ABSTRACT
In this multicentric study performed in 12 French hospitals, we reported that 26.9% (14/52) of the amoxicillin-clavulanate-resistant Proteus mirabilis isolates produced the OXA-23 carbapenemase. We found that an inhibition zone diameter of <11 mm around the amoxicillin-clavulanate disc was an accurate screening cutoff to detect these OXA-23 producers. We confirmed by whole-genome sequencing that these OXA-23-producers all belonged to the same lineage that has been demonstrated to disseminate OXA-23 or OXA-58 in P. mirabilis.
KEYWORDS: OXA-23, epidemiology, Proteae, Acinetobacter, carbapenemase, infectious clones
INTRODUCTION
Proteus mirabilis are Gram-negative rods belonging to the Morganellaceae family inside the Enterobacterales order. This species is widespread in the environment but is also part of the gastrointestinal tract (GIT) microbiota. P. mirabilis clinical isolates are mainly responsible for urinary tract infections (UTIs), including health care-associated infections (1). Intrinsically, P. mirabilis is resistant to polymyxins, nitrofurantoin and tetracyclines. It does not produce any β-lactamase and remains susceptible to all β-lactams except imipenem. Decreased susceptibility to imipenem (but not to the other carbapenems such as meropenem and ertapenem) corresponds to the expression of PBPs (penicillin-binding proteins) of low affinity for this molecule (2). Acquired resistance to β-lactams is mainly due to the acquisition of extended-spectrum β-lactamases (ESBLs), cephalosporinases and sporadically carbapenemases (3). These carbapenemases are those usually identified in Enterobacterales such as KPC (Ambler class A), metallo-β-lactamases of NDM-, VIM-, or IMP-type (Ambler class B) and carbapenem-hydrolyzing Ambler class D β-lactamases (CHDLs) of OXA-48 type. In addition, as opposed to other Enterobacterales species, the most prevalent carbapenemases reported in Acinetobacter spp. (i.e., OXA-23, OXA-24/40, and OXA-58) have also been reported in P. mirabilis: OXA-23 in France and Finland, OXA-24/40 in Algeria, and OXA-58 in Belgium and Germany (4–9). Recently, a global phylogenetic analysis demonstrated that a unique clone of P. mirabilis is responsible for the dissemination OXA-23 or OXA-58 carbapenemases in humans and animals since 1996 (10).
Despite OXA-23 and OXA-58 are carbapenemases, the production of these enzymes surprisingly does not lead to multidrug resistance in P. mirabilis. Usually, OXA-23-producing P. mirabilis isolates exhibits an AST profile with only resistance to amoxicillin, ticarcillin, and piperacillin with no recovery of susceptibility when combined with clavulanate or tazobactam. They remain susceptible to third-generation cephalosporins, and resistance to carbapenems (meropenem and ertapenem) is difficult to detect due to the poor carbapenem-hydrolyzing activity of OXA-23, OXA-24/-40, and OXA-58 and the common chromosomal localization of the carbapenemase-encoding genes in this major clone of OXA-23/OXA-58-producing P. mirabilis (3). Accordingly, this phenotype is very lucky to be confused with the high-level production of a penicillinase or the expression of a narrow-spectrum oxacillinase (e.g., OXA-1) (see Fig. S1 in the supplemental material). In addition, the frequency of the acquisition of carbapenemase from Acinetobacter in P. mirabilis remains unknown.
Here, we aimed to determine the prevalence of Acinetobacter main carbapenemases (i.e., OXA-23, OXA-58, and OXA-24/40) in P. mirabilis clinical isolates resistant to amoxicillin-clavulanate collected in a French multicentric cohort.
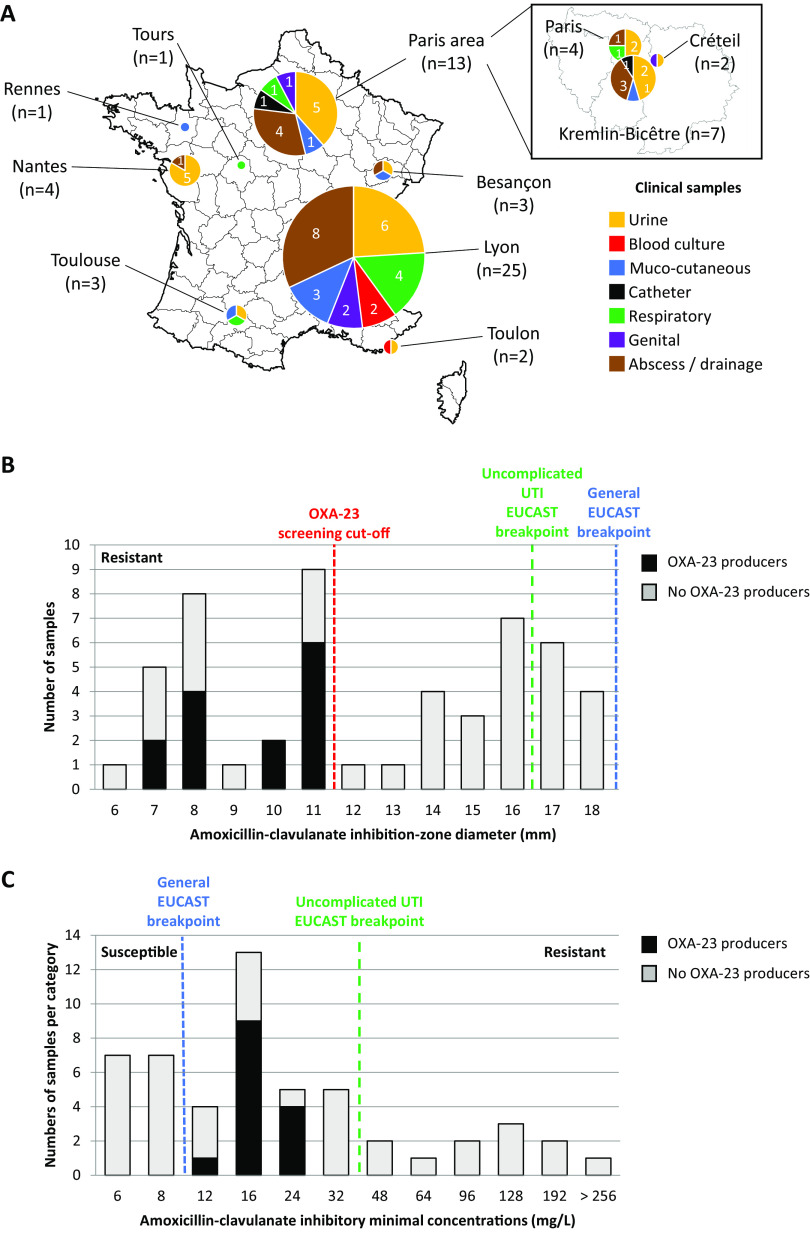
From 1 January to 31 December 2019, 139 P. mirabilis isolates recovered from human clinical samples (no screening sample) collected in 12 French hospitals were analyzed. Antimicrobial susceptibility testing was performed using the disc diffusion method on Mueller-Hinton (MH) agar (Bio-Rad, Marnes-La-Coquette, France) and interpreted according to EUCAST guidelines. Among these P. mirabilis isolates, 52 strains with an inhibition diameter zone below 16 mm for urinary samples or below 19 mm for all other clinical samples were included as amoxicillin-clavulanate-resistant isolates, according to EUCAST breakpoints. These strains were isolated from urine samples (n = 19), blood cultures (n = 3), mucocutaneous samples (n = 7), catheters (n = 1), respiratory samples (n = 7), genital samples (n = 3), and abscesses and drainage (n = 14) (Fig. 1A). One additional ESBL-producing isolate was excluded from the study.
FIG 1.
Characteristics of the 52 amoxicillin-clavulanate-resistant P. mirabilis isolates. (A) Geographic distribution and clinical samples. (B) Distribution of the amoxicillin-clavulanate zone inhibition diameters. (C) Distribution of the amoxicillin-clavulanate minimal inhibition concentrations.
All 52 amoxicillin-clavulanate-resistant P. mirabilis isolates were then screened by conventional PCR for the presence of blaOXA-23, blaOXA-24/40, or blaOXA-58 genes, as previously described (5, 11). No strain was found to be positive for blaOXA-24/40 or blaOXA-58 gene, whereas 26.9% (14/52) of amoxicillin-clavulanate-resistant P. mirabilis isolates gave a positive signal for blaOXA-23. The production of the OXA-23 carbapenemase was confirmed with an OXA-23 K-SeT immunochromatographic detection assay (Coris Bioconcept) performed as previously described (12). These OXA-23-producing P. mirabilis were isolated from urine samples (n = 7), respiratory samples (n = 2), and abscesses and drainage (n = 5). Of note, all OXA-23-producing P. mirabilis isolates had an amoxicillin-clavulanate inhibition zone diameter between 7 and 11 mm (Fig. 1B). This suggests that the screening cutoff for OXA-23 production in P. mirabilis might be a ≤11-mm inhibition zone diameter around an amoxicillin-clavulanate-containing disc (20 mg amoxicillin plus 10 mg clavulanate). The determination of amoxicillin-clavulanate MICs by Etest (bioMérieux) confirmed that all OXA-23-producing P. mirabilis isolates had MICs comprised between 12 and 24 mg/L. According to EUCAST breakpoints, these OXA-23-producing P. mirabilis isolates are categorized as susceptible (MIC ≤32 mg/L) if they are responsible for uncomplicated UTIs. Indeed, clavulanate can concentrate in urine, leading to a higher breakpoint (32 mg/L) for uncomplicated UTI compared to strains responsible for other infections (breakpoint at 8 mg/L). However, since the OXA-23 enzyme is not inhibited by clavulanate, it might be possible that such susceptible categorization leads to treatment failure. Unfortunately, we could not have access to clinical data to confirm if such treatment failure occurred. Of note, among the 38 amoxicillin-clavulanate-resistant P. mirabilis isolates that were negative for blaOXA-23, 76.3% (29/38) expressed the TEM-1 penicillinase and the narrow-spectrum oxacillinase OXA-1 (assessed by PCR and sequencing), 13.2% (5/38) were positive only for blaOXA-1, 7.9% (3/38) were positive only for blaTEM-1, and only one isolate (2.6%) was negative for both blaTEM-1 and blaOXA-1.
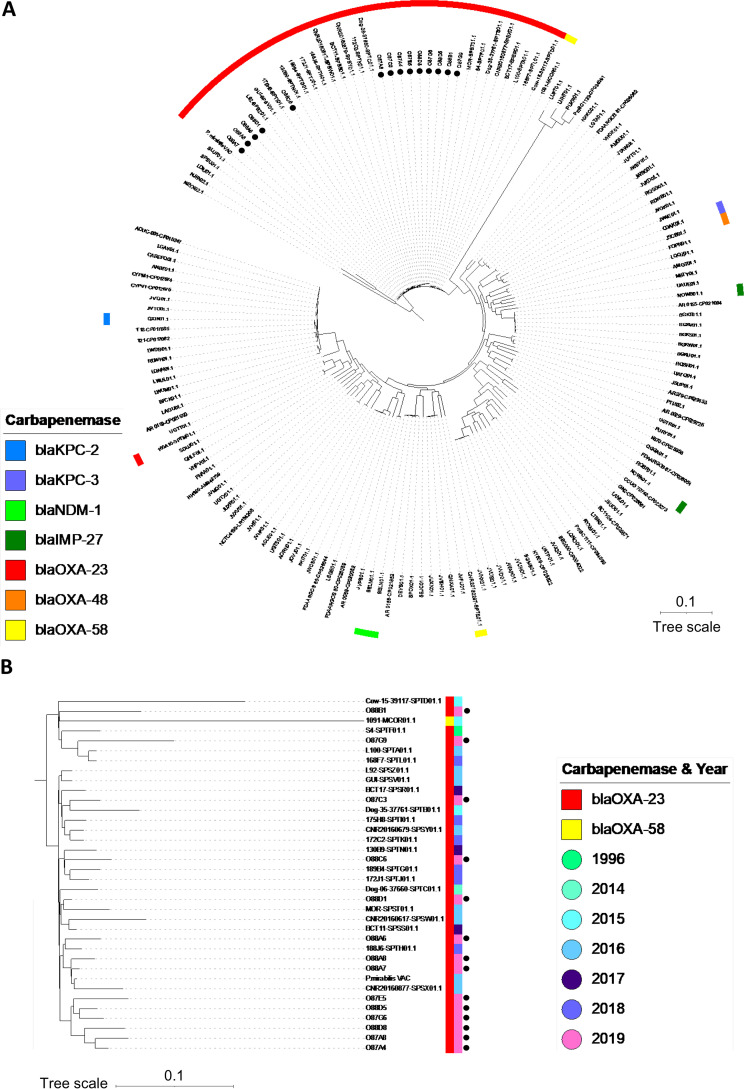
No obvious epidemiological link could be identified between the 14 OXA-23-producing P. mirabilis isolates since they were recovered in different areas. However, to assess the clonal relationship between these 14 P. mirabilis isolates, we performed a whole-genome sequencing and comparison as previously described (10). The genomes of OXA-23-producing P. mirabilis were submitted to GenBank (BioProject PRJNA780406). As previously reported (10), all OXA-23-producing P. mirabilis isolates were part of the major lineage that disseminated in France and Belgium at least since 1996 (Fig. 2).
FIG 2.
(A) Phylogenetic relationship of the 14 OXA-23-producing P. mirabilis isolates with the 145 reference genomes of P. mirabilis reported by Bonnin et al. (10). This comparison was performed on 17.83% of the genome of OXA-23-producing P. mirabilis VAC used as a reference. (B) Phylogenetic relationship of the OXA-23/OXA-58-producing P. mirabilis isolates. This comparison was performed on 57.36% of the genome of OXA-23-producing P. mirabilis VAC used as a reference. OXA-23-producing P. mirabilis from this study are marked by a black point. Scale bar on tree indicates the number of single-nucleotide polymorphisms per position of common sequences.
In conclusion, we demonstrated that nearly one-quarter of the P. mirabilis clinical isolates with an amoxicillin-clavulanate zone inhibition of <19 mm were OXA-23-producing strains. We established that an amoxicillin-clavulanate zone inhibition of <11 mm is an efficient screening cutoff to detect these OXA-23 producers. However, since currently only one clone of P. mirabilis has been reported to vehiculate blaOXA-23, such a screening cutoff might have to be adapted if the emergence of another clone is further reported. As previously reported, we demonstrated that the immunochromatographic assay OXA-23 K-Set is a useful tool to rapidly identify the production of OXA-23 by P. mirabilis. The amoxicillin-clavulanate MICs for these OXA-23-producing P. mirabilis strains are between 12 and 24 mg/L. Accordingly, these strains might be categorized as susceptible if they were considered to be responsible for uncomplicated UTIs. Since OXA-23 is not inhibited by clavulanate, clinical failure might occur. However, several therapeutic options are often possible outside the β-lactams family (fluoroquinolones, aminoglycosides, and sulfamethoxazole-trimethoprim), particularly for UTIs. In addition, OXA-23 do not hydrolyze third-generation cephalosporins and only leads to a low-level resistance that is often remain undetectable for carbapenems (ertapenem or meropenem) or piperacillin-tazobactam, suggesting a potential use of these molecules. However, complementary studies are needed to decipher this hypothesis.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P, European Network on Carbapenemases. 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 18:413–431. 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 2.Neuwirth C, Siebor E, Duez JM, Pechinot A, Kazmierczak A. 1995. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J Antimicrob Chemother 36:335–342. 10.1093/jac/36.2.335. [DOI] [PubMed] [Google Scholar]
- 3.Girlich D, Bonnin RA, Dortet L, Naas T. 2020. Genetics of acquired antibiotic resistance genes in Proteus spp. Front Microbiol 11:256. 10.3389/fmicb.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet R, Marchandin H, Chanal C, Sirot D, Labia R, De Champs C, Jumas-Bilak E, Sirot J. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob Agents Chemother 46:2004–2006. 10.1128/AAC.46.6.2004-2006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, Glaser P, Glupczynski Y, Naas T. 2017. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 61:e01697-16. 10.1128/AAC.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange F, Pfennigwerth N, Gerigk S, Gohlke F, Oberdorfer K, Purr I, Wohanka N, Roggenkamp A, Gatermann SG, Kaase M. 2017. Dissemination of blaOXA-58 in Proteus mirabilis isolates from Germany. J Antimicrob Chemother 72:1334–1339. [DOI] [PubMed] [Google Scholar]
- 7.Leulmi Z, Kandouli C, Mihoubi I, Benlabed K, Lezzar A, Rolain JM. 2019. First report of blaOXA-24 carbapenemase gene, armA methyltransferase and aac(6′)-Ib-cr among multidrug-resistant clinical isolates of Proteus mirabilis in Algeria. J Glob Antimicrob Resist 16:125–129. 10.1016/j.jgar.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Osterblad M, Karah N, Halkilahti J, Sarkkinen H, Uhlin BE, Jalava J. 2016. Rare detection of the Acinetobacter class D carbapenemase blaOXA-23 gene in Proteus mirabilis. Antimicrob Agents Chemother 60:3243–3245. 10.1128/AAC.03119-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potron A, Hocquet D, Triponney P, Plesiat P, Bertrand X, Valot B. 2019. Carbapenem-susceptible OXA-23-producing Proteus mirabilis in the French community. Antimicrob Agents Chemother 63:e00191-19. 10.1128/AAC.00191-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnin RA, Girlich D, Jousset AB, Gauthier L, Cuzon G, Bogaerts P, Haenni M, Madec JY, Couve-Deacon E, Barraud O, Fortineau N, Glaser P, Glupczynski Y, Dortet L, Naas T. 2020. A single Proteus mirabilis lineage from human and animal sources: a hidden reservoir of OXA-23 or OXA-58 carbapenemases in Enterobacterales. Sci Rep 10:9160. 10.1038/s41598-020-66161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnin RA, Nordmann P, Potron A, Lecuyer H, Zahar JR, Poirel L. 2011. Carbapenem-hydrolyzing GES-type extended-spectrum beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 55:349–354. 10.1128/AAC.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riccobono E, Bogaerts P, Antonelli A, Evrard S, Giani T, Rossolini GM, Glupczynski Y. 2019. Evaluation of the OXA-23 K-SeT immunochromatographic assay for the rapid detection of OXA-23-like carbapenemase-producing Acinetobacter spp. J Antimicrob Chemother 74:1455–1457. 10.1093/jac/dkz001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Download aac.01983-21-s0001.pdf, PDF file, 0.9 MB (938.7KB, pdf)