PURPOSE
The optimal characteristics among patients with breast cancer to recommend neoadjuvant chemotherapy is an active area of clinical research. We developed and compared several approaches to developing prediction models for pathologic complete response (pCR) among patients with breast cancer in Alberta.
METHODS
The study included all patients with breast cancer who received neoadjuvant chemotherapy in Alberta between 2012 and 2014 identified from the Alberta Cancer Registry. Patient, tumor, and treatment data were obtained through primary chart review. pCR was defined as no residual invasive tumor at surgical excision in breast or axilla. Two types of prediction models for pCR were built: (1) expert model: variables selected on the basis of oncologists' opinions and (2) data-driven model: variables selected by trained machine. These model types were fit using logistic regression (LR), random forests (RF), and gradient-boosted trees (GBT). We compared the models using area under the receiver operating characteristic curve and integrated calibration index, and internally validated using bootstrap resampling.
RESULTS
A total of 363 cases were included in the analyses, of which 86 experienced pCR. The RF and GBT fits yielded higher optimism-corrected area under the receiver operating characteristic curves compared with LR for the expert (RF: 0.70; GBT: 0.69; LR: 0.65) and data-driven models (RF: 0.71; GBT: 0.68; LR: 0.64). The LR fit yielded the lowest integrated calibration indices for the expert (LR: 0.037; GBT: 0.05; RF: 0.10) and data-driven models (LR: 0.026; GBT: 0.06; RF: 0.099).
CONCLUSION
Our models demonstrated predictive ability for pCR using routinely collected clinical and demographic variables. We show that machine learning fit methods can be used to optimize models for pCR prediction. We also show that additional variables beyond clinical expertise do not considerably improve predictive ability and may not be of value on the basis of the burden of data collection.
BACKGROUND
Preoperative or neoadjuvant chemotherapy (NAC) was previously reserved for locally advanced and inflammatory breast cancer with the goal of enabling resection. NAC is now used more widely in clinical practice for earlier-stage, operable breast cancer following trial data that demonstrated its equivalency to adjuvant chemotherapy in terms of event-free survival and overall survival.1-5 Pathologic complete response (pCR) has been shown to be predictive for superior event-free survival and overall survival, especially in human epidermal growth factor receptor 2 (HER2)-positive and triple-negative disease, and has therefore been used as the primary end point for many NAC trials.6-11 NAC confers several potential advantages including tumor downstaging to reduce the extent of local surgery, using tumor response for prognostication, and determination of need for postoperative therapies.12
CONTEXT
Key Objective
To compare regression-based and machine learning model fitting, as well as a priori and data-driven feature selection approaches, to predict pathologic complete response (pCR) in patients with breast cancer following neoadjuvant chemotherapy.
Knowledge Generated
Using clinical and pathologic variables, models fit with random forest algorithms showed better discrimination of patients with and without pCR, whereas models fit with logistic regression showed better calibration. A priori and data-driven feature selection did not result in differences in model discrimination or calibration.
Relevance
As artificial intelligence continues to emerge in cancer research, there is a need to compare traditional regression and novel machine learning approaches to pCR prediction. This may improve the ability to identify patients who are more likely to benefit from neoadjuvant chemotherapy and facilitate precision-based treatment decisions.
Despite its potential advantages, only 20%-40% of patients with breast cancer in observational and trial settings achieve pCR following NAC.6,13-17 Thus, it is important to develop decision support tools to choose the right patients who will be more likely to benefit from NAC. Many studies in the current literature have developed models for pCR prediction among patients on NAC13-17; however, no clear superior models have been identified because key questions on the development of these models remain unanswered. These questions include whether machine learning is superior to traditional regression modeling and whether routinely collected widely available data are sufficient to accurately predict pCR. These questions are imperative in terms of the predictivity and utility of prediction models.
We aimed to address these gaps by comparing traditional regression-based and novel machine learning approaches to developing prognostic prediction models for pCR following NAC among patients with breast cancer in Alberta. We also aimed to quantify the gain in predictive performance associated with increasing the number of candidate variables in the statistical model to identify the optimal balance between measurement burden and predictive ability.
METHODS
This study reports the details of the development of prediction models in accordance with the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis guidelines.18 The models developed in this study constitute type 2 models according to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis guidelines.
Study Cohort and Data Collection
This was a population-based retrospective cohort study, which included all patients who received NAC and underwent surgery for invasive breast cancer between January 1, 2012, and June 30, 2014 in Alberta, Canada. Patients were identified through our provincial, synoptic, web-based surgical medical record database, which is described in detail elsewhere.19
Web-based surgical medical record and individual chart review were used to obtain detailed information regarding patient age at surgery, treatment facility, use of neoadjuvant treatment, pathologic response to chemotherapy, types of surgery performed, surgery side, the surgeon's perception of appropriateness for breast-conserving surgery (BCS), adjuvant therapy (chemotherapy, radiation therapy, and hormonal), and pretreatment tumor characteristics, including tumor (T) and node (N) stage. Estrogen receptor (ER) status, progesterone receptor (PR) status, and HER2 status were obtained from the Alberta Cancer Registry database, which captures all malignancies in Alberta. pCR was confirmed by microscopic assessment of tissue samples by a pathologist and defined as the absence of invasive cancer in the breast and axillary nodes, irrespective of in situ carcinoma (ypT0/is ypN0).
Modeling Approaches
Only variables gathered before the administration of NAC were considered for inclusion in the prediction models. We examined different methods for selection of predictors to build two types of prediction models for pCR. (1) Expert model: One surgeon (M.L.Q.) and two medical oncologists (S.L. and W.Y.C.) in our study team listed all the likely factors that may be used to predict pCR on the basis of clinical knowledge and experience. Then, on the basis of our data availability, we established our list of predictors for the expert model including age at surgery (in years), T stage (T1-T4), BCS candidate (yes v no), ER status (positive v negative), PR status (positive v negative), HER2 status (positive v negative), and treatment facility (academic v community). (2) Data-driven model: In this model, an automated (data-driven) variable selection procedure was used to select a final list of predictors from all candidate variables available with complete data. These automated variable selection procedures were dependent on the approach that was used to fit the model, which is discussed in detail in the next section.
Statistical Analyses
Each of the two types of models (expert and data-driven) were fit using multivariable logistic regression (LR), random forests (RF), and gradient-boosted trees (GBT), resulting in six prediction models. The associations between pCR and predictors were presented using odds ratios (ORs) with 95% CI for expert model when fit using LR. The data-driven model built on LR was penalized using least absolute shrinkage and selection operator regularization. The method aims to minimize the prediction error by shrinking the coefficient of some variables to zero and keep those variables with nonzero coefficients after shrinking. Ten-fold cross-validation was used to determine the penalization (λ) parameter that minimized the mean cross-validated error.
For the expert and data-driven models fit with RF, the number of trees was 500 and the number of randomly sampled predictors chosen as split candidates in each tree was equal to the square root of number of predictors included in the model. The tree depth used in the RF models was tuned to minimizing the out-of-bag error. For models fit with GBT, the number of trees, learning rate, and tree depth were tuned to minimize test classification error with 10-fold cross-validation.
The automated variable selection process for the data-driven model when fit using RF and GBT was based on feature importance. Feature importance for RF was based on permutation importance measured by out-of-bag accuracy and for GBT was based on gain, which represents fractional contribution of each feature to the model on the basis of the total gain of this feature's splits. Higher percentage means a more important predictive feature.20 The automated variable selection procedure for the data-driven models built on RF and GBT occurred in a backward elimination fashion. First, a model was fit with pCR as the outcome variable and all candidate variables with complete data as predictors of pCR. Then, feature importance statistics were extracted and ranked from highest to lowest. Finally, the least important predictor was removed. This process iterated until eight predictors remained, indicating the eight most important predictors. Details regarding tuning parameters and feature sets for the data-driven models are provided in the Data Supplement.
Predictive performance of the models was compared using discrimination and calibration measures. Discrimination was measured by area under the receiver operating characteristic curve (AUC). The DeLong algorithm21,22 was used to compute 95% CIs of each AUC. Calibration was measured using Emax,23 the maximum difference between predicted and observed probabilities, and integrated calibration index (ICI),24 the average difference between a smooth calibration curve and the diagonal line of perfect calibration. Two thousand bootstrap samples were used to compute 95% CIs for calibration measures. Internal validation was performed using bootstrap resampling in which 200 bootstrap samples were used to quantify the optimism-corrected AUC, ICI, and Emax values.25 We performed 0.632 bootstrap resampling, developed by Bradley Efron, for internal validation.26 The adequacy of the sample size was evaluated using 10 events-per-variable, which is widely used in prediction research.18
RESULTS
Cohort Characteristics
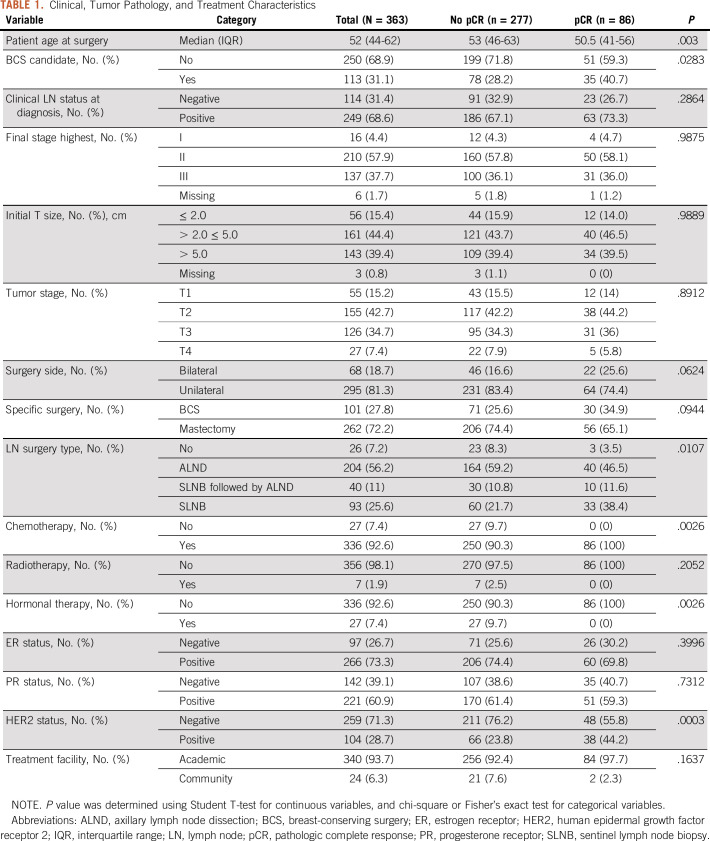
The study cohort consisted of 363 patients, with a median age of 52 (interquartile range, 44-62) years. One hundred thirteen (31%) patients were deemed as a candidate for BCS, which is slightly higher than the proportion of the patients who actually received BCS (28%, n = 101). Sixty-eight (18%) patients received bilateral surgery and the majority of the patients (n = 337, 93%) underwent lymph node surgery. Overall, 86 patients (24%) patients in this cohort achieved pCR. The univariate analysis showed that the patient group with pCR was significantly different from the patient group without pCR regarding age, BCS candidates, lymph node surgery, chemotherapy, hormone therapy, and HER2 status (Table 1).
TABLE 1.
Clinical, Tumor Pathology, and Treatment Characteristics
Predictors of pCR
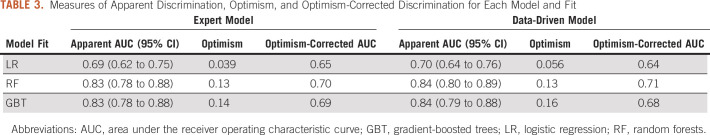
When fit using LR, patient age at time of surgery (OR = 0.97; 95% CI, 0.95 to 0.99, per 1-year increase in age), BCS candidate (OR = 2.01; 95% CI, 1.14 to 3.55), and HER2 status (OR = 2.72; 95% CI, 1.59 to 4.67) remained significant predictors of pCR in the expert model (Table 2).
TABLE 2.
The Associations Between Patient Characteristics and Pathologic Complete Response in the Expert Model
Predictive Performance
Expert model.
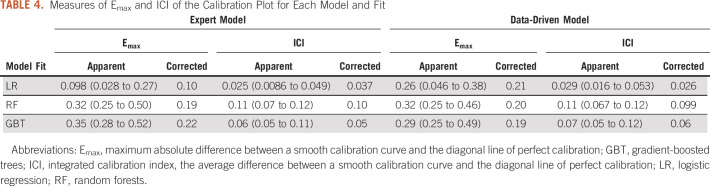
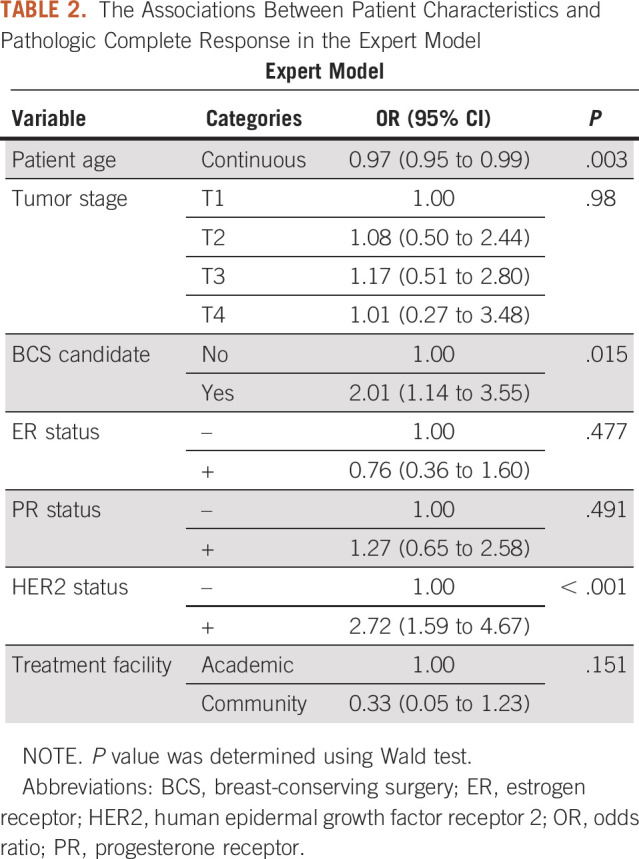
The receiver operating characteristic curves illustrating the discriminative performance of the expert model for the three fitting approaches are presented in Figures 1A-1C. The RF and GBT fit yielded optimism-corrected AUCs of 0.70 and 0.69, respectively, whereas the LR fit yielded an optimism-corrected AUC of 0.65 (Table 3). Figures 1D-1F illustrate calibration plots for the expert models. The LR fit resulted in an Emax of 0.10 and ICI of 0.037. The RF resulted in an Emax of 0.19 and ICI of 0.10. The GBT resulted in an Emax of 0.22 and ICI of 0.05 (Table 4).
FIG 1.
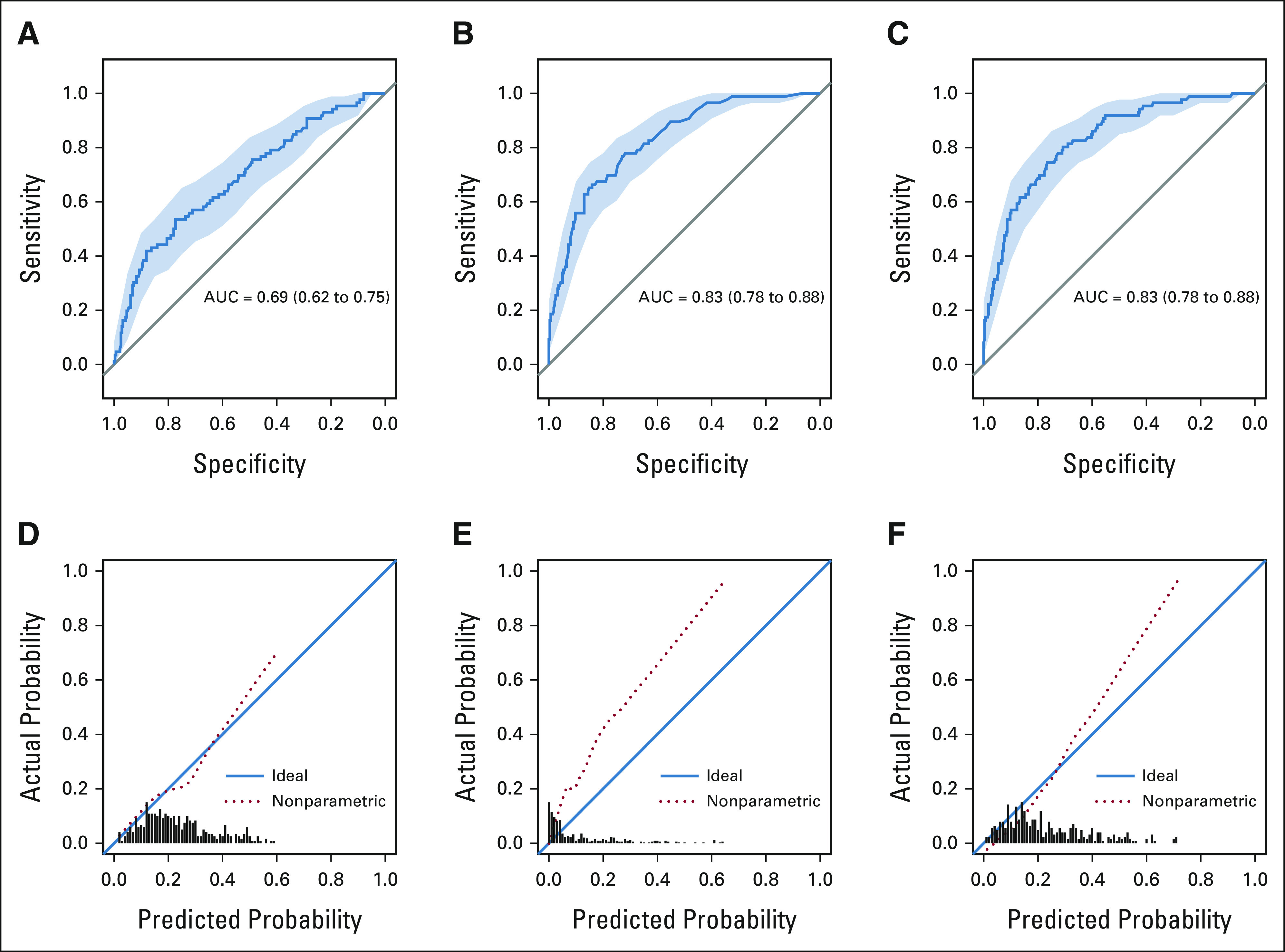
(A-F) Receiver operating characteristic and calibration plot for expert models. AUC, area under the receiver operating characteristic curve.
TABLE 3.
Measures of Apparent Discrimination, Optimism, and Optimism-Corrected Discrimination for Each Model and Fit
TABLE 4.
Measures of Emax and ICI of the Calibration Plot for Each Model and Fit
Data-driven model.
Figures 2A-2C present the receiver operating characteristic curves of the data-driven model fit with LR, RF, and GBT. The optimism-corrected AUCs for the RF, GBT, and LR were 0.71, 0.68, and 0.64, respectively (Table 3). The calibration plots for data-driven models are presented in Figure 2D-2F. The Emax was 0.21 and ICI was 0.026 for the LR fit. For GBT fit, the Emax and ICI were 0.19 and 0.06, respectively. The Emax was 0.20 and ICI was 0.099 for the RF fit (Table 4).
FIG 2.
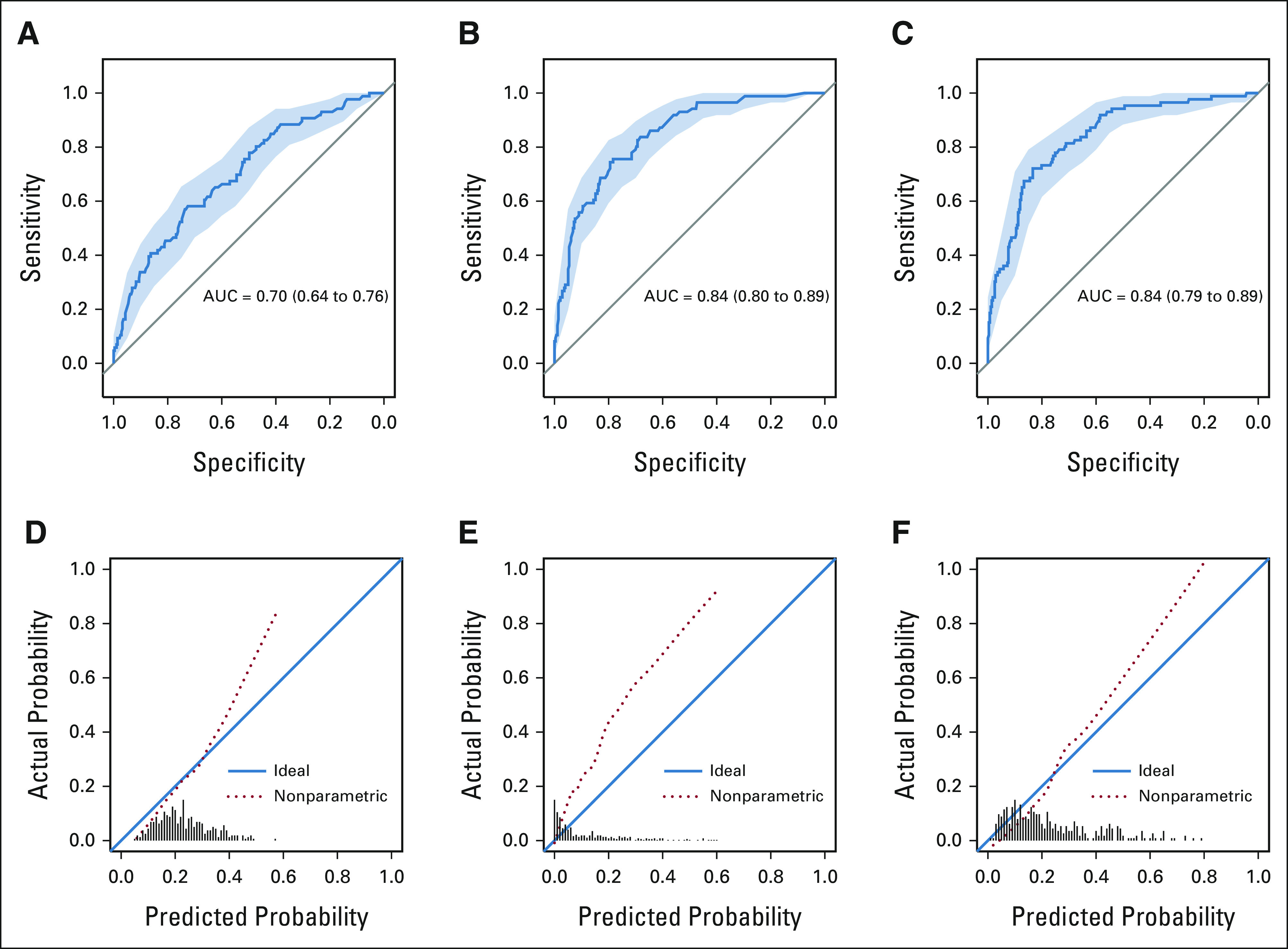
(A-F) Receiver operating characteristic and calibration plot for data-driven models. AUC, area under the receiver operating characteristic curve.
DISCUSSION
In this study, we analyzed 363 patients with breast cancer in Alberta and demonstrated that traditional regression and novel machine learning models on the basis of routinely collected patient data are able to meaningfully predict pCR following NAC. Following internal validation, we observed that the performance of each type of model when fit using RF and GBT varied the most, that is, had the highest optimism. Despite this high optimism, the RF and GBT fits produced higher optimism-corrected AUCs than LR fit for the two types of models. The internally validated calibration of the models fit with LR was superior compared with RF and GBT. However, the sample size for this study was likely not sufficient for the RF or GBT algorithms, resulting in statistical overfitting, miscalibration, and requiring cautious interpretation.
Rates of pCR have ranged between 13%-65% in randomized trials of NAC, depending on molecular subtype.1-5,27-29 In our population-based cohort, 24% of patients achieved pCR. Patients in the study displayed similar tumor and demographic characteristics as those in the previously published literature.7,13 There is substantial evidence that HER2-positive tumors are more likely to achieve pCR and similar associations were observed in our study cohort. However, predictors found to be significant in other studies, such as negative ER and/or PR status, clinical stage, and lymph node status, were not observed in our analysis.7,30-33
Several different approaches have been used to develop models for predicting pathologic response to NAC in patients with breast cancer. LR is among the most common owing to its interpretability to a clinical audience, with insight into the relative effects of predictors by ORs and in displays, such as nomograms.34 A nomogram proposed by Hennessy et al13 predicts pCR on the basis of patient age, ER status, grade, and stage; however, no discriminative performance is reported. Another built by Pu et al35 using Ki-67 index, NAC regimen, lymphovascular invasion, hemoglobin level, and ER status produced an AUC of 0.76. Other nomograms built on LR using clinicopathologic variables had AUCs that range from 0.77 to 0.8530,36; however, these models were built in node-positive patient populations.
To our knowledge, our analyses are among the first to compare regression-based and machine learning methods to fit prediction models for pCR in patients with breast cancer following NAC. LR is considered to be the default modeling approach to probability estimation in medical risk prediction.34 Although it is easily interpretable, LR is parametric in nature and the model will not produce consistent probability estimates if it is mis-specified, such as ignoring nonlinearity and interactions.37 Even after applying more modern modeling techniques to increase the flexibility of LR, such as least absolute shrinkage and selection operator penalization, no significant improvement in model calibration was observed in our study. Machine learning techniques are a more flexible alternative to probability estimation as they are able to learn more directly from data without assuming an underlying statistical model.23 Tree-based methods do not make the same additive and linear assumption as LR. RF generates predictions by running a subject through multiple decision trees built in bootstrap data sets, with averaging of the result. At each split in a tree, only a subset of randomly selected predictors is considered. This process decorrelates the trees, thereby making the average of the resulting trees less variable and more reliable.38 Boosting does not involve bootstrap resampling; instead, trees are grown sequentially using information from previously grown trees. Boosting learns more slowly and is thought to be more robust to overfitting.38 It appeared that the nonparametric nature of machine learning techniques was more suitable for pCR discrimination in our study cohort; however, whether this is true in independent data requires further investigation.
Despite promising discriminative ability, the models fit with RF were the most miscalibrated. The RF models had ICI values of approximately 0.1, suggesting that predicted probabilities of pCR from these models are on average 10% off from the true probability of pCR.24 Comparatively, the models fit with LR had ICI values range between 0.026 and 0.037, suggesting that predicted probabilities of pCR from these models are on average 2.6%-3.7% off from the true probability of pCR.24 Therefore, improvements in calibration are needed for these RF models to be clinically useful. Machine learning offers more flexible algorithms for outcome prediction and thus requires large amounts of data.34 The size of our sample was likely not sufficient enough to adequately fit the prediction models using RF, resulting in overly optimistic discrimination measures and miscalibration owing to statistical overfitting. The GBT approach yielded better calibration than RF and a wider range of predicted probabilities. It is likely that the slower learning approach of boosting makes it more robust to overfitting in smaller data sets and better calibration than RF. Validation of these models on a larger independent data set is required to accurately assess their performance and clinical utility.
Our study was also novel in that we compared different variable selection techniques to find an optimal balance between predictive ability and measurement burden. The performance between expert and data-driven models was similar regardless of model fit. On the basis of our data-driven models, training a machine to automatically select a feature set did not result in improvements in predictive performance to offset the burden of collecting data for these additional variables. We also report that the feature sets selected by the trained machine was similar to that of expert opinion for each model fit. To further explore how model performance changed on the basis of the number of features included in the data-driven RF model, we continued the backward deletion procedure until one feature remained. The primary observation from this procedure was that as variables were removed from the model, calibration decreased because of high counts of probability predictions of 0. Therefore, we show that a priori variable selection on the basis of subject knowledge allows for sufficient predictive performance on the basis of limited predictors. This may be desirable in clinical practice as little data collection is required to provide reliable probability estimates of pCR following NAC. Steyerberg23 claims it would be ideal to prespecify a prediction model completely as it implies that candidate predictors are selected without studying the predictor-outcome relation in the data under study. Subject knowledge is viewed as a useful technique to restrict the number of candidate predictors and increase the robustness and validity of prediction models, particularly in smaller samples.23 The predictor variables considered in this analysis were those routinely collected in clinical practice to facilitate the uptake of such prediction tools among oncologists. As these variables are included in the standard initial breast cancer workup, these models may be applied to each patient before surgery, avoiding the need for additional diagnostic procedures and their associated costs.
There were several strengths to this analysis. This was a population-based study—the sample of patients was derived from a cohort that captures nearly all (> 93%) breast cancer cases in Alberta, Canada, which provides real-world evidence of current practice. These findings may be inferred to other patients with breast cancer on NAC in this province and potentially others, because of the universal health system and similar patterns of care, and the similarities in administrative data between provinces in Canada. This study was not without limitations. First, there was no independent study sample to perform external validation on these models. Several predictive variables identified in the literature were not available for analysis, including body mass index,39,40 tumor infiltration lymphocytes,41,42 p53 status,43,44 Ki-67 status,45,46 and BRCA1 and BRCA2 status.47 Finally, the sample size may not have been sufficient for the data-hungry machine learning approaches, resulting in overfitting of the models built on RF and GBT.
In conclusion, we developed and compared several approaches to modeling pCR prediction in patients with breast cancer following NAC. As the use of NAC increases in patients with lower burden of disease, it becomes increasingly important to identify characteristics best suited for NAC. These preliminary models are promising and may assist clinical decision making by determining patients with a high probability of achieving pCR to improve prognostication, limit surgical morbidity, dictate further adjuvant treatment, and provide optimal cancer care. External validation studies are needed to investigate how the models developed in this study perform in independent data. Furthermore, net benefit approaches, described by Vickers et al,48 may provide a clearer understanding of the clinical utility of these models. Our study group is collaborating with additional external collaborators to examine external validation and implementation to accelerate the significance and impact of these prediction models. We also plan to abstract more chart review data to include information regarding type and duration of NAC received in our prediction models. We have developed an online prediction tool to demonstrate proof of concept where patients or clinicians can input readily available patient data to estimate probability of pCR.49
ACKNOWLEDGMENT
The authors would like to thank and acknowledge the Oncology Outcomes (O2) research team and Strategies for Precision Health in Breast Cancer (SPHERE) research team for their valuable contribution to the preparation of this manuscript.
Devon Boyne
Employment: Cytel
Darren Brenner
Employment: Cytel
No other potential conflicts of interest were reported.
AUTHOR CONTRIBUTIONS
Conception and design: Robert B. Basmadjian, Tamer N. Jarada, Yuan Xu, Winson Y. Cheung, Darren R. Brenner
Administrative support: Yuan Xu
Provision of study materials or patients: Winson Y. Cheung, May Lynn Quan
Collection and assembly of data: Yuan Xu, Winson Y. Cheung
Data analysis and interpretation: Robert B. Basmadjian, Shiying Kong, Devon J. Boyne, Tamer N. Jarada, Winson Y. Cheung, Sasha Lupichuk, May Lynn Quan
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Devon Boyne
Employment: Cytel
Darren Brenner
Employment: Cytel
No other potential conflicts of interest were reported.
REFERENCES
- 1.Bear HD, Anderson S, Smith RE, et al. : Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol 24:2019-2027, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Mauri D, Pavlidis N, Ioannidis JP: Neoadjuvant versus adjuvant systemic treatment in breast cancer: A meta-analysis. J Natl Cancer Inst 97:188-194, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Mieog JS, van der Hage JA, van de Velde CJ: Preoperative chemotherapy for women with operable breast cancer. Cochrane Database Syst Rev 2007:CD005002, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rastogi P, Anderson SJ, Bear HD, et al. : Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol 26:778-785, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Wolmark N, Wang J, Mamounas E, et al. : Preoperative chemotherapy in patients with operable breast cancer: Nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J Natl Cancer Inst Monogr:96-102, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Cortazar P, Zhang L, Untch M, et al. : Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 384:164-172, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Kuerer HM, Newman LA, Smith TL, et al. : Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol 17:460-469, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Kümmel S, Holtschmidt J, Loibl S: Surgical treatment of primary breast cancer in the neoadjuvant setting. Br J Surg 101:912-924, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Liedtke C, Mazouni C, Hess KR, et al. : Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26:1275-1281, 2008 [DOI] [PubMed] [Google Scholar]
- 10.von Minckwitz G, Blohmer JU, Costa SD, et al. : Response-guided neoadjuvant chemotherapy for breast cancer. J Clin Oncol 31:3623-3630, 2013 [DOI] [PubMed] [Google Scholar]
- 11.von Minckwitz G, Untch M, Blohmer JU, et al. : Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 30:1796-1804, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Graham PJ, Brar MS, Foster T, et al. : Neoadjuvant chemotherapy for breast cancer, is practice changing? A population-based review of current surgical trends. Ann Surg Oncol 22:3376-3382, 2015 [DOI] [PubMed] [Google Scholar]
- 13.Hennessy BT, Hortobagyi GN, Rouzier R, et al. : Outcome after pathologic complete eradication of cytologically proven breast cancer axillary node metastases following primary chemotherapy. J Clin Oncol 23:9304-9311, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Kantor O, Sipsy LM, Yao K, et al. : A predictive model for axillary node pathologic complete response after neoadjuvant chemotherapy for breast cancer. Ann Surg Oncol 25:1304-1311, 2018 [DOI] [PubMed] [Google Scholar]
- 15.Parker JS, Mullins M, Cheang MCU, et al. : Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27:1160-1167, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Y-H, Zhu H-T, Cao K, et al. : Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac Cancer 11:651-658, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vila J, Mittendorf EA, Farante G, et al. : Nomograms for predicting axillary response to neoadjuvant chemotherapy in clinically node-positive patients with breast cancer. Ann Surg Oncol 23:3501-3509, 2016 [DOI] [PubMed] [Google Scholar]
- 18.Moons KG, Altman DG, Reitsma JB, et al. : Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): Explanation and elaboration. Ann Intern Med 162:W1-W73, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Graham PJ, Brar MS, Foster T, et al. : Neoadjuvant chemotherapy for breast cancer, is practice changing? A population-based review of current surgical trends. Ann Surg Oncol 22:3376-3382, 2015 [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Guestrin C: XGBoost: A scalable tree boosting system. Presented at the Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, August 13-17, 2016
- 21.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44:837-845, 1988 [PubMed] [Google Scholar]
- 22.Sun X, Xu W: Fast implementation of DeLong's algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process Lett 21:1389-1393, 2014 [Google Scholar]
- 23.Steyerberg EW: Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York, NY, Springer, 2009 [Google Scholar]
- 24.Austin PC, Steyerberg EW: The Integrated Calibration Index (ICI) and related metrics for quantifying the calibration of logistic regression models. Stat Med 38:4051-4065, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steyerberg EW, Harrell FE, Borsboom GJJM, et al. : Internal validation of predictive models: Efficiency of some procedures for logistic regression analysis. J Clin Epidemiol 54:774-781, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Efron B: Estimating the error rate of a prediction rule: Improvement on cross-validation. J Am Stat Assoc 78:316-331, 1983 [Google Scholar]
- 27.Baselga J, Bradbury I, Eidtmann H, et al. : Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet 379:633-640, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang HR: Trastuzumab-based neoadjuvant therapy in patients with HER2-positive breast cancer. Cancer 116:2856-2867, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Robidoux A, Tang G, Rastogi P, et al. : Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): An open-label, randomised phase 3 trial. Lancet Oncol 14:1183-1192, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Choi HJ, Ryu JM, Kim I, et al. : Nomogram for accurate prediction of breast and axillary pathologic response after neoadjuvant chemotherapy in node positive patients with breast cancer. Ann Surg Treat Res 96:169-176, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JY, Park HS, Kim S, et al. : Prognostic nomogram for prediction of axillary pathologic complete response after neoadjuvant chemotherapy in cytologically proven node-positive breast cancer. Medicine 94:e1720, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasanpour P, Sandoughdaran S, Mosavi-Jarrahi A, et al. : Predictors of pathological complete response to neoadjuvant chemotherapy in Iranian breast cancer patients. Asian Pac J Cancer Prev 19:2423-2427, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan MC, Al Mushawah F, Gao F, et al. : Predictors of complete pathological response after neoadjuvant systemic therapy for breast cancer. Am J Surg 198:520-525, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steyerberg EW, van der Ploeg T, Van Calster B: Risk prediction with machine learning and regression methods. Biometrical J 56:601-606, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Pu S, Wang K, Liu Y, et al. : Nomogram-derived prediction of pathologic complete response (pCR) in breast cancer patients treated with neoadjuvant chemotherapy (NCT). BMC Cancer 20:1120, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schipper R-J, Moossdorff M, Nelemans PJ, et al. : A model to predict pathologic complete response of axillary lymph nodes to neoadjuvant chemo(immuno)therapy in patients with clinically node-positive breast cancer. Clin Breast Cancer 14:315-322, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Kruppa J, Liu Y, Biau G, et al. : Probability estimation with machine learning methods for dichotomous and multicategory outcome: Theory. Biometrical J 56:534-563, 2014 [DOI] [PubMed] [Google Scholar]
- 38.James G, Witten D, Hastie T, et al. : An Introduction to Statistical Learning. New York, NY, Springer, 2013 [Google Scholar]
- 39.Raman R, Mott SL, Schroeder MC, et al. : Effect of body mass index- and actual weight-based neoadjuvant chemotherapy doses on pathologic complete response in operable breast cancer. Clin Breast Cancer 16:480-486, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litton JK, Gonzalez-Angulo AM, Warneke CL, et al. : Relationship between obesity and pathologic response to neoadjuvant chemotherapy among women with operable breast cancer. J Clin Oncol 26:4072-4077, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang HW, Jung H, Hyeon J, et al. : A nomogram to predict pathologic complete response (pCR) and the value of tumor-infiltrating lymphocytes (TILs) for prediction of response to neoadjuvant chemotherapy (NAC) in breast cancer patients. Breast Cancer Res Treat 173:255-266, 2019 [DOI] [PubMed] [Google Scholar]
- 42.Ingold Heppner B, Untch M, Denkert C, et al. : Tumor-infiltrating lymphocytes: A predictive and prognostic biomarker in neoadjuvant-treated HER2-positive breast cancer. Clin Cancer Res 22:5747-5754, 2016 [DOI] [PubMed] [Google Scholar]
- 43.Kim T, Han W, Kim MK, et al. : Predictive significance of p53, Ki-67, and Bcl-2 expression for pathologic complete response after neoadjuvant chemotherapy for triple-negative breast cancer. J Breast Cancer 18:16-21, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Xu Y, Chen J, et al. : TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int J Cancer 138:489-496, 2016 [DOI] [PubMed] [Google Scholar]
- 45.Chen R, Ye Y, Yang C, et al. : Assessment of the predictive role of pretreatment Ki-67 and Ki-67 changes in breast cancer patients receiving neoadjuvant chemotherapy according to the molecular classification: A retrospective study of 1010 patients. Breast Cancer Res Treat 170:35-43, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain P, Doval DC, Batra U, et al. : Ki-67 labeling index as a predictor of response to neoadjuvant chemotherapy in breast cancer. Jpn J Clin Oncol 49:329-338, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Dumont A, Pannier D, Ducoulombier A, et al. : ERCC1 and CYP1B1 polymorphisms as predictors of response to neoadjuvant chemotherapy in estrogen positive breast tumors. SpringerPlus 4:327, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vickers AJ, Van Calster B, Steyerberg EW: Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 352:i6, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oncology Outcomes (O2). Neoadjuvant Chemotherapy for Breast Cancer in Alberta - Decision Support Tool. http://tools.oncoutcomes.com/pcr.aspx