Abstract
Over the past decade, several Helicobacter species have been isolated from rodents. With the advent of PCR for the diagnosis of infectious agents, it has become clear that several previously uncharacterized Helicobacter species also colonize rodents. In this report, we describe a novel urease-negative helicobacter, Helicobacter typhlonius sp. nov., which was isolated from colonies of laboratory mice independently by two laboratories. Infection of immunodeficient mice by this bacterium resulted in typhlocolitis similar to that observed with other helicobacter infections. H. typhlonius is genetically most closely related to H. hepaticus. Like H. hepaticus, it is a spiral bacterium with bipolar sheathed flagella. However, this novel species contains a large intervening sequence in its 16S rRNA gene and is biochemically distinct from H. hepaticus. Notably, H. typhlonius does not produce urease or H2S nor does it hydrolize indoxyl-acetate. Compared to other Helicobacter species that commonly colonize rodents, H. typhlonius was found to be less prevalent than H. hepaticus and H. rodentium but as prevalent as H. bilis. H. typhlonius joins a growing list of helicobacters that colonize mice and are capable of inducing enteric disease in various strains of immunodeficient mice.
The genus Helicobacter has rapidly expanded since the discovery of the type species, Helicobacter pylori, in 1982. This genus currently contains 24 named species and numerous provisionally named species. It is likely that several novel Helicobacter species await discovery. Members of this genus are microaerobic, have a fusiform or curved to spiral rod morphology and are motile by flagella that vary in number and location among different species (44). All known helicobacters live in human and animal hosts, where colonization occurs primarily in the gastrointestinal tract. The type species, H. pylori, was isolated from the stomach of humans and has been associated with a variety of gastric anomalies including gastritis, peptic ulcer disease, gastric carcinoma, and gastric mucosa-associated lymphoma (25, 26, 28, 29, 48). Like H. pylori, other species of Helicobacter have also been shown to colonize the stomach and cause disease in animals. Gastric colonizers include H. felis, H. mustelae, H. acinonychis, H. bizzozeronii, H. heilmannii, H. salomonis, and a recently isolated novel Helicobacter sp. of dolphins (5, 19–21, 30). Several species of Helicobacter have been identified in rodents, including the species H. hepaticus, H. bilis, H. muridarum, H. aurati, H. cinaedi, H. cholecystus, H. trogontum, H. rodentium, and a bacterium morphologically resembling Helicobacter Flexispira taxon 8 (formerly Flexispira rappini) (9, 13, 14, 17, 24, 27, 31, 32, 36, 37). Of these, only H. muridarum and H. aurati have been demonstrated to colonize the rodent stomach. Instead, most rodent helicobacters colonize the large intestine and in some cases translocate to the liver and colonize the biliary system.
Several rodent Helicobacter spp., including H. hepaticus, H. bilis, and H. rodentium, have been associated with enterohepatic disease. H. hepaticus has been shown to cause inflammatory bowel disease and chronic active hepatitis in both immunocompetent and immunodeficient mice (2, 12, 45–47). Conversely, H. bilis and H. rodentium have only been convincingly associated with disease in immunodeficient rodents (15, 18, 38). Lesions caused by these rodent Helicobacter spp. often mimic those seen in human idiopathic enterohepatic diseases, especially inflammatory bowel diseases. Therefore, rodent helicobacter infections have attracted the attention of scientists interested in the study of these diseases. Not only have these infections been used as models for disease, but inadvertent infections by rodent Helicobacter spp. have confounded interpretation of results from established models of inflammatory bowel disease (2, 7, 10, 23). As a result, it has become increasingly important to ascertain the Helicobacter status of experimental animals prior to the initiation of studies using rodent inflammatory bowel disease models.
With the advent of molecular diagnostic techniques such as PCR designed to detect known Helicobacter species, it is becoming increasingly evident that rodents are colonized by numerous novel Helicobacter species. Our laboratories have been screening rodents by PCR and culture for several years. As a result, several uncharacterized Helicobacter spp. have been identified (L. K. Riley, unpublished results; J. G. Fox, unpublished results). Whether or not these novel species of Helicobacter cause disease awaits further systematic study of infected animals.
Recently, Fox et al. described a novel urease-negative helicobacter, designated Helicobacter sp. strain MIT 97-6810, that was isolated from interleukin-10 (IL-10)-deficient mice with typhlocolitis (10). IL-10-deficient and SCID/NCr mice experimentally infected with this Helicobacter strain also developed typhlitis, colitis, and proctitis, and experimentally infected A/JCr mice developed minimal to mild typhlitis. Shortly thereafter, Franklin et al. described a disease syndrome characterized by proliferative typhlocolitis in C.B-17 scid/scid mice naturally and experimentally infected with a novel species of Helicobacter that was provisionally named “Helicobacter typhlonicus” (16). H. typhlonicus was genetically and morphologically identical to Helicobacter sp. strain MIT 97-6810. In this report, we propose to use the more appropriate neo-Latin adjective “typhlonius” and formally name this novel helicobacter Helicobacter typhlonius. Furthermore, we demonstrate that H. typhlonius is a common intestinal colonizer of research mice.
MATERIALS AND METHODS
Bacterial isolates.
Isolates MIT 97-6810 and MIT 97-6811 were isolated from IL-10−/− knockout mice on a C57BL6/129-Ola background (10). One mouse had a rectal prolapse, while the second was clinically normal. Isolates MU 96-1 and MU 96-2 were cultured from feces of BALB/c mice (16). There was no history of clinical signs, abnormal necropsy findings, or significant histologic lesions in mice from this colony. Isolate MU 96-3 was recovered from the feces of a FOX CHASE SCID C.B-17/IcrCrl-scidBR (SCID) mouse that had been inoculated with MU 96-1 (16). Isolation procedures were similar to those previously described for the isolation of H. hepaticus and H. bilis (9, 13, 14). Briefly, fecal material was homogenized in either phosphate-buffered saline (PBS) or brain heart infusion broth containing horse serum and yeast extract. The fecal slurries were filtered through either a 0.45- or 0.80-μm (pore-size) filter and placed on plates of Trypticase soy or brucella agar that contained 5% sheep blood. In some cases, blood agar plates also contained trimethoprim, vancomycin, and polymyxin. Cultures were incubated in a microaerobic environment at 37 and 42°C for up to 2 weeks.
DNA sequencing.
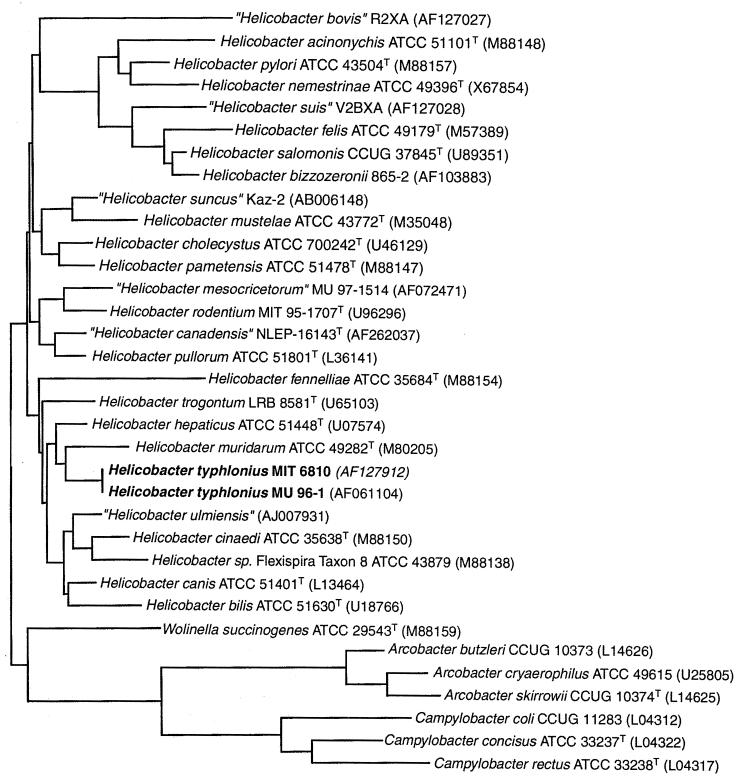
Sequence analyses of bacterial 16S rRNA genes were performed as previously described (13, 14). Briefly, bacterial DNA was extracted from samples, and sequencing templates were prepared by PCR with one of the following primer sets: (i) C70f and B37r, (ii) H35f and H587r, (iii) C70f and H676r, (iv) H276f and cg4r, and (v) H276f and p13Br (3, 13, 33, 34). Templates were purified on 3.5% polyacrylamide gels, and the gene sequence was determined by using either the Taq Dye Deoxy Termantory Cycle Sequencing Kit (Applied BioSystems, Inc., Foster City, Calif) or the TAQuence Cycle Seq Kit (U.S. Biochemicals, Cleveland, Ohio) (10, 16). Sequence analyses were performed by using the Sequence Analysis Software Package (Wisconsin Package, version 10.0; Genetics Computer Group, Inc., Madison, Wis.). The sequences (1,307 bp) of the five novel Helicobacter isolates were aligned with sequences of 32 bacteria representing formally and provisionally named species of Helicobacter, and select species of Wolinella, Campylobacter, and Arcobacter. A matrix of pairwise evolutionary distances between aligned sequences (similarity matrix) was constructed; both uncorrected distances and corrected distances made using the Jukes-Cantor method (22) were calculated. A phylogenetic tree was created by the neighbor-joining method (35, 42, 43). Sequence data used for comparisons were obtained from GenBank (accession numbers are provided in Fig. 1).
FIG. 1.
Phylogenetic tree of members of the genera Helicobacter, Campylobacter, “Flexispira,” Wolinella, and Arcobacter based on 16S ribosomal DNA sequences and prepared by using the neighbor-joining method. Phylogenetic distances between bacteria are calculated by totaling horizontal branches between bacteria. Bar, 1 nucleotide substitution per 100 nucleotides.
Specific identification of H. typhlonius by PCR.
Two PCR primer sets were generated from sequence data. Primer set 1 consisted of the forward primer JGF-F1 (5′-GAA ACT ATC ACT CTA GAG TAT G-3′) and the reverse primer JGF-R1 (5′-TGC TCC TCA TTG TAT GCC-3′). This primer set amplified a 622-bp fragment from nucleotide positions 639 to 1260 (Escherichia coli numbering). The following conditions were used: 50 μl of reaction mixture with 50 ng of template, 200 μg of bovine serum albumin ml−1, 5 pmol of primer, 1× reaction buffer (100 mM Tris-HCl, 15 mM MgCl2, 500 mM KCl; pH 8.3; Roche Molecular Biochemicals, Indianapolis, Ind.), and 2.5 U of Taq, with 40 cycles of 1-min denaturation at 94°C, 1-min annealing at 52°C, and 1-min extension at 72°C. Primer set 2 consisted of the forward primer Ht184f (5′-TTA AAG ATA TTC TAG GGG TAT AT-3′) and the reverse primer Ht640r (5′-TCT CCC ATA CTC TAG AGT GA-3′). The forward primer was designed from the intervening sequence. This primer set amplified a 455-bp fragment from base position 131 of the intervening sequence to nucleotide position 665 (E. coli numbering). The following conditions were used for this PCR: 50 μl of reaction mixture with 1.25 μg of template, 2.5 pmol of primer, 10× reaction buffer, and 1.25 U of Taq, with 45 cycles of 2-s denaturation at 94°C, 10-s annealing at 53°C, and 30-s extension at 72°C. Samples were analyzed by PCR as previously described (1, 10, 34).
Biochemical characterization.
To further characterize isolates, phenotypic tests commonly used to characterize helicobacters were performed (14). Growth was examined at 25, 37, and 42°C. Tolerances to 1% (wt/vol) glycine and 1.5% NaCl were determined as previously described (9). Bacteria were tested for urease activity by a selective rapid urea test (Remel, Lenexa, Kans.) and examined for oxidase and catalase activity by standard microbiological methods (6). Alkaline phosphatase activity and hydrolysis of indoxyl-acetate were analyzed by using the Ani-Ident system (bioMerieux Vitek, Inc., Hazelwood, Mo.). Gamma-glutamyltransferase and alkaline phosphatase activities, hydrolysis of hippurate, reduction of nitrate to nitrite, and production of hydrogen sulfide were analyzed by using the Campy identification system (bioMerieux Vitek). Reduction of nitrate was also determined by using nitrate impregnated disks (Remel). Susceptibilities to cephalothin and nalidixic acid were determined by culturing isolates in the presence of antibiotic impregnated disks (Remel).
Electron microscopy.
Bacteria were collected from blood agar plates, placed in PBS (pH 7.4), and centrifuged at 12,000 × g. Bacteria were washed, resuspended, stained with phosphotungstic acid for 20 to 30 s, and examined with either a JEOL model JEM-1 200EX or an Hitachi H-6700 transmission electron microscope.
Nucleotide sequence accession numbers.
The type strains of H. typhlonius sp. nov., MIT 97-6810 and MU 96-1, isolated from the intestinal contents of an IL-10−/− knockout mouse and a BALB/c mouse, respectively, have been deposited with The American Type Culture Collection (ATCC) and are designated strains ATCC BAA-367T and ATCC BAA-368. The 16S rRNA sequences of the type strains are available for electronic retrieval from GenBank under accession numbers AF127912 and AF061104.
RESULTS
Bacterial analyses.
Five isolates of the H. typhlonius (MIT 97-6810, MIT 97-6811, MU 96-1, MU 96-2, and MU 96-3), were examined by 16S rRNA gene sequence analysis. Sequences were obtained from a 1,614-bp fragment or a 1,448-bp fragment internal to the 1,614-bp fragment. The 1,448-bp fragments from all five isolates were 100% identical. A 166-bp intervening sequence (IVS) was identified following position 198 in all isolates. This IVS occupied the area normally occupied by a seven-base stem-loop centered on position 210 in several other species of Helicobacter. Because not all helicobacters contain an IVS, this sequence was removed for genetic comparisons. The consensus sequence from the novel Helicobacter sp. was aligned with sequences from bacteria of the genera Helicobacter, Campylobacter, and Wolinella as previously described (13, 14). A similarity matrix was generated (data not shown), and a phylogenetic tree was constructed (Fig. 1). Sequences of all known rodent helicobacters were included in this comparison. On the basis of this comparison, H. typhlonius was most closely related to H. hepaticus but was a distinct species exhibiting 97.64% similarity and containing the aforementioned intervening sequence.
Six additional isolates obtained from mice inoculated with MIT 97-6810 were analyzed by PCR with primer set 1. Isolates 98-6781 and 98-6782 were from 4-week postinoculation mice, and isolates 98-784, 98-6785, 98-7686, and 98-6787 were from 18-week postinoculation mice. All isolates were found to be positive by this PCR. DNA from H. rodentium, another urease-negative Helicobacter sp., did not amplify with this primer set.
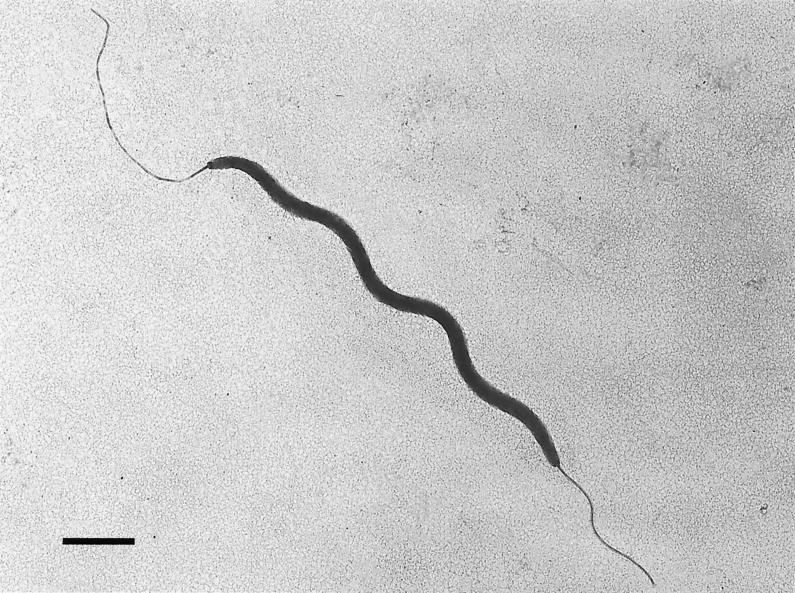
Phenotypic tests commonly used to characterize helicobacters were performed on MU 96-1, MU 96-2, MIT 97-6810, and MIT 97-6811. All isolates had identical biochemical phenotypes (Table 1). Cells were motile and helical and grew on moist blood agar as transparent pinpoint colonies. Upon ultrastructural examination, isolates MIT 97-6810 and MU 96-1 were identical. The majority of bacteria were 0.3-μm by 2- to 3-μm curved to spiral rods, with single sheathed bipolar flagella and no periplasmic fibers (Fig. 2).
TABLE 1.
Biochemical and morphologic characteristics of H. typhlonius compared to closely related and other rodent Helicobacter spp.
| Characteristic | Results witha:
|
|||||
|---|---|---|---|---|---|---|
| H. typhloniusb | H. hepaticus | H. trogontum | H. muridarum | H. rodentium | H. bilis | |
| Enzyme activities | ||||||
| Catalase | 4/4 | + | + | + | + | + |
| Urease | 0/4 | + | + | + | − | + |
| Oxidase | 4/4 | + | + | + | + | + |
| Alkaline phosphatase | 0/2 | − | − | + | − | − |
| Gamma-glutamyltransferase | 0/2 | − | + | + | − | + |
| H2S production | 0/4 | + | − | + | ND | + |
| Indoxyl-acetate hydrolysis | 0/2 | + | − | + | − | − |
| Hippurate hydrolysis | 0/4 | − | − | − | ND | ND |
| Nitrate reduction | 2/4 | + | + | − | + | + |
| Flagellar morphology | ||||||
| Periplasmic fibers | 0/2 | − | + | + | − | + |
| Number | 2 | 2 | 5–7 | 10–14 | 2 | 3–14 |
| Location | Bipolar | Bipolar | Bipolar | Bipolar | Bipolar | Bipolar |
| Growth conditions | ||||||
| 1% Glycine | 4/4 (wk)c | + | − | − | + | + |
| 1.5% NaCl | 0/4 | + | − | − | + | − |
| 25°C | 0/4 | − | − | − | − | − |
| 37°C | 4/4 | + | + | + | + | + |
| 42°C | 4/4 | − | + | − | + | + |
| Anaerobic | 0/4 | + | ND | + | + | ND |
| Aerobic | 0/2 | − | ND | − | − | ND |
| Antibiotic sensitivities | ||||||
| Nalidixic acid | S (4/4) | R | R | R | R | R |
| Cephalothin | R (4/4) | R | R | R | R | R |
Symbols: +, positive result; −, negative result; ND, not determined; R, resistant; S, susceptible. Information on H. hepaticus is based on references 4 and 9; information on H. trogontum is based on references 4 and 27; information on H. muridarum is based on references 13 and 24; information on H. rodentium is based on reference 37; and information on H. bilis is based on references 4 and 13.
Number of positive strains/number tested.
Most of the cells from isolates MU 96-1 and MU 96-2 harvested in these conditions had coccoid morphology.
FIG. 2.
Negatively stained preparation of H. typhlonius MU 96-1 demonstrating single sheathed bipolar flagella and no periplasmic fibers. Bar, 1 μm.
H. typhlonius is common in research mice.
Fecal samples submitted to the University of Missouri Research Animal Diagnostic and Investigative Laboratory for helicobacter testing were examined by PCR with primer sets designed to detect (i) all species of Helicobacter (generic test), (ii) H. hepaticus, (iii) H. bilis, (iv) H. rodentium, and (v) H. typhlonius (with primer set 2 [Ht184f and Ht640r]). Samples that were found to be positive by the generic helicobacter PCR but negative by all species tests were designated as positive for Helicobacter spp. Of 1,271 samples tested from November 1999 through April 2000, 4.88% were positive for H. typhlonius, 16.44% were positive for H. hepaticus, 4.33% were positive for H. bilis, 15.11% were positive for H. rodentium, and 10.54% were positive for Helicobacter spp.
DISCUSSION
In this report we describe the phylogenetic characterization of a novel urease-negative spiral bacterium that was isolated independently by two laboratories. This bacterium merits a formal name as a novel Helicobacter species because it is genetically distinct from other Helicobacter spp. and exhibits important biochemical differences compared to its closest relatives. We propose the name H. typhlonius because this bacteria was isolated from inflamed intestinal contents of naturally infected IL-10−/− mice and has, under defined conditions, produced inflammation of the ceca and colon of both IL-10−/− and SCID mice (10, 16).
H. typhlonius is most closely related to H. hepaticus (97.44%), and it clusters with H. muridarum and H. trogontum. While H. typhlonius is on a genetic basis closely related to these species, it also possesses a unique IVS in its 16S rRNA sequence, and it is morphologically and/or biochemically distinguishable from its closest phylogenetic relatives. Unlike H. muridarum and H. trogontum, H. typhlonius lacks the periplasmic fibers and multiple flagella and does not have gamma-glutamyltransferase activity. H. typhlonius is morphologically similar to H. hepaticus; however, unlike H. hepaticus, it does not produce H2S or hydrolyze indoxyl-acetate. Perhaps the most dramatic difference between H. typhlonius and its three closest relatives is its lack of urease activity. Preliminary data obtained by both of our laboratories suggest that H. typhlonius lacks the gene for urease (10; C. S. Beckwith, unpublished results).
The role of urease in species of Helicobacter that colonize the large intestine is unknown. Both urease-positive (H. hepaticus and H. bilis) and two urease-negative helicobacters have been shown to cause proliferative disease of the cecum and colon (10, 15, 16, 39, 46). Because the novel urease-negative helicobacter described by Shomer et al. produces significant hepatic and intestinal disease in A/JCr mice, as well as in immunodeficient mice, it is unlikely that urease plays a major role in the pathogenesis of enteric disease (40). Interestingly, only mild portal inflammation in the liver has been seen in H. typhlonius-infected immunocompromised mice (10, 16). This is in contrast to the severe necrotizing liver disease seen in SCID mice infected with either H. hepaticus and H. bilis. There was no evidence of colonization of the liver by H. typhlonius, as evidenced by uniformly negative PCR results (16). Also, H. rodentium, another urease-negative helicobacter, has not been isolated from livers nor is it associated with liver disease (37). This may be due to a lack of undetermined virulence factors in both of these urease-negative Helicobacter spp.
Several other species of Helicobacter also lack urease activity. These include H. pullorum, H. canis, H. fennelliae, H. cinaedi, and H. canadensis (8, 11, 41, 44). Interestingly, all of have been isolated from the feces of diarrheic humans, supporting the premise that urease is not critical to the pathogenesis of enteric disease. Also, H. pullorum, H. cinaedi, and H. canis have been isolated from inflamed livers (11, 41). The only other known rodent helicobacters that lack urease are an unnamed novel species of Helicobacter that can cause cholangiohepatitis and inflammatory bowel disease (40) and H. rodentium (37). Coinfection with H. rodentium and H. bilis has been associated with diarrhea in SCID mice, suggesting that, like H. typhlonius, H. rodentium may be pathogenic to immunocompromised mice (38). Although H. typhlonius is genetically distinct from H. rodentium (94.28% identity), these bacteria are biochemically and phenotypically very similar. Both agents produce catalase, reduce nitrate, and have bipolar flagella, and both are negative for indoxyl-acetate hydrolysis and gamma-glutamyltranspeptidase and alkaline phosphatase activities. Like H. rodentium, H. typhlonius grows in the presence of 1% glycine, but the majority of the bacteria grown in these conditions are degenerative coccoid forms. The only other known phenotypic differences are growth in anaerobic conditions, susceptibility to nalidixic acid, and flagellar morphology; H. rodentium grows in anaerobic conditions, is resistant to nalidixic acid, and has nonsheathed flagella, whereas H. typhlonius does not grow in anaerobic conditions, is susceptible to nalidixic acid, and has sheathed flagella. Another previously described, unnamed novel urease-negative Helicobacter sp. is also characterized by single polar sheathed flagella (40).
Screening of over 1,000 fecal samples submitted to the University of Missouri Research Animal Diagnostic and Investigative Laboratory demonstrated H. typhlonius colonization in 4.88% of the samples. This finding suggests that H. typhlonius is prevalent in laboratory rodent colonies in the United States. H. typhlonius was not as prevalent as H. hepaticus (16.44%) or H. rodentium (15.11%) but was slightly more prevalent than H. bilis (4.33%). While these data must be interpreted cautiously because some samples were likely submitted from colonies known or suspected to be contaminated by Helicobacter spp., they highlight that multiple Helicobacter spp. are common in research mouse colonies. Furthermore, the IL-10 knockout mice from which the MIT isolates were obtained originated in Germany, suggesting that H. typhlonius has a worldwide distribution (10).
In conclusion, geographically disparate laboratories independently identified a novel Helicobacter sp. that is capable of causing enteric disease in immunodeficient mice (10, 16). We propose to name this species H. typhlonius. Routine screening of rodent colonies throughout the United States suggests that H. typhlonius is prevalent in rodent colonies. Its importance in causing naturally occurring gastrointestinal disease in immunocompetent mice, as well as enteric disease in other hosts, will require further studies.
Description of H. typhlonius sp. nov.
Helicobacter typhlonius (ti.flo′ ni.us Gr. n. typhlon, cecum; N.L. adj. typhlonius, pertaining to the cecum). Filamentous cells are 0.3- by 2- to 3-μm curved to spiral rods, with no periplasmic fibers. Cells do not form spores and are motile by single sheathed bipolar flagella. Colonies are pinpoint. Cells grow in microaerobic but not in anaerobic or aerobic conditions. There is growth at 37 or 42°C but not at 25°C nor in the presence of 1.5% NaCl. There is growth in the presence of 1% glycine; however, the majority of cells harvested in these conditions have coccoid morphology. H. typhlonius is oxidase and catalase positive but does not have urease, alkaline phosphatase, or gamma-glutamyltransferase activities. Nitrate is reduced to nitrite. Indoxyl-acetate and hippurate are not hydrolyzed. Cells are resistant to cephalothin and sensitive to nalidixic acid.
ACKNOWLEDGMENTS
We thank H. G. Trüper of the Institut for Mikrobiology and Biotechnology in Bonn, Germany, for assistance with bacterial nomenclature and Beth Livingston, Samantha McCasland, Sue Bingaman, Sandy Xu, and Allen Maddy for excellent technical support.
This research was supported in part by a grant from the Pittsburgh Supercomputing Center through the NIH National Center for Research Resources resource grants P41RR0600 (C.L.F.) and R01CA067529 (J.G.F.).
REFERENCES
- 1.Beckwith C S, Franklin C L, Hook R R, Jr, Besch-Williford C L, Riley L K. Fecal PCR assay for diagnosis of Helicobacter infection in laboratory rodents. J Clin Microbiol. 1997;35:1620–1623. doi: 10.1128/jcm.35.6.1620-1623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahill R J, Foltz C J, Fox J G, Dangler C A, Powrie F, Schauer D B. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox R A, Kempsell K, Fairclough L, Colston M J. The 16S ribosomal RNA of Mycobacterium leprae contains a unique sequence which can be used for identification by the polymerase chain reaction. J Med Microbiol. 1991;35:284–290. doi: 10.1099/00222615-35-5-284. [DOI] [PubMed] [Google Scholar]
- 4.Dewhirst F E, Fox J G, On S L. Recommended minimal standards for describing new species of the genus Helicobacter. Int J Syst Evol Microbiol. 2000;50(Pt. 6):2231–2237. doi: 10.1099/00207713-50-6-2231. [DOI] [PubMed] [Google Scholar]
- 5.Eaton K A, Dewhirst F E, Radin M J, Fox J G, Paster B J, Krakowka S, Morgan D R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 6.Finegold S M, Martin W J. Diagnostic microbiology. 6th ed. St. Louis, Mo: C. V. Mosby Co.; 1982. [Google Scholar]
- 7.Foltz C J, Fox J G, Cahill R C, Murphy J C, Yan L, Shames B, Schauer D B. Spontaneous inflammatory bowel diseae in multiple mutant mouse lines: association with colonization by Helicobacter hepaticus. Helicobacter. 1998;3:69–78. doi: 10.1046/j.1523-5378.1998.08006.x. [DOI] [PubMed] [Google Scholar]
- 8.Fox J G, Chien C C, Dewhirst F E, Paster B J, Shen Z, Melito P L, Woodward D L, Rodgers F G. Helicobacter canadensis sp. nov. isolated from humans with diarrhea: an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox J G, Gorelick P L, Kullberg M C, Ge Z, Dewhirst F E, Ward J M. A novel urease-negative Helicobacter species associated with colitis and typhlitis in IL-10-deficient mice. Infect Immun. 1999;67:1757–1762. doi: 10.1128/iai.67.4.1757-1762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J G, Handt L, Sheppard B J, Xu S, Dewhirst F E, Motzel S, Klein H. Isolation of Helicobacter cinaedi from the colon, liver and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J Clin Microbiol. 2001;39:1580–1585. doi: 10.1128/JCM.39.4.1580-1585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox J G, Yan L, Shames B, Campbell J, Murphy J C, Li X. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect Immun. 1996;64:3673–3681. doi: 10.1128/iai.64.9.3673-3681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin C L, Beckwith C S, Livingston R S, Riley L K, Gibson S V, Besch-Williford C L, Hook R R., Jr Isolation of a novel Helicobacter species, Helicobacter cholecystus sp. nov., from the gallbladders of Syrian hamsters with cholangiofibrosis and centrilobular pancreatitis. J Clin Microbiol. 1996;34:2952–2958. doi: 10.1128/jcm.34.12.2952-2958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin C L, Riley L K, Livingston R S, Beckwith C S, Besch-Williford C L, Hook J R R. Enterohepatic lesions in SCID mice infected with Helicobacter bilis. Lab Anim Sci. 1998;48:334–339. [PubMed] [Google Scholar]
- 16.Franklin C L, Riley L K, Livingston R S, Beckwith C S, Hook R R, Jr, Besch-Williford C L, Hunziker R, Gorelick P L. Enteric lesions in SCID mice infected with “Helicobacter typhlonicus,” a novel urease-negative Helicobacter species. Lab Anim Sci. 1999;49:496–505. [PubMed] [Google Scholar]
- 17.Gebhart C J, Fennell C L, Murtaugh M P, Stamm W E. Campylobacter cinaedi is normal intestinal flora in hamsters. J Clin Microbiol. 1989;27:1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haines D C, Gorelick P L, Battles J K, Pike K M, Anderson R J, Fox J G, Taylor N S, Shen Z, Dewhirst F E, Anver M R, Ward J M. Inflammatory large bowel disease in immunodeficient rats naturally and experimentally infected with Helicobacter bilis. Vet Pathol. 1998;35:202–208. doi: 10.1177/030098589803500305. [DOI] [PubMed] [Google Scholar]
- 19.Hanninen M L, Happonen I, Saari S, Jalava K. Culture and characteristics of Helicobacter bizzozeronii, a new canine gastric Helicobacter sp. Int J Syst Bacteriol. 1996;46:160–166. doi: 10.1099/00207713-46-1-160. [DOI] [PubMed] [Google Scholar]
- 20.Harper C M, Dangler C A, Xu S, Feng Y, Shen Z, Sheppard B, Stamper A, Dewhirst F E, Paster B J, Fox J G. Isolation and characterization of a Helicobacter sp. from the gastric mucosa of dolphins, Lagenorhynchus acutus and Delphinus delphis. Appl Environ Microbiol. 2000;66:4751–4757. doi: 10.1128/aem.66.11.4751-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalava K, Kaartinen M, Utriainen M, Happonen I, Hanninen M L. Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeronii. Int J Syst Bacteriol. 1997;47:975–982. doi: 10.1099/00207713-47-4-975. [DOI] [PubMed] [Google Scholar]
- 22.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 23.Kullberg M C, Ward J M, Gorelick P L, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee A, Phillips M W, J. L. O, Paster B J, Dewhirst F E, Fraser G J, Fox J G, Sly L I, Romaniuk P J, Trust T J, et al. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol. 1992;42:27–36. doi: 10.1099/00207713-42-1-27. [DOI] [PubMed] [Google Scholar]
- 25.Marshall B J. Campylobacter pylori: its link to gastritis and peptic ulcer disease. Rev Infect Dis. 1990;12:S87–S93. doi: 10.1093/clinids/12.supplement_1.s87. [DOI] [PubMed] [Google Scholar]
- 26.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 27.Mendes E N, Queiroz D M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 28.Parsonnet J. Bacterial infection as a cause of cancer. Environ Health Perspect. 1995;103(Suppl. 8):263–268. doi: 10.1289/ehp.95103s8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsonnet J. Helicobacter pylori. Infect Dis Clin N Am. 1998;12:185–197. doi: 10.1016/s0891-5520(05)70417-7. [DOI] [PubMed] [Google Scholar]
- 30.Paster B J, Lee A, Fox J G, Dewhirst F E, Tordoff L A, Fraser G J, O'Rourke J L, Taylor N S, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 31.Patterson M M, Schrenzel M D, Feng Y, Fox J G. Gastritis and intestinal metaplasia in Syrian hamsters infected with Helicobacter aurati and two other microaerobes. Vet Pathol. 2000;37:589–596. doi: 10.1354/vp.37-6-589. [DOI] [PubMed] [Google Scholar]
- 32.Patterson M M, Schrenzel M D, Feng Y, Xu S, Dewhirst F E, Paster B J, Thibodeau S A, Versalovic J, Fox J G. Helicobacter aurati sp. nov., a urease-positive Helicobacter species cultured from gastrointestinal tissues of Syrian hamsters. J Clin Microbiol. 2000;38:3722–3728. doi: 10.1128/jcm.38.10.3722-3728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 34.Riley L K, Franklin C L, Hook R R, Jr, Besch-Williford C. Identification of murine helicobacters by PCR and restriction enzyme analyses. J Clin Microbiol. 1996;34:942–946. doi: 10.1128/jcm.34.4.942-946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Z, Fox J G, Dewhirst F E, Paster B J, Foltz C J, Yan L, Shames B, Perry L. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol. 1997;47:627–634. doi: 10.1099/00207713-47-3-627. [DOI] [PubMed] [Google Scholar]
- 38.Shomer N H, Dangler C A, Marini R P, Fox J G. Helicobacter bilis/Helicobacter rodentium co-infection associated with diarrhea in a colony of scid mice. Lab Anim Sci. 1998;48:455–459. [PubMed] [Google Scholar]
- 39.Shomer N H, Dangler C A, Schrenzel M D, Fox J G. Helicobacter bilis-induced inflammatory bowel disease in scid mice with defined flora. Infect Immun. 1997;65:4858–4864. doi: 10.1128/iai.65.11.4858-4864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shomer N H, Dangler C A, Schrenzel M D, Whary M T, Paster B J, Dewhirst F E, Fox J G. Cholangiohepatitis and inflammatory bowel disease (IBD) induced by a novel urease-negative Helicobacter species in A/J and Tac:ICR:Hascid mice. Biol Exp Med. 2001;226:420–428. doi: 10.1177/153537020122600505. [DOI] [PubMed] [Google Scholar]
- 41.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 42.Studier J A, Keppler K J. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988;5:729–731. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- 43.Swofford D L, Olsen G J. Phylogeny reconstruction. In: Hillis D M, Moritz C, editors. Molecular systematics. Sunderland, Mass: Sinauer Association, Inc.; 1990. pp. 411–501. [Google Scholar]
- 44.Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990;28:1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward J M, Anver M R, Haines D C, Benveniste R E. Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 46.Ward J M, Anver M R, Haines D C, Melhorn J M, Gorelick P, Yan L, Fox J G. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46:15–20. [PubMed] [Google Scholar]
- 47.Ward J M, Fox J G, Anver M R, Haines D C, George C V, Collins M J, Jr, Gorelick P L, Nagashima K, Gonda M A, Gilden R V, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 48.Witherell H L, Hansen S, Jellum E, Orentreich N, Vogelman J H, Parsonnet J. Risk for gastric lymphoma in persons with CagA+ and CagA−Helicobacter pylori infection. J Infect Dis. 1997;176:1641–1644. doi: 10.1086/517346. [DOI] [PubMed] [Google Scholar]