Abstract
A method for detection and genotyping of genital Chlamydia trachomatis infections based on omp1 gene amplification and sequencing was developed. DNA was extracted from urogenital or urine samples using a Chelex-based method, and an approximately 1,100-bp-long fragment from the omp1 gene was directly amplified and sequenced. Genotyping was performed by BLAST similarity search, and phylogenetic tree analysis was used to illustrate the evolutionary relationships between clinical isolates and reference strains. The method was used to determine the genotypes of C. trachomatis in 237 positive urogenital and/or urine specimens collected at a Swedish sexually transmitted disease clinic during 1 year. The most common genotypes corresponded to serotypes E (47%) and F (17%). The omp1 gene was highly conserved for genotype E (106 of 112 samples without any mutation) and F (41 of 42 samples without any mutation) strains but appear slightly less conserved for genotypes G (n = 6) and H (n = 6), where the sequences displayed one to four nucleotide substitutions relative to the reference sequence. Genotyping of samples collected at the follow-up visit indicated that two patients had become reinfected, while three other patients suffered treatment failure or reinfection. One woman appeared to have a mixed infection with two different C. trachomatis strains. This omp1 genotyping method had a high reproducibility and could be used for epidemiological characterization of sexually transmitted Chlamydia infections.
Sexually transmitted infection with Chlamydia trachomatis is a common treatable urogenital infection in young adults worldwide and is associated with a spectrum of clinical diseases, including urethritis and epididymitis among men and cervicitis, salpingitis, and pelvic inflammatory disease among women (4, 10, 21). In Sweden, reporting gonorrhea and syphilis has been mandatory under the Communicable Diseases Act since 1919, and reporting genital infection with C. trachomatis has been mandatory since April 1988. The incidence of genital chlamydial infections declined in all Swedish counties until 1996. It has been suggested that this decline was due to contact tracing, screening, and treatment of asymptomatic men and women (10), but data from the Swedish Institute for Infectious Disease Control show that genital infections with C. trachomatis are now again increasing. The incidence was 172 cases per 100,000 inhabitants in 1998 and 217 cases per 100,000 inhabitants in 2000 (27).
Characterization of C. trachomatis strains can provide valuable information about the variants circulating in the community, and with better knowledge of the epidemiology of Chlamydia infection, efforts against spread can probably be more effective. Serotyping with monoclonal antibodies recognizing antigenic determinants located on the major outer membrane protein (MOMP) is the reference method for typing C. trachomatis isolates (12, 29, 30). The MOMP is the immunodominant antigen of C. trachomatis and contains four variable domains (VDI to VDIV) that are flanked and interspaced by five constant domains (30). In order to study the epidemiology of C. trachomatis infections, new methods—such as PCR, restriction fragment length polymorphism, and sequencing of the omp1 gene, which encodes the MOMP protein—have recently been described (7, 9, 11, 12, 17, 20, 23, 25).
The aim of the present study was to establish a PCR method for genetic characterization of clinical C. trachomatis isolates in a Swedish population by sequence analysis of the omp1 gene.
MATERIALS AND METHODS
Clinical samples and strains.
Urogenital and/or urine samples for diagnosis of C. trachomatis were prospectively obtained from all new attendees (n = 2,195) of the Outpatient Sexually Transmitted Disease (STD) Clinic, Örebro Medical Centre Hospital, Örebro, Sweden, during 1 year (1 March 1999 to 29 February 2000). The mean age for men (n = 1,141) was 28.5 (range, 14 to 68) years, and the mean age for women (n = 1,054) was 25.7 (range, 13 to 59) years.
Urethral or endocervical specimens for tissue culture were obtained from males and females, respectively, using sterile Dacron swabs. Swabs were placed into transport medium containing sucrose-phosphate buffer, 5% fetal bovine serum, and antibiotics (2SP medium) and were directly transported to the laboratory and stored at −70°C until processed for tissue culture. At the same examination, first-void urine samples (5 to 10 ml) from both men and women were collected and stored at 2 to 8°C in a sterile screw-cap plastic tube. The first 779 urine samples were tested by the Chlamydia trachomatis Amplicor PCR (Roche Diagnostic Systems, Inc., Branchburg, N.J.), and the remaining samples were tested by the COBAS Amplicor Chlamydia trachomatis Test (Roche Diagnostic Systems) due to changed diagnostic screening PCR methods during the period of the study. All C. trachomatis-positive patients were treated with appropriate antibiotics and requested to come for a checkup visit 4 to 5 weeks after the initial sample was obtained.
A total of 240 specimens were found to be C. trachomatis positive by tissue culture and/or by the Amplicor PCR or COBAS Amplicor test. Two samples were lost, while 238 were stored at −20°C until used in the study. The first choice for the omp1 PCR was the urogenital specimen (n = 190), and the second choice was the urine sample (n = 48), if the urogenital sample was negative or not obtained. One C. trachomatis-negative patient sample per every C. trachomatis-positive sample was randomly selected each day, and these were used as negative controls in the study. Twenty-four patients who were epidemiologically highly suspected of having a C. trachomatis infection, but whose diagnostic tests for C. trachomatis were negative, were also included in the study.
The following C. trachomatis reference strains were used for optimization of PCR and DNA sequencing: serotypes A/HAR-1/OT, B/TW-5, Ba/AP-2/OT, C/UW-1/OT, D/ICCAL-8/ON, E/DK-20/ON, F/MRC-301/GU, G/IOL-238/R, H/UW-4/GCx, I/UW-12/GU, J/UW-36/GCx, K/UW-31/GCx, L1/440 Bu, L2/434 Bu, and L3/404 Bu, as well as Chlamydia pneumoniae strain IOL-207 and Chlamydia psittaci strain 6BC. A C. trachomatis serotype E strain, used as a control in the diagnostic tissue culture and PCR analyses above (provided from Department of Microbiology, Halmstad Hospital, Halmstad, Sweden), was used as a positive control in each PCR run. All the reference strains were originally from the Institute of Ophthalmology, London, United Kingdom.
Isolation of DNA.
DNA was isolated directly from the clinical samples and the reference strains using Chelex 100 resin (catalog no. CA 94547; Bio-Rad Laboratories) (28). A volume of 100 μl from the clinical specimens (in 2SP medium) for tissue culture or a pellet from 1,000 μl of urine was washed in distilled water for 30 min at room temperature and microcentrifuged for 5 min at 18,000 × g. The pellet was resuspended to a final volume of 200 μl in distilled water and mixed with 2 μl of 10-mg/ml proteinase K (catalog no. P6556; Sigma) and incubated for 1 h at 37°C. The tubes were then microcentrifuged for 5 min at 18,000 × g, and the pellet was resuspended in 200 μl of 5% Chelex 100 resin, thoroughly mixed and incubated at 56°C for 30 min, mixed again, and incubated in boiling water for 8 min. The cell debris was pelleted by centrifugation at 10,000 × g for 3 min, and the supernatant containing the DNA was withdrawn and stored at 4°C until used.
omp1 PCR.
The optimized omp1 PCR was carried out as follows. The DNA preparation (10 μl) was added to the PCR mixture (40 μl) containing 1.5 U of AmpliTaqGold DNA polymerase (PE Biosystems, Branchburg, N.J.), 1× PCR buffer (PE Biosystems), 2.0 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate (PE Biosystems), and a 1.0 μM concentration of each primer (Scandinavian Gene Synthesis AB, Köping, Sweden): primer P1 (5′-ATG AAA AAA CTC TTG AAA TCG G-3′) (7) and primer OMP 2 (5′-ACT GTA ACT GCG TAT TTG TCT G-3′) (designed from C. trachomatis strains sequenced in reference 25). The PCR mixture was overlaid with 50 μl of mineral oil and was run in a GeneAmp PCR System 9600 (PE Biosystems) at 94°C for 10 min followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min. At the end of the final cycle, an extension at 72°C for 7 min was included before storage at 4°C. A positive control (DNA of C. trachomatis serotype E) and a negative reagent control (distilled water), as well as the randomly selected negative sample controls, were included in each PCR run. The amplification product, which consisted of approximately 1,100 bp from the omp1 gene, was visualized after electrophoresis through a 1.5% agarose gel containing ethidium bromide. The DNA Molecular Weight Marker VI (Boehringer Mannheim, GmbH, Mannheim, Germany) was included in each electrophoresis.
DNA sequencing.
The PCR products were purified using the High Pure PCR product purification kit (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer's instructions. Sufficient amounts of purified DNA (1 to 4 μl) were mixed with 2 μl of a 50 μM concentration of one of the primers, S1 (5′-TTG AGT TCT GCT TCC TCC T-3′) or OMP 2 (5′-ACT GTA ACT GCG TAT TTG TCT G-3′) (Scandinavian Gene Synthesis AB), in separate reaction mixtures and sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Biosystems, Warrington, United Kingdom). The DyeEx Spin kit (catalog no. 63106; Qiagen, Hilden, Germany) was used to remove the dye terminators in the cycle sequencing reaction mixtures according to the manufacturer's instructions. The sequence of the omp1 gene was determined by using an ABI PRISM 310 Genetic Analyser (PE Biosystems). Each PCR product was sequenced twice in each direction, and this gave an overlap of about 200 bp in the middle of the omp1 gene.
BLAST and phylogenetic analyses.
In a first analysis, the individual consensus sequence (about 1,100 nucleotides) of the clinical isolates was determined by comparison to omp1 nucleotide sequences of known serovars of C. trachomatis strains using the BLAST search tool at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Phylogenetic tree analyses were used to illustrate the evolutionary relationships between clinical isolates and the following reference strains of C. trachomatis obtained from GenBank (accession numbers in parentheses): A/SA1/OT (M58938), Ba/AP-2 (AF063194), B/TW-5 (M33636), C/TW-3/OT (M17343), D/B-120 (X62918), D/B-185 (X62919), D/IC-CAL-8 (X62920), E/DK-20/ON (X52557), F/MRC-301/GU (X52080), G/UW57/Cx (AF063199), H/Wash (X16007), I/UW-12 (AF063200), J/UW36/Cx (AF063202), K/UW31/Cx (AF063204), L1/440 Bu (M36533), L2/434 Bu (M14738), and L3/404 Bu (X55700). MOMP sequences representing C. psittaci (AF131889), C. pneumoniae (L25436), and a murine variant of C. trachomatis (MoPn [M64171]) were used as outgroup sequences to root the tree. The sequences were manually aligned using BioEdit (version 5.0.0) software. Preliminary phylogenetic trees with all sequences were constructed using the DNADIST and NEIGHBOR programs in the PHYLIP (version 3.52c) package (6). A final tree with selected sequences was constructed using a parallelized version of DNAml (5, 6; A. Holmberg et al., unpublished data). Bootstrapping was performed using the SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE programs in the PHYLIP (version 3.52c) package (6). All programs were run under Linux on a custom-built Beowulf cluster, consisting of one master and four slaves (all five were 350-MHz 586 AMD-K6 PCs).
Ethics.
The research ethics committee at Örebro County Council, Örebro, Sweden, approved the study.
RESULTS
omp1 PCR.
All tested prototype isolates of serovars A to L3 of C. trachomatis were successfully amplified in the omp1 PCR, whereas the C. pneumoniae and C. psittaci strains were PCR negative. The omp1 PCR showed high concordance with the diagnostic tests for C. trachomatis, and out of 238 C. trachomatis-positive clinical samples analyzed with the diagnostic tests, 235 were found to be positive in the omp1 PCR. Consequently, three clinical samples were found to be negative in the omp1 PCR and cell culture but positive in the Amplicor PCR. Retesting with the COBAS Amplicor test showed that two of the three samples were negative, and thus these were considered false positives in the Amplicor PCR. The remaining COBAS Amplicor-positive sample was repeatedly negative in the omp1 PCR.
Samples from 2 of the 24 C. trachomatis-exposed but -negative partners were found to be positive in the omp1 PCR. When these two samples were retested with the COBAS Amplicor test, one was positive and the other was negative.
All clinical samples included in the study as negative controls (one per positive sample) were negative in the omp1 PCR.
Sequence and phylogenetic analyses.
Optimization of the DNA sequence analysis was performed using reference isolates for C. trachomatis serovars A, B, Ba, C, D, E, F, G, H, I, J, K, L1, L2, and L3. All isolates were successfully amplified and sequenced.
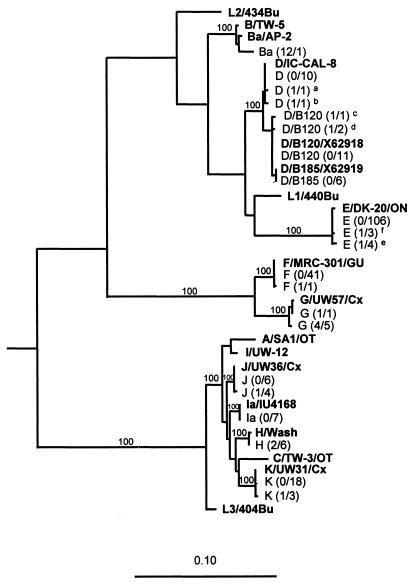
Sequence analysis of the omp1 gene from amplified DNA from the 237 clinical strains revealed that the most prevalent genotypes corresponded to C. trachomatis serovar E (47.3%), followed by F, K, D/B-120, D, J, Ia, D/B-185, G, H, and 1 strain of Ba. All nucleotide sequences were easy to read and interpret when compared by BLAST similarity search. The results are summarized in Table 1, and the evolutionary relationships between clinical isolates and reference strains are shown in Fig. 1.
TABLE 1.
omp1 genotype distribution of 237 urogenital C. trachomatis strains isolated in Örebro, Sweden, from March 1999 to February 2000
| Genotype | No. of persons infected with genotype
|
|||
|---|---|---|---|---|
| Men (n = 138) | Women (n = 99) | Total (n = 237) | %a | |
| Ba | 1 | 0 | 1 | 0.4 |
| D | 9 | 3 | 12 | 5.1 |
| D/B-120 | 8 | 6 | 14 | 5.9 |
| D/B-185 | 3 | 3 | 6 | 2.5 |
| E | 71 | 41 | 112 | 47.3 |
| F | 24 | 17 | 41 | 17.3 |
| G | 2 | 4 | 6 | 2.5 |
| H | 3 | 3 | 6 | 2.5 |
| Ia | 4 | 3 | 7 | 3.0 |
| J | 6 | 4 | 10 | 4.2 |
| K | 7 | 14 | 21 | 8.9 |
| Mixed (E + F) | 0 | 1 | 1 | 0.4 |
Values are percentages of total numbers of C. trachomatis-positive specimens (n = 237).
FIG. 1.
Phylogenetic tree showing the evolutionary relationship between the C. trachomatis nucleotide sequences of the omp1 gene from one representative of each genetic variant in the clinical material and 18 serovars of C. trachomatis available from GenBank. Sequences representing C. psittaci, C. pneumoniae, and a murine variant of C. trachomatis (MoPn) were used as outgroups to root the tree. The tree was calculated by the maximum-likelihood method (transition/transversion ratio, 1.4; global rearrangement option on) from an alignment of 1,032 aligned nucleotides. Relevant bootstrap values (as a percentage of 1,000 replicates) are also given. Reference strains are shown in boldface text. The clinical strains (n = 237) are illustrated with genotype and number of mutations per number of samples (within parentheses). Footnote letters: a, genotype D with one mutation at position 331 compared to D/IC-CAL-8; b, genotype D with one mutation at position 1092 compared to D/IC-CAL-8; c, genotype D/B120 with one mutation at position 975 compared to the strain X62918 (GenBank); d, genotype D/B120 with one mutation at position 1045 compared to the strain X62918 (GenBank); e, genotype E with one mutation at position 420 compared to E/DK20/ON; f, genotype E with one mutation at position 997 compared to E/DK20/ON.
The phylogenetic analysis of the omp1 gene showed that the serovars of C. trachomatis are segregated into three main clusters. Two clusters were characterized by small genetic distances within each cluster; one consisted of genotypes F and G, and the other consisted of A, I, J, H, C, K, and L3. The third cluster consisted of genotypes B, Ba, D, D/B-120, D/B-185, E, L1, and L2 and displayed larger genetic distances (Fig. 1).
Detailed analysis revealed that there were limited sequence differences within genotypes (Table 2 and Fig. 1). However, some differences from the reference sequences were observed, and these are listed in Table 2. Some of these sequence variants were observed in a single sample, whereas others were observed in several samples. Notably, all of the sequences within genotypes G and H differed from the respective reference sequence by one or more nucleotides (Table 2). Some of these nucleotide substitutions resulted in amino acid replacements and could thus potentially alter the function and antigenicity of the MOMP. For genotypes D, D/B-120, D/B-185, E, F, J, and K a majority of the omp1 gene sequences from the clinical samples had sequences that were identical to those reported for the respective reference isolates. However, in genotypes E, J, and K several clinical isolates displayed identical nucleotide differences relative to the reference sequence. Thus, 4 of 113 genotype E strains displayed an A→G substitution at position 420, and 3 strains displayed a G→A substitution at position 997 (compared to E/DK-20/ON [19]). The latter substitution resulted in an alanine-to-threonine amino acid replacement, whereas the former was silent. Similarly, 4 of 10 genotype J sequences displayed a silent C→T substitution at position 369 (compared to J/UW36), and 3 of 21 genotype K sequences displayed a silent C→T substitution at position 132 (compared to K/UW 31/Cx).
TABLE 2.
Mutations found in 237 clinical specimens compared to strains in GenBank
| Genotype | No.a | No. of mutations | Mutation
|
Amino acid change | GenBank accession no. | |
|---|---|---|---|---|---|---|
| Nucleotide | Position | |||||
| D | 1 (12) | 1 | G→A | 331 | R→H | X62920 |
| D | 1 | 1 | G→A | 1092 | A→T | X62920 |
| D/B-120 | 1 (14) | 1 | A→T | 975 | Silent | X62918 |
| D/B-120 | 2 | 1 | T→C | 1045 | Silent | X62918 |
| E | 4 (113) | 1 | A→G | 420 | Silent | X52557 |
| E | 3 | 1 | G→A | 997 | A→T | X52557 |
| F | 1 (42) | 1 | A→T | 1409 | Silent | X52080 |
| J | 4 (10) | 1 | C→T | 369 | Silent | AF063202 |
| K | 3 (21) | 1 | C→T | 132 | Silent | AF063204 |
| H | 6 (6) | 2 | A→G | 440 | D→S | X16007 |
| C→T | 1018 | Silent | X16007 | |||
| G | 1 (6) | 1 | T→G | 1003 | S→A | AF063199 |
| G | 5 | 4 | T→A | 228 | Silent | AF063199 |
| G→A | 487 | G→S | AF063199 | |||
| G→C | 700 | E→Q | AF063199 | |||
| T→A | 1003 | S→T | AF063199 | |||
Number of clinical strains with mutations. The values in parentheses are the total numbers of strains of the same genotype.
As shown in Table 3, three women and two men were found to be C. trachomatis positive at follow-up visits 1 to 5 months after treatment of their initial infections. The omp1 sequence of isolates obtained from the initial sample was compared to those obtained from the follow-up samples. The sequence analysis suggested that a reinfection had occurred in two of these five individuals. Thus, one woman was found to have C. trachomatis genotype K sequence in the initial endocervical sample and a genotype D/B-185 sequence 1 month later. Similarly, in the initial urethral specimen from one man and also in the second sample (received 2 months later) a genotype E sequence was found, while the third urethral sample, 5 months after the initial sample, contained genotype D/B 120. The remaining three individuals showed no change in genotype, which suggests either treatment failure or reinfection with the same genotype. Further on, one woman showed evidence of double infection, since she was found to have C. trachomatis genotype E in an endocervical sample and genotype F in the urine sample (Table 1).
TABLE 3.
Patients still Chlamydia positive at follow-up visit after initial antibiotic treatment
| Patient sex | Genotype in initial sample | Genotype after:
|
||
|---|---|---|---|---|
| 1 mo | 2 mo | 5 mo | ||
| Male | E | NDa | E | D/B-120 |
| Female | K | D/B-185 | Negative | ND |
| Male | E | E | ND | ND |
| Female | K | K | Negative | ND |
| Female | D/B-120 | D/B-120 | Negative | ND |
ND, not done (diagnostic samples not obtained).
The epidemiological information suggested that eight patients had been infected abroad. One woman and one man, both infected with genotype E, were likely to have acquired their infections in Bulgaria. Like most other genotype E sequences these two omp1 gene sequences were identical to that of the reference strain. The other five patients infected abroad included one genotype Ia infection from the United States, one genotype F infection from Greece, one genotype D/B-120 infection from Austria, one genotype D infection from Switzerland, one genotype D infection from Norway, and one genotype Ba infection from Thailand. Interestingly, this genotype Ba infection was the only genotype Ba sample in the entire study.
DISCUSSION
In this study we have established a method for PCR amplification and sequence analysis of the omp1 gene of C. trachomatis from clinical specimens. Our omp1 PCR worked successfully with both urogenital samples and urine samples and showed no cross-reactions with C. pneumoniae or C. psittaci or other false-positive reactions. All but 1 of 238 samples that were positive by the diagnostic Chlamydia tests were positive in the omp1 PCR. One reason for this false-negative result could be that the omp1 gene is only present in one copy per organism, whereas the plasmid that is targeted by the diagnostic tests is present in approximately 10 copies per organism (13, 18, 24). However, the omp1 PCR was also positive in 2 of 24 diagnostic test negative samples from individuals epidemiologically highly suspected of having a C. trachomatis infection. One of these two samples was positive after retesting by the diagnostic test, perhaps due to the freeze-thawing, as recently described (3). The remaining sample, which was found positive in the omp1 PCR only, could contain a plasmid-free variant of C. trachomatis (1).
C. trachomatis in these Swedish clinical isolates was most frequently genotyped as E (47%) or F (17%). Also relatively prevalent were omp1 genotypes D, D/B-120, J, and K (5 to 10%), whereas the remaining genotypes (Ba, D/B-185, G, H, and Ia) were more infrequently encountered. The genotype distribution in our 237 clinical specimens was broadly similar to the serovar distributions reported previously (15, 20). There was a high level of conservation of the omp1 gene in infections caused by genotype E, of which 106 out of 112 had 100% similarity to the strain E/DK-20/ON. These findings are in agreement with those of Rodriguez et al. (22), who reported a high level of conservation of E strains from different geographic origins. However, we found minor sequence variations within the different genotypes in our clinical material. Some of these genetic variants were detected only in a single sample, which makes it difficult to exclude the possibility that they represent sequencing artifacts, but most of them were independently detected in two or more samples, which strongly indicates that they were accurate. Thus, 4 of 10 clinical genotype J strains were found to have one identical substitution, but at a different position than that described by Morré et al. (14). Similarly, five of six of genotype G sequences had identical substitutions that were different from the substitutions reported previously (14, 16), and all six genotype H sequences harbored two identical mutations. It has been speculated that such omp1 genovariants occur as a result of point mutations and recombination events selected by immune pressure (8). However, several of the nucleotide substitutions that we detected were synonymous (i.e., silent), which suggests that they were evolutionarily neutral.
The phylogenetic tree analysis of all the genotype sequences found indicates that there is no simple correlation between the disease manifestation and omp1 gene phylogeny. The omp1 sequences of genotype H, I, J, and K, which were represented in our material from patients with symptomatic or subclinical genital infections, were closely related to each other. However, they were also closely related to genotypes A and C, which are associated with trachoma, and genotype L3, which is associated with lymphogranuloma venerum. These observations are broadly the same as those reported by Stothard et al. when they compared omp1 sequences from clinical strains from the United States to sequences registered in GenBank (26). Few studies have addressed the possible correlation between specific serovars or genotypes and disease manifestations and severity of disease. However, in a recent Finnish serological study it was stated that serovar G could be associated with subsequent development of cervical squamous cell carcinoma (2). This finding is interesting and requires further study, especially due to the fact that some genotype G sequences in our study differ from the prototype sequence by three amino acids (Table 2).
Genotyping was also useful in the follow-up of the Chlamydia-infected patients, and five patients were still C. trachomatis positive at the follow-up visit. In two of these patients, the initial sample and follow-up sample showed different omp1 genotypes, which indicates that new sexual partners had reinfected these patients. In the other three patients the genotypes were identical in the initial and follow-up samples, suggesting treatment failure or reinfection with the same genotype. Furthermore, one woman was found to have a mixed infection with two genotypes of C. trachomatis, and she had had two sexual partners, each carrying one of these different genotypes.
In conclusion, we have established a sensitive and relatively simple method for the genotyping of C. trachomatis strains in clinical samples based on sequencing of the omp1 gene. Genotypes E and F dominated in our Swedish material, and the individual sequences were stable and showed limited variation. This omp1 genotyping method provided interesting results concerning double infections and reinfections and could be useful for epidemiological characterization of circulating C. trachomatis strains in the community.
ACKNOWLEDGMENTS
We thank the staff at the Outpatient Sexually Transmitted Disease Clinic and at the Department of Clinical Microbiology and Immunology for all effort and positive attitude shown during the study.
This work was supported by grants from the Örebro Medical Research Foundation, Örebro Medical Centre Hospital, and The National Institute for Public Health, Stockholm, Sweden.
REFERENCES
- 1.An Q, Radcliffe G, Vassallo R, Buxton D, O'Brien W J, Pelletier D A, Weisburg W G, Klinger J D, Olive D M. Infection with a plasmid-free variant Chlamydia related to Chlamydia trachomatis identified by using multiple assays for nucleic acid detection. J Clin Microbiol. 1992;30:2814–2821. doi: 10.1128/jcm.30.11.2814-2821.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antilla T, Saikku P, Koskela P, Bloigu A, Dillner J, Ikäheimo I, Jellum E, Lehtinen M, Lenner P, Hakulinen T, Närvänen A, Pukkala E, Thoresen S, Youngman L, Paavonen J. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA. 2001;285:47–51. doi: 10.1001/jama.285.1.47. [DOI] [PubMed] [Google Scholar]
- 3.Berg E S, Ånestad G, Moi H, Storvold G, Skaug K. False-negative results of a ligase chain reaction assay to detect Chlamydia trachomatis due to inhibitors in urine. Eur J Clin Microbiol Infect Dis. 1997;16:727–731. doi: 10.1007/BF01709252. [DOI] [PubMed] [Google Scholar]
- 4.Cates W J, Jr, Wassenheit J N. Genital chlamydial infections: epidemiology and reproductive sequelae. Am J Obstet Gynecol. 1991;164:1771–1781. doi: 10.1016/0002-9378(91)90559-a. [DOI] [PubMed] [Google Scholar]
- 5.Ceron C, Dopazo J, Zapata E L, Carazo J M, Trelles O. Parallel implementation for DNAml program on message-passing architectures. Parallel Comput. 1998;24:701–716. [Google Scholar]
- 6.Felsenstein J. PHYLIP: phylogenetic inference package, version 3.52.c. Seattle: University of Washington; 1993. [Google Scholar]
- 7.Frost E H, Deslandes S, Veilleux S, Bourgaux-Ramoisy D. Typing Chlamydia trachomatis by detection of restriction fragment length polymorphism in the gene encoding the major outer membrane protein. J Infect Dis. 1991;163:1103–1107. doi: 10.1093/infdis/163.5.1103. [DOI] [PubMed] [Google Scholar]
- 8.Hayes L J, Yearsley P, Treharne J D, Ballard R A, Fehler G H, Ward M E. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venerum isolates of Chlamydia trachomatis. Infect Immun. 1994;62:5659–5663. doi: 10.1128/iai.62.12.5659-5663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaltenboeck B, Kousoulas K G, Storz J. Two-step polymerase chain reactions and restriction endonuclease analyses detect and differentiate ompA DNA of Chlamydia spp. J Clin Microbiol. 1992;30:1098–1104. doi: 10.1128/jcm.30.5.1098-1104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamwendo F, Forslin L, Bodin L, Danielsson D. Programmes to reduce pelvic inflammatory disease—the Swedish experience. Lancet. 1998;351(Suppl. 3):25–28. doi: 10.1016/s0140-6736(98)90008-3. [DOI] [PubMed] [Google Scholar]
- 11.Lampe M F, Wong K G, Stamm W E. Sequence conservation in the major outer membrane protein gene among Chlamydia trachomatis strains isolated from the upper and lower urogenital tract. J Infect Dis. 1995;172:589–592. doi: 10.1093/infdis/172.2.589. [DOI] [PubMed] [Google Scholar]
- 12.Lampe M F, Suchland R J, Stamm W E. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun. 1993;61:213–219. doi: 10.1128/iai.61.1.213-219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahony J B, Luinstra K E, Sellors J W, Chernesky M A. Comparison of plasmid- and chromosome-based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J Clin Microbiol. 1993;31:1753–1758. doi: 10.1128/jcm.31.7.1753-1758.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morré S A, Ossewaarde J M, Lan J, van Doornum G J J, Walboomers J M M, MacLaren D M, Meijier C J L M, van den Brule A J C. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J Clin Microbiol. 1998;36:345–351. doi: 10.1128/jcm.36.2.345-351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morré S A, Moesvan R, Valkengoed I, Boeke J P, van Eijk J T M, Meijer C J L, van den Brule A J C. Genotyping of Chlamydia trachomatis in urine specimens will facilitate large epidemiological studies. J Clin Microbiol. 1998;36:3077–3078. doi: 10.1128/jcm.36.10.3077-3078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norum Pedersen L, Kjaer O H, Kjolseth Moller J, Falck Orntoft T, Ostergaard L. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J Clin Microbiol. 2000;38:3068–3071. doi: 10.1128/jcm.38.8.3068-3071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostergaard L. Diagnosis of urogenital Chlamydia trachomatis infection by use of DNA amplification. APMIS. 1999;107(Suppl. 89):5–36. [PubMed] [Google Scholar]
- 18.Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986;16:52–62. doi: 10.1016/0147-619x(86)90079-x. [DOI] [PubMed] [Google Scholar]
- 19.Peterson E M, Markhoff B A, de la Maza L M. The major outer membrane protein nucleotide sequence of Chlamydia trachomatis, serovar E. Nucleic Acids Res. 1990;18:3414. doi: 10.1093/nar/18.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole E, Lamont I. Chlamydia trachomatis serovar differentiation by direct sequence analyses of the variable segment 4 region of the major outer membrane protein gene. Infect Immun. 1992;60:1089–1094. doi: 10.1128/iai.60.3.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn T C, Gaydos C, Shepherd M, Bobo L, Hook III E W, Viscidi R, Rompalo A. Epidemiologic and microbiologic correlates of Chlamydia trachomatis infection in sexual partnerships. JAMA. 1996;276:1737–1742. [PubMed] [Google Scholar]
- 22.Rodriguez P, de Barbeyrac B, Persson K, Dutilh B, Bebear C. Evaluation of molecular typing for epidemiological study of Chlamydia trachomatis genital infections. J Clin Microbiol. 1993;31:2238–2240. doi: 10.1128/jcm.31.8.2238-2240.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayada C, Denamur E, Elion J. Complete sequence of the major outer membrane protein-encoding gene of Chlamydia trachomatis serovar Da. Gene. 1992;120:129–130. doi: 10.1016/0378-1119(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 24.Sriprakash K S, Macavoy E S. Characterization and sequence of a plasmid from the trachoma biovar of Chlamydia trachomatis. Plasmid. 1987;18:205–214. doi: 10.1016/0147-619x(87)90063-1. [DOI] [PubMed] [Google Scholar]
- 25.Stephens R S, Sanchez-Pescador R, Wagar E A, Inouye C, Urdea M S. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169:3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stothard D R, Boguslawski G, Jones R B. Phylogenic analyses of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swedish Institute for Infectious Disease Control. Annual report. Stockholm, Sweden: Swedish Institute for Infectious Disease Control; 2000. [Google Scholar]
- 28.Walsh S P, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 29.Wang S P, Kuo C C, Barnes R C, Stephens R S, Grayson J T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 1985;152:791–800. doi: 10.1093/infdis/152.4.791. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Y, Zhang Y X, Watkins N G, Caldwell H D. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect Immun. 1989;57:1040–1049. doi: 10.1128/iai.57.4.1040-1049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]