ABSTRACT
The WalR-WalK two component signaling system in Bacillus subtilis functions in the homeostatic control of the peptidoglycan (PG) hydrolases LytE and CwlO that are required for cell growth. When the activities of these enzymes are low, WalR activates transcription of lytE and cwlO and represses transcription of iseA, a secreted inhibitor of LytE. Conversely, when PG hydrolase activity is too high, WalR-dependent expression of lytE and cwlO is reduced and iseA is derepressed. In a screen for additional factors that regulate this signaling pathway, we discovered that overexpression of the membrane-anchored PG deacetylase PdaC increases WalR-dependent gene expression. We show that increased expression of PdaC, but not catalytic mutants, prevents cell wall cleavage by both LytE and CwlO, explaining the WalR activation. Importantly, the pdaC gene, like iseA, is repressed by active WalR. We propose that derepression of pdaC when PG hydrolase activity is too high results in modification of the membrane-proximal layers of the PG, protecting the wall from excessive cleavage by the membrane-tethered CwlO. Thus, the WalR-WalK system homeostatically controls the levels and activities of both elongation-specific cell wall hydrolases.
IMPORTANCE Bacterial growth and division requires a delicate balance between the synthesis and remodeling of the cell wall exoskeleton. How bacteria regulate the potentially autolytic enzymes that remodel the cell wall peptidoglycan remains incompletely understood. Here, we provide evidence that the broadly conserved WalR-WalK two-component signaling system homeostatically controls both the levels and activities of two cell wall hydrolases that are critical for cell growth.
KEYWORDS: peptidoglycan; two component system; d,l-endopeptidase; deacetylation; deacetylation; peptidoglycan
INTRODUCTION
The cell wall peptidoglycan (PG) is composed of glycan strands that are cross-linked together by short peptides. This three-dimensional exoskeleton protects the cell from osmotic lysis and specifies cell shape. To grow, cells must enlarge this covalently closed macromolecule. This requires both the synthesis of new material and hydrolysis of the existing meshwork to allow for PG expansion (1). How these potentially autolytic enzymes are regulated remains incompletely understood. Bacillus subtilis contains two functionally redundant d,l-endopeptidases (CwlO and LytE) that cleave peptide cross-links to allow expansion of the PG. LytE is a secreted factor that interacts with the cell wall via its LysM domains (2), while CwlO is regulated by a membrane complex composed of FtsEX and SweDC and therefore acts on membrane-proximal layers of the PG (3–5). The genes encoding these enzymes are under the transcriptional control of the essential and broadly conserved WalR-WalK (WalRK) two component system (6). We have previously shown that this regulatory pathway monitors the activity of LytE and CwlO by sensing their cleavage products and, in response, modulates their levels (7). Specifically, when d,l-endopeptidase activity is too high, WalRK responds by decreasing the production of LytE and CwlO; conversely, when d,l-endopeptidase activity is low, WalRK increases their transcription in response (7).
In addition to adjusting the expression of these enzymes, WalRK modulates the activity of LytE. Specifically the WalRK system reciprocally controls IseA, a secreted inhibitor of LytE (8, 9). Accordingly, when d,l-endopeptidase activity is too high and WalRK signaling drops, not only is transcription of cwlO and lytE reduced but iseA is de-repressed allowing the cell to rapidly adjust PG cleavage activity (7). A second gene, pdaC, is negatively regulated by activated WalR (6) and is therefore also derepressed when PG hydrolase activity is high and WalRK signaling is reduced. pdaC encodes a membrane-anchored PG deacetylase that removes the N-acetyl group from the MurNAc sugars in the PG matrix (10). Previous studies suggest that deacetylation by PdaC reduces LytE activity (11), providing a second mechanism to modulate D,L-endopeptidase activity. Thus, the WalRK system controls the levels of LytE and CwlO and reciprocally modulates LytE activity.
The sensor histidine kinase, WalK, is thought to monitor the cleavage products produced by LytE and CwlO through its extracellular sCache domain (7, 12). However, WalK also contains an intracellular Per-ARNT-Sim (PAS) sensing domain (13) that was found to be essential for WalK function (14), suggesting it might respond to additional inputs. Intriguingly, a paralog of WalK, involved in phosphate homeostasis called PhoR, is thought to monitor intracellular precursors of wall teichoic acid synthesis via its PAS domain (15). These data raise the possibility that the conserved intracellular PAS domain of WalK could modulate kinase activity in response to a cytoplasmic signal, like intermediates in the PG precursor pathway (16).
In a screen aimed at identifying signals sensed by WalK's intracellular PAS domain, we discovered that increasing levels of PdaC activates WalRK signaling. Our data indicate that WalRK activation is due to inhibition of PG hydrolysis and therefore is related to the signal sensed by the extracellular sCache domain. Strikingly, our analysis revealed that PdaC-mediated deacetylation of the cell wall impaired the activity of both LytE and CwlO. Our data support a model in which derepression of pdaC when PG hydrolase activity is too high results in deacetylation of the membrane-proximal layers of the PG, which reduces cleavage by the membrane-tethered CwlO enzyme. More broadly, our findings indicate that the WalRK system is finely tuned to homeostatically control the levels and activities of both elongation-specific cell wall hydrolases.
RESULTS
A genetic screen for regulators of WalRK signaling identifies pdaC.
To identify perturbations that could be sensed by the intracellular PAS domain on WalK, we performed a genetic screen for activators of WalRK signaling. We used a WalR-responsive transcriptional reporter that contains the promoter of the WalR-activated gene yocH (PyocH) fused to lacZ (7). yocH codes for a putative lytic transglycosylase and is one of most highly expressed genes under WalR control (6, 17). A B. subtilis strain harboring this fusion was mutagenized with a transposon containing an outward-facing promoter (Ppen) (18) that can generate both loss-of-function insertions and overexpression of adjacent genes. The library was plated on Luria-Bertani (LB) agar plates containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Approximately 200,000 colonies were screened and ∼1,000 appeared dark blue, indicative of increased WalRK signaling. We picked and pooled these colonies and mapped their insertion sites and the orientation of the transposon by transposon-sequencing (Tn-seq) (see Materials and Methods). In validation of our screen, we isolated transposon insertions in the genes encoding CwlO and its regulators FtsEX (see Fig. S1 in the supplemental material). Both reduce PG hydrolase activity and increase WalRK signaling (7). Similarly, we isolated insertions in walH and walI (Fig. S1) that encode negative regulators of WalK, and thus result in high WalR activity (19).
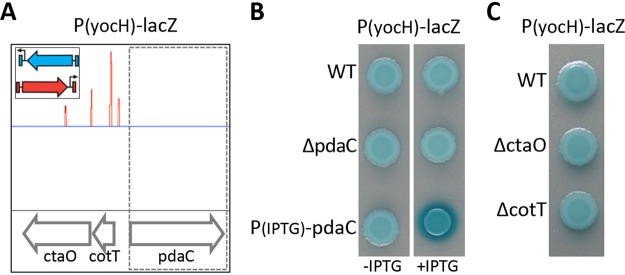
Our screen identified several additional insertions predicted to cause loss-of-function or overexpression of genes previously unknown to impact WalRK signaling (Table S1). To validate these hits, we generated in-frame deletions, insertion-deletions, or IPTG-inducible promoter fusions in our PyocH-lacZ reporter strain and analyzed colony color on LB agar plates containing X-Gal. Surprisingly, most hits exhibited modest or no change in PyocH-lacZ compared to wild-type (see Discussion). However, overexpression of the gene encoding the PG deacetylase PdaC (10), identified by oriented transposons inserted upstream of its coding sequence (Fig. 1A), reproducibly increased WalR-dependent transcription from PyocH-lacZ (Fig. 1B). Importantly, deletions of cotT and ctaO, the genes upstream of pdaC that contained the transposon insertions, did not affect PyocH-lacZ activity (Fig. 1C).
FIG 1.
Transposon-sequencing screen for activators of WalR-WalK signaling identifies pdaC. To identify mutants that increase WalRK signaling, a transposon containing a strong outward-facing promoter (insert) was used to mutagenize a strain harboring the WalR activity reporter PyocH-lacZ. (A) Transposon insertion profile from a region of the B. subtilis genome containing pdaC. Each line indicates a transposon insert site, its height represents the number of sequencing reads, and its color (red or blue) indicates its orientation. Oriented insertions upstream of pdaC suggest overexpression of pdaC activates WalR-WalK signaling. (B) Increased expression of pdaC in a strain harboring an IPTG-regulated promoter fused to pdaC results in high PyocH-lacZ transcription. (C) Strains harboring deletions of the genes into which the transposons had inserted did not affect WalRK signaling. Cells harboring the PyocH-lacZ reporter were spotted on LB agar plates containing 100 μg/ml X-Gal with and without IPTG (500 μM). Images were taken after overnight incubation at 37°C.
PG deacetylase activity of pdaC is required to activate WalRK signaling.
To investigate the impact of pdaC overexpression on WalR activity during exponential growth, we used a fluorescent PyocH transcriptional reporter (PyocH-venus) (7) and monitored the cells by fluorescence microscopy. We induced pdaC expression at an optical density at 600 nm (OD600) of 0.15 and imaged the cells before and 60 min after IPTG addition. Consistent with our population-based plate assay, PyocH-venus fluorescence increased by ∼2.5-fold after pdaC induction (Fig. 2A and C).
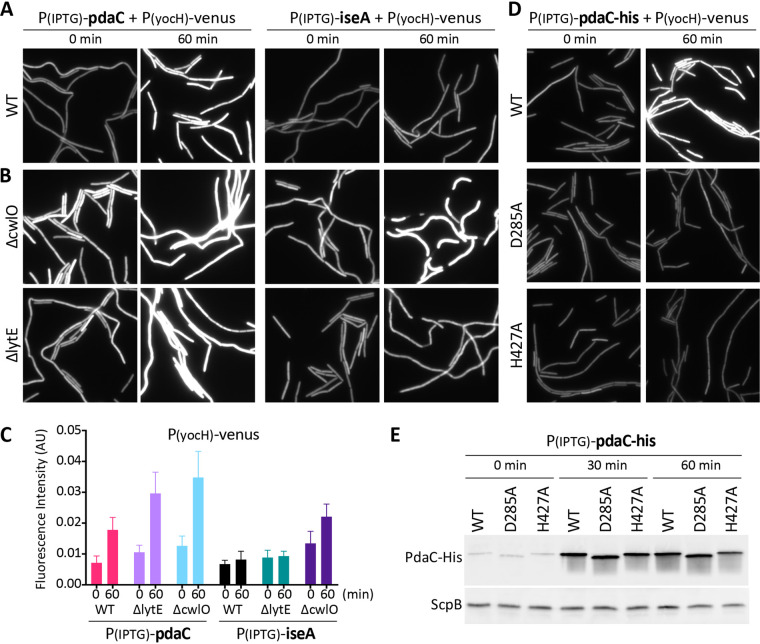
FIG 2.
Overexpression of pdaC, but not catalytic mutants, activates WalRK signaling. (A) Overexpression of pdaC during exponential growth activates WalR-dependent transcription. Representative images of YFP fluorescence in the indicated strains harboring the WalR activity reporter PyocH-venus and IPTG-regulated promoter fusions to pdaC or iseA. Cells were grown to an OD600 of ∼0.15 in LB medium and analyzed by fluorescence microscopy before (0 min) and after (60 min) IPTG addition (500 μM). (B) Overexpression of pdaC activates WalR in the absence of CwlO or LytE while activation of WalR by overexpression of iseA requires LytE. Representative fluorescent images of cells grown to an OD600 of ∼0.15 in LB medium before (0 min) and after (60 min) induction with 500 μM IPTG. (C) Quantification of average fluorescence intensity, normalized to cell area, of strains shown in panels A and B. More than 300 cells were analyzed for each strain. (D) Overexpression of pdaC catalytic mutants does not affect WalRK signaling. The indicated strains with His-tagged PdaC variants were grown to an OD600 of ∼0.15 in LB medium, and images were taken before (0 min) and after (60 min) addition of IPTG (500 μM). (E) Immunoblot analysis of PdaC-His protein levels from cultures shown in panel D. ScpB was used to control for loading. Images are from one of three biological replicates. Representative immunoblots are from one of two biological replicates.
PdaC has an N-terminal transmembrane segment followed by a PG deacetylase domain that belongs to the carbohydrate esterase family 4 (CE4) (20, 21). Biochemical characterization of the soluble domain indicates that it catalyzes the removal of the N-acetyl group of N-acetylmuramic acid (MurNAc) in PG (10). To investigate whether PdaC’s effect on WalRK signaling requires deacetylase activity, we generated previously characterized catalytically inactive mutants of PdaC (D285A and H427A) (22) and fused them to the identical IPTG-regulated promoter. Both mutants and a matched wild-type control were expressed as His-tagged fusions to enable assessment of protein levels using anti-His antibodies. Overexpression of either catalytic mutant had little to no impact on PyocH-venus transcription compared to the matched wild-type control (Fig. 2D). Importantly, the expression levels of the mutants were similar to wild-type PdaC (Fig. 2E). These data argue that PdaC's deacetylase activity is required for WalRK activation.
Overexpression of pdaC impacts LytE and CwlO.
Overexpression of pdaC was previously reported to prevent cell death in a strain with hyperactive WalR activity (11). Furthermore, suppression of walRK essentiality by overexpressing lytE required deletion of pdaC (11). Based on these findings, Takada and coworkers proposed that the deacetylation of the PG by PdaC reduces the affinity of LytE for its substrate, thereby lowering LytE activity. Accordingly, we suspected that the increase in WalRK signaling we observed when pdaC was overexpressed was due to reduced LytE activity. To test this, we used a strain lacking CwlO (ΔcwlO) in which the only elongation-specific d,l-endopeptidase was LytE. In this background, overexpression of pdaC for 60 min increased WalR-dependent expression of PyocH-venus by ∼3-fold (Fig. 2B and C). These data are consistent with the idea PdaC-mediated deacetylation of the PG reduces LytE activity resulting in WalRK activation. As a positive control for these experiments, we overexpressed the LytE inhibitor IseA. In wild-type, PyocH-venus transcription was largely unchanged. However, in the ΔcwlO mutant, WalR-dependent transcription dramatically increased (Fig. 2B and C).
To investigate whether overexpression of pdaC also impaired CwlO activity, we analyzed WalRK signaling in a ΔlytE mutant in which CwlO was the only elongation-specific PG hydrolase. Consistent with the idea that PdaC-mediated deacetylation of the PG impacts not only the activity of LytE but also the activity of CwlO, overexpression of pdaC increased PyocH-venus transcription in this background (Fig. 2B and C). As a control for these experiments, we analyzed WalR-dependent transcription in the ΔlytE mutant when iseA was overexpressed. As anticipated, PyocH-venus fluorescence was largely unchanged (Fig. 2B and C), consistent with previous findings (9).
Overexpression of pdaC reduces LytE and CwlO activity.
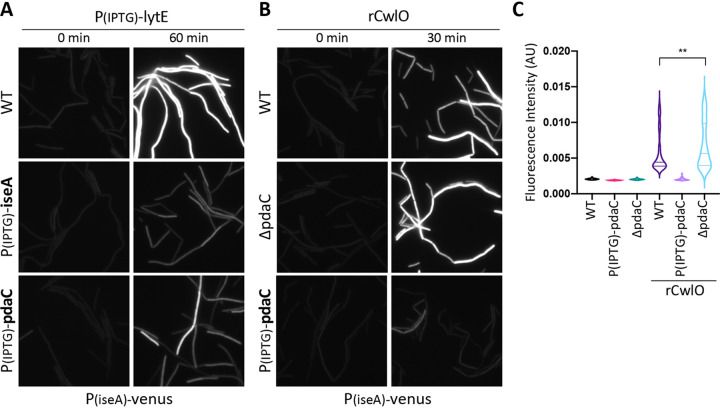
To more directly investigate whether PdaC inhibits LytE activity, we took advantage of the fact that high levels of LytE activity reduce WalRK signaling and derepress a PiseA-venus transcriptional reporter (7) (Fig. 3A). If PdaC activity impairs LytE's ability to cut the cell wall, then overexpressing pdaC at the same time as overexpressing lytE should prevent or reduce de-repression of PiseA-venus. As can be seen in Fig. 3A, this was indeed the case. Importantly, this effect required the deacetylation activity of PdaC, as a catalytic mutant did not alter the WalRK response to lytE overexpression (Fig. S2A in the supplemental material). Similarly, co-overexpression of iseA and lytE also prevented PiseA-venus derepression (Fig. 3A). These data argue that both IseA and PdaC inhibit LytE, one by directly inhibiting LytE enzymatic activity (9), the other by modifying its substrate.
FIG 3.
Overexpression of pdaC counteracts inhibition of WalRK signaling in response to high d,l-endopeptidase activity. (A) Representative fluorescent images of the PiseA-venus transcriptional reporter in the indicated strains containing an IPTG-regulated promoter fusion to lytE. Cells were grown to an OD600 of ∼0.15 and images were taken before (0 min) and after (60 min) addition of IPTG (50 μM). Overexpression of both iseA and pdaC prevents inhibition of WalRK signaling in response to overexpression of lytE. (B) Representative fluorescent images of the PiseA-venus transcriptional reporter in the indicated strains before (0 min) and after (30 min) the addition of recombinant CwlO (rCwlO) (70 μg/ml final concentration). rCwlO inhibited WalRK signaling and derepressed PiseA-venus in wild-type (WT) and the ΔpdaC mutant. Overexpression of pdaC (25 μM IPTG) for 60 min prior to addition of rCwlO prevents inhibition of WalRK signaling. (C) Quantification of average fluorescence intensity, normalized to cell area, of strains shown in panel B. >300 cells were analyzed for each strain.
Next, we investigated whether PdaC impacts CwlO's PG hydrolase activity. Unlike overexpression of lytE, overexpression of cwlO does not affect WalRK signaling, likely due to its postranslational regulation by FtsEX and SweDC (5). Instead, we took advantage of the fact that recombinantly produced CwlO (rCwlO), when added exogenously, reduces WalRK signaling and derepresses PiseA-venus (Fig. 3B). Importantly, when rCwlO was added to a strain lacking PdaC (ΔpdaC), we detected a subtle but reproducible increase in PiseA-venus derepression (Fig. 3B and C). Reciprocally, when we added rCwlO to a strain overexpressing pdaC, WalRK activity remained unchanged and PiseA-venus expression remained low (Fig. 3B and C). Finally, as anticipated, overexpression of iseA had no impact on derepression of PiseA-venus caused by rCwlO (Fig. S2B in the supplemental material).
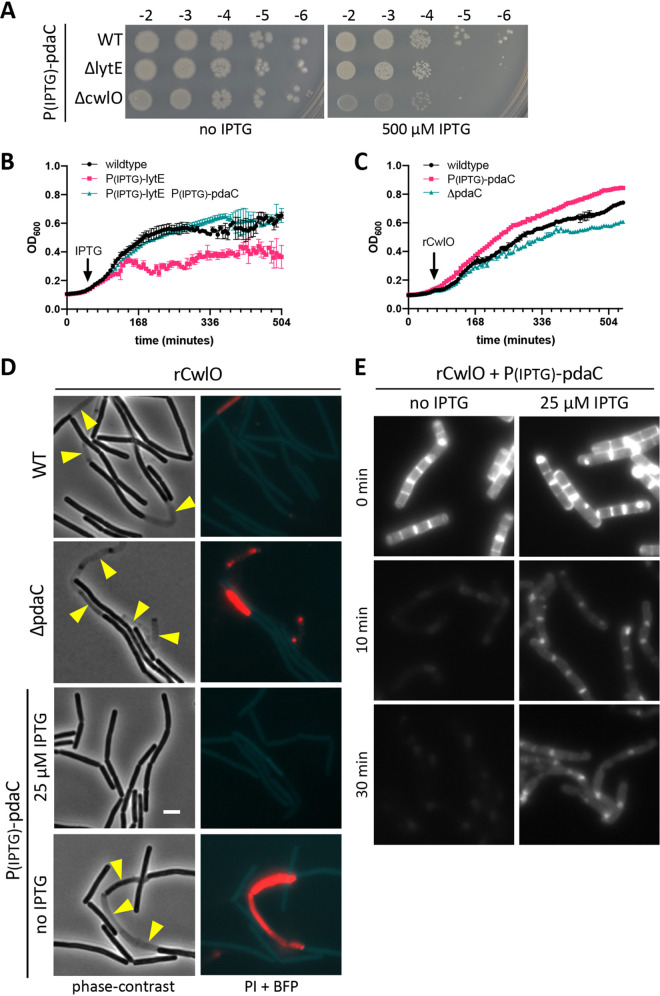
Derepression of PiseA-venus is an indirect assay for CwlO and LytE activity. To more directly assess whether PdaC-mediated PG deacetylation inhibits LytE and CwlO, we took advantage of the fact that high levels of either enzyme impair cell growth and/or cause lysis. As can be seen in Fig. 4B, chronic overexpression of lytE caused a cessation of growth. However, this defect could be suppressed by overexpression of wild-type pdaC (Fig. 4B) but not a catalytic mutant (Fig. S3 in the supplemental material). Similarly, addition of rCwlO to wild-type cells reduces growth and causes lysis (Fig. 4C and D). As anticipated, cells lacking PdaC were even more growth-impaired and susceptible to lysis by rCwlO (Fig. 4C and D). Furthermore, overexpression of pdaC (but not iseA) protected cells from rCwlO (Fig. 4C and D; Fig. S4). Consistent with these findings, we found that overexpression of pdaC in cells lacking CwlO or LytE impaired growth on LB agar (Fig. 4A). Altogether, these results provide strong genetic evidence that PdaC deacetylase activity inhibits both LytE and CwlO activity.
FIG 4.
Overexpression of pdaC inhibits PG hydrolase activity of LytE and CwlO. (A) Spot dilutions of the indicated strains harboring an IPTG-regulated promoter fusion to pdaC on LB agar plates with and without 500 μM IPTG. Strains were grown to mid-log phase in LB medium and normalized to an OD600 of 0.5. Serial 10-fold dilutions were spotted on LB agar plates with or without IPTG, and imaged after overnight incubation at 37°C. (B) Growth curves of the indicated strains. Cells were grown to mid-log phase in LB medium, normalized to an OD600 of 0.02 in LB, and grown at 37°C. After 1 h, IPTG was added (500 μM), and growth resumed. The OD600 was measured every 6 min for 8 h. (C) Growth curves of the indicated strains after addition of recombinant CwlO (rCwlO). The indicated strains were grown to mid-log phase in LB medium, normalized to an OD600 of 0.02 in LB containing 25 μM IPTG to overexpress pdaC, and grown at 37°C. After 1 h, 70 μg/ml (final) rCwlO was added, and growth was resumed. OD600 measurements were taken every 6 min for 8 h. (D) Representative fluorescent images of the PiseA-venus transcriptional reporter, propidium iodide (PI) staining, and a BFP cytoplasmic marker in the indicated strains. Strains were grown to an OD600 of ∼0.15 in LB medium with IPTG, if indicated, at 37°C. After 1 h of growth, rCwlO was added (70 μg/ml final) and cultures were imaged after 30 min. Scale bar indicates 3 μm. (E) Representative images of fluorescently labeled sacculi incubated with rCwlO. Sacculi were purified from cultures harboring an IPTG-regulated promoter fusion to pdaC that were grown for 1.5 h in the presence of HADA and concurrently for 1 h in the presence (25 μM IPTG) or absence (no IPTG) of IPTG. Fluorescent sacculi were resuspended in 25 mM MES [pH 5.5] at 2.5 mg/ml, and rCwlO was added to 5 μg/ml (final). Reactions were incubated with agitation at 37°C, and images were taken at the indicated time. Representative data from one of two biological replicates.
PdaC-deacetylated sacculi are inefficiently cleaved by recombinant CwlO.
To directly test whether CwlO activity is influenced by deacetylation of its substrate, we analyzed rCwlO cleavage of sacculi isolated from a strain harboring an IPTG-regulated allele of pdaC that was grown in the presence or absence of inducer. The fluorescent d-amino acid HADA was included in the growth medium to fluorescently label the cell wall. We then incubated the fluorescently labeled purified sacculi with rCwlO and monitored the loss of fluorescence over time. As can be seen in Fig. 4E, sacculi derived from cells in which pdaC was not induced lost fluorescence within 10 min. By contrast, sacculi derived from cells in which pdaC was overexpressed were less efficiently cleaved by rCwlO and retained fluorescent signal even after 30 min. These results indicate that PdaC-mediated deacetylation of the PG impairs CwlO's ability to cleave it.
DISCUSSION
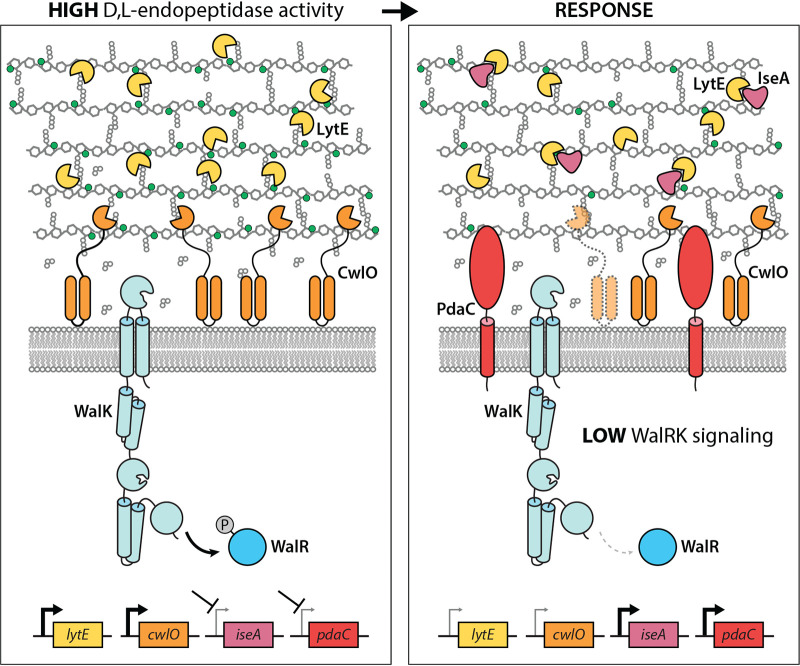
Our data support a model in which the WalRK two-component system can maintain precise PG hydrolase activity during cell growth through both transcriptional and post-transcriptional control of the elongation-specific cell wall hydrolases, CwlO and LytE (Fig. 5). We previously demonstrated that when d,l-endopeptidase activity is too high, WalRK signaling decreases lowering transcription of these enzymes (7). However, a purely transcriptional response is likely to be too slow to effectively decrease hydrolytic activity mediated by these enzymes. To reduce d,l-endopeptidase activity at short time scales, a reduction in WalRK activity also leads to derepression of the secreted protein IseA that directly inhibits LytE activity (8, 9, 11). Furthermore, we have shown that CwlO has a half-life of 5 to 7 min and this likely contributes to reducing PG cleavage activity on short time scales (7). Here, we add an additional level of posttranscriptional control to this picture. A reduction in WalRK signaling due to high d,l-endopeptidase activity causes derepression of the PG deacetylase PdaC (Fig. 5). Deacetylation of the MurNAc sugars reduces PG cleavage by both d,l-endopeptidases. Since PdaC is anchored in the cytoplasmic membrane, we hypothesize that it acts on the membrane-proximal layers of the peptidoglycan, and therefore exerts much of its immediate influence on membrane-associated CwlO (Fig. 5).
FIG 5.
Schematic model for the homeostatic control of CwlO and LytE by WalRK. Phosphorylated WalR (WalR∼P) activates transcription of lytE and cwlO and represses iseA and pdaC (left panel). When d,l-endopeptidase activity gets too high, the cleavage products generated by these enzymes are thought to inhibit the sensor kinase WalK. The drop in WalR∼P leads to a reduction in lytE and cwlO expression and derepression of iseA and pdaC (right panel). IseA directly inhibits LytE activity, while PdaC deacetylates the N-acetyl groups (green balls) of the MurNAc sugars in the PG matrix. Deacetylation impairs cleavage by both CwlO and LytE. Finally, the short half-life of CwlO (shown schematically as translucent with dashed lines) further ensures a rapid adjustment in CwlO levels. CwlO is controlled by the FtsEX/SweDC membrane complex (not shown). Since PdaC and CwlO act on the membrane-proximal layers of the PG, it is hypothesized that PdaC principally modulates CwlO activity.
The conclusion that PdaC-mediated deacetylation of the cell wall prevents its cleavage by CwlO and LytE principally comes from experiments involving overexpression of the deacetylase. However and importantly, even when we expressed PdaC at levels similar to those observed when pdaC is derepressed due to increased LytE activity, we could detect WalR-dependent activation of PyocH-venus (see Fig. S5 in the supplemental material). Furthermore, cells lacking PdaC exhibited increased sensitivity to rCwlO compared to wild-type (Fig. 4C and D). Based on these findings, we propose that the levels of PdaC produced in response to high PG hydrolase activity likely function to modulate or reduce cell wall cleavage, rather than to completely inhibit it.
Our original goal was to identify mutants that would provide information about the role of the intracellular PAS domain of WalK (14). However, we did not identify any hits related to its function. One possibility is that our screen was not saturating. Alternatively, the signal that is sensed by the intracellular PAS domain could cause inhibition of signaling rather than activation. If so, a screen that monitors a reduction in WalRK signaling using the PiseA promoter that is inhibited by active WalR would be more sensitive. A third possibility is that the intracellular PAS domain is not involved in signal sensing but instead functions in transducing the signal sensed by the extracellular sCache domain to the histidine kinase domain.
We note that many of the hits from our Tn-seq screen did not confirm when we tested insertion-deletions, in-frame deletions, or overexpression. When picking blue colonies in our screen, we were very liberal in assessing color change, and likely included colonies with more modest increases in PyocH-lacZ transcription. A work-intensive improvement to this screen would be to streak all potential hits before pooling them. Alternatively, one could use a fluorescent reporter and screen by fluorescence activated cell sorting. This latter method has the additional advantage of eliminating false-positives that result from envelope permeability defects that increase the ability of X-Gal to access β-galactosidase in the cell cytoplasm.
Previous studies on the effects of PG deacetylation have focused on how removal of acetyl groups from GlcNAc or MurNAc sugars affect enzymes that cleave glycan strands (23). Accordingly, we were surprised to find that MurNAc deacetylation by PdaC had inhibitory effects on the d,l-endopeptidase activities of LytE and CwlO. LytE contains three PG-binding LysM domains (2), that have been modeled to interact with acetyl groups in the glycan strand (24) and could explain how PdaC modulates LytE's ability to cleavage PG (11). However, CwlO lacks domains that are known to be influenced by the acetylation state of its PG substrate. Instead we hypothesize that MurNAc deacetylation alters the conformation of the PG such that cleavage of the peptide cross-link by CwlO is less efficient. Establishing the molecular basis for this regulation awaits future structural studies.
In their seminal study of WalRK essentiality, Takada and coworkers suggested that PdaC may play a role in inhibiting CwlO when d,l-endopeptidase cleavage is too high but speculated that this could be indirect (11). Here, we confirm and extend their findings and provide evidence that PG deacetylation by PdaC directly impairs cleavage by both CwlO and LytE. Thus, homeostatic control of d,l-endopeptidases by WalRK involves multiple layers of regulation, both transcriptional and posttranslational, to continually adjust hydrolase activity during cell growth (Fig. 5). Deacetylation of the cell wall is a well-established mechanism of protection from the innate immune response (25, 26). Our data add to the growing body of evidence that PG deacetylases may play equally important roles in controlling cell shape, growth, and cell separation (27–29).
MATERIALS AND METHODS
General methods.
All B. subtilis strains were derived from the prototrophic strain PY79 (30). Cells were grown in LB medium at 37°C. Unless otherwise indicated, B. subtilis strains were constructed using genomic DNA and a 1-step competence method. Antibiotic concentrations were used at: 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 10 μg/ml tetracycline, 10 μg/ml kanamycin, 1 μg/ml erythromycin and 25 μg/ml lincomycin. A list of strains, plasmids, and oligonucleotide primers used in this study can be found in Table S2, Table S3, and Table S4 in the supplemental material, respectively.
Transposon-sequencing and screen for activators of WalRK signaling.
Tn-seq was performed as described previously (31) with modifications described below. The transposon library was generated using a modified B. subtilis mariner-based transposon plasmid (32). The E. coli-B. subtilis shuttle vector contained a temperature-sensitive replicon for B. subtilis, a hyperactive allele of mariner-Himar1 transposase, and a spectinomycin resistance cassette flanked by inverted repeats recognized by the transposase. An MmeI restriction site was engineered within one of the inverted repeats and an outward-facing Ppen promoter was inserted adjacent to it (18). The plasmid was transformed into a B. subtilis strain harboring PyocH-lacZ. Transformants were selected at 30°C and grown in liquid medium at 22°C overnight selecting for resistance to spectinomycin. Transposants were selected on LB agar plates contain spectinomycin at 42°C. Approximately 750,000 transposants were pooled, aliquoted, and frozen at −80°C. An aliquot was thawed and plated on LB agar containing 100 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Dark blue colonies were picked and pooled, and genomic DNA was isolated followed by cleavage with MmeI. After adapter-ligation chromosome-transposon junctions were amplified using 16 cycles of PCR. The PCR products were purified and sequenced using an Illumina MiSeq v3 (150-cycle) reagent kit, and transposon insertion sites were mapped to the B. subtilis 168 genome (NCBI NC_00964.3).
Fluorescence microscopy.
Exponentially growing cells were harvested and concentrated by centrifugation at 6,800 × g for 1.5 min, resuspended in 1/10 volume growth medium, and then immobilized on 2% (wt/vol) agarose pads containing growth medium. Fluorescence microscopy was performed on a Nikon Ti inverted microscope equipped with a Plan Apo 100×/1.4 Oil Ph3 DM phase contrast objective, an Andor Zyla 4.2 Plus sCMOS camera, and Lumencore SpectraX LED Illumination. Images were acquired using Nikon Elements 4.3 acquisition software. Propidium iodide (PI) was added at a final concentration of 5 μM. Venus was imaged using a Chroma ET filter cube for yellow fluorescent protein (49003) with an exposure time of 800 ms; mTagBFP was visualized using a Chroma ET filter cube for 4′,6-diamidino-2-phenylindole (DAPI) (49000) with an exposure time of 800 ms; PI was visualized using a Chroma ET filter cube for mCherry (49008) with an exposure time of 100 ms; HADA was visualized using a Chroma ET filter cube for DAPI (49000) with an exposure time of 500 ms. Image processing was performed using Metamorph software (version 7.7.0.0) and Oufti (33) was used for quantitative image analysis.
Immunoblot analysis.
Immunoblot analysis was performed as described previously (34). Briefly, for each culture, 1 ml of cells, normalized to OD600 = 0.5, was collected and the cell pellet resuspended in 50 μl lysis buffer (20 mM Tris [pH 7.0], 10 mM MgCl2, 1 mM EDTA, 1 mg/ml lysozyme, 10 μg/ml DNase I, 100 μg/ml RNase A, 1× protease inhibitor cocktail (Roche)). The cells were incubated at 37°C for 10 min followed by addition of an equal volume sample buffer (0.25 M Tris [pH 6.8], 4% SDS, 20% glycerol, 10 mM EDTA) containing 10% β-mercaptoethanol. Samples were heated for 15 min at 65°C prior to loading. Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto nitrocellulose membranes (Thermo) and blocked in 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) with 0.5% Tween 20. The blocked membranes were probed with anti-His (1:4,000) (Genscript) or anti-ScpB (1:10,000) (34) diluted into 3% BSA in 1× PBS with 0.05% Tween 20. The anti-His antibody was detected using anti-mouse IgG (Bio-Rad); anti-ScpB was detected using anti-rabbit IgG (Bio-Rad), and the Clarity Western ECL Blotting Substrate chemiluminescence reagent as described by the manufacturer (Bio-Rad). Signal was detected using an Azure 400 Imager (Azure Biosystems).
Growth curves.
Exponential cultures of indicated strains grown in LB medium at 37°C were back-diluted to OD600 = 0.02 in 96-well, clear, flat-bottom plates (Corning). Plates were incubated in an Infinite M Plex microplate reader (Tecan) at 37°C with shaking, and growth was monitored by measuring OD600 every 5 min. After 1 h, rCwlO was added (70 μg/ml final) or lytE was induced (50 μM IPTG), and incubation was resumed. The figures that report growth curves are representative of experiments that were performed on at least two independent samples.
Purification of recombinant CwlO (rCwlO).
rCwlO lacking its N-terminal coiled coil domain was expressed in E. coli BL21(DE3) ΔfhuA (NEB) using pJM63. Cells were grown in 1 L Terrific Broth (TB) supplemented with 100 μg/ml ampicillin at 37°C to OD600 = 0.5. Cultures were allowed to equilibrate to room temperature for 30 min and then transferred to 18°C. His6-SUMO-CwlOΔcc expression was induced with 1 mM IPTG for 18 h. Cells were collected by centrifugation, resuspended in 50 ml Buffer A (20 mM Tris-HCl [pH 7.4], 500 mM NaCl, 20 mM imidazole, and 2× complete protease inhibitor tablets; Roche), and stored at −80°C. The cell suspension was thawed on ice and lysed by two passes through a cell disruptor (Constant Systems Ltd.) at 25,000 lb/in2. The lysate was clarified by ultracentrifugation at 35,000 rpm for 30 min at 4°C. The supernatant was mixed with 1 ml Ni2+-NTA resin (Qiagen) and rolled for 1 h at 4°C. The suspension was loaded into a 10 ml column (Bio-Rad), washed twice with 4 ml Buffer A, and eluted with 2.5 ml Buffer B (20 mM Tris-HCl [pH 7.4], 500 mM NaCl, 300 mM imidazole). 10 μl of purified His6-Ulp1 (1.25 mg/ml) was added to the eluate, and the mixture was dialyzed into storage buffer (20 mM Tris-HCl [pH 8], 250 mM NaCl, 10% glycerol, 1 mM 1,4-dithiothreitol [DTT]) overnight at 4°C. The next morning 10 μl more His6-Ulp1 was added to the dialysate and incubated for 1 h at 30°C. The dialysate was mixed with 1 ml of Ni2+-NTA resin for 1 h at 4°C to remove free His6-Ulp1 and His6-SUMO. The suspension was loaded onto a column and the CwlOΔcc-containing flowthrough was collected, aliquoted, and stored at −80°C.
Sacculus preparation.
B. subtilis cells harboring Phyperspank-pdaC were grown in 100 ml LB at 37°C to an OD600 of ∼0.2 and HADA (Tocris Bioscience) was added to a final concentration of 25 μM. After 30 min of growth, the culture was split between two flasks and one was induced with 25 μM IPTG. After 1 h, cultures were normalized to OD600 = 0.5, pelleted, resuspended in 1 ml 0.1 M Tris-HCl [pH 7.5], with 2% SDS (wt/vol) and boiled for 2 h. The samples were cooled to room temperature and incubated with proteinase K solution (Invitrogen) at a final concentration of 0.4 mg/ml at 50°C for 1 h. Sacculi were pelleted at 20,000 × g and washed five times with 1 ml ddH2O until free of SDS. The sacculi were then subjected to acid hydrolysis by suspension in 1 ml 1 M HCl at 37°C for 4 h. The sacculi were then washed five times with 1 ml ddH2O. Sacculus pellets, each containing 100 mg of peptidoglycan, were stored at −80°C.
Sacculus cleavage assay.
B. subtilis sacculus aliquots were resuspended in cleavage buffer (25 mM morpholineethanesulfonic acid [MES; pH 5.5], 1 mM DTT) to a final concentration of 2.5 mg/ml and dispersed in a water bath sonicator for 30 min. rCwlO (CwlOΔcc) was added to sonicated sacculi at a final concentration of 5 μg/ml and incubated while shaking at 37°C. Samples were removed at the indicated time points and imaged by fluorescence microscopy.
Strain and plasmid construction: insertion-deletion mutants.
Insertion-deletion mutants were generated in PY79 using genomic DNA from the Bacillus knockout (BKE) collection (35) or by isothermal assembly (36) of PCR products followed by direct transformation into B. subtilis. All BKE mutants were back-crossed twice into B. subtilis PY79. All deletions were confirmed by PCR. Oligonucleotide primers were used to make strains as follows: ΔcwlO::cat and ΔcwlO::kan strains, oJM36/37, oJM28/29, and oJM38/39; ΔftsEX::cat strain, oJM54/55, oJM28/29, and oJM56/57; ΔlytE::kan strain, oJM40/41, oJM28/29 and oJM42/53; ΔpdaC::erm, ΔctaO::erm, ΔcotT::erm, ΔwalH::erm, ΔwalI::erm and ΔwalJ::erm strains, BKE collection). Antibiotic cassettes were amplied with oJM3/4 or oJM28/29.
bGD919 [ΔpdaC::erm yhdG::PpdaC-pdaC-His6 (kan)]. yhdG::PpdaC-pdaC-His6 could not be propagated in E. coli due to toxicity. The strain was generated by direction transformation of a 2-piece isothermal assembly reaction containing (i) a PCR product of the pdaC gene and its native promoter (oligonucleotide primers oGD521/525 and PY79 genomic DNA) and (ii) pCB37 digested with EcoRI and XhoI. pCB37 is an ectopic integration vector for insertions at the yhdG locus (R. Barajas and D. Z. Rudner, unpublished data). The resulting reaction product was directly transformed into bGD170 to generate strain bGD919. The construct was PCR amplified and sequence confirmed. bGD965 [pdaC-His6 (kan)] was generated by direct transformation of a 2-piece isothermal assembly reaction containing (i) a PCR product from bGD919 that contained the 3′ half of the pdaC gene including a C-terminal 6-histidine tag and the kanamycin cassette (oligonucleotide primers oGD517/571 and genomic DNA from bGD919) and (ii) the 1.5 kb region directly downstream of pdaC (oligonucleotide primers oGD572/573 and PY79 genomic DNA). The resulting reaction was directly transformed into PY79 to generate bGD965. The construct was PCR amplified and sequence confirmed. bGD950 [yvbJ::Phyperspank-(optRBS)-pdaC-His6 (spec)] was generated by direct transformation of a 2-piece isothermal assembly reaction containing (i) a PCR product containing an optimized RBS and the pdaC gene with a C-terminal 6-histidine tag (oligonucleotide primers oGD509/565 from genomic DNA of bGD919) and (ii) pCB126 digested with HindIII and SpeI. pCB126 is an ectopic integration vector for insertions at the ycgO locus (Barajas and Rudner, unpublished). The resulting reaction was directly transformed into BDR3899 (yvbJ::cat) selecting for Specr and screening for Cms. The resulting construct was PCR amplified and sequence confirmed. bGD729 [ycgO::PyocH-(optRBS)-lacZ (kan)] was generated by direct transformation of a two-piece isothermal assembly reaction containing (i) a PCR product containing the PyocH promoter, an optimized RBS, and the lacZ gene (oligonucleotide primers oGD487/486 from genomic DNA of bYB1344 [7] and (ii) pCB41 [ycgO::kan] digested with EcoRI and BamHI). pCB41 is an ectopic integration vector for insertions at the ycgO locus (Barajas and Rudner, unpublished). The resulting reaction was directly transformed into bDR2036 [ycgO::cat] selecting for Kanr and screened for Cms. The resulting construct was PCR amplified and sequence confirmed. pGD179 [yvbJ::Phyperspank-(optRBS)-pdaC (spec amp)] was generated in a two-piece isothermal assembly reaction containing (i) a PCR product containing pdaC and an optimized RBS (oligonucleotide primers oGD509/510 and PY79 genomic DNA) and (ii) pCB126 [yvbJ::Phyperspank (spec)] digested with HindIII and SpeI. pCB126 is an ectopic integration vector with Phyperspank for insertions at the yvbJ locus (Barajas and Rudner, unpublished). The resulting construct was sequence confirmed. pGD187 [ycgO::Phyperspank-(optRBS)-pdaC (erm amp)] was generated in a two-piece isothermal assembly reaction containing (i) a PCR product containing pdaC and an optimized RBS (oligonucleotide primers oGD509/510 and PY79 genomic DNA) and (ii) pCB089 digested with HindIII and SpeI. pCB089 is an ectopic integration vector with Phyperspank for insertions at the yvbJ locus (Barajas and Rudner, unpublished). The resulting construct was sequence confirmed. pGD196 [ycgO::Phyperspank-(optRBS)-pdaC(D285A) (erm amp)] was generated in a three-piece isothermal assembly reaction containing (i) a PCR product containing an optimized RBS and amino acids 1 to 285 of pdaC including a point mutation to create D285A (oligonucleotide primers oGD509/541 and PY79 genomic DNA), (ii) a PCR product containing amino acids 285 to 474 of pdaC including a point mutation to create D285A (oligonucleotide primers oGD540/510 and PY79 genomic DNA), and (iii) pC0B89 digested with HindIII and SpeI. The resulting construct was sequence confirmed. pGD197 [ycgO::Phyperspank-(optRBS)-pdaC(H427A) (erm amp)] was generated in a three-piece isothermal assembly reaction containing (i) a PCR product containing an optimized RBS and amino acids 1 to 427 of pdaC including a point mutation to create H427A (oligonucleotide primers oGD509/543 and PY79 genomic DNA), (ii) a PCR product containing amino acids 427 to 474 of pdaC including a point mutation to create H427A (oligonucleotide primers oGD542/510 and PY79 genomic DNA), and (iii) pCB089 digested with HindIII and SpeI. The resulting construct was sequence confirmed. pGD203 [yvbJ::Phyperspank-(optRBS)-pdaC(D285A)-6×His (spec amp)] was generated in a three-piece isothermal assembly reaction containing (i) a PCR product containing an optimized RBS and amino acids 1 to 285 of pdaC including a point mutation to create D285A (oligonucleotide primers oGD509/541 and PY79 genomic DNA), (ii) a PCR product containing amino acids 285 to 474 of pdaC including a point mutation to create D285A (oligonucleotide primers oGD540/565 from genomic DNA of bGD919), and (iii) pCB126 digested with HindIII and SpeI. The resulting construct was sequence confirmed. pGD204 [yvbJ::Phyperspank-(optRBS)-pdaC(H427A)-6×His (spec amp)] was generated in a three-piece isothermal assembly reaction containing (i) a PCR product containing an optimized RBS and amino acids 1 to 427 of pdaC including a point mutation to create H427A (oligonucleotide primers oGD509/543 and PY79 genomic DNA), (ii) a PCR product containing amino acids 427 to 474 of pdaC including a point mutation to create H427A (oligonucleotide primers oGD542/565 from genomic DNA of bGD919), and (iii) pCB126 digested with HindIII and SpeI. The resulting construct was sequence-confirmed.
Strains containing [ytoI::Pveg-(optRBS)-mTagBFP (kan)] were generated using genomic DNA from BDR2678. This strain was built by direct transformation of a two-piece ligation reaction containing (i) an EcoRI-BamHI digestion product from pER083 [sacA::Pveg-(optRBS)-mTagBFP (phleo)] containing the Pveg promoter, an optimized RBS, and the mTagBFP gene and (ii) pBB289 digested with EcoRI and BamHI. pBB289 [ytoI::kan] is an ectopic integration vector for insertions at the ytoI locus (B. Burton and D. Z. Rudner, unpublished data). The resulting reaction was directly transformed into BDR2257 [ytoI::cat] selecting for Kanr and screening for Cms. pIR242 [Himar1C9 IR-spec Ppen-IR terminators (erm amp)] was generated in a two-way ligation with pWX642 digested with PstI and SpeI, and a gBlock (IDT) containing the Ppen promoter, a mariner inverted repeat insertion element, the rrnB T1 and T2 terminators, and the lambda t0 terminator, amplified using oIR541 and oIR542. The resulting plasmid was sequence confirmed. pWX642 [Himar1C9 IR-spec-IR (erm) (amp)] contains the repG(Ts) origin, ermr cassette and Himar1C9 from pMarA (32) and also contains the Specr cassette flanked by inverted repeats with one harboring an MmeI site. pWX642 [pACYC MmeI-TnKRM (spec erm amp)] was generated in three steps. First, an MmeI site was introduced into the mariner-based transposon TnKRM (37) to generate pWX634. This was achieved by annealing oWX1154 and oWX1155 and ligating the linker into pDP383 (37) cut with EagI and HindIII. The insert was sequence confirmed using oML78 and oWX1156. Second, the fragment containing the HiMar transposase, repG(Ts), and erm from pKB176 (37) were cloned into a pACYC-based low-copy-number plasmid by isothermal assembly of two PCR products: (i) the HiMar transposase, repG(Ts), erm fragment was amplified from pKB176 with oWX1159 and oWX1160; (ii) the pACYC replication origin and the ampicillin resistance gene were amplified from pWX294 with oWX1157 and oWX1158. Third, the MmeI-TnKRM transposon from pWX634 was liberated using HindIII and PstI and cloned into pWX638 cut with HindIII and PstI. The resulting plasmid was pWX642.
ACKNOWLEDGMENTS
We thank all members of the Bernhardt-Rudner super-group past and present for helpful advice, discussions, and encouragement; Paula Montero Llopis and the HMS Microscopy Resources on the North Quad (MicRoN) core for advice on microscopy; Jeff Meisner for deletion strain construction; Eammon Riley for Pveg-mTagBFP; Marek Basler for mTagBFP plasmid.
Support for this work comes from the NIH grants GM086466, GM127399, U19 AI109764 (D.Z.R.), F32AI36431 (JFK), 5T32AI132120 (G.S.D.), GM141242 (X.W.).
Footnotes
Supplemental material is available online only.
Contributor Information
David Z. Rudner, Email: rudner@hms.harvard.edu.
Elizabeth Anne Shank, University of Massachusetts Medical School.
REFERENCES
- 1.Rohs PDA, Bernhardt TG. 2021. Growth and division of the peptidoglycan matrix. Annu Rev Microbiol 75:315–336. 10.1146/annurev-micro-020518-120056. [DOI] [PubMed] [Google Scholar]
- 2.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez-Cuevas P, Mercier R, Leaver M, Kawai Y, Errington J. 2012. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol Microbiol 83:52–66. 10.1111/j.1365-2958.2011.07920.x. [DOI] [PubMed] [Google Scholar]
- 4.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet YR, Wang X, Rudner DZ. 2019. SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet 15:e1008296. 10.1371/journal.pgen.1008296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65:180–200. 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 7.Dobihal GS, Brunet YR, Flores-Kim J, Rudner DZ. 2019. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 8. 10.7554/eLife.52088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzberg LI, Helmann JD. 2007. An antibiotic-inducible cell wall-associated protein that protects Bacillus subtilis from autolysis. J Bacteriol 189:4671–4680. 10.1128/JB.00403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto H, Hashimoto M, Higashitsuji Y, Harada H, Hariyama N, Takahashi L, Iwashita T, Ooiwa S, Sekiguchi J. 2008. Post-translational control of vegetative cell separation enzymes through a direct interaction with specific inhibitor IseA in Bacillus subtilis. Mol Microbiol 70:168–182. 10.1111/j.1365-2958.2008.06398.x. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, Sudiarta IP, Kodama T, Fukushima T, Ara K, Ozaki K, Sekiguchi J. 2012. Identification and characterization of a novel polysaccharide deacetylase C (PdaC) from Bacillus subtilis. J Biol Chem 287:9765–9776. 10.1074/jbc.M111.329490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takada H, Shiwa Y, Takino Y, Osaka N, Ueda S, Watanabe S, Chibazakura T, Su'etsugu M, Utsumi R, Yoshikawa H. 2018. Essentiality of WalRK for growth in Bacillus subtilis and its role during heat stress. Microbiology (Reading) 164:670–684. 10.1099/mic.0.000625. [DOI] [PubMed] [Google Scholar]
- 12.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukushima T, Furihata I, Emmins R, Daniel RA, Hoch JA, Szurmant H. 2011. A role for the essential YycG sensor histidine kinase in sensing cell division. Mol Microbiol 79:503–522. 10.1111/j.1365-2958.2010.07464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botella E, Devine SK, Hubner S, Salzberg LI, Gale RT, Brown ED, Link H, Sauer U, Codee JD, Noone D, Devine KM. 2014. PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol Microbiol 94:1242–1259. 10.1111/mmi.12833. [DOI] [PubMed] [Google Scholar]
- 16.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 17.Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, Devine K. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol 49:1639–1655. 10.1046/j.1365-2958.2003.03661.x. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin JR, Chang SY, Chang S. 1982. Transcriptional analyses of the Bacillus licheniformis penP gene. Nucleic Acids Res 10:3905–3919. 10.1093/nar/10.13.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szurmant H, Nelson K, Kim EJ, Perego M, Hoch JA. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J Bacteriol 187:5419–5426. 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–8. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukushima T, Tanabe T, Yamamoto H, Hosoya S, Sato T, Yoshikawa H, Sekiguchi J. 2004. Characterization of a polysaccharide deacetylase gene homologue (pdaB) on sporulation of Bacillus subtilis. J Biochem 136:283–291. 10.1093/jb/mvh151. [DOI] [PubMed] [Google Scholar]
- 22.Grifoll-Romero L, Sainz-Polo MA, Albesa-Jove D, Guerin ME, Biarnes X, Planas A. 2019. Structure-function relationships underlying the dual N-acetylmuramic and N-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC. J Biol Chem 294:19066–19080. 10.1074/jbc.RA119.009510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev 32:287–306. 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 24.Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, Foster SJ, Williamson MP. 2014. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun 5:4269. 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L, Ellermeier CD. 2016. The anti-sigma factor RsiV Is a bacterial receptor for lysozyme: Co-crystal structure determination and demonstration that binding of lysozyme to RsiV is required for sigmaV activation. PLoS Genet 12:e1006287. 10.1371/journal.pgen.1006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brott AS, Clarke AJ. 2019. Peptidoglycan O-acetylation as a virulence factor: Its effect on lysozyme in the innate immune system. Antibiotics (Basel) 8:94. 10.3390/antibiotics8030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balomenou S, Fouet A, Tzanodaskalaki M, Couture-Tosi E, Bouriotis V, Boneca IG. 2013. Distinct functions of polysaccharide deacetylases in cell shape, neutral polysaccharide synthesis and virulence of Bacillus anthracis. Mol Microbiol 87:867–883. 10.1111/mmi.12137. [DOI] [PubMed] [Google Scholar]
- 28.Bonnet J, Durmort C, Jacq M, Mortier-Barriere I, Campo N, VanNieuwenhze MS, Brun YV, Arthaud C, Gallet B, Moriscot C, Morlot C, Vernet T, Di Guilmi AM. 2017. Peptidoglycan O-acetylation is functionally related to cell wall biosynthesis and cell division in Streptococcus pneumoniae. Mol Microbiol 106:832–846. 10.1111/mmi.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rismondo J, Wamp S, Aldridge C, Vollmer W, Halbedel S. 2018. Stimulation of PgdA-dependent peptidoglycan N-deacetylation by GpsB-PBP A1 in Listeria monocytogenes. Mol Microbiol 107:472–487. 10.1111/mmi.13893. [DOI] [PubMed] [Google Scholar]
- 30.Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci USA 80:2305–2309. 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci USA 112:6437–6442. 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Breton Y, Mohapatra NP, Haldenwang WG. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl Environ Microbiol 72:327–333. 10.1128/AEM.72.1.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. 2015. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev 29:1661–1675. 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann AB, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291–305. 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozsgai ER, Blair KM, Kearns DB. 2012. Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl Environ Microbiol 78:778–785. 10.1128/AEM.07098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Tables S1 to S4. Download jb.00533-21-s0001.pdf, PDF file, 6.8 MB (6.8MB, pdf)