Abstract
While COVID-19 research has seen an explosion in the literature, the impact of pandemic-related societal and lifestyle disruptions on brain health among the uninfected remains underexplored. However, a global increase in the prevalence of fatigue, brain fog, depression and other “sickness behavior”-like symptoms implicates a possible dysregulation in neuroimmune mechanisms even among those never infected by the virus.
We compared fifty-seven ‘Pre-Pandemic’ and fifteen ‘Pandemic’ datasets from individuals originally enrolled as control subjects for various completed, or ongoing, research studies available in our records, with a confirmed negative test for SARS-CoV-2 antibodies. We used a combination of multimodal molecular brain imaging (simultaneous positron emission tomography / magnetic resonance spectroscopy), behavioral measurements, imaging transcriptomics and serum testing to uncover links between pandemic-related stressors and neuroinflammation.
Healthy individuals examined after the enforcement of 2020 lockdown/stay-at-home measures demonstrated elevated brain levels of two independent neuroinflammatory markers (the 18 kDa translocator protein, TSPO, and myoinositol) compared to pre-lockdown subjects. The serum levels of two inflammatory markers (interleukin-16 and monocyte chemoattractant protein-1) were also elevated, although these effects did not reach statistical significance after correcting for multiple comparisons. Subjects endorsing higher symptom burden showed higher TSPO signal in the hippocampus (mood alteration, mental fatigue), intraparietal sulcus and precuneus (physical fatigue), compared to those reporting little/no symptoms. Post-lockdown TSPO signal changes were spatially aligned with the constitutive expression of several genes involved in immune/neuroimmune functions.
This work implicates neuroimmune activation as a possible mechanism underlying the non-virally-mediated symptoms experienced by many during the COVID-19 pandemic. Future studies will be needed to corroborate and further interpret these preliminary findings.
Keywords: PET, Neuroimaging, Stress, Pandemic, Mental health, MRS
1. Introduction
Beyond the staggering number of infections and deaths, the Coronavirus Disease 2019 (COVID-19) pandemic (Cucinotta and Vanelli, 2020) has caused lifestyle, societal, and other disruptions, impacting the lives of a large swath of the world population in multiple ways. For instance, behavioral data show that the severity and/or prevalence of symptoms of psychological distress have increased considerably in the United States since the pandemic onset (CDC, 2020, Abbott, 2021). Likewise, an increased prevalence of fatigue, dyscognition (i.e., “brain fog”) and other symptoms has been reported, including among the non-infected. As such, the scientific and medical communities are urgently calling for studies promoting a better understanding of the effects of the pandemic on brain and mental health (Holmes et al., 2020, Gonçalves de Andrade et al., 2021, Kar Ray et al., 2021).
While the mechanisms underlying the non-virally mediated effects of the COVID-19 pandemic on brain health are currently unknown, we hypothesized that an elevation in neuroinflammatory responses might play a role. In fact, exposure to social stressors, including social isolation (a state experienced by many during the pandemic), has been previously linked to elevations in serum levels of pro-inflammatory cytokines (Smith et al., 2020) and activation of brain glial cells (Stein et al., 2017, Calcia et al., 2016), involving mechanisms largely overlapping with those observed during infection-induced inflammation (Eisenberger et al., 2017). Such “sterile” forms of (neuro)inflammation -much like their pathogen-associated counterparts- have been linked to produce a constellation of “sickness behaviors”, including fatigue, depressive symptoms, social withdrawal, etc (Dantzer, 2009, Fleshner et al., 2017). Indeed, sterile-(neuro)inflammation is thought to be a key neurobiological process in the pathophysiology of mood disorders (Setiawan et al., 2018, Albrecht et al., 2021, Troubat et al., 2021). Thus, results from both clinical and preclinical literature raise the possibility that (neuro)inflammation might be a potential mechanism underlying the symptoms experienced during the pandemic by healthy individuals that were not infected by SARS-CoV-2.
Here we have hypothesized that subjects examined after the onset of the pandemic and the implementation of lockdown/stay-at-home measures rendered necessary to limit its spread would demonstrate increased (neuro)inflammatory markers. To test this hypothesis, we conducted a retrospective analysis of advanced, multimodal [11C]PBR28 Positron Emission Tomography/Magnetic Resonance (PET/MR) imaging data collected at the A. A. Martinos Center for Biomedical Imaging in the last decade. Specifically, we investigated brain levels of the 18 kDa translocator protein (TSPO) (Cosenza-Nashat et al., 2009); and myoinositol (mIns) (Datta et al., 2017), in subjects evaluated after pandemic onset, compared to subjects scanned pre-pandemic. TSPO and mIns are two putative glial markers that can be detected with PET and MR spectroscopy, respectively. To assess the clinical significance of our findings, we performed a preliminary investigation of the link between neuroinflammatory signals and scores on a questionnaire assessing mental and physical impacts of the pandemic. Then, to understand the genetic underpinnings of our imaging results, we evaluated their spatial association with constitutive brain gene expression in post-mortem brains (Allen Human Brain Atlas) (Hawrylycz et al., 2012). Finally, in order to evaluate possible links between central and circulating inflammatory markers (Wang et al., 2015), we quantified serum levels of cytokines and chemokines from venous blood measurements.
Because the focus of this study was on “sterile”, and not virally-mediated, neuroimmune activation, only individuals with a confirmed negative test for SARS-CoV-2 antibodies at the time of the scan (indicating a remote probability of infection prior to the study (Choe et al., 2021) were included among those tested after the onset of the pandemic.
2. Methods
This research study was conducted at the Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital. Data were pooled for several protocols, which were approved by the Partners Healthcare / Mass General Brigham Institutional Review Board. All participants gave written informed consent at the time of their screening.
2.1. Study design and participants
We conducted a retrospective brain imaging study of healthy individuals, originally enrolled as control subjects for various completed or ongoing, research studies investigating the role of neuroinflammation in various disorders.
In total, this study included fifty-seven ‘Pre-Pandemic’ datasets (acquired between 04/2012 and 02/2020) and fifteen ‘Pandemic’ datasets (acquired between 08/2020 and 07/2021) (Supplementary Fig. 1). See Supplementary materials for details on study design. At the time of the study, subjects from the Pandemic group were all residents of Massachusetts, where the lockdown lasted approximately for two months (March-May 2020). Several social-distancing restrictions (such as limited social gathering) remained active in the following year, even after the stay-at-home order was lifted. Over the time-period of this study, the average COVID-19 positivity rate in Massachusetts ranged between 0.5% (July 2021) and ∼ 9% (January 2021).
All subjects in the Pandemic cohort had a negative COVID-19 antibody test (Elecsys® Anti-SARS-CoV-2, Roche Diagnostics; 99.81% specificity; 100% sensitivity 14 + days post infection) at the time of the scan. This broad-spectrum test is sensitive to both IgM, IgA and IgG antibodies, allowing the detection of SARS-CoV-2 infections occurred over a period of at least several months prior the scan (Choe et al., 2021). No COVID-19 antibody testing was available for any of the participants within the Pre-Pandemic group.
Demographics for the participants used in the PET and 1H-MRS analyses are displayed in Table 1 .
Table 1.
Demographics from both main and matched cohorts. Categorical variables are summarized as frequencies (proportions) and continuous variables are summarized as mean (standard deviation). HAB = high affinity binders; MAB = mixed affinity binders; mCi = millicuries. *=p < 0.05; **=p < 0.01.
| [11C]PBR28 PET cohorts | ||||
|---|---|---|---|---|
| Pre-Pandemic | Pre-PandemicMatched | Pandemic | ||
| N | 57 | 29 | 15 | |
| Sex | Male Female |
30 (52.6) 27 (47.4) |
19 (65.5) 10 (35.5) |
12 (80) 3 (20) |
| Age [y] | 42.85(15.36) ** | 55.72(9.49) | 60.73(10.7) | |
| TSPO genotype | HAB MAB |
33(57.9) 24(42.1) |
16(55.1) 13(44.9) |
7 (46.6) 8 (53.4) |
| Scanner | Scanner 1 Scanner 2 |
44(77.2) 13(22.8) |
21(72.4) 8(27.6) |
9 (60) 6 (40) |
| Weight [kg] | 73.71(16.0) | 73.86(16.94) | 78.04(19.7) | |
| Inj. dose [mCi] | 13.28(1.57) | 13.26(1.66) | 13.86(1.69) | |
| Blood samples cohorts | ||||
| Pre-Pandemic | Pre-PandemicMatched | Pandemic | ||
| N | 11 | 9 | 13 | |
| Sex | Male Female |
6 (54.5) 5 (45.5) |
5 (55.5) 4 (44.5) |
11 (84.6) 2 (15.4) |
| Age [y] | 47.2(17.7) * | 51.33(17.7) | 60.61(11.47) | |
| Weight [kg] | 78.24(11.53) | 77.38(12.7) | 78.78(21.10) | |
| 1H-MRS cohorts | ||||
| Pre-Pandemic | Pre-PandemicMatched | Pandemic | ||
| N | 13 | 9 | 11 | |
| Sex | Male Female |
7 (53.8) 6 (46.2) |
6 (66.7) 3 (33.3) |
8 (72.7) 3 (27.3) |
| Age [y] | 46.34(13.51) * | 55.25(7.95) | 60.36(12.2) | |
| Scanner | Scanner 1 Scanner 2 |
2(15.4) * 11(84.6) * |
2(22.2) 7(77.8) |
6 (54.5) 5 (45.5) |
| Weight [kg] | 77.4(11.69) | 78.03(13.76) | 76.73(22.61) | |
2.2. Data acquisition and processing
During an initial screening visit, participants were consented, and genotyped for the Ala147Thr polymorphism in the TSPO gene, which is known to affect binding affinity for several TSPO radioligands, including [11C]PBR28 (Owen et al., 2015, Kreisl et al., 2013). Individuals with the Ala/Ala or Ala/Thr genotypes (predicted high- and mixed-affinity binders -HABs and MABs-, respectively) were included, and the genotype was modeled as a covariate in the statistical design (see below). Individuals with the Thr/Thr genotype (predicted “low-affinity binders”) were excluded at the time of the screening and therefore not represented in our dataset.
At the beginning of the imaging visit, a subset of subjects (n = 24; 11 Pre-Pandemic; 13 Pandemic; Table 1) had venous blood collected to measure the level of circulating inflammatory cytokines (IFN-ɣ, IL-15, IL-16, IL-1RA, IL-6, IL-8, MCP-1, MDC, MIP-1ɑ, and TNF-ɑ). See Supplementary materials for further details.
Dynamic PET/MR scans were performed with two different Siemens scanners: a 3 T Tim Trio whole-body MRI with a dedicated brain PET insert (BrainPET; Scanner 1) (Kolb et al., 2012) and a 3 T Verio whole-body, MRI whole-body PET tomograph (Biograph mMR; Scanner 2) (Delso et al., 2011). Participants were injected with up to ∼ 15 mCi (mCi) [11C]PBR28 as an intravenous bolus and dynamic PET was acquired as described in previous studies (Albrecht et al., 2021, Alshelh et al., 2020). [11C]PBR28 is a second-generation radioligand commonly used to image the glial marker TSPO for the study of neuroinflammation in various conditions (Albrecht et al., 2016). All participants were scanned for a time-period that included ∼ 60–90 min post-injection (the framing window used in our PET analyses; see below). See Supplementary materials for further details on PET data acquisition and processing.
SUV ratio (SUVR) images were obtained via intensity-normalization using the occipital cortex as a pseudo-reference region, as done in previous research (Alshelh et al., 2020, Albrecht et al., 2019). This region demonstrated no statistically significant group differences in mean SUV (p = 0.18; GLM, correcting for age, scanner and genotype; see below), indicating that the use of this signal as a normalizing factor did not bias our results. To support findings from SUVR measurements, distribution volume (VT) and ratio (DVR) outcomes were determined using kinetic modeling in a subset of subjects for whom a radiometabolite-corrected arterial input function was available (n = 24; 16 Pre-Pandemic; 8 Pandemic), as described in the Supplementary materials.
Simultaneously to the PET data collection, a subset (13 Pre-Pandemic; 11 Pandemic) was also scanned with 1H-MRS, allowing us to quantify the brain concentration of myoinositol (mIns), another putative glial marker (Datta et al., 2017), and -for reference- Creatine (Cr; a cellular energetic marker) and N-Acetyl Aspartate (NAA; a marker of neuronal integrity). See Supplementary materials for details on 1H-MRS data acquisition and processing.
To interrogate the possible clinical significance of our results, we retrospectively administered a questionnaire assessing the impact of the COVID-19 pandemic on mental and physical well-being, specifically assessing mental/physical fatigue, dyscognition and mood alterations (n = 11; See Supplementary Table 3).
2.3. Statistical analysis
Descriptive summaries were computed by group (Pre-Pandemic, Pandemic). Continuous variables were summarized as mean and standard deviation (SD) and categorical variables were summarized as frequencies and percentages (Table 1). Group differences in demographics were assessed with Student’s t-tests for continuous variables (age, injected dose, weight) and Chi-Square () tests for categorical variables (sex, TSPO polymorphism, scanner).
Covariate imbalance was assessed in all the analyses by adjusting for confounding variables but was also accounted for via matching. In particular, given the significant difference in age across Pre-Pandemic and Pandemic groups, we re-ran all group analyses using smaller subsets of the Pre-Pandemic group who were better demographically matched to the Pandemic cohort (p’s ≥ 0.11) (Table 1). Specifically, all Pre-Pandemic subjects with an age range 20–40 (n = 28) were removed, resulting in a non-statistical difference in age (p > 0.1) between groups.
Pre-Pandemic and Pandemic [11C]PBR28 signals were compared in both a-priori region-of-interest (ROI) analysis (in areas previously associated with stress or depression: hippocampus (Calcia et al., 2016, DiSabato et al., 2020), thalamus (Kang et al., 2018), and anterior cingulate cortex (ACC) (Albrecht et al., 2021), using a General Lineal Model (GLM), including TSPO genotype, scanner type and age as covariates. The same analyses were performed in a non-parametric voxel-wise analysis of the whole brain. The significant cluster obtained from the whole brain analysis was parcellated by intersecting it with anatomical labels in standard space. From these parcels, the mean PET signal was extracted for visualization purposes, as well as correlational and support/sensitivity analyses (see Supplementary materials). Coefficient of variation (CV=; =standard deviation; =mean), was computed to compare inter-individual variability across various PET metrics (SUVR, DVR, VT).
Further, clinical indicators from behavioral measurements and serum testing, as well as imaging transcriptomics statistical analyses were used to uncover links between pandemic-related stressors and neuroinflammation. 1H-MRS data were also tested for group differences using a GLM, including scanner type and age as covariates in the model. See Supplementary materials for further details.
3. Results
Participants from the Pandemic cohort reported experiencing various symptoms since the onset of the pandemic, including mood alterations (54% of respondents), mental (36%) and physical (27%) fatigue, and dyscognition (18%). Paralleling these behavioral findings, [11C]PBR28 signal was higher in the Pandemic vs the Pre-Pandemic group, as was apparent in both ROI and voxelwise analyses (including in all support/sensitivity analyses performed).
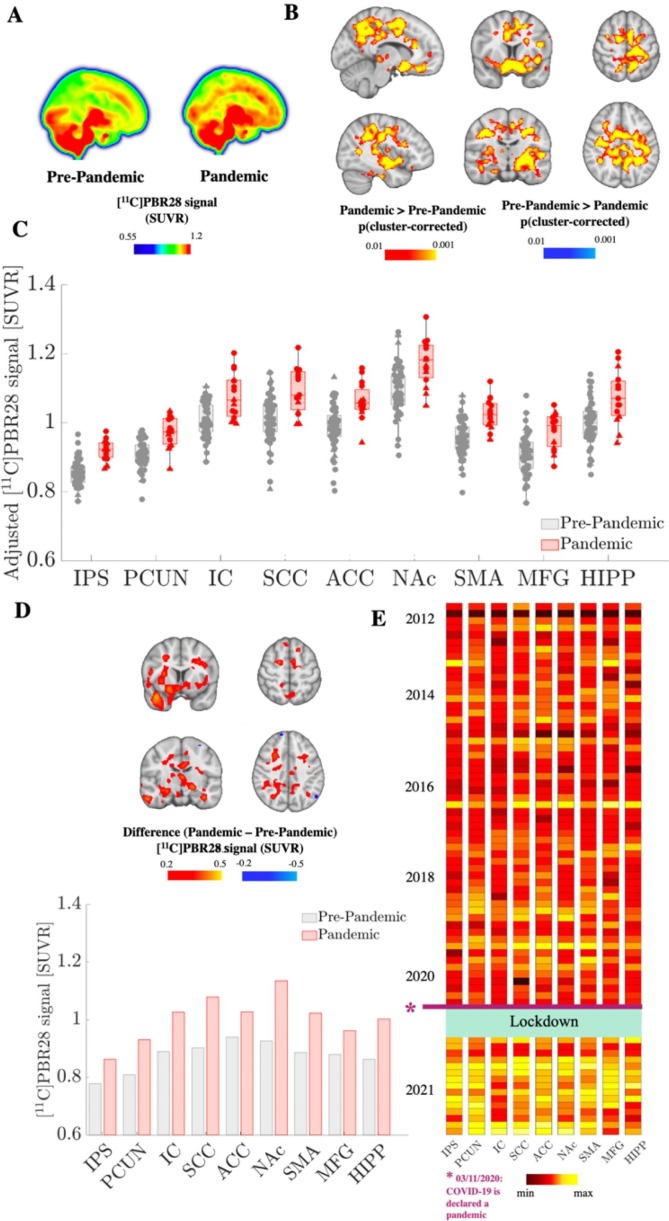
The a priori ROI analyses revealed that, compared to the Pre-Pandemic group, the Pandemic group demonstrated higher [11C]PBR28 signal in all the four ROI evaluated (p’s < 0.05). Whole-brain voxelwise analyses (Fig. 1 , Supplementary Fig. 2) confirmed the [11C]PBR28 signal elevation in the Pandemic cohort, identifying a large cluster encompassing both grey and white matter regions, including portions of the anatomical regions used as our a-priori ROIs as well as additional regions (precental, postcentral, superior, middle and inferior frontal gyri, precuneus, superior parietal lobule, anterior and posterior insula, amygdala, putamen, supplementary motor cortex, anterior, middle, posterior cingulate and subcallosal cortex; see Table 2 ).
Fig. 1.
[11C]PBR28 PET signal elevations in the Pandemic group. (A) Mean images computed from 57 Pre-pandemic and 15 Pandemic subjects are displayed as maximum intensity projections. (B) Significant cluster from the Pandemic > Pre-Pandemic voxel-wise contrast is shown in a red–yellow color scale. There were no significant regions for the Pre-Pandemic > Pandemic contrast. (C) Visualization of mean [11C]PBR28 SUVR extracted from sub-portions of the cluster statistically significant in A. IPS = Intraparietal Sulcus, PCUN = Precuneus, IC = Insular Cortex, SCC = Subcallosal Cortex, ACC = Anterior Cingulate Cortex, NAc = Nucleus Accumbens, SMA = Supplementary Motor Area, MFG = Middle Frontal Gyrus, HIPP = Hippocampus. Error bars denote 25th to 75th inter-quartile range. Triangles denote data from Scanner 1 and circles denote data from Scanner 2. (D) Case study of one subject scanned before- (September 2016) and during the pandemic (October 2020) in Scanner 1. Bar graph of mean [11C]PBR28 SUVR are extracted from the cluster sub-portions in (C) and difference image (post-pre) are reported in red-yellow and cyan-blue color bars. E) Mean [11C]PBR28 SUVR extracted from sub-clusters in (C) and sorted by scan date. The range of the color scale was set for each region independently to best illustrate the pandemic PET signal increase, for visualization purposes.
Table 2.
Peak coordinates from brain regions significant in whole-brain [11C]PBR28 PET voxel-wise group analyses.
| Cluster size (# voxels) | Cluster p-value | Peak | Anatomical Location | |||
|---|---|---|---|---|---|---|
| Z | x | y | z | |||
| Pandemic > Pre-Pandemic | ||||||
| 31,504 | 0.0006 | 3.54 | −20 | −36 | −1 | L Hippocampus |
| 3.54 | −6 | 2 | 57 | L Supplementary Motor Cortex | ||
| 3.54 | 10 | 5 | 36 | R Anterior Cingulate Cortex | ||
| 3.54 | −40 | –23 | 37 | L Postcentral Gyrus | ||
| 3.54 | –32 | −7 | 46 | L Precentral Gyrus | ||
| 3.54 | −29 | −47 | 43 | L Superior Parietal Lobe | ||
| 3.54 | −40 | 4 | −6 | L Anterior Insula | ||
| 3.54 | −19 | −27 | 54 | L White Matter | ||
| 3.54 | −9 | 11 | −12 | L Nucleus Accumbens | ||
| 3.54 | 7 | 23 | −16 | R Subcallosal Cortex | ||
| 3.54 | −46 | 6 | 17 | L Inferior Frontal Gyrus | ||
| 3.54 | −25 | 3 | 0 | L Putamen | ||
| 3.35 | −20 | 8 | 48 | L Superior Frontal Gyrus | ||
| 3.23 | −16 | 6 | –22 | L Frontal Orbital Cortex | ||
| 3.23 | −37 | –22 | 1 | L Posterior Insula | ||
| 3.23 | 4 | −5 | 0 | R Thalamus | ||
| 3.09 | 35 | −1 | 55 | L Middle Frontal Gyrus | ||
| 3.03 | 22 | −4 | −24 | R Amygdala | ||
| 2.98 | 18 | −53 | 25 | R Precuneus Cortex | ||
| Pre-Pandemic > Pandemic | ||||||
| n.s. | ||||||
Notably, [11C]PBR28 signal elevation in the Pandemic group could be observed irrespective of scanner, age, genotype, sex, injected dose and body weight (Supplementary Fig. 3). In all the evaluated sub-clusters, [11C]PBR28 signal elevations in the Pandemic group were also confirmed by DVR, computed from the small sample (∼30%) of participants with available arterial blood data (n = 24) (0.009 ≤ p ≤ 0.04; Supplementary Fig. 4A). In all regions, DVR was strongly correlated with SUVR (r ≥ 0.77; p ≤ 0.001; Supplementary Fig. 4B). The VT values, computed from the same subsample of participants, demonstrated larger interindividual variability (CV ranging from 34.6% (IPS) to 39.2% (HIPP), as opposed to 5.9–9.1% for SUVR and 5.8–12.1% for DVR (see Supplementary Table 4) and did not statistically differ across groups (p’s > 0.6). No significant correlation was found between SUVR or DVR and VT (p’s > 0.1).
The results of the group analyses were replicated within-subject in the single participant scanned before and after the pandemic onset in the same scanner. Indeed, during the pandemic this subject demonstrated widespread PET signal elevation in all brain regions significant in the group analyses (Fig. 1C), ranging up to 22.6% (Nucleus Accumbens), compared to the Pre-Pandemic scan.
Fig. 1D displays the PET signal across subjects, indicating relatively low between-subject variability for nearly a decade (as corroborated by time-stability and scan-re-scan reliability tests; Supplementary Fig. 5A, Fig. 5B), followed by a noticeable increase during the pandemic.
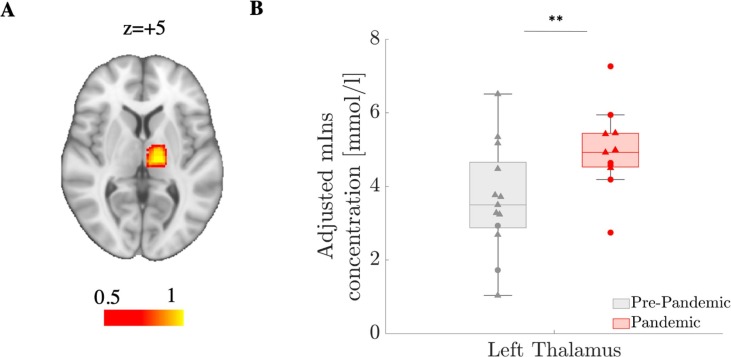
Fig. 5.
Elevations in the Pandemic group in 1H-MRS-measured myoinositol concentration. (A) Probabilistic map of thalamic voxel placement in standard space, calculated via non-linear transformation between each subjects’ MPRAGE volume and MNI151 template, then applied to the MRS voxel. (B) Group comparison in mIns concentration (left thalamus) between Pre-pandemic (n = 13) and Pandemic (n = 11) cohorts. Error bars denote 25th to 75th inter-quartile range. Triangles denote data from Scanner 1 and circles denote data from Scanner 2.
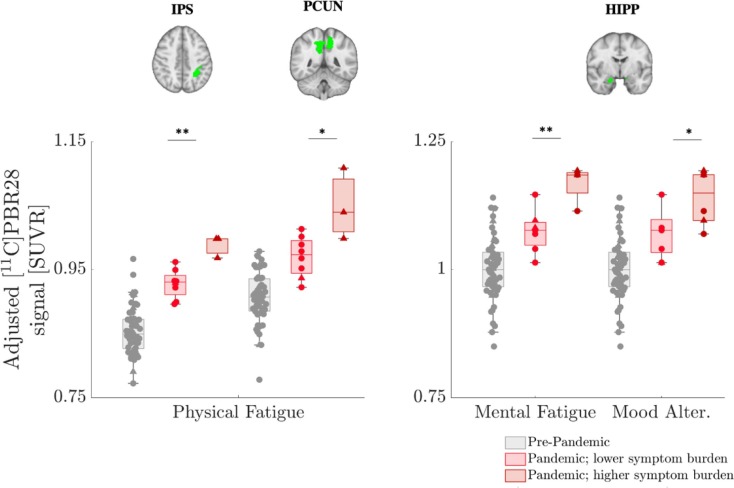
When categorizing the Pandemic group subjects based on their symptom burden (higher vs lower), we found the PET signal in the IPS and precuneus to be associated with physical fatigue (p < 0.01 and p < 0.05 uncorrected, respectively), whereas the hippocampal PET signal was more closely linked to mental fatigue and mood alterations (p < 0.01 and p < 0.05 uncorrected, respectively) (see Fig. 2 ). Of these results, the IPS-physical fatigue association remained statistically significant after correction for multiple comparisons. No association was found with dyscognition symptoms (although the very small sample size of participants reporting an increase in this symptom, n = 2, rendered this analysis inconclusive).
Fig. 2.
[11C]PBR28 PET signal elevations in the Pandemic group are proportional to symptom burden. [11C]PBR28 signal in the subset of Pandemic subjects who completed the questionnaire on the impact of the pandemic con physical and mental health (n = 11) shows elevations in the IPS, PCUN and HIPP for those individuals who showed higher symptom burden (physical fatigue, mental fatigue and/or mood alterations). Pre-pandemic data (n = 57) are also displayed as reference, for visualization purposes only. Data are adjusted for TSPO genotype. Error bars denote 25th to 75th inter-quartile range, and the horizontal line represents the median. Triangles denote data from Scanner 1 and circles denote data from Scanner 2. *=p < 0.05; **= p < 0.01. Abbreviations: Mood Alter. = Mood Alterations. See Fig. 2 caption for abbreviations.
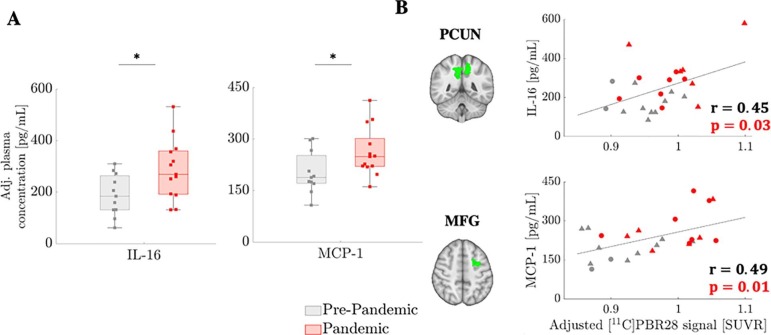
Fig. 3 shows that IL-16 and MCP-1 were significantly higher in the Pandemic cohort than in the Pre-Pandemic cohort (p’s < 0.03) and that higher levels of those inflammatory markers were correlated with increased [11C]PBR28 uptake, when tested across the entire dataset (although these correlations did not reach significance when tested only in the Pandemic group). Elevations in IL-16 and MCP-1 did not reach statistical significance after correcting for the number of cytokines/chemokines evaluated (n = 5) (p’s = 0.075 and 0.13, corrected). Results were consistent when using cohorts matched in demographics (Supplementary Fig. 8).
Fig. 3.
Plasma inflammatory marker elevations in the Pandemic group. Plasma inflammatory marker (IL-16 and MCP-1) elevation in the Pandemic group (n = 13) compared to Pre-Pandemic group (n = 11) (A), and their correlation with [11C]PBR28 signal (B). Error bars denote 25th to 75th inter-quartile range, and the horizontal line represents the median. GLM group difference accounts for age as regressor of no interest. Partial correlations are computed adjusting for TSPO genotype. Triangles denote data from Scanner 1 and circles denote data from Scanner 2. *=p < 0.05.
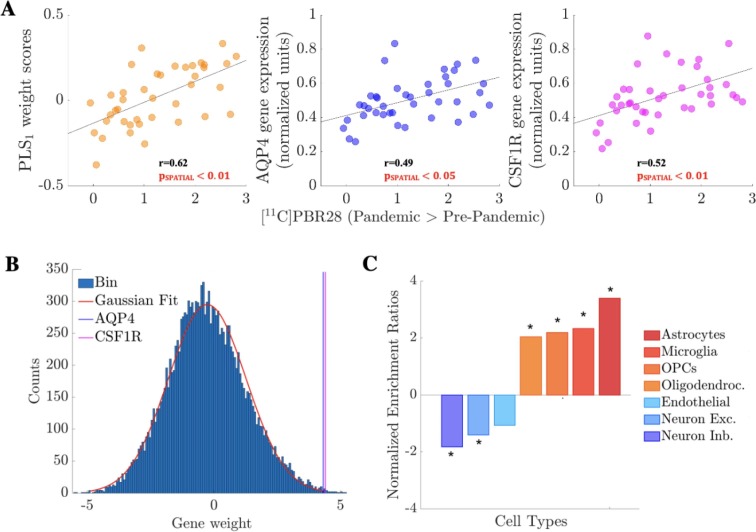
Regional vulnerability to changes in [11C]PBR28 signal between Pre- and Pandemic groups was predicted by regional variability in the human brain transcriptome: PLS1 alone explained 37.33% of variance in [11C]PBR28 T-statistics and did so above chance (pSPATIAL < 0.01; see Supplementary materials for details). As Fig. 4 A illustrates, PLS1 regional scores correlated positively with [11C]PBR28 contrast map, i.e. genes with positive weights in PLS1 have higher-than-average expression where [11C]PBR28 signal showed the largest increases and lower-than-average expression in regions with minimal changes. Among the genes with the highest PLS1 weights were AQP4 (24th out of 15,633) and CSF1R (32th), two genes strongly expressed in glial cells (Fig. 4B), as well as CCR1, a monocyte marker. Indeed, our analyses revealed significant enrichment for genes: i) highly expressed in glial cells, including astrocytes and microglia (first and second hits respectively; Fig. 4C and Supplementary Table 1); ii) belonging to several biological pathways related to immune/neuroimmune functions, including “macrophage activation”, “leukocyte activation involved in inflammatory response”, “purinergic receptor signaling pathway”, “neuroinflammatory response”, “gliogenesis”, “mast cell activation”, “T cell activation”, “chemokine production”, “acute inflammatory response”, “cytokine secretion” and “interleukin-1 production” (Supplementary Table 2).
Fig. 4.
Imaging transcriptomics analyses. (A) First component of the PLS analyses (PLS1; weight scores), AQP4 (astrocyte marker) and CSF1R (microglia marker) gene expression (normalized units) plotted against [11C]PBR28 contrast t-stat (Pandemic > Pre-pandemic; see Fig. 1) in the 41 regions of left hemisphere regions (34 cortical plus 7 subcortical regions) of the Desikan-Killiany atlas. Genes with positive weights in PLS1 have higher-than-average expression where [11C]PBR28 showed the largest increases and lower-than-average expression in regions with minimal changes. Of note, pSPATIAL is obtained via spatial permutation testing (spin test) to account for the inherent spatial autocorrelation of the imaging data (see Supplementary materials). (B) Histogram (150 bins) of gene weights in PLS1 with the highly-ranked positions of AQP4 and CSF1R highlighted (scores 4.31 and 4.40 respectively). (C) Brain cell-type gene set enrichment analysis. Positive normalized enrichment ratios (in orange-red shades) indicate enrichment for genes of a certain cell-type among those genes with high expression in regions with the largest increases in [11C]PBR28 signal (positive weights in PLS1). Negative normalized enrichment ratios (in blue shades) indicate enrichment for genes of a certain cell-type among genes with high expression in regions with minimal or negligible increases in [11C]PBR28 signal (negative weights in PLS1). *=p < 0.05, after FDR correction for the total number of cell-types tested. Abbreviations: OPCs = Oligodendrocyte Precursor Cells; Oligodendroc. = Oligodendrocytes; Neuron Exc. = Excitatory Neurons; Neuron Inb. = Inhibitory Neurons.
Paralleling the [11C]PBR28 PET results, the subset of Pandemic cohort with 1H-MRS data also demonstrated significantly higher thalamic concentration of the glial metabolite mIns, measured using 1H-MRS, compared to the Pandemic cohort (p < 0.01; Fig. 5 ). The increases in mIns concentration in the Pandemic group appeared to be metabolite-specific, since group differences in Cr (a cellular energetic marker) and NAA (a marker of neuronal integrity) concentrations were not statistically significant (p > 0.2). Notably, the elevations in mIns remained statistically significant after correcting for the number of metabolites evaluated (n = 3). No significant correlation was found between thalamic mIns and SUVR signal (p = 0.31).
Ancillary analyses replicated both [11C]PBR28 and 1H-MRS results when 1) including only a subset of participants from the Pre-Pandemic group better demographically matched to the Pandemic cohort (Table 1, Fig. 6A, 7A); 2) using alternate datasets (i.e., datasets collected in individuals scanned twice and not used in the primary analyses; Supplementary Fig. 6B, 7B); or 3) excluding subjects who had received the vaccine against the SARS-CoV-2 virus (Supplementary Fig. 6C, 7C)..
4. Discussion
This study provides novel evidence of elevated neuroinflammatory markers in healthy, non-infected individuals during the COVID-19 pandemic. Subjects evaluated after the onset of the pandemic onset and the implementation of necessary lockdown/stay-at-home measures showed elevations in the brain levels of [11C]PBR28 (measured PET) and mIns (measured using 1H-MRS), thus providing multimodal evidence for neuroinflammation in cortical and subcortical regions including sensory, motor and higher order association areas, and white matter. [11C]PBR28 signal elevations were positively associated with physical fatigue (IPS/precuneus), mental fatigue and mood alterations (hippocampus) and with pro-inflammatory blood markers (IL-16 and MCP-1). Collectively, these findings provide support to neuroimmune responses as mechanisms underlying stress, depression and other symptoms of psychological distress (Calcia et al., 2016, DiSabato et al., 2020). Further, the regional variability in increased [11C]PBR28 signal could be predicted by constitutive expression of genes related to glial neuroimmune response in healthy post-mortem human brains. Overall, our results indicate a possible link between pandemic-associated stressors and neuroimmune responses.
While several studies have recently raised the urgent need for research to quantitatively address consequences of COVID-19-related disruptions on human health, (Holmes et al., 2020, Gonçalves de Andrade et al., 2021) neuroinflammatory responses in non-infected individuals during the pandemic so far have not been explored. However, a wealth of pre-clinical studies has shown that inflammatory processes and social behaviors are deeply connected: as inflammatory processes can affect social behaviors, exposure to chronic stressors can also upregulate inflammation (Liu et al., 2017). Specifically, it has been recently proposed that chronic stress can trigger the sympathetic nervous system to release catecholamine and glucocorticoids (Sapolsky et al., 2000), and that microglia can respond to those elevations by altering their density and triggering upregulation of pro-inflammatory molecules (Tian et al., 2017), ultimately promoting sickness behaviors (Dantzer, 2009, Konsman et al., 2002). Further, clinical research has provided evidence for neuroinflammation in subjects with major depression (Setiawan et al., 2018) or depressive symptoms comorbid to chronic pain (using the same simplified ratio metric employed in the current study (Albrecht et al., 2021). Likewise, glial activation has been recently implicated in the pathophysiology of fibromyalgia (Albrecht et al., 2019) and Gulf War Illness (Alshelh et al., 2020), both conditions characterized by several symptoms including excessive fatigue and “brain fog”, which are symptoms reported by many during the pandemic. As such, preclinical and clinical work provide support the investigation of neuroinflammation as a mechanism of the symptoms observed with increasing frequency during the COVID-19 pandemic, -importantly- even among the non-infected.
TSPO is a five-transmembrane domain protein mostly expressed on the outer membrane of mitochondria which is dramatically overexpressed in activated microglia and astrocytes (Cosenza-Nashat et al., 2009), in the context of neuroinflammatory responses. Indeed, a strong colocalization between TSPO upregulation and activated glial cells has been found across multiple preclinical and human studies of various disorders, including in rodent and human studies of multiple sclerosis, (Airas et al., 2015, Sucksdorff et al., 2020) human immunodeficiency encephalitis (Cosenza-Nashat et al., 2009), ischemia (Li et al., 2017), depression (Richards et al., 2018), and Alzheimer’s disease (Cosenza-Nashat et al., 2009, Albrecht et al., 2016, Richards et al., 2018). However, TSPO is also expressed by circulating immune cells and, as such, it is possible that some of the observed central TSPO signal may be explained by TSPO-rich peripheral immune cells, such as monocytes and macrophages, infiltrating the brain (Zhao et al., 2020). Indeed, the elevation of MCP-1 and IL-16, both molecules regulating migration and infiltration of monocyte/macrophages (Deshmane et al., 2009, Hridi et al., 2021), may provide some support to this view. For instance, data in knock-out mice suggest that MCP-1 may be required for glia to mount an effective neuroinflammatory response, and that MCP-1 may be critical in transferring inflammatory signals from the periphery to the brain (Thompson et al., 2008).
It should be noted that the specificity of TSPO upregulation to (neuro)inflammatory responses has been questioned (Nutma et al., 2019). However, the co-occurrence of elevations in the Pandemic cohort also in the brain concentration of mIns (a metabolite that is more abundant in glial cells rather than other cell types) (Datta et al., 2017), the elevation in serum levels of pro-inflammatory cytokines, as well as our imaging transcriptomics analyses (which map the TSPO signal increases to the constitutive architecture of genes implicated in immune/neuroimmune signaling) corroborate a neuroinflammatory interpretation of our imaging findings.
Interestingly, our results have shown that the mean thalamic levels of mIns and [11C]PBR28 were not correlated, albeit we did previously observe an association between the two glial markers in patients with amyotrophic lateral sclerosis (Ratai et al., 2018). It is possible that changes in [11C]PBR28 and mIns levels observed in our study may reflect distinct neuroinflammatory processes or two elements of a common process with different time courses (as previously suggested) (Datta et al., 2017).
When interpreting the results of our study, the reader should be mindful of several limitations. First, the Pandemic cohort had a relatively small sample size (a limitation largely imposed by the COVID-19 pandemic-related disruption on clinical research) (Mitchell et al., 2020). However, multiple factors provide high confidence in the solidity of our observations, including the consistency of our observations across individuals, scanners, age groups, genotypes, and sexes (see Supplementary Fig. 3), the relative stability of the markers prior to the pandemic onset, the observed elevation of two separate imaging markers measured using independent imaging modalities and the finding of significant enrichment in genes highly expressed in glial / immune cells.
Second, our analysis was largely based on unpaired comparisons of groups of individuals scanned either before or after the pandemic onset. While the data from our single subject scanned in both periods demonstrated PET signal elevations in the same regions observed in the group comparison, thus supporting a causal relationship between pandemic-related factors and neuroinflammation, longitudinal assessments of more individuals scanned in the same manner will be needed to better understand the impact of the COVID-19 pandemic on brain health.
Third, the insight offered by our pandemic-specific questionnaire was limited, for several reasons. For instance, these data were not collected at the time of the scan visits, but retrospectively, and only from a subset of subjects. Furthermore, these data were focused on physical and mental symptoms, but did not allow us to isolate the effect of specific factors contributing to those symptoms (e.g., social isolation, financial strain, overall changes in lifestyle, etc). We also note that some of the items might have been difficult to tease apart by participants (e.g., brain fog vs. mental fatigue). As such the link between neuroinflammation and clinical symptoms presented in this report should be regarded as preliminary and will need validation in larger studies, and with additional behavioral assessments.
Fourth, only five out of ten of the collected cytokines/chemokines were detectable, limiting the number our ability to evaluate to what extent circulating inflammatory markers can be linked to neuroinflammatory processes.
One additional limitation is given by the fact that only ∼ 30% of the subjects included in this study had available arterial blood data, yielding limited power to detect significant differences in absolute metrics such as VT. Future studies including a larger number of participants with arterial blood available for VT calculation, will be needed to provide further support to our conclusions.
Finally, in spite of a negative SARS-Cov-2 antibody test in all the subjects in the Pandemic group, it is worth mentioning that we cannot completely exclude the possibility that at least some of our participants might still have been exposed to the virus in the past. In fact, there remains considerable controversy about the persistence of antibodies as a marker of previous infection. However, recent studies confirming the persistence of neutralizing antibodies several months after of an infection (Choe et al., 2021) would exclude this scenario with some confidence. At the same time, none of our Pre-pandemic subjects completed a SARS-Cov-2 antibody test at the time of their participation in our study, which means that we cannot categorically exclude that perhaps a small number of subjects scanned in early 2020 might have been infected. This scenario would suggest that the difference in neuroinflammatory signals between Pre-pandemic and Pandemic groups we described might actually have been somewhat underestimated, although the large sample size of the Pre-pandemic group would likely allay this concern.
This study presents preliminary evidence of pandemic-related neuroinflammation in non-infected participants, providing an example of how broad the impact of the pandemic has been on human health, extending beyond the morbidity directly induced by the virus itself. As prolonged inflammation can be implicated in the pathogenesis of a variety of conditions, including breakdown of immune tolerance (Furman et al., 2019), future studies are needed to assess the long-term implications of COVID-19-pandemic related neuroinflammatory responses. Extending these observations to a larger cohort of healthy subjects followed up longitudinally and/or subjects with pre-existing medical conditions (De Marchi et al., 2021, Colizzi et al., 2020) would represent a valuable objective of future research. Moreover, comparing our findings with those from other studies conducted in world regions with different lockdown lengths, positivity and death rates would be a valuable way to better understand the relationship between pandemic-related stressors and neuroinflammation.
5. Contributors
LB conceived the idea for the study, conducted the main study design and validation, undertook the data analysis, wrote and revised the manuscript. ZA contributed to PET data collection, data pre-processing and manuscript revision. DM performed the imaging transcriptomics analyses, contributed to data interpretation and manuscript content. MK contributed to PET and clinical data collection, data analysis and manuscript revision. AW contributed to 1H-MRS data collection and pre-processing, and manuscript content. HH contributed to data analysis and manuscript revision. EJM, PCK, KACB, OJA, NRZ and DSA contributed to data collection. MM contributed to the development of the main pipeline used for data reconstruction. EMR contributed to the study funding, data collection and manuscript revision. CET contributed to manuscript revision. CC, MV, FT and NH contributed to manuscript revision and data interpretation. BRR contributed to data interpretation. The statistician (NDM) contributed to data analysis. JMH contributed to study funding. The corresponding author (MLL) contributed to study funding, data collection, supervised data analysis and data interpretation and contributed to writing the manuscript. All authors approved the manuscript and had final responsibility for the decision to submit for publication. LB and MLL had full access to and verified the underlying data. The study results were shared with all study team members through online presentations. All authors had the opportunity to contribute to the data interpretation and manuscript content through helpful discussions and written communications. All authors have read and approved the final manuscript.
6. Data sharing
Following publication of the study results, data will be made available upon reasonable request.
7. Ethics approval and consent to participate
The Mass General Brigham (MGB) institutional review board (IRB) gave ethical approval to the study. Written informed consent was obtained from all participants before enrollment, according to the ethical standards of the 1964 Helsinki declaration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to thank Dr. Shibani Mukerji and Dr. Julie Price for helpful discussion on data and quality control, and Angel Torrado-Carvajal, Atreyi Saha, Courtney Bergan and Yang Lin for help with data collection. We thank Dr. Pia Kivisakk and Bianca Trombetta for their work on serum biomarkers. We also thank Grae Arabasz, Regan Butterfield and Shirley Hsu and all the A.A. Martinos Radiochemistry team for producing and administering the radioligand. The authors acknowledge that a preprint of this manuscript is available on medRxiv (https://www.medrxiv.org/content/10.1101/2021.09.21.21263740v1).
Funding sources
This work was funded by the NIH grants R01-NS094306-01A1, R01-NS095937-01A1, R01-DA047088-01 and by The Landreth Family Foundation. The funder had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbi.2022.02.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, National Center for Health Statistics. Indicators of anxiety or depression based on reported frequency of symptoms during the last 7 days. Household Pulse Survey. US Department of Health and Human Services, CDC, National Center for Health Statistics, 2020.
- Abbott A. COVID's mental-health toll: how scientists are tracking a surge in depression. Nature. 2021;590(7845):194–195. doi: 10.1038/d41586-021-00175-z. [DOI] [PubMed] [Google Scholar]
- Holmes E.A., O'Connor R.C., Perry V.H., Tracey I., Wessely S., Arseneault L., Ballard C., Christensen H., Cohen Silver R., Everall I., Ford T., John A., Kabir T., King K., Madan I., Michie S., Przybylski A.K., Shafran R., Sweeney A., Worthman C.M., Yardley L., Cowan K., Cope C., Hotopf M., Bullmore E.d. Multidisciplinary research priorities for the COVID-19 pandemic: a call for action for mental health science. Lancet Psych. 2020;7(6):547–560. doi: 10.1016/S2215-0366(20)30168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves de Andrade E., Šimončičová E., Carrier M., Vecchiarelli H.A., Robert M.-È., Tremblay M.-È. Microglia Fighting for Neurological and Mental Health: On the Central Nervous System Frontline of COVID-19 Pandemic. Front. Cell. Neurosci. 2021;15(12) doi: 10.3389/fncel.2021.647378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar Ray M., Chow K.K., Theodoros T., Wyder M., Steginga A., Sorrensen R., Hickey P., Lau G., Kinsella K., Lombardo C. LOVE in the time of Covid-19: a brief mental health intervention to overcome loneliness. Australas Psychiatry. 2021;29(5):529–534. doi: 10.1177/10398562211010806. [DOI] [PubMed] [Google Scholar]
- Smith K.J., Gavey S., RIddell N.E., Kontari P., Victor C. The association between loneliness, social isolation and inflammation: a systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2020;112:519–541. doi: 10.1016/j.neubiorev.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Stein D.J., Vasconcelos M.F., Albrechet-Souza L., Ceresér K.M., De Almeida R.M. Microglial over-activation by social defeat stress contributes to anxiety-and depressive-like behaviors. Front. Behav. Neurosci. 2017;11:207. doi: 10.3389/fnbeh.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcia M.A., Bonsall D.R., Bloomfield P.S., Selvaraj S., Barichello T., Howes O.D. Stress and neuroinflammation: a systematic review of the effects of stress on microglia and the implications for mental illness. Psychopharmacology. 2016;233(9):1637–1650. doi: 10.1007/s00213-016-4218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberger N.I., Moieni M., Inagaki T.K., Muscatell K.A., Irwin M.R. In Sickness and in Health: The Co-Regulation of Inflammation and Social Behavior. Neuropsychopharmacology. 2017;42(1):242–253. doi: 10.1038/npp.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R. Cytokine, sickness behavior, and depression. Immunol. Allergy Clin. North Am. 2009;29(2):247–264. doi: 10.1016/j.iac.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleshner M., Frank M., Maier S.F. Danger Signals and Inflammasomes: Stress-Evoked Sterile Inflammation in Mood Disorders. Neuropsychopharmacology. 2017;42(1):36–45. doi: 10.1038/npp.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan E., Attwells S., Wilson A.A., Mizrahi R., Rusjan P.M., Miler L., Xu C., Sharma S., Kish S., Houle S., Meyer J.H. Association of translocator protein total distribution volume with duration of untreated major depressive disorder: a cross-sectional study. Lancet Psychiatry. 2018;5(4):339–347. doi: 10.1016/S2215-0366(18)30048-8. [DOI] [PubMed] [Google Scholar]
- Albrecht D.S., Kim M., Akeju O., Torrado-Carvajal A., Edwards R.R., Zhang Y., Bergan C., Protsenko E., Kucyi A., Wasan A.D., Hooker J.M., Napadow V., Loggia M.L. The neuroinflammatory component of negative affect in patients with chronic pain. Mol. Psychiatry. 2021;26(3):864–874. doi: 10.1038/s41380-019-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troubat R., Barone P., Leman S., Desmidt T., Cressant A., Atanasova B., Brizard B., El Hage W., Surget A., Belzung C., Camus V. Neuroinflammation and depression: A review. Eur. J. Neurosci. 2021;53(1):151–171. doi: 10.1111/ejn.14720. [DOI] [PubMed] [Google Scholar]
- Cosenza-Nashat M., Zhao M.L., Suh H.S., Morgan J., Natividad R., Morgello S., et al. Expression of the translocator protein of 18 kDa by microglia, macrophages and astrocytes based on immunohistochemical localization in abnormal human brain. Neuropathol. Appl. Neurobiol. 2009;35(3):306–328. doi: 10.1111/j.1365-2990.2008.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta G., Violante I.R., Scott G., Zimmerman K., Santos-Ribeiro A., Rabiner E.A., Gunn R.N., Malik O., Ciccarelli O., Nicholas R., Matthews P.M. Translocator positron-emission tomography and magnetic resonance spectroscopic imaging of brain glial cell activation in multiple sclerosis. Mult Scler. 2017;23(11):1469–1478. doi: 10.1177/1352458516681504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., Abajian C., Beckmann C.F., Bernard A., Bertagnolli D., Boe A.F., Cartagena P.M., Chakravarty M.M., Chapin M., Chong J., Dalley R.A., Daly B.D., Dang C., Datta S., Dee N., Dolbeare T.A., Faber V., Feng D., Fowler D.R., Goldy J., Gregor B.W., Haradon Z., Haynor D.R., Hohmann J.G., Horvath S., Howard R.E., Jeromin A., Jochim J.M., Kinnunen M., Lau C., Lazarz E.T., Lee C., Lemon T.A., Li L., Li Y., Morris J.A., Overly C.C., Parker P.D., Parry S.E., Reding M., Royall J.J., Schulkin J., Sequeira P.A., Slaughterbeck C.R., Smith S.C., Sodt A.J., Sunkin S.M., Swanson B.E., Vawter M.P., Williams D., Wohnoutka P., Zielke H.R., Geschwind D.H., Hof P.R., Smith S.M., Koch C., Grant S.G.N., Jones A.R. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.Y., Tan M.S., Yu J.T., Tan L. Role of pro-inflammatory cytokines released from microglia in Alzheimer's disease. Ann Transl Med. 2015;3(10):136. doi: 10.3978/j.issn.2305-5839.2015.03.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe P.G., Kim K.-H., Kang C.K., Suh H.J., Kang E., Lee S.Y., Kim N.J., Yi J., Park W.B., Oh M.-D. Antibody Responses 8 Months after Asymptomatic or Mild SARS-CoV-2 Infection. Emerg. Infect. Dis. 2021;27(3):928–931. doi: 10.3201/eid2703.204543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Guo Q., Rabiner E.A., Gunn R.N. The impact of the rs6971 polymorphism in TSPO for quantification and study design. Clin. Transl. Imag. 2015;3(6):417–422. [Google Scholar]
- Kreisl W.C., Jenko K.J., Hines C.S., Lyoo C.H., Corona W., Morse C.L., Zoghbi S.S., Hyde T., Kleinman J.E., Pike V.W., McMahon F.J., Innis R.B. A genetic polymorphism for translocator protein 18 kDa affects both in vitro and in vivo radioligand binding in human brain to this putative biomarker of neuroinflammation. J. Cereb. Blood Flow Metab. 2013;33(1):53–58. doi: 10.1038/jcbfm.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Wehrl H.F., Hofmann M., Judenhofer M.S., Eriksson L., Ladebeck R., Lichy M.P., Byars L., Michel C., Schlemmer H.-P., Schmand M., Claussen C.D., Sossi V., Pichler B.J. Technical performance evaluation of a human brain PET/MRI system. Eur. Radiol. 2012;22(8):1776–1788. doi: 10.1007/s00330-012-2415-4. [DOI] [PubMed] [Google Scholar]
- Delso G., Fürst S., Jakoby B., Ladebeck R., Ganter C., Nekolla S.G., Schwaiger M., Ziegler S.I. Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. J. Nucl. Med. 2011;52(12):1914–1922. doi: 10.2967/jnumed.111.092726. [DOI] [PubMed] [Google Scholar]
- Alshelh Z., Albrecht D.S., Bergan C., Akeju O., Clauw D.J., Conboy L., Edwards R.R., Kim M., Lee Y.C., Protsenko E., Napadow V., Sullivan K., Loggia M.L. In-vivo imaging of neuroinflammation in veterans with Gulf War illness. Brain Behav. Immun. 2020;87:498–507. doi: 10.1016/j.bbi.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D.S., Granziera C., Hooker J.M., Loggia M.L. In Vivo Imaging of Human Neuroinflammation. ACS Chem. Neurosci. 2016;7(4):470–483. doi: 10.1021/acschemneuro.6b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D.S., Forsberg A., Sandström A., Bergan C., Kadetoff D., Protsenko E., Lampa J., Lee Y.C., Höglund C.O., Catana C., Cervenka S., Akeju O., Lekander M., Cohen G., Halldin C., Taylor N., Kim M., Hooker J.M., Edwards R.R., Napadow V., Kosek E., Loggia M.L. Brain glial activation in fibromyalgia - A multi-site positron emission tomography investigation. Brain Behav. Immun. 2019;75:72–83. doi: 10.1016/j.bbi.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSabato D.J., Nemeth D.P., Liu X., Witcher K.G., O'Neil S.M., Oliver B., et al. Interleukin-1 receptor on hippocampal neurons drives social withdrawal and cognitive deficits after chronic social stress. Mol. Psychiatry. 2020 doi: 10.1038/s41380-020-0788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L., Zhang A., Sun N., Liu P., Yang C., Li G., Liu Z., Wang Y., Zhang K. Functional connectivity between the thalamus and the primary somatosensory cortex in major depressive disorder: a resting-state fMRI study. BMC Psychiatry. 2018;18(1) doi: 10.1186/s12888-018-1913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.Z., Wang Y.X., Jiang C.L. Inflammation: The Common Pathway of Stress-Related Diseases. Front. Hum. Neurosci. 2017;11:316. doi: 10.3389/fnhum.2017.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky R.M., Romero L.M., Munck A.U. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 2000;21(1):55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- Tian L.i., Hui C.W., Bisht K., Tan Y., Sharma K., Chen S., Zhang X., Tremblay M.-E. Microglia under psychosocial stressors along the aging trajectory: Consequences on neuronal circuits, behavior, and brain diseases. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2017;79:27–39. doi: 10.1016/j.pnpbp.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Konsman J.P., Parnet P., Dantzer R. Cytokine-induced sickness behaviour: mechanisms and implications. Trends Neurosci. 2002;25(3):154–159. doi: 10.1016/s0166-2236(00)02088-9. [DOI] [PubMed] [Google Scholar]
- Airas L., Rissanen E., Rinne J.O. Imaging neuroinflammation in multiple sclerosis using TSPO-PET. Clin. Transl. Imaging. 2015;3(6):461–473. doi: 10.1007/s40336-015-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucksdorff M, Matilainen M, Tuisku J, Polvinen E, Vuorimaa A, Rokka J, et al. Brain TSPO-PET predicts later disease progression independent of relapses in multiple sclerosis. Brain. 2020;143(11):3318-30. [DOI] [PMC free article] [PubMed]
- Li H.-D., Li M., Shi E., Jin W.-N., Wood K., Gonzales R., Liu Q. A translocator protein 18 kDa agonist protects against cerebral ischemia/reperfusion injury. J. Neuroinflamm. 2017;14(1) doi: 10.1186/s12974-017-0921-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.M., Zanotti-Fregonara P., Fujita M., Newman L., Farmer C., Ballard E.D., Machado-Vieira R., Yuan P., Niciu M.J., Lyoo C.H., Henter I.D., Salvadore G., Drevets W.C., Kolb H., Innis R.B., Zarate Jr C.A. PET radioligand binding to translocator protein (TSPO) is increased in unmedicated depressed subjects. EJNMMI Res. 2018;8(1) doi: 10.1186/s13550-018-0401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Tuo H., Wang S., Zhao L. The Roles of Monocyte and Monocyte-Derived Macrophages in Common Brain Disorders. Biomed Res. Int. 2020;2020:1–11. doi: 10.1155/2020/9396021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane S.L., Kremlev S., Amini S., Sawaya B.E. Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hridi S.U., Barbour M., Wilson C., Franssen A.J., Harte T., Bushell T.J., Jiang H.-R. Increased Levels of IL-16 in the Central Nervous System during Neuroinflammation Are Associated with Infiltrating Immune Cells and Resident Glial Cells. Biology (Basel) 2021;10(6):472. doi: 10.3390/biology10060472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W.L., Karpus W.J., Van Eldik L.J. MCP-1-deficient mice show reduced neuroinflammatory responses and increased peripheral inflammatory responses to peripheral endotoxin insult. J. Neuroinflamm. 2008;5(1):1–13. doi: 10.1186/1742-2094-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutma E, Stephenson JA, Gorter RP, de Bruin J, Boucherie DM, Donat CK, et al. 2019. A quantitative neuropathological assessment of translocator protein expression in multiple sclerosis. Brain. 142 (11), 3440-55. [DOI] [PMC free article] [PubMed]
- Ratai E.-M., Alshikho M.J., Zürcher N.R., Loggia M.L., Cebulla C.L., Cernasov P., Reynolds B., Fish J., Seth R., Babu S., Paganoni S., Hooker J.M., Atassi N. Integrated imaging of [(11)C]-PBR28 PET, MR diffusion and magnetic resonance spectroscopy (1)H-MRS in amyotrophic lateral sclerosis. Neuroimage Clin. 2018;20:357–364. doi: 10.1016/j.nicl.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell E.J., Ahmed K., Breeman S., Cotton S., Constable L., Ferry G., Goodman K., Hickey H., Meakin G., Mironov K., Quann N., Wakefield N., McDonald A. It is unprecedented: trial management during the COVID-19 pandemic and beyond. Trials. 2020;21(1) doi: 10.1186/s13063-020-04711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D., Campisi J., Verdin E., Carrera-Bastos P., Targ S., Franceschi C., Ferrucci L., Gilroy D.W., Fasano A., Miller G.W., Miller A.H., Mantovani A., Weyand C.M., Barzilai N., Goronzy J.J., Rando T.A., Effros R.B., Lucia A., Kleinstreuer N., Slavich G.M. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25(12):1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi F., Gallo C., Sarnelli M.F., De Marchi I., Saraceno M., Cantello R., Mazzini L. Accelerated Early Progression of Amyotrophic Lateral Sclerosis over the COVID-19 Pandemic. Brain Sci. 2021;11(10):1291. doi: 10.3390/brainsci11101291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colizzi M., Bortoletto R., Silvestri M., Mondini F., Puttini E., Cainelli C., Gaudino R., Ruggeri M., Zoccante L. Medically unexplained symptoms in the times of COVID-19 pandemic: a case-report. Brain, Behav, Immun - Health. 2020;5:100073. doi: 10.1016/j.bbih.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.