ABSTRACT
We described and characterized Shiga-toxin-producing Escherichia coli (STEC) strains with high levels of resistance to azithromycin isolated in France between 2004 and 2020. Nine of 1,715 (0.52%) STEC strains were resistant to azithromycin, with an increase since 2017. One isolate carried a plasmid-borne mef(C)-mph(G) gene combination, described here for the first time for E. coli. Azithromycin resistance, although rare, needs consideration, as this treatment may be useful in cases of STEC infection.
KEYWORDS: EHEC, azithromycin resistance, fosfomycin, macrolide resistance, macrolides, mef(C)-mph(G), mph(A), mph(B), phosphorylases, rifaximin
INTRODUCTION
Shiga-toxin-producing Escherichia coli (STEC) strains are major foodborne pathogens responsible for gastrointestinal diseases, ranging from diarrhea to hemolytic uremic syndrome (HUS) (1). Typical STEC isolates carry the intimin (eae gene) and the prophage-encoded Shiga-toxin (Stx). Although no clear risk factors for HUS development, except for young age, have been identified, higher levels of Stx production could be an important trigger of HUS occurrence (2). Unlike quinolones or beta-lactams, macrolides and especially azithromycin do not modify or increase Stx production in vitro (3, 4) and decrease bacterial shedding after HUS (5).
Azithromycin is one of the most prescribed antibiotics worldwide, among adults (6) and children (7), and could participate in the selection of resistant strains (8). Macrolide resistance is mainly observed in E. coli strains with phosphorylases [mph(A) to mph(O) genes (9, 10)], especially mph(A) (11). However, in STEC strains, resistance to azithromycin is poorly studied. Two studies address this subject; one, carried out in France, reported a prevalence of 0.3% (2/508 isolates in 2004 to 2014) (12), and another one, in England in 2017, reported a prevalence of 0.2% (1/430) (13).
Here, we aimed to describe and characterize STEC strains with high-level resistance to azithromycin, isolated in France between 2004 and 2020, and to assess alternative drugs in that case. To this end, we included all STEC strains collected in France between January 2004 and June 2020 by the French National Reference Center for E. coli (NRC). For azithromycin-resistant isolates, antimicrobial susceptibility testing was performed either by disk diffusion (rifaximin [40 μg; MAST] and spiramycin [100 μg; Bio-Rad]) or by using Etest (bioMérieux, France) (erythromycin, clindamycin, clarithromycin, fosfomycin, and rifampicin) (Table 1). We used the reference strain E. coli ATCC 25922 as a control. Fosfomycin MICs were interpreted using the European Committee on Antimicrobial Susceptibility Testing (EUCAST; https://mic.eucast.org/) breakpoints (14). The breakpoint of >16 mg/L has been proposed to define resistance to azithromycin in Salmonella and Shigella, and therefore, it is also often used for intestinal pathogenic E. coli (11). No resistance breakpoints are established for the other molecules (rifaximin, erythromycin, clindamycin, clarithromycin, spiramycin, and rifampicin) in E. coli.
TABLE 1.
Antibiotic susceptibility of azithromycin-resistant Shiga-toxin producing E. coli strains isolated between 2004 and 2020 in France
| Isolate | MIC (mg/L) |
Inhibition zone diam (mm) in disk diffusion |
||||||
|---|---|---|---|---|---|---|---|---|
| Azithromycin | Erythromycin | Clindamycin | Clarithromycin | Fosfomycin | Rifampicin | Rifaximin | Spiramycin | |
| 34396 | >256 | >256 | >256 | >256 | 0.75 | 16 | 12 | 6 |
| 36493 | >256 | >256 | >256 | >256 | 0.75 | 16 | 16 | 8 |
| 42514 | 256 | >256 | >256 | >256 | 0.38 | 16 | 13 | 9 |
| 43037 | 96 | >256 | >256 | >256 | 1 | 16 | 16 | 9 |
| 45195 | 256 | >256 | >256 | >256 | 0.5 | 16 | 13 | 6 |
| 45381 | 48 | >256 | >256 | >256 | 1 | 16 | 16 | 6 |
| 45466 | 64 | >256 | >256 | >256 | 12 | 16 | 17 | 8 |
| 47439 | 48 | >256 | >256 | >256 | 0.75 | 12 | 15 | 10 |
| 47750 | 48 | >256 | >256 | >256 | 0.5 | 12 | 17 | 6 |
| ATCC 25922 | 4 | 32 | >256 | 48 | 1,5 | 12 | 12 | 10 |
Azithromycin-resistant isolates were subjected to whole-genome sequencing (WGS) using Illumina, and sequences were analyzed as previously described (3, 15).
The genetic support of mef(C)-mph(G) genes was studied in E. coli isolate 45466 using long-read sequencing with Nanopore (Oxford, UK). It was performed with the rapid sequencing kit SQK-RAD and a MinION device with a R9.4 flow cell. We used Unicycler software (v0.4.0) to carry out hybrid assembly of the Illumina and Nanopore sequences (16). Sequences were analyzed with PlasmidFinder (17) and annotated with RAST software (18). Finally, we searched both for homologous plasmids from Enterobacterales and for mef(C)-mph(G) genes with 100% coverage and homology in all E. coli/Shigella sequences in the NCBI database.
Between 2004 and 2020, 1,715 STEC strains were isolated by the NRC. Azithromycin MIC50 and MIC90 were 4 mg/L and 6 mg/L, respectively (range, 0.5 to >256 mg/L; median, 4 mg/L). Nine isolates were resistant to azithromycin (MIC range, 48 to >256 mg/L; median, 64 mg/L), resulting in 0.52% resistance. Of note, only 2/807 (0.25%) azithromycin-resistant isolates were found before 2017 (in 2012 and in 2013 [12]), and the other 7 resistant isolates (7/908; 0.77%) were collected between January 2017 and June 2020 (Tables 1 and 2).
TABLE 2.
Whole-genome sequencing results of azithromycin-resistant Shiga toxin-producing E. coli strains isolated between 2004 and 2020 in France
| Isolate | Collection yr | Patient age (yr) | Clinical diseasea | Quality sequencing data |
Serotype | Sequence type | Virulence factor(s) | Macrolide resistance gene(s) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| N50 (bp) | Contig no. | Mean genome size (bp) | ||||||||
| 34396 | 2012 | 62 | HUS | 278,669 | 279 | 5,300,106 | O17/O44:H18 | 69 | Stx2d | mph(A), erm(B) |
| 36493 | 2013 | <1 | HUS | 94,290 | 408 | 5,562,733 | O26:H11 | 29 | Stx2d, Eaeβ1 | mph(A) |
| 42514 | 2017 | 28 | BD | 29,905 | 469 | 5,171,176 | O117:H7 | 6880 | Stx1a | mph(A) |
| 43037 | 2017 | 5 | HUS | 48,855 | 301 | 5,125,621 | O146:H8 | 8356 | Stx1c | mph(A) |
| 45195 | 2018 | 24 | BD | 39,047 | 332 | 5,095,835 | ND:H7 | 5292 | Stx1a | mph(A), erm(B) |
| 45381 | 2019 | 29 | BD | 52,202 | 318 | 5,272,679 | O146:H8 | 8356 | Stx1c | mph(A), mph(B) |
| 45466 | 2019 | 1 | BD | 64,220 | 472 | 5,942,917 | O26:H11 | 21 | Stx1a, Eaeβ1, EhxA | mph(G), mef(C) |
| 47439 | 2020 | 1 | HUS | 79,390 | 382 | 5,411,518 | O111:H8 | 16 | Stx1a, Stx2a, Eaeγ2, EhxA | mph(A) |
| 47750 | 2020 | 44 | BD | 70,538 | 333 | 5,642,051 | O157:H7 | 11 | Stx1a, Stx2c, Eaeγ1, EhxA | mph(B), erm(B) |
HUS, hemolytic uremic syndrome; BD, bloody diarrhea.
For azithromycin-resistant isolates, MICs of rifampicin and fosfomycin ranged from 12 to 16 mg/L (median, 16 mg/L) and from 0.5 to 12 mg/L (median, 1.25 mg/L), respectively. MICs of other macrolides (erythromycin, clindamycin, and clarithromycin) were >256 mg/L, and spiramycin zone diameter inhibition was 6 to 10 mm (median, 8 mm). Zone diameter inhibition of rifaximin was between 12 and 17 mm (median, 16 mm). Of note, 45466 had the highest fosfomycin MIC (12 mg/L; however, this MIC remains susceptible according to EUCAST criteria) (Table 1).
Resistance to azithromycin was mostly mediated by mph(A) (n = 7), either alone (n = 4) or associated with mph(B) (n = 1) or erm(B) (n = 2). One isolate carried the association mph(B)/erm(B), and one carried the rare couple mef(C)/mph(G) (strain 45466) (Table 2).
For the latter, long-read sequencing allowed us to identify a 202,201-bp plasmid (p45466-R) harboring the mef(C)-mph(G) genes with IncHI1A, IncHI1B(R27), and IncFIA(HI1) incompatibility groups (17). These genes are localized in a cassette carrying several other resistance genes conferring resistance to penicillins (blaTEM), sulfamethoxazole/trimethoprim (dhfr1 and sul2), or aminoglycosides [aph(3)-Ib and aph(6)-I]. The mef(C)-mph(G) genes are organized in tandem-pair arrangement only 5 nucleotides apart (Fig. 1 and Table 2) and are surrounded by IS6-like element IS26 family transposases and an IS91 family transposase.
FIG 1.
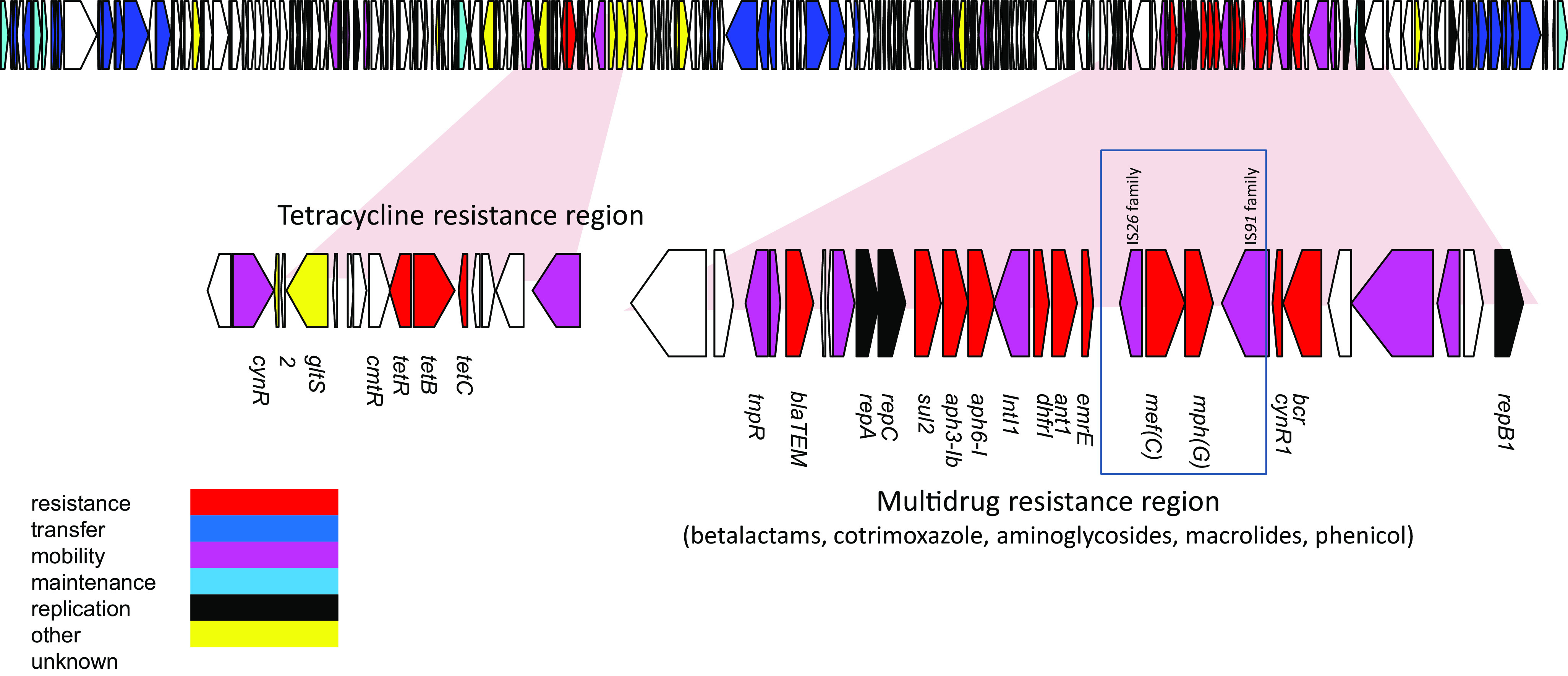
Annotation of the 202-kb plasmid p45466 from E. coli strain 45466, harboring the mef(C)-mph(G) gene pair involved in macrolide resistance.
After subjecting this plasmid to a BLAST search in the NCBI database, we found that it was close to several plasmids described in Enterobacterales, with the closest being p14ODMR, described in E. coli 14OD0056 with a 99.98% identity and a coverage of 94% (accession number MG904992.1). Interestingly, none of these plasmids carry the mef(C)-mph(G) genes.
Finally, we identified six isolates carrying the combination mef(C)-mph(G) with 100% identity and coverage that were E. coli/Shigella in the NCBI database. Of note, 2 of these also carried stx genes (Table 3).
TABLE 3.
Whole-genome sequence data (from public databases) of E.coli/Shigella isolates carrying mef(C)-mph(G) genes
| Strain | Biosample ID | Country | Source | Origin | Serotype | Sequence type | Virulence factor(s) |
|---|---|---|---|---|---|---|---|
| IHIT32077 | SAMN14279035 | Spain | Environmental | O128:H26 | 2197 | ||
| 978891 | SAMN15933666 | United Kingdom | Human | Clinical | O166:H28 | 1819 | Stx1c, EhxA |
| As Lw Down3-2 | SAMN11124839 | Germany | River sediment | Environmental | O93:H28 | 4038 | |
| B P Zu-1 | SAMN11125139 | Germany | Raw sewage | Environmental | O8:H19 | 201 | Stx2e |
| S18-17 | SAMN11125178 | Germany | River sediment | Environmental | O149:H1 | 5748 | |
| Win2012_WWKa_NEU_19 | SAMN06641869 | Germany | Wastewater inflow | Environmental | O143:H4 | 117 |
While exploring antibiotic resistance among STEC strains in France, we observed low but increasing rates of high-level resistance to azithromycin. Isolates identified in our study were from various serotypes and sequence types, showing that the diffusion is not due to the emergence of a particular clone.
Interestingly, we identified an azithromycin-resistant E. coli strain (strain 45466) carrying the gene combination mef(C)-mph(G), encoding the Mph(G) phosphotransferase associated with the Mef(C) efflux pump. To our knowledge, this is the first description of an azithromycin-resistant E. coli strain carrying this combination. The mef(C)-mph(G) gene association was first described in Photobacterium damselae, an indigenous marine bacterium known as a zoonotic pathogen (19, 20). The combination of both genes is synergic and is associated with a high azithromycin MIC (19).
The mef(C)-mph(G) genes are located on a plasmid of 202 kb whose main structure was found in several other Enterobacterales. The presence of the mef(C)-mph(G) genes in the whole-genome sequences of six E. coli/Shigella in the NCBI database supports the idea that the diffusion of these genes is not an isolated event. However, as there are only genomic data, it is not possible to determine the azithromycin MICs for these isolates and thus be sure of the expression of these genes.
The triggering role of antibiotics in HUS is still debated (21). Some bactericidal antibiotics (co-trimoxazole and fluoroquinolones, for example) increase Stx production in vitro and therefore could increase the risk of progression to HUS (22, 23). Inversely, azithromycin is associated with a decrease in the STEC inoculum and in Stx release and therefore probably decreases the risk of developing HUS (3, 22). The use of azithromycin is currently being assessed in a clinical trial concerning patients with HUS (NCT02336516). Therapeutic alternatives to azithromycin were proposed in some countries, such as rifamycins (especially rifaximin) in the United States and salts of fosfomycin in Japan (24). Both are associated with a favorable clinical outcome when administered early after the first symptoms of bloody diarrhea (24–26). Although no critical inhibition zone diameters exist for these two antibiotics, all isolates had inhibition zone diameters or MICs similar to those of wild-type E. coli ATCC 25922, suggesting susceptibility, except for the 45466 isolates, which had a higher fosfomycin MIC than other isolates.
Together, these results show that resistance to azithromycin in STEC strains remains rare but tends to increase in France; hence the need to test sensitivity before therapeutic use. Here, we describe a new combination of genes, mef(C)-mph(G), in E. coli that was initially described in a marine bacterium. In vitro data suggest that rifaximin or fosfomycin could be interesting therapeutic alternatives in STEC infections with azithromycin-resistant E. coli.
Data availability.
Raw reads have been deposited in GenBank under BioProject ID PRJNA735027.
ACKNOWLEDGMENTS
This study was supported by internal funding.
We have no conflicts of interest to declare.
REFERENCES
- 1.Williams DM, Sreedhar SS, Mickell JJ, Chan JCM. 2002. Acute kidney failure: a pediatric experience over 20 years. Arch Pediatr Adolesc Med 156:893–900. 10.1001/archpedi.156.9.893. [DOI] [PubMed] [Google Scholar]
- 2.Joseph A, Cointe A, Mariani Kurkdjian P, Rafat C, Hertig A. 2020. Shiga toxin-associated hemolytic uremic syndrome: a narrative review. Toxins 12:67. 10.3390/toxins12020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cointe A, Birgy A, Bridier-Nahmias A, Mariani-Kurkdjian P, Walewski V, Lévy C, Cohen R, Fach P, Delannoy S, Bidet P, Bonacorsi S. 2020. Escherichia coli O80 hybrid pathotype strains producing Shiga toxin and ESBL: molecular characterization and potential therapeutic options. J Antimicrob Chemother 75:537–542. 10.1093/jac/dkz484. [DOI] [PubMed] [Google Scholar]
- 4.Ohara T, Kojio S, Taneike I, Nakagawa S, Gondaira F, Tamura Y, Gejyo F, Zhang H-M, Yamamoto T. 2002. Effects of azithromycin on shiga toxin production by Escherichia coli and subsequent host inflammatory response. Antimicrob Agents Chemother 46:3478–3483. 10.1128/AAC.46.11.3478-3483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nitschke M, Sayk F, Härtel C, Roseland RT, Hauswaldt S, Steinhoff J, Fellermann K, Derad I, Wellhöner P, Büning J, Tiemer B, Katalinic A, Rupp J, Lehnert H, Solbach W, Knobloch JK-M. 2012. Association between azithromycin therapy and duration of bacterial shedding among patients with Shiga toxin-producing enteroaggregative Escherichia coli O104:H4. JAMA 307:1046–1052. 10.1001/jama.2012.264. [DOI] [PubMed] [Google Scholar]
- 6.Hicks LA, Taylor TH, Hunkler RJ. 2013. U.S. outpatient antibiotic prescribing, 2010. N Engl J Med 368:1461–1462. 10.1056/NEJMc1212055. [DOI] [PubMed] [Google Scholar]
- 7.Fleming-Dutra KE, Demirjian A, Bartoces M, Roberts RM, Taylor TH, Hicks LA. 2018. Variations in antibiotic and azithromycin prescribing for children by geography and specialty—United States, 2013. Pediatr Infect Dis J 37:52–58. 10.1097/INF.0000000000001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidman JC, Coles CL, Silbergeld EK, Levens J, Mkocha H, Johnson LB, Muñoz B, West SK. 2014. Increased carriage of macrolide-resistant fecal E. coli following mass distribution of azithromycin for trachoma control. Int J Epidemiol 43:1105–1113. 10.1093/ije/dyu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, Ruiz J. 2017. Macrolide resistance mechanisms in Enterobacteriaceae: focus on azithromycin. Crit Rev Microbiol 43:1–30. 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 10.Pawlowski AC, Stogios PJ, Koteva K, Skarina T, Evdokimova E, Savchenko A, Wright GD. 2018. The evolution of substrate discrimination in macrolide antibiotic resistance enzymes. Nat Commun 9:112. 10.1038/s41467-017-02680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes C, Ruiz-Roldán L, Mateu J, Ochoa TJ, Ruiz J. 2019. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci Rep 9:6089. 10.1038/s41598-019-42423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jost C, Bidet P, Carrère T, Mariani-Kurkdjian P, Bonacorsi S. 2016. Susceptibility of enterohaemorrhagic Escherichia coli to azithromycin in France and analysis of resistance mechanisms. J Antimicrob Chemother 71:1183–1187. 10.1093/jac/dkv477. [DOI] [PubMed] [Google Scholar]
- 13.Day M, Doumith M, Jenkins C, Dallman TJ, Hopkins KL, Elson R, Godbole G, Woodford N. 2017. Antimicrobial resistance in Shiga toxin-producing Escherichia coli serogroups O157 and O26 isolated from human cases of diarrhoeal disease in England, 2015. J Antimicrob Chemother 72:145–152. 10.1093/jac/dkw371. [DOI] [PubMed] [Google Scholar]
- 14.Leclercq R, Cantón R, Brown DFJ, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy C-J, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 19:141–160. 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones G, Pardos de la Gandara M, Herrera-Leon L, Herrera-Leon S, Varela Martinez C, Hureaux-Roy R, Abdallah Y, Nisavanh A, Fabre L, Renaudat C, Mossong J, Mattheus W, Huard C, Le Borgne C, de Valk H, Weill F-X, Jourdan-Da Silva N. 2019. Outbreak of Salmonella enterica serotype Poona in infants linked to persistent Salmonella contamination in an infant formula manufacturing facility, France, August 2018 to February. Euro Surveill 24:2019. 10.2807/1560-7917.ES.2019.24.13.1900161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, Hasman H. 2020. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2075:285–294. 10.1007/978-1-4939-9877-7_20. [DOI] [PubMed] [Google Scholar]
- 18.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonaka L, Maruyama F, Miyamoto M, Miyakoshi M, Kurokawa K, Masuda M. 2012. Novel conjugative transferable multiple drug resistance plasmid pAQU1 from Photobacterium damselae subsp. damselae isolated from marine aquaculture environment. Microbes Environ 27:263–272. 10.1264/jsme2.me11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto Y, Suzuki S, Nonaka L, Boonla C, Sukpanyatham N, Chou H-Y, Wu J-H. 2017. The novel mef(C)–mph(G) macrolide resistance genes are conveyed in the environment on various vectors. J Glob Antimicrob Resist 10:47–53. 10.1016/j.jgar.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Safdar N, Said A, Gangnon RE, Maki DG. 2002. Risk of hemolytic uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 enteritis: a meta-analysis. JAMA 288:996–1001. 10.1001/jama.288.8.996. [DOI] [PubMed] [Google Scholar]
- 22.Bielaszewska M, Idelevich EA, Zhang W, Bauwens A, Schaumburg F, Mellmann A, Peters G, Karch H. 2012. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob Agents Chemother 56:3277–3282. 10.1128/AAC.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassar FJ, Rahal EA, Sabra A, Matar GM. 2013. Effects of subinhibitory concentrations of antimicrobial agents on Escherichia coli O157:H7 Shiga toxin release and role of the SOS response. Foodborne Pathog Dis 10:805–812. 10.1089/fpd.2013.1510. [DOI] [PubMed] [Google Scholar]
- 24.Kakoullis L, Papachristodoulou E, Chra P, Panos G. 2019. Shiga toxin-induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. J Infect 79:75–94. 10.1016/j.jinf.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Ochoa TJ, Chen J, Walker CM, Gonzales E, Cleary TG. 2007. Rifaximin does not induce toxin production or phage-mediated lysis of Shiga toxin-producing Escherichia coli. Antimicrob Agents Chemother 51:2837–2841. 10.1128/AAC.01397-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang ZD, Ke S, Palazzini E, Riopel L, Dupont H. 2000. In vitro activity and fecal concentration of rifaximin after oral administration. Antimicrob Agents Chemother 44:2205–2206. 10.1128/AAC.44.8.2205-2206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads have been deposited in GenBank under BioProject ID PRJNA735027.


