Electrophoretic mobility shift assays completed with the indicated promoter DNA and increasing concentrations of untreated BfmR and BfmR treated with the phosphomimic BeF3− (BfmR-BeF3−). The negative-control promoter fragment was an internal fragment from gene I5593_11150 of strain ATCC 17961. Results are representative of at least two independent experiments.
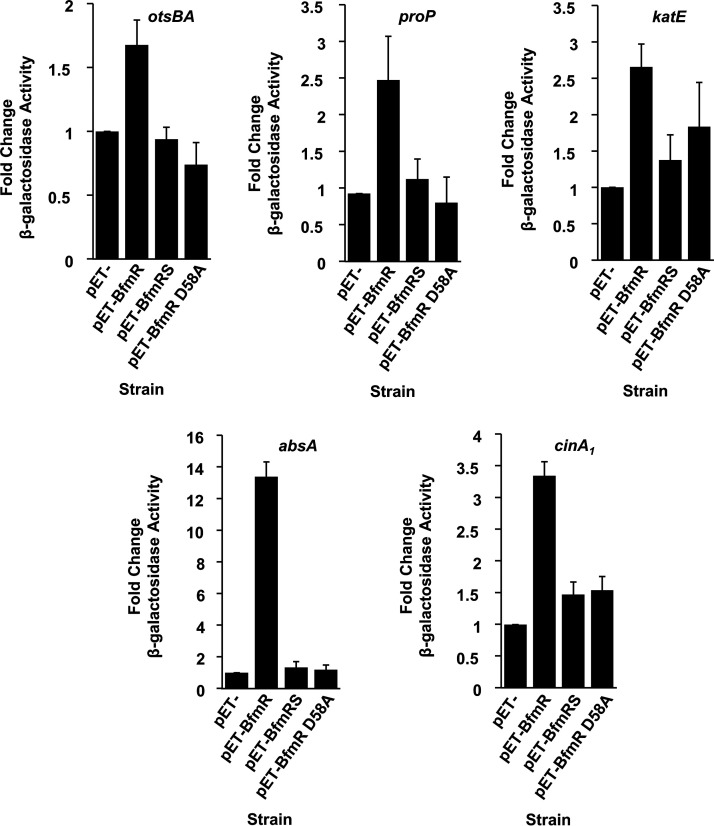
To determine if DNA binding leads to the activation of transcription at these promoters, and to clarify a role for BfmS in this regulation, we utilized a two-plasmid system in Escherichia coli. In this system, one plasmid carried the promoter region of a stress-related gene fused to a lacZ-transcriptional reporter, and the second plasmid was designed to express either BfmR (pET-BfmR), BfmR and BfmS (pET-BfmRS) or BfmR with the D58A mutation, which prevents phosphorylation of BfmR (pET-BfmR D58A). We found that expression of BfmR alone was able to induce lacZ activity from all five of the stress-related promoter fusions tested in E. coli (Fig. 3). However, we observed little to no induction of lacZ activity when BfmS was coexpressed with BfmR. These data indicate that BfmS represses BfmR’s regulatory activity, which agrees with previous observations (19–21). We also found that expression of BfmR D58A was unable to activate expression from the otsBA, proP, and absA promoters, and it had a reduced ability to activate transcription for katE and cinA1 promoters compared with expression of the wild type BfmR protein. These results agree with our analysis of reporter fusions in A. baumannii (Fig. 1) that showed that the BfmR D58A mutation appeared to inactivate BfmR and imply that phosphorylation of BfmR is required to induce the expression of stress-related genes.
FIG 3.
BfmR directly activates stress-related transcripts in E. coli. E. coli strain NovaBlue(DE3) carrying the indicated promoters on lacZ-transcriptional fusion plasmids and either the empty expression vector (pET-), pET-BfmR, pET-BfmRS, or pET-BfmR D58A was grown in LB at 37°C, and protein expression was induced as described in Materials and Methods. β-Galactosidase activity was assayed and is presented as the fold change; data are means and SD of results from at least three independent experiments.
Together, these data show that phosphorylation appears to alter BfmR’s interactions with DNA, allowing direct induction of protective genes. The data also confirm that BfmS inhibits BfmR activity. Since BfmR requires phosphorylation to directly activate these genes in E. coli, and since sensor kinases can have both kinase and phosphatase functions (52), we hypothesized that BfmS alters the BfmR phosphorylation state to regulate BfmR’s activity.
BfmS dephosphorylates BfmR to negatively regulate stress gene activation.
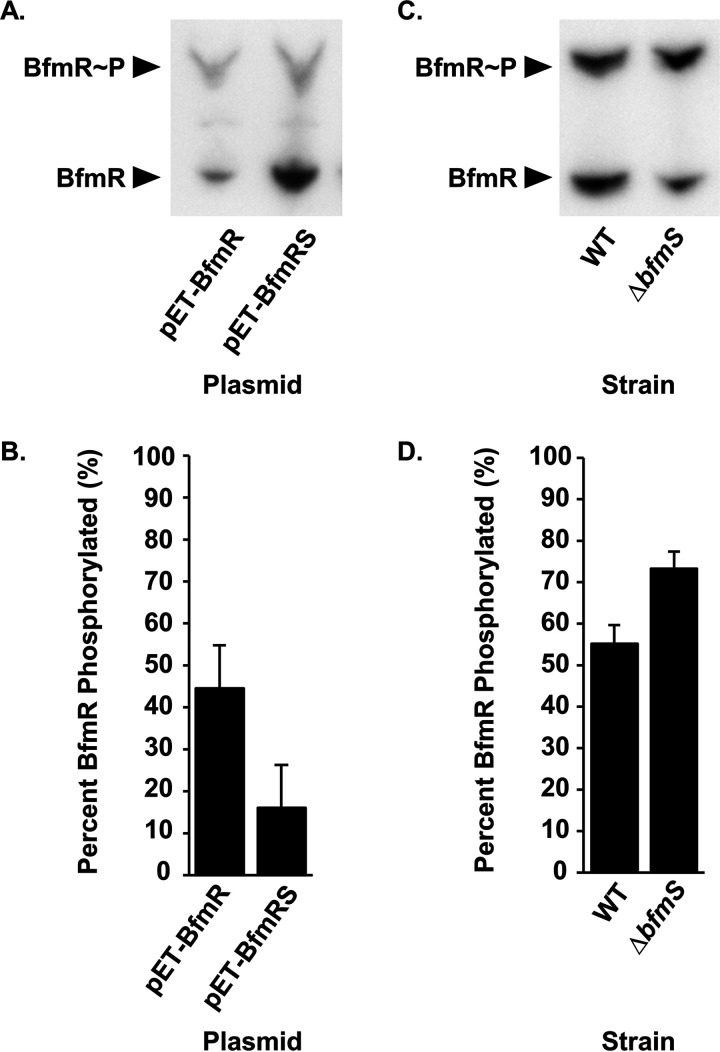
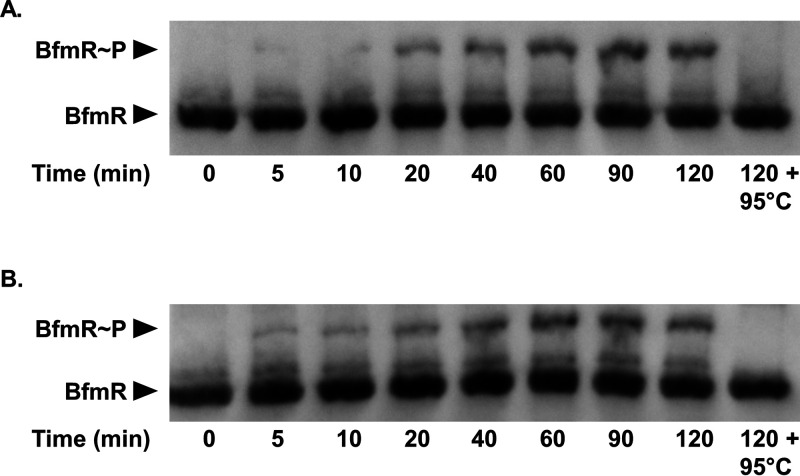
To explore the idea that BfmS affects the BfmR phosphorylation state, we performed Phos-tag gel analysis. The Phos-tag reagent is a phosphate-binding molecule that, in the presence of divalent metal ions, traps phosphorylated proteins during migration through an SDS-PAGE gel. Hence, phosphorylated proteins appear “shifted” due to slower migrations (53). We used Phos-tag gel electrophoresis to separate proteins in whole-cell lysates from E. coli expressing either BfmR alone or BfmR and BfmS. Proteins were then analyzed by Western blotting using anti-BfmR antibodies. Two distinct bands representing both BfmR∼P and BfmR were detected, where BfmR∼P migrated more slowly than BfmR (Fig. 4A). These results confirm that BfmR is phosphorylated in E. coli. The Phos-tag gel also showed that expression of BfmS greatly reduced phosphorylation of BfmR. We quantified the relative amounts of BfmR∼P and BfmR present in the blots using densitometry and found a significant decrease in BfmR∼P in the presence of BfmS (from 44.5% phosphorylated down to 16.1%) (Fig. 4B). These data provide evidence that BfmS dephosphorylates BfmR.
FIG 4.
BfmS dephosphorylates BfmR. (A and C) Phos-tag gel and Western blot analysis of the BfmR phosphorylation state in E. coli NovaBlue(DE3) carrying either pET-BfmR or pET-BfmRS (A) or in the wild-type A. baumannii strain ATCC 17961 and the ΔbfmS mutant strain (C). (B and D) Quantification of phosphorylated BfmR as a percentage of total BfmR protein in E. coli strains from panel A (B) or in A. baumannii strains from panel C (D). Data are means and SD from at least three independent experiments.
To examine the effects of BfmS on the BfmR phosphorylation state in A. baumannii, we repeated the Phos-tag gel analysis using whole-cell lysates from the ATCC 17961 wild-type and ΔbfmS mutant strains grown to stationary phase (Fig. 4C). The wild-type strain showed approximately equal amounts of BfmR∼P and BfmR. In the ΔbfmS mutant strain, there was a significant increase in BfmR∼P compared to the wild-type strain (73.2% versus 55.1%, respectively) (Fig. 4D). Notably, we did not observe any BfmR∼P when we examined a cell lysate from the bfmR D58A mutant strain using Phos-tag analysis (Fig. S1). This supports our conclusion that BfmS dephosphorylates BfmR at aspartate 58 to negatively regulate BfmR activity.
BfmR autophosphorylates in vitro using small phosphodonors.
In the prototypical two-component system, the role of the sensor kinase is to phosphorylate its cognate response regulator, resulting in activation (54). However, since our data showed that BfmS dephosphorylates BfmR (Fig. 4), we questioned how BfmR becomes phosphorylated and hence activated to regulate transcription. It has been demonstrated that response regulators, including OmpR and PhoB, can autophosphorylate in vitro using small phosphodonors such as acetylphosphate, carbamoylphosphate, and phosphoramidate (55, 56). Moreover, acetylphosphate can influence a number of response regulators in vivo (55). Therefore, we performed in vitro phosphorylation assays by incubating purified BfmR protein with either acetylphosphate or carbamoylphosphate. At indicated time points, the BfmR phosphorylation state was analyzed using Phos-tag gel analysis. Phospho-aspartate bonds are extremely heat labile (56). Thus, as a negative control, one reaction mixture was heat shocked at 95°C for 5 min. We observed that over a period of 120 min, a population of BfmR became phosphorylated, as evidenced by the appearance of an upshifted band (Fig. 5). Since phospho-aspartate bonds are heat labile, the absence of the slower-migrating band in the samples that were heat shocked at 95°C confirms that this higher band represents BfmR∼P. These assays showed that BfmR can autophosphorylate in vitro using acetylphosphate (Fig. 5A) and carbamoylphosphate (Fig. 5B) as phosphodonors. Therefore, not only can BfmR activate its own expression (Fig. 1), but it can also induce its own activity through autophosphorylation.
FIG 5.
Autophosphorylation of BfmR in vitro by small phosphodonors. Purified BfmR protein (3 μM) was incubated with 10 mM acetylphosphate (A) or 10 mM carbamoyl phosphate (B) at 37°C for the indicated times, at which point reactions were stopped by the addition of SDS loading buffer. As a control, the last sample was heat shocked at 95°C for 5 min after 120 min incubation with the phosphodonor, prior to the addition of SDS loading buffer. Samples were then analyzed by Phos-tag gel electrophoresis and Western blotting.
DISCUSSION
Our overall goal was to clarify the function of the BfmRS two-component regulatory system in A. baumannii. Previously, it was observed that BfmS exerted a negative regulatory effect on BfmR activity (19–21). This did not fit with the typical two-component regulatory mechanism, where the sensor kinase usually activates the response regulator (15). Mutations that were predicted to inactivate the histidine kinase domain of BfmS blocked repression of BfmR activity (20), suggesting that BfmS controlled BfmR via phosphoregulation. However, the details behind the BfmS inhibitory mechanism remained unclear, because it was unknown whether BfmR required phosphorylation to activate transcription. Previous EMSA analysis demonstrated that unphosphorylated BfmR, BfmR activated with the phosphomimic BeF3−, and the nonphosphorylatable BfmR D58A mutated protein could all bind the bfmRS promoter region (22). Our current results strongly suggest that phosphorylated BfmR (BfmR∼P) is the active form of this response regulator that directly influences gene expression (Fig. 1 to 3). Our data also provide evidence that BfmS acts as a BfmR phosphatase to inhibit activation of BfmR (Fig. 4) and thus repress BfmR-controlled transcriptional responses. Furthermore, we showed that BfmR can autophosphorylate in vitro using small phosphodonors such as acetylphosphate (Fig. 5), and we observed that phosphorylated BfmR was present in E. coli in the absence of BfmS or other A. baumannii proteins. These data showed that BfmS is not necessary to convert BfmR to its active, phosphorylated form.
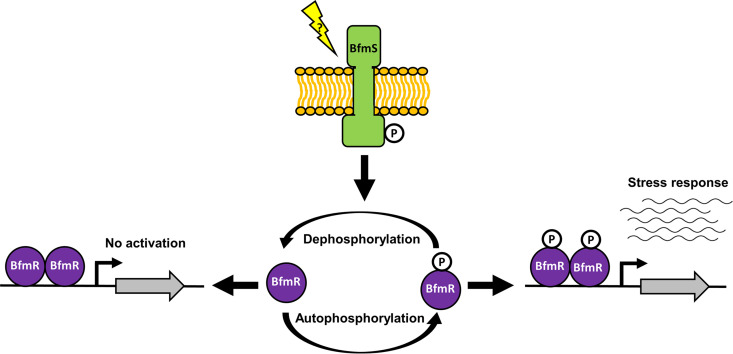
Overall, our data have allowed us to propose a working model (Fig. 6) of how the BfmRS two-component regulatory system functions to control gene expression. BfmR∼P induces the expression of A. baumannii genes, including those involved in stress responses, by directly binding to promoter regions and initiating transcription. BfmR activation potentially occurs through autophosphorylation using small phosphodonors, such as acetylphosphate. However, the signal(s) or condition(s) that stimulates BfmR autophosphorylation in A. baumannii remain to be elucidated. While the levels of acetylphosphate in E. coli have been shown to fluctuate depending on growth phase, carbon source, oxygen availability, nitrogen availability, phosphate availability, temperature, pH, and extracellular acetate concentrations (55), acetylphosphate metabolism in A. baumannii is yet to be explored. It is possible that similar fluctuations of acetylphosphate or other small molecule phosphodonors occur in response to stress in A. baumannii and that BfmR autophosphorylates in response to these fluctuations to activate a transcriptional response. Alternatively, BfmR may be the target of other cellular kinases. In either case, phosphorylation of BfmR leads to increased expression of stress-related genes and the bfmRS operon.
FIG 6.
Working model for the BfmRS two-component system regulatory circuit. Phosphorylated BfmR (BfmR∼P) binds to promoter regions to activate transcription of the general stress response genes. BfmR can autophosphorylate using small molecule phosphodonors, such as acetylphosphate and carbamoylphosphate. BfmS represses BfmR∼P activation of stress-related genes by dephosphorylating BfmR. The signal that BfmS responds to is unknown. Unphosphorylated BfmR can also bind DNA, but this does not induce transcription of the genes we tested, and thus the role of unphosphorylated BfmR is yet to be determined.
The activation of BfmR is limited by BfmS. BfmS dephosphorylates BfmR∼P (Fig. 4), preventing the induction of stress-related genes (Fig. 6). Our data also suggest that cross talk occurs between BfmS and other noncognate response regulators to activate bauD and fimT expression in the absence of BfmR (Fig. 1). This could occur, for instance, when the concentration of phosphodonor inside the cell decreases and thus the rate of BfmR autophosphorylation decreases. If BfmR does not interact with BfmS, competition for BfmS is reduced, allowing interactions between BfmS and other response regulators. Alternatively, since expression of these transcripts is increased in the ΔbfmRS strain compared to the wild-type strain (Fig. 1), it is also possible that BfmR could be responsible for some of the inhibitory effects on bauD and fimT transcription. Notably, bauD transcription is likely also repressed by the ferric uptake regulator Fur (44). We did not observe any change in fur expression in our RNA-seq analysis, and it is unclear whether the BfmRS-mediated effects on bauD expression occur through interactions with Fur or through an alternative mechanism, such as cross talk.
While it is possible that the function of BfmS relies on the concentrations of BfmR∼P, the signal(s) that BfmS responds to remains to be identified. BfmS is predicted to have two membrane-spanning regions that are common to sensor kinases (23). The BfmS C-terminal, cytoplasmic domain contains the histidine kinase catalytic domain found in bacterial sensor proteins (54). However, the N-terminal, extracellular region of BfmS has no known sequence conservation to a putative domain. This makes it difficult to predict the types of stimuli that BfmS responds to. A previous study found that the BfmRS system regulated genes that are important for defense against cell envelope damage (20), suggesting that cell envelope stress could be a signal that affects BfmS activity. However, the BfmRS system is also important for defense against multiple other stresses and appears to be important for some starvation-induced responses (23, 31). This suggests that other signals could also influence BfmS’s phosphatase activity.
The activity of BfmS leads to increased levels of unphosphorylated BfmR (Fig. 4). Unphosphorylated BfmR can bind DNA (Fig. 2) (22), although our data suggest that it could not activate transcription from the promoters that we tested (Fig. 1 and 3). It is not unusual for unphosphorylated response regulators to bind to DNA. For instance, the Salmonella enterica serovar Typhimurium response regulator SsrB binds to DNA in both the phosphorylated and unphosphorylated forms. Unphosphorylated SsrB can activate transcription of csgD, the master regulator of biofilms in S. Typhimurium (57). Furthermore, the S. Typhimurium response regulator ArcA forms a multimer composed of a 1:1 ratio of ArcA and ArcA∼P, demonstrating a requirement for both states for DNA binding (58). Interestingly, we observed similar amounts of phosphorylated and unphosphorylated BfmR in wild-type ATCC 17961 (55.1% and 44.9%, respectively) (Fig. 4D). Therefore, it may be beneficial to further investigate the role, if any, of unphosphorylated BfmR.
In addition to understanding the mechanistic details of the BfmRS control circuit, we wanted to elucidate how the BfmRS system functions in controlling different stress responses, with a particular focus on the general stress response (also known as the stationary phase response). Multiple lines of evidence suggest that the BfmRS system has a significant role in controlling this response. Studies show that the BfmRS system controls stress and stationary phase-induced phenotypes, including increased resistance to desiccation, oxidative stress, and osmotic stress, increased tolerance to antibiotics, increased survival during long-term nutrient starvation, and biofilm formation (20, 23, 31). Another characteristic of the general stress response is cross-protection, whereby exposure to one stress stimulates protection against different stresses (32). BfmR was required for starvation-induced cross-protection against desiccation (31). During stationary phase, bacterial cells often decrease in size due to changes in replication and cell wall composition, allowing increased survival in unfavorable conditions (59). Deletion of bfmR causes elongation of A. baumannii cells (23), and it has been shown that the BfmRS system has a role in cell shortening (20). Finally, in agreement with a previous analysis (20), our RNA-seq analysis revealed large decreases in expression for multiple stress response genes upon mutation of bfmR, and we showed that BfmR can directly control a selection of these genes (Fig. 1 to 3). Together, these data indicate that the BfmRS system has a major role in controlling the general stress response in A. baumannii.
In most gammaproteobacteria, and some beta- and deltaproteobacteria, the RpoS sigma factor coordinates the general stress response and the transition to stationary phase (59–61). Multiple regulatory signals can alter the cellular levels of RpoS, which competes with other sigma factors to bind and recruit RNA polymerase core enzyme to specific promoter sites (60). Alphaproteobacteria do not encode a RpoS homolog (32), but some bacteria in this class regulate their general stress responses via the response regulator PhyR and the extracytoplasmic function sigma factor EcfG (62). In response to phosphorylation, PhyR sequesters the anti-sigma factor NepR from binding to EcfG, allowing EcfG to direct transcription of stress-related genes (32, 62). Interestingly, unlike other members of the gammaproteobacteria, A. baumannii does not encode an RpoS homolog. Instead, the BfmRS system appears to fulfill a similar function. Also, unlike the system present in alphaproteobacteria, we found that BfmR can directly regulate multiple stress response genes without the help of other A. baumannii proteins (Fig. 3). Therefore, the BfmRS two-component system seems to be part of a unique scheme of general stress response control that is different from previously described systems.
A number of questions still remain about the coordination of stress responses in A. baumannii. In addition to BfmRS, several other regulators appear to broadly affect A. baumannii’s ability to survive in stressful conditions, and it is currently unclear if stress responses in this species are subject to overlapping control by multiple systems, or if a single factor acts as a signal integration point. An analysis of multiple A. baumannii genomes identified only five sigma factor homologs: RpoD, RpoH, RpoN, RpoE, and FecI (63). Two of these factors, RpoN and FecI, do not appear to be involved in stress response regulation, but there is some evidence that RpoE may take part in coordinating stress responses along with the GigA/GigB regulatory system. GigA and GigB are part of a phosphorelay, along with the components of the nitrogen phosphotransferase system (PTSNtr). Mutation of GigA or GigB causes A. baumannii to become less virulent and more sensitive to antibiotics, acid stress, and temperature stress. Inactivation of RpoE caused similar phenotypes, but it is unclear if GigA/GigB/PTSNtr directly affect RpoE’s function or if other factors are involved (36).
In addition to the GigA/GigB regulatory system, multiple other regulators are known to be involved in controlling stress responses, resistance, and virulence in A. baumannii. These include the GacSA two-component regulatory system (37, 38), the AdeRS two-component regulatory system (38, 39), the RNA chaperone Hfq (41), and the regulatory proteins involved in phase variation (64–66). Likewise, it is unclear if these systems act directly or indirectly to influence the expression of stress-related genes, or if there are interactions that link these regulatory systems together.
To conclude, it is clear that the BfmRS system has multiple roles in A. baumannii’s ability to endure unfavorable environments. Overall, our data support the idea that this two-component regulatory system harbors characteristics of a master regulator of the general stress response in A. baumannii. Hence, the BfmRS system represents an attractive target for the development of novel antimicrobials to combat prolonged survival of A. baumannii in nosocomial environments.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 2. Stocks of each strain containing 15% glycerol (vol/vol) were stored at −80°C. Bacteria were freshly plated prior to each experiment and cultured in lysogeny broth (LB; Lennox formulation). Unless otherwise specified, cultures were incubated at 37°C with shaking at 260 to 280 rpm. When necessary to maintain plasmids, medium was supplemented with 100 μg/mL carbenicillin, 50 μg/mL kanamycin, 30 μg/mL chloramphenicol, 10 μg/mL tetracycline, or 10 μg/mL gentamicin for Escherichia coli and 150 μg/mL carbenicillin or 10 μg/mL gentamicin for A. baumannii.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α strains | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U196 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 77 |
| BL21(DE3) | F− dcm ompT hsdSB(rB− mB−) gal (λDE3) | Millipore |
| NovaBlue(DE3) | endA1 hsdR17(rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac(DE3) [F′ proA+B+ lacIqZΔM15::Tn10(Tetr)] | Novagen |
| A. baumannii strains | ||
| ATCC 17961 | Clinical isolate from blood | ATCC |
| 17961-ΔbfmR | bfmR deletion mutant derived from strain ATCC 17961 | 31 |
| ABGW-ΔbfmS | bfmS deletion mutant derived from strain ATCC 17961 | This study |
| ABGW-ΔbfmRS | bfmRS deletion mutant derived from strain ATCC 17961 | This study |
| ABGW-bfmR D58A | ATCC 17961 derivative with a single nucleotide substitution at bp 174 (T to A) of the bfmR coding sequence, resulting in an amino acid change from Asp to Ala at position 58 of BfmR | This study |
| Plasmids | ||
| pEX18Ap | Suicide vector | 78 |
| pGW-bfmS-entire-suc | Suicide plasmid carrying bfmS region | This study |
| pGW-ΔbfmS-suc | Suicide plasmid carrying an in-frame deletion that removed the bfmS coding sequence from +180 to +1473 relative to the translational start site | This study |
| pGW-bfmRS-entire-suc | Suicide plasmid carrying bfmRS region | This study |
| pGW-ΔbfmRS | Suicide plasmid carrying ΔbfmRS deletion | This study |
| pBfmR-suc | Suicide plasmid carrying bfmR region | 31 |
| pGW-bfmR D58A | Suicide plasmid carrying bfmR with a substitution at bp 174 (T to A) of the bfmR coding sequence, resulting in an amino acid change from Asp to Ala at position 58 of BfmR | This study |
| pLPV3Z | A. baumannii GFP transcriptional reporter vector | 67 |
| pJF330 | A. baumannii absA-GFP transcriptional fusion vector | This study |
| pJF331 | A. baumannii katE-GFP transcriptional fusion vector | This study |
| pJF332 | A. baumannii cinA1-GFP transcriptional fusion vector | This study |
| pJF333 | A. baumannii otsBA-GFP transcriptional fusion vector | This study |
| pJF334 | A. baumannii proP-GFP transcriptional fusion vector | This study |
| pJF338 | A. baumannii fimT-GFP transcriptional fusion vector | This study |
| pJF339 | A. baumannii bauD-GFP transcriptional fusion vector | This study |
| pJF340 | A. baumannii bfmRS-GFP transcriptional fusion vector | This study |
| pUC18T-mini-Tn7T-Gm | Mini-Tn7 lacZ transcriptional fusion vector | 68 |
| pACYC184 | E. coli cloning vector | 79 |
| pACYC-lacZ | E. coli lacZ transcriptional fusion vector | This study |
| pSP-cinA1lacZ | E. coli cinA1-lacZ transcriptional fusion vector | This study |
| pSP-otsBAlacZ | E. coli otsBA-lacZ transcriptional fusion vector | This study |
| pSP-proPlacZ | E. coli proP-lacZ transcriptional fusion vector | This study |
| pJF320 | E. coli absA-lacZ transcriptional fusion vector | This study |
| pJF321 | E. coli katE-lacZ transcriptional fusion vector | This study |
| pET28b | pET28b expression vector | Novagen |
| pET-BfmR | pET28b expression vector containing bfmR from ATCC 17961, lacI | This study |
| pET-BfmRS | pET28b expression vector containing bfmRS from ATCC 17961, lacI | This study |
| pET-BfmR D58A | pET28b expression vector containing bfmR with a D58A point mutation from ATCC 17961, lacI | This study |
| pMU360 | pET200 expression vector containing bfmR with an amino-terminal His6 affinity tag, lacI | 23 |
| pET28a-bfmR | pET28a expression vector containing bfmR with an amino-terminal His6 affinity tag, lacI | This study |
Construction of plasmids and mutant strains.
Plasmids and primers used in this study are listed in Tables 2 and 3, respectively. To construct strain ABGW-ΔbfmS, an approximately 2.1-kb DNA fragment containing bfmS was amplified by PCR using A. baumannii strain ATCC 17961 chromosomal DNA as a template. Primers (bfmS del 1 and bfmS del 4) were designed to contain a PstI site. The PCR fragment and vector plasmid pEX18Ap were digested with PstI, purified from an agarose gel, and ligated to produce pGW-bfmS-entire-suc. This plasmid was then used as a template for inverse PCRs using 5′ phosphorylated primers (bfmS INV 1 and 2). The resulting DNA fragment was recircularized by ligation to produce plasmid pGW-ΔbfmS-suc. This plasmid contained an in-frame deletion in bfmS that removed the sequence encoding amino acids 60 to 491 (78.7% of the protein sequence).
TABLE 3.
Primers used in this study
| Purpose and primer | Sequence (5′→3′)a |
|---|---|
| Generation of mutant strains | |
| bfmS del 1 | AAAAACTGCAGGGTTGTCATGTATCAGTTTG |
| bfmS del 4 | AAAAACTGCAGACAGTCAGTCCGCCTACT |
| bfmS INV 1 | [Phos]TTGTCGAGCGACTCCTTCACT |
| bfmS INV 2 | [Phos] GCATCTGGCGGTTATGGTTTG |
| bfmRS entire 2 F | AAAAAACTGCAGGCGCACTCCATTCTGAATTAA |
| bfmRS entire 3 R | AAAAAACTGCAGATTGCTTGAACATCAATACCTT |
| INV bfmR F | [Phos] ATCATTGCCCCTATAAATCTCATTC |
| INV bfmS R | [Phos] GGTGCTTTTTTATTGGTTTATTTATAATTG |
| bfmR asp ala 1 | TTGGCTGTCATGTTGCCGGGTGCA |
| bfmR asp 2 | GACCACAAGATCCGGTTGCTCACT |
| Cloning transcriptional reporter plasmids | |
| PabsA gfpF | AAAAAAACTGCAGTGTGTATGAATAGGTTGCATCATTC |
| PabsA gfpR2 | AAAAAAAATCTAGACAGGATCCATACTTGCAAATCC |
| cinA_lacZF | AAAAACTGCAGTTTTCACTACAACAGGCATTTTC |
| cinA gfpR | AAAAAAAATCTAGACATTTTAAATCTCCCTACAATTG |
| katE_lacZF | AAAAACTGCAGCATTTTAAATCTCCCTACAATTG |
| PkatE gfpR2 | AAAAAAAATCTAGATTCACTACAACAGGCATTTTC |
| otsBA gfpF | AATTAATTGGTACCAACCTAATTTTAACGATTGCAAAG |
| otsBA gfpR | AAAAAAAATCTAGATCTCCCATCAGATATTATACTTTG |
| proP_lacZF | AAAAACTGCAGGATTATTTAGAGTCAATCCCTG |
| proP gfpR | AAAAAAAATCTAGAGCGCATATCTAGCCTAAGAATTG |
| lacZ-F | AAAAAAAATCTAGAAAGTATAGGAACTTCAGA |
| lacZ-R | GCCGATTCATTAATGCAGC |
| cinA_lacZR | AAAAAAAGCTTCATTTTAAATCTCCCTACAATTG |
| otsBA_lacZF2 | AAAAAAAGCTTAACCTAATTTTAACGATTGCAAAG |
| otsBA_lacZR | AAAAAAAGCTTTCTCCCATCAGATATTATACTTTG |
| proP_lacZR | AAAAAAAGCTTGCGCATATCTAGCCTAAGAATTG |
| PabsA gfpR | AAAAAAAGGTACCTTCAGGATCCATACTTGCAAATCC |
| PkatE gfpR | AAAAAAAAGGTACCTTCACTACAACAGGCATTTTC |
| Cloning expression plasmids | |
| bfmR exR | TTTTTTTTACATGTCGCAAGAAGAAAAGTTACCAAAGATTCTG |
| bfmR exF | TTTTTTTTGGATCCGGAAGTTTAATCAGATTTTACAATCCATTGG |
| BfmSexp_R2 | AAAAAGAATTCCAATAAAAAAGCACCTTATGCAGGTG |
| EMSA analysis | |
| cinA_outF2 | AAAAAAAGCTTGATGATAATACTCGTCTTC |
| bfmR_negR | TCTTGCTAAAGAGATAGATAAACTAGAG |
| absA_lacZF | AAAAAGAATTCTGTGTATGAATAGGTTGCATCATTC |
| absA_lacZR | AAAAAAAGCTTTTCAGGATCCATACTTGCAAATCC |
| cinA_proF | AAGCTAGAAACATCAGGATCAAGG |
| cinA_proR | AAGAGAAACGATAAGCAAGGTAGC |
| otsBA_proF | AACCTAATTTTAACGATTGCAAAG |
| otsBA_proR | TCTCCCATCAGATATTATACTTTG |
| proP_inF | GCGCATATCTAGCCTAAGAATTGAATC |
| proP_proR | GATTATTTAGAGTCAATCCCTGAAGC |
Underlining indicates restriction sites.
To construct the ABGW-ΔbfmRS double-deletion mutant strain, an approximately 4.4-kb DNA fragment containing bfmRS was amplified by PCR using A. baumannii strain ATCC 17961 chromosomal DNA as a template. Primers (bfmRS entire 2 F and bfmRS entire 3 R) were designed to contain a PstI site. The PCR fragment and vector plasmid pEX18Ap were digested with PstI, purified from an agarose gel, and ligated to produce pGW-bfmRS-entire-suc. This plasmid was then used as a template for inverse PCRs using 5′ phosphorylated primers (INV bfmR F and INV bfmS R) to remove the bfmRS region. The resulting DNA fragment was recircularized by ligation to produce plasmid pGW-ΔbfmRS. This plasmid contained a complete deletion of the bfmRS DNA sequence encoding amino acids 1 to 238 of BfmR and amino acids 1 to 549 of BfmS (100% of the protein sequences for both).
To construct the ABGW-bfmR D58A strain, the plasmid pBfmR-suc (31) was used as a template for inverse PCRs, using primers with 5′ phosphate groups (bfmR asp ala 1 and bfmR asp2). The resulting DNA fragment was recircularized by ligation to produce plasmid pGW-bfmR D58A.
All mutant strains were generated by first transferring suicide plasmids (carrying mutant alleles) into E. coli strain SM10. Next, suicide plasmids were transferred from E. coli to A. baumannii via conjugation. This was followed by selection on sucrose, as described previously (31). Potential mutant strains were confirmed using PCR amplification and DNA sequencing of the mutated chromosomal DNA region.
The GFP reporter plasmids were generated using the vector plasmid pLPV3Z (67). Promoter fragments upstream from absA, katE, cinA1, otsBA, and proP were amplified by PCR using A. baumannii strain ATCC 17961 chromosomal DNA as a template. DNA fragments corresponding to the fimT, bauD, and bfmRS promoter regions were synthesized by Integrated DNA Technologies. Each promoter DNA fragment was digested with PstI and XbaI except for bfmRS and otsBA promoters, which were digested with KpnI and PstI and with KpnI and XbaI, respectively. Digested fragments were purified from an agarose gel and ligated with plasmid pLPV3Z that had been digested with the appropriate restriction enzymes. Promoter fragments were cloned into plasmid pLPV3Z directly upstream from the promoterless GFP reporter gene. This produced plasmids pJF330, pJF331, pJF332, pJF333, pJF334, pJF337, pJF338, pJF339, and pJF340.
To generate lacZ-transcriptional fusion plasmids, we constructed plasmid pACYC-lacZ by amplifying the multicloning site (MCS) and lacZ reporter gene from plasmid pUC18T-mini-Tn7T-Gm (68) by PCR using primers lacZ-F and lacZ-R. The primer upstream from the MCS contained a XbaI restriction site. Plasmid pACYC184 was digested with AvaI, and the 3′ overhangs were filled in with Pfu Turbo DNA polymerase (Agilent Technologies). Next, the plasmid was digested with XbaI, and the 2,750-bp fragment carrying the p15A origin of replication and the cat gene was purified from an agarose gel. This fragment was ligated with the lacZ-containing PCR fragment, which had also been digested with XbaI, to generate plasmid pACYC-lacZ.
To generate transcriptional reporter fusions in plasmid pACYC-lacZ, the cinA1, otsBA, and proP promoter inserts were amplified by PCR using A. baumannii strain ATCC 17961 chromosomal DNA as a template. Primers for cinA1 and proP promoters were designed to contain PstI and HindIII sites. These inserts, in addition to the plasmid pACYC-lacZ, were digested with PstI and HindIII and purified from an agarose gel. The resulting fragments were ligated to produce plasmids pSP-cinA1lacZ and pSP-proPlacZ. The same process was used to generate plasmid pSP-otsBAlacZ using the HindIII restriction enzyme only. Plasmids pJF320 and pJF321 were also generated by this method, using the restriction enzymes PstI and KpnI.
The expression plasmids pET-BfmR and pET-BfmRS were generated by amplifying the bfmR and bfmRS DNA regions by PCR using A. baumannii strain ATCC 17961 chromosomal DNA as a template. The primers for these reactions (bfmR exR, bfmR exF, and BfmSexp_R2) were designed to create a PciI site overlapping the bfmR translational start site and either a BamHI site or EcoRI site downstream from the bfmR or bfmRS coding region, respectively. These PCR fragments were digested with PciI and either BamHI or EcoRI, purified from an agarose gel, and ligated with plasmid pET28b, which had been digested with NcoI and either BamHI or EcoRI, to produce plasmids pET-BfmR and pET-BfmRS. To introduce a point mutation into the bfmR coding sequence, plasmid pET-BfmR was used as a template for inverse PCR using primers bfmR asp ala 1 and bfmR asp2. The resulting product was purified from an agarose gel and recircularized by ligation to produce plasmid pET-BfmR D58A.
All plasmids were confirmed by restriction digest analysis and DNA sequencing.
Protein expression and purification.
The expression vector pET28a-bfmR was used to express BfmR with an amino-terminal His6 affinity tag. To generate pET28a-bfmR, the bfmR gene was cloned out of pMU360 (23) by PCR using primers designed to contain BamHI and XhoI restriction sites. The PCR fragment and pET28a were digested with BamHI and XhoI and ligated to produce plasmid pET28a-bfmR. This plasmid was then transformed into BL21(DE3), and overnight cultures were used to subculture into 1 L LB medium at a 1:80 dilution. Cultures were grown at 37°C with shaking at 160 rpm until the optical density at 600 nm (OD600) reached 0.7. Then, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and cultures were shifted to 25°C overnight with shaking at 120 rpm. Cells were harvested and pellets were resuspended in 30 mL lysis buffer (25 mM Tris [pH 7.9], 500 mM NaCl, 1 mM dithiothreitol [DTT], 5 mM imidazole). Cells were lysed by sonication and lysates were then centrifuged at 15,000 × g for 15 min. The resulting clarified supernatants were loaded onto a nickel-nitrilotriacetic acid (Ni-NTA)–agarose column (Qiagen) by gravity flow. Next, the column was washed with 100 mL lysis buffer, followed by 100 mL lysis buffer containing 1 M NaCl. Proteins were eluted using an imidazole gradient (0 to 300 mM) in lysis buffer. Fractions containing BfmR-His6 were pooled and dialyzed into 20 mM Tris (pH 7.9) and 400 mM NaCl. The His6 affinity tag was cleaved by addition of 100 U thrombin for 2 h at room temperature, and the reaction was quenched with 20 μM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) for 20 min at room temperature. Dialysis was continued in 20 mM Tris (pH 7.9) and 400 mM NaCl, and then protein was concentrated using Millipore 10 K spin columns according to the manufacturer’s protocol.
RNA-sequencing analysis.
To isolate RNA for primer extension analyses, overnight A. baumannii cultures were subcultured in LB medium to an OD600 of 0.05 and grown at 37°C for 6 h. Samples from cultures were treated with RNAprotect bacteria reagent (Qiagen), and then cells were harvested by centrifugation and stored at −80°C until RNA extraction. To isolate RNA, cell pellets were thawed and incubated in lysis buffer (30 mM Tris [pH 8], 1 mM EDTA, 10 mg/mL lysozyme, 2 mg/mL proteinase K) for 5 min at room temperature, and RNA was purified using TRIzol reagent (Invitrogen). RNA samples were treated with Turbo DNase (Invitrogen) to remove any remaining DNA. Sequencing library preparation, Illumina sequencing, and gene expression analyses were performed by LC Sciences, LLC (Houston, TX). Sequencing libraries were prepared using the Illumina TruSeq total RNA library protocol, including rRNA depletion with the Ribo-Zero kit (Illumina). Sequencing was performed using the Illumina HiSeq system to generate 150-bp paired-end reads. The raw reads were trimmed and filtered for quality, and adapter sequences were removed using Trimmomatic v0.32 (69). The processed reads were mapped to the strain ATCC 17961 draft genome sequence (http://www.patricbrc.org; genome ID 470.2202 [70]), and differential gene expression analysis was performed using Rockhopper v2.0.3 (71). Gene names and locus tags in the text have been updated to the identifiers found in the completed genome sequence for strain ATCC 17961 (GenBank accession numbers CP065432 [chromosome], CP065433 [pAB17961-1], and CP065434 [pAB17961-2]) (72). Functional enrichment analysis was performed using ShinyGO v0.66 (73).
GFP reporter assays.
To assay transcriptional fusions for absA, katE, cinA1, otsBA, proP, and fimT (plasmids pJF330, pJF331, pJF332, pJF333, pJF334, and pJF338, respectively), initial A. baumannii cultures (harboring relevant plasmids) were grown in LB medium at 37°C for 6 to 7 h and then subcultured to an OD600 of 0.05. Subcultures were grown at 37°C for 16 h, at which point aliquots were diluted either 1:5 or 1:10 in sterile phosphate-buffered saline, pH 7. Three 100-μL samples of diluted culture were transferred to a 96-well plate, and fluorescence was measured using a Tecan Spark microplate reader (excitation, 480 nm; emission, 520 nm). Fluorescence values were adjusted by subtracting the background fluorescence value generated by uninoculated LB medium diluted in phosphate-buffered saline (PBS). The reported values were calculated by dividing the average of three replicate measurements of fluorescence units (RFU) by the average A600 value of the three replicates (RFU/A600) to account for differences in bacterial growth. The data reported are the averages and standard deviations (SD) from at least three independent experiments. The same assay procedure was used for bauD and bfmR transcriptional fusions (plasmids pJF339 and pJF340, respectively), except initial cultures were grown overnight 37°C, and then subcultured to an OD600 of 0.05 for 6 h prior to assays for fluorescence.
Chemical activation of BfmR protein.
Protein samples were activated using beryllium fluoride as previously descried (22, 50, 51). Briefly, 1 mg/mL purified protein was activated by addition of 7 mM MgCl2, 5 mM BeCl2, and 35 mM NaF. The solution was mixed and incubated at room temperature for at least 1 h, then transferred to 4°C.
EMSAs.
For EMSAs, PCR was used to synthesize DNA promoter fragments from A. baumannii strain ATCC 17961 chromosomal DNA. These DNA fragments corresponded to the region from −435 to +33 relative to the otsBA translational start site (468 bp), from −433 to +6 for proP (439 bp), from −419 to +97 for cinA1 (516 bp), from −507 to +78 for absA (585 bp), and from +132 to +543 (412 bp) for the internal fragment of gene I5593_11150, which was used as a negative control. DNA probes were radiolabeled using [γ-32P]ATP (PerkinElmer, Inc.) and T4 polynucleotide kinase (New England Biolabs). Binding reaction mixtures containing 0.3 μg salmon sperm DNA, 30,000 cpm of radiolabeled DNA probe, and indicated concentrations of purified BfmR protein were incubated in a mixture containing 20 mM Tris (pH 7.3), 50 mM NaCl, 5 mM MgCl2, and 0.5 mM DTT for 20 min at room temperature. Binding reaction mixtures were then mixed with loading dye and loaded onto native 6% Tris-borate-EDTA (TBE)–polyacrylamide gels that had been prerun for 20 min in chilled 0.5× TBE buffer at 80 V. Samples were electrophoresed at 100 V at 0°C, and gels were visualized by autoradiography.
β-Galactosidase assays.
To assay transcriptional fusions for absA, katE, and cinA1 (plasmids pJF320, pJF321, and pSP-cinA1lacZ, respectively), cells from overnight E. coli strain NovaBlue(DE3) cultures (harboring both the transcriptional fusion plasmid and the expression plasmid or vector control) were used to subculture to an OD600 of 0.05. Subcultures were grown at 37°C for 3 h (OD600 of ∼0.3), and then expression plasmids were induced by addition of 5 μM IPTG for 6 h. To assay transcriptional fusions for proP and otsBA (pSP-proPlacZ and pSP-otsBAlacZ, respectively), initial cultures were grown at 37°C for 6 to 7 h and then subcultured to an OD600 of 0.05 for growth at 37°C for 16 h. At indicated time points, aliquots were collected to assay for β-galactosidase activity in duplicate. Activity is reported as the mean fold change and standard deviation from at least three independent experiments (74).
Sample preparation for in vivo detection of BfmR and Phos-tag acrylamide gel analysis.
For samples collected from E. coli, overnight E. coli strain NovaBlue(DE3) cells expressing either BfmR alone or BfmR and BfmS were subcultured to an OD600 of 0.1 in LB medium. Cultures were then grown at 37°C for 3 h, after which expression plasmids were induced with 5 μM IPTG for a further 2 h before harvesting by centrifugation. For samples collected from A. baumannii, overnight A. baumannii strains were subcultured to an OD600 of 0.05. Cultures were grown in LB medium at 37°C for 6 h and were then harvested by centrifugation. All E. coli and A. baumannii cell pellets were weighed, flash frozen on dry ice/ethanol, and stored at −80°C until assays were performed. Pellets were thawed at room temperature and then resuspended in 10 μL/mg (wet pellet weight) of Bug Buster protein extraction reagent (EMD Millipore) supplemented with 2 mg/mL lysozyme and 1 μL/mL Benzonase nuclease (EMD Millipore). Aliquots were removed from each sample and used in a Bradford assay (Bio-Rad) (75) to estimate total protein concentrations in each lysate, while remaining lysates were flash frozen on dry ice/ethanol and stored at −80°C until ready to be loaded onto gels. Lysates were then thawed at room temperature, mixed with SDS-PAGE loading buffer, and kept on ice until all samples had been prepared. Samples were separated by SDS-PAGE on a 12% (wt/vol) polyacrylamide gel containing 50 μM Phos-tag acrylamide (Wako-Chem) and 100 μM ZnCl2, as per the manufacturer’s instructions. Samples were electrophoresed at 150 V and 4°C.
Phos-tag gels were fixed by incubating in transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol, 1.3 mM SDS) containing 1 mM EDTA to chelate Zn2+ ions. A total of three incubations were carried out for 10 min each at room temperature. This was followed by incubation for 10 min at room temperature in transfer buffer without EDTA. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane using semidry transfer. BfmR protein was detected by Western blot analysis using polyclonal rabbit antiserum raised against purified His-tagged BfmR (23). Prior to use, the anti-BfmR antibody was preadsorbed with an acetone powder (76) derived from either an E. coli strain BL21(DE3) pET28b cell lysate or an A. baumannii strain ATCC 17961 ΔbfmR cell lysate. Blots were incubated with a goat anti-rabbit IgG–horseradish peroxidase (HRP) conjugate (Invitrogen) as a secondary antibody, and blots were visualized with chemiluminescence using SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific).
Phosphorylated BfmR protein was quantified using densitometry as a percentage of total (phosphorylated plus unphosphorylated) BfmR protein. This was calculated using Image Lab software v6.1.0 volume quantity tools (Bio-Rad Laboratories, Inc.).
In vitro phosphorylation of BfmR.
Purified BfmR protein (3 μM) was incubated with 10 mM lithium potassium acetyl phosphate (Sigma) or lithium carbamoyl phosphate dibasic hydrate (Sigma) in 20 mM Tris-HCl (pH 7.3), 50 mM NaCl, 5 mM MgCl2, and 1 mM DTT in a total volume of 15 μL, for the indicated times at 37°C. As a negative control, one sample was heat shocked at 95°C for 5 min after incubation at 37°C. Samples were analyzed using Phos-tag gel electrophoresis and Western blotting, as described above.
Statistical analysis.
For gfp and lacZ reporter assays, statistical significance was determined by analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test (P ≤ 0.05). For quantification of phosphorylated BfmR, statistical significance was determined using Student's t test (P ≤ 0.05).
Data availability.
Raw Illumina sequencing reads were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA780533, with accession numbers SRX13141184, SRX13141185, and SRX13141186 for strain ATCC 17961 and SRX13141187, SRX13141188, and SRX13141189 for strain 17961-ΔbfmR.
ACKNOWLEDGMENTS
We thank P. Visca, University Roma Tre, for providing the Acinetobacter baumannii GFP reporter plasmid.
This work was supported by NIH grant 1R21AI59072 (to E.C.P.), National Science Foundation (NSF) grant 0420479 (to L.A.A.), Miami University research funds (to L.A.A.), and NIH grant R01AI136904 (to J.C.).
Footnotes
Supplemental material is available online only.
Contributor Information
Everett C. Pesci, Email: pescie@ecu.edu.
Laurie E. Comstock, Duchossois Family Institute
REFERENCES
- 1.Almasaudi SB. 2018. Acinetobacter spp. as nosocomial pathogens: epidemiology and resistance features. Saudi J Biol Sci 25:586–596. 10.1016/j.sjbs.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayobami O, Willrich N, Harder T, Okeke IN, Eckmanns T, Markwart R. 2019. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg Microbes Infect 8:1747–1759. 10.1080/22221751.2019.1698273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard A, O'Donoghue M, Feeney A, Sleator RD. 2012. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3:243–250. 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris FC, Dexter C, Kostoulias X, Uddin MI, Peleg AY. 2019. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol 10:1601. 10.3389/fmicb.2019.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodríguez-Villodres Á, Martín-Gandul C, Peñalva G, Guisado-Gil AB, Crespo-Rivas JC, Pachón-Ibáñez ME, Lepe JA, Cisneros JM. 2021. Prevalence and risk factors for multidrug-resistant organisms colonization in long-term care facilities around the world: a review. Antibiotics (Basel) 10:680. 10.3390/antibiotics10060680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurihara MNL, de Sales RO, da Silva KE, Maciel WG, Simionatto S. 2020. Multidrug-resistant Acinetobacter baumannii outbreaks: a global problem in healthcare settings. Rev Soc Bras Med Trop 53:e20200248. 10.1590/0037-8682-0248-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 8.Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. 2021. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10:373. 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. 2021. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens 10:387. 10.3390/pathogens10040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, Roberts JA, Sime FB. 2019. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: a systematic review and meta-analysis. J Infect 79:593–600. 10.1016/j.jinf.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Sherertz RJ, Sullivan ML. 1985. An outbreak of infections with Acinetobacter calcoaceticus in burn patients: contamination of patients’ mattresses. J Infect Dis 151:252–258. 10.1093/infdis/151.2.252. [DOI] [PubMed] [Google Scholar]
- 12.van den Broek PJ, Arends J, Bernards AT, De Brauwer E, Mascini EM, van der Reijden TJK, Spanjaard L, Thewessen EAPM, van der Zee A, van Zeijl JH, Dijkshoorn L. 2006. Epidemiology of multiple Acinetobacter outbreaks in The Netherlands during the period 1999–2001. Clin Microbiol Infect 12:837–843. 10.1111/j.1469-0691.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- 13.Cefai C, Richards J, Gould FK, McPeake P. 1990. An outbreak of Acinetobacter respiratory tract infection resulting from incomplete disinfection of ventilatory equipment. J Hosp Infect 15:177–182. 10.1016/0195-6701(90)90128-b. [DOI] [PubMed] [Google Scholar]
- 14.Papon N, Stock AM. 2019. Two-component systems. Curr Biol 29:R724–R725. 10.1016/j.cub.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Bourret RB, Silversmith RE. 2010. Two-component signal transduction. Curr Opin Microbiol 13:113–115. 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai SK, Kenney LJ. 2017. To ∼P or not to ∼P? Non-canonical activation by two-component response regulators. Mol Microbiol 103:203–213. 10.1111/mmi.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. 2010. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol 13:232–239. 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisinger E, Mortman NJ, Vargas-Cuebas G, Tai AK, Isberg RR. 2018. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog 14:e1007030. 10.1371/journal.ppat.1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, Shuman HA. 2015. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. mBio 6:e01660-15. 10.1128/mBio.01660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draughn GL, Milton ME, Feldmann EA, Bobay BG, Roth BM, Olson AL, Thompson RJ, Actis LA, Davies C, Cavanagh J. 2018. The structure of the biofilm-controlling response regulator BfmR from Acinetobacter baumannii reveals details of its DNA-binding mechanism. J Mol Biol 430:806–821. 10.1016/j.jmb.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology (Reading) 154:3398–3409. 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 24.Pakharukova N, Tuittila M, Paavilainen S, Malmi H, Parilova O, Teneberg S, Knight SD, Zavialov AV. 2018. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci USA 115:5558–5563. 10.1073/pnas.1800961115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo TA, Manohar A, Beanan JM, Olson R, MacDonald U, Graham J, Umland TC. 2016. The response regulator BfmR is a potential drug target for Acinetobacter baumannii. mSphere 1:e00082-16. 10.1128/mSphere.00082-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umland TC, Wayne Schultz L, MacDonald U, Beanan JM, Olson R, Russo TA. 2012. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. mBio 3:e00113-12. 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crépin S, Ottosen EN, Peters K, Smith SN, Himpsl SD, Vollmer W, Mobley HLT. 2018. The lytic transglycosylase MltB connects membrane homeostasis and in vivo fitness of Acinetobacter baumannii. Mol Microbiol 109:745–762. 10.1111/mmi.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5:e01163-14. 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marr CM, MacDonald U, Trivedi G, Chakravorty S, Russo TA. 2020. An evaluation of BfmR-regulated antimicrobial resistance in the extensively drug resistant (XDR) Acinetobacter baumannii strain HUMC1. Front Microbiol 11:2688. 10.3389/fmicb.2020.595798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liou M-L, Soo P-C, Ling S-R, Kuo H-Y, Tang CY, Chang K-C. 2014. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect 47:275–281. 10.1016/j.jmii.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Farrow JM, III, Wells G, Pesci EC. 2018. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One 13:e0205638. 10.1371/journal.pone.0205638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storz G, Hengge R. 2010. Bacterial stress responses. ASM Press, Washington, DC. [Google Scholar]
- 33.Robinson A, Brzoska AJ, Turner KM, Withers R, Harry EJ, Lewis PJ, Dixon NE. 2010. Essential biological processes of an emerging pathogen: DNA replication, transcription, and cell division in Acinetobacter spp. Microbiol Mol Biol Rev 74:273–297. 10.1128/MMBR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCann MP, Kidwell JP, Matin A. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol 173:4188–4194. 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange R, Hengge-Aronis R. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol 5:49–59. 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 36.Gebhardt MJ, Shuman HA. 2017. GigA and GigB are master regulators of antibiotic resistance, stress responses, and virulence in Acinetobacter baumannii. J Bacteriol 199:e00066-17. 10.1128/JB.00066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerqueira GM, Kostoulias X, Khoo C, Aibinu I, Qu Y, Traven A, Peleg AY. 2014. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J Infect Dis 210:46–55. 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 38.Kröger C, Kary SC, Schauer K, Cameron ADS. 2016. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes (Basel) 8:12. 10.3390/genes8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, Chua KL, Webber MA, Sutton JM, Peterson ML, Piddock LJV. 2021. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7:e00430-16. 10.1128/mBio.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbé J, Bou G. 2011. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol 193:3740–3747. 10.1128/JB.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo H-Y, Chao H-H, Liao P-C, Hsu L, Chang K-C, Tung C-H, Chen C-H, Liou M-L. 2017. Functional characterization of Acinetobacter baumannii lacking the RNA chaperone Hfq. Front Microbiol 8:2068. 10.3389/fmicb.2017.02068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elhosseiny NM, Amin MA, Yassin AS, Attia AS. 2015. Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int J Med Microbiol 305:114–123. 10.1016/j.ijmm.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 43.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheldon JR, Skaar EP. 2020. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog 16:e1008995. 10.1371/journal.ppat.1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bijlsma JJE, Groisman EA. 2003. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol 11:359–366. 10.1016/S0966-842X(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 47.Bourret RB. 2010. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 13:142–149. 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem 266:8348–8354. 10.1016/S0021-9258(18)92982-0. [DOI] [PubMed] [Google Scholar]
- 49.Goulian M. 2010. Two-component signaling circuit structure and properties. Curr Opin Microbiol 13:184–189. 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachhawat P, Swapna GVT, Montelione GT, Stock AM. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13:1353–1363. 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. 2001. BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase. Proc Natl Acad Sci USA 98:8525–8530. 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao R, Stock AM. 2013. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc Natl Acad Sci USA 110:672–677. 10.1073/pnas.1214587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757. 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 55.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbieri CM, Stock AM. 2008. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem 376:73–82. 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desai SK, Winardhi RS, Periasamy S, Dykas MM, Jie Y, Kenney LJ. 2016. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. Elife 5:e10747. 10.7554/eLife.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem 276:40873–40879. 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- 59.Pletnev P, Osterman I, Sergiev P, Bogdanov A, Dontsova O. 2015. Survival guide: Escherichia coli in the stationary phase. Acta Naturae 7:22–33. 10.32607/20758251-2015-7-4-22-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong T, Schellhorn HE. 2010. Role of RpoS in virulence of pathogens. Infect Immun 78:887–897. 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang SM, Schellhorn HE. 2010. Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J Mol Evol 70:557–571. 10.1007/s00239-010-9352-0. [DOI] [PubMed] [Google Scholar]
- 62.Gourion B, Francez-Charlot A, Vorholt JA. 2008. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J Bacteriol 190:1027–1035. 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casella LG, Weiss A, Pérez-Rueda E, Antonio Ibarra J, Shaw LN. 2017. Towards the complete proteinaceous regulome of Acinetobacter baumannii. Microb Genom 3:mgen000107. 10.1099/mgen.0.000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tipton KA, Dimitrova D, Rather PN. 2015. Phase-variable control of multiple phenotypes in Acinetobacter baumannii strain AB5075. J Bacteriol 197:2593–2599. 10.1128/JB.00188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chin CY, Tipton KA, Farokhyfar M, Burd EM, Weiss DS, Rather PN. 2018. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat Microbiol 3:563–569. 10.1038/s41564-018-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmad I, Karah N, Nadeem A, Wai SN, Uhlin BE. 2019. Analysis of colony phase variation switch in Acinetobacter baumannii clinical isolates. PLoS One 14:e0210082. 10.1371/journal.pone.0210082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lucidi M, Visaggio D, Prencipe E, Imperi F, Rampioni G, Cincotti G, Leoni L, Visca P. 2019. New shuttle vectors for real-time gene expression analysis in multidrug-resistant Acinetobacter species: in vitro and in vivo responses to environmental stressors. Appl Environ Microbiol 85:e01334-19. 10.1128/AEM.01334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farrow JM, 3rd, Pesci EC, Slade DJ. 2021. Genome sequences for two Acinetobacter baumannii strains obtained using the Unicycler Hybrid Assembly Pipeline. Microbiol Resour Announc 10:e00017-21. 10.1128/MRA.00017-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ge SX, Jung D, Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36:2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller JH. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 75.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 76.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 77.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 79.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jb.00494-21-s0001.pdf, PDF file, 0.09 MB (89.1KB, pdf)
Table S1. Download jb.00494-21-s0002.xlsx, XLSX file, 0.2 MB (161.4KB, xlsx)
Data Availability Statement
Raw Illumina sequencing reads were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA780533, with accession numbers SRX13141184, SRX13141185, and SRX13141186 for strain ATCC 17961 and SRX13141187, SRX13141188, and SRX13141189 for strain 17961-ΔbfmR.