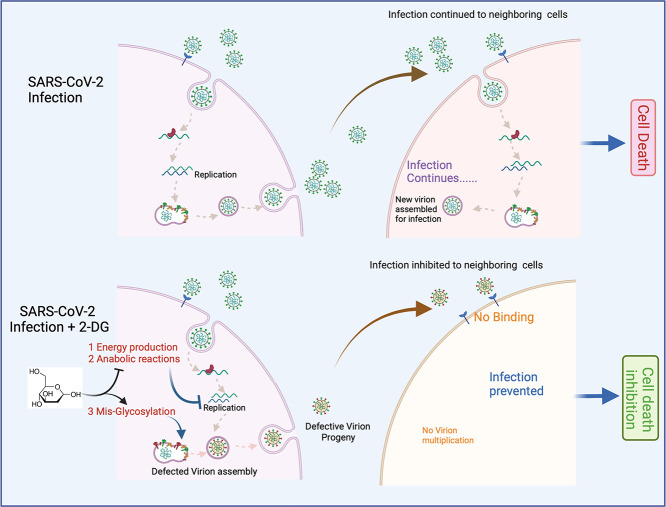
Abstract
Aims
Virus-infected host cells switch their metabolism to a more glycolytic phenotype, required for new virion synthesis and packaging. Therefore, we investigated the effect and mechanistic action of glycolytic inhibitor 2-Deoxy-d-glucose (2-DG) on virus multiplication in host cells following SARS-CoV-2 infection.
Main methods
SARS-CoV-2 induced change in glycolysis was examined in Vero E6 cells. Effect of 2-DG on virus multiplication was evaluated by RT-PCR (N and RdRp genes) analysis, protein expression analysis of Nucleocapsid (N) and Spike (S) proteins and visual indication of cytopathy effect (CPE), The mass spectrometry analysis was performed to examine the 2-DG induced change in glycosylation status of receptor binding domain (RBD) in SARS-CoV-2 spike protein.
Key findings
We observed SARS-COV-2 infection induced increased glucose influx and glycolysis, resulting in selectively high accumulation of the fluorescent glucose analog, 2-NBDG in Vero E6 cells. 2-DG inhibited glycolysis, reduced virus multiplication and alleviated cells from virus-induced cytopathic effect (CPE) in SARS-CoV-2 infected cells. The progeny virions produced from 2-DG treated cells were found unglycosylated at crucial N-glycosites (N331 and N343) of the receptor-binding domain (RBD) in the spike protein, resulting in production of defective progeny virions with compromised infective potential.
Significance
The mechanistic study revealed that the inhibition of SARS-COV-2 multiplication is attributed to 2-DG induced glycolysis inhibition and possibly un-glycosylation of the spike protein, also. Therefore, based on its previous human trials in different types of Cancer and Herpes patients, it could be a potential molecule to study in COVID-19 patients.
Keywords: SARS-CoV-2, Glycolysis, 2-DG, Unglycosylation, COVID-19
Graphical abstract
1. Introduction
The coronavirus pandemic is an ongoing global outbreak of COVID-19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and recognized as Public Health Emergency of International Concern by the World Health Organization (WHO). The highly infectious virus pathogenesis caused by SARS-CoV-2 is a respiratory disease occurring in three Phases 1) Virus multiplication, 2) Immune hyper-reactivity, and 3) Pulmonary destruction [1]. The faster multiplication of COVID-19 virus-driven damage to epithelial cells results in pulmonary destruction and cytokine storm [2]. An immense effort has already been made, and many are underway worldwide to develop effective anti-virus therapeutic against this fast mutating virus to control the disease severity and mortality [3]. Currently, newly identified therapeutics are under investigation, and there is no established therapy for the treatment of COVID-19. Treatment is primarily based on supportive and symptomatic care. Therefore, the development of effective therapeutic against COVID-19 is urgently needed.
Upon infection, the virus hijacks and reprograms the host cell metabolism for its own rapid multiplication as new virion assembly involves very high demand and turnover of nucleotides and lipids [4]. This enhanced demand of virus-infected host cells is solely fulfilled by induced anabolic reactions to synthesize more nucleotide and lipid using glucose and glutamine as substrate [5]. Like cancer cells, the enhanced aerobic glycolysis, popularly known as the Warburg effect, was also observed in SARS-CoV-2 infected host cells, satisfying the elevated anabolic demand [6], [7], [8]. In addition to providing direct substrates for virion assembly, adjustments to metabolic pathways are also required to provide ATP at a rapid rate for the high energy costs of virus genome replication and packaging in host cells [5]. While oxidative phosphorylation provides significantly more ATP per glucose, glycolysis is a much faster process for providing ATP rapidly [9]. However, utilizing glycolysis as the main metabolic pathway requires an increased influx of extracellular glucose using enhanced expression of glucose transporters viz. GLUT1, GLUT4, etc., to match the increased metabolic rate [9]. Unlike normal cells, which show more plasticity in their metabolism by relying on both glycolytic and mitochondrial pathways, SARS-CoV-2 infected cells operate predominantly on glycolysis for bioenergetic and anabolic demand due to compromised mitochondrial respiratory function [6]. Based on these facts, the intervention at the level of SARS-CoV-2 induced host cell metabolism is a promising target to exploit as a potential strategy for developing anti-virus therapy to control the progression of the disease and thereby the pandemic.
The glycolytic inhibitor, 2-Deoxy-d-glucose (2-DG), is a synthetic analog of glucose, which blocks glycolysis at the initial stage and causes depletion of ATP, anabolic intermediates required for virus synthesis, and glucose derivatives used in protein glycosylation [10]. It is demonstrated in earlier studies that inhibition of reprogrammed metabolism of virus-infected host cells using 2-DG potently impairs virus multiplication by reverting anabolic reprogramming [6], [7], [8], [11], [12], [13], [14]. Hence, we used 2-DG to target the selective and predominant metabolic pathway of the virus-infected host cell, glycolysis, to inhibit virus multiplication.
The present study was undertaken in the Vero E6 cell line used as an in-vitro host model system for SARS-CoV-2 infection and multiplication. Since Vero E6 cells support viral replication to high titers and moreover high expression levels of ACE-2 receptor used by SARS-CoV-2 virus for internalization and/or their inability to produce interferon makes this cell system as a suitable in-vitro model to isolate, propagate and study SARS-CoV-like viruses [15]. In this study, we studied the effect of SARS-CoV-2 infection on the alterations of host cell metabolism with respect to enhanced expression of transporters and enzymes responsible for glucose uptake and glycolysis pathway. We found 2-DG treatment not only inhibited the virus multiplication but also led to the formation of defective virions, which have reduced ability to infect the newer cells resulting in overall effective inhibition of virus growth.
2. Results
2.1. SARS-CoV-2 infection induces cellular glucose influx and glycolysis
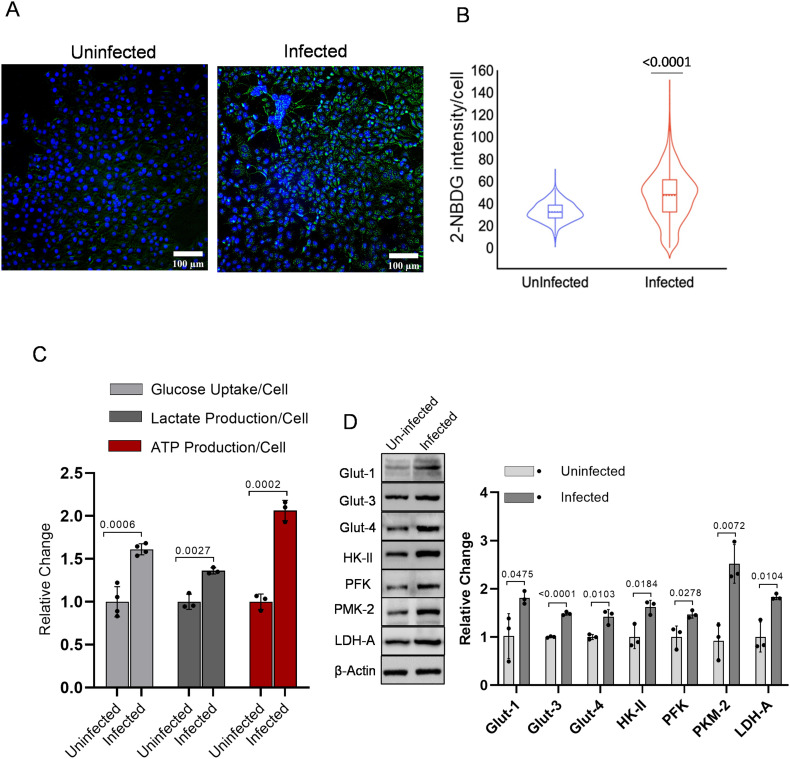
To evaluate the effect of SARS-CoV-2 infection mediated reprogramming of the host cell metabolism towards enhanced glucose utilization, we estimated the glucose uptake using 2-NBDG, a fluorescent glucose/2-DG analog. Using fluorescence imaging of cells, we found SARS-CoV-2 infection in Vero E6 cell leads to profound accumulation of 2-NBDG as compared to uninfected cells (Fig. 1A), which was confirmed by image analysis and quantitative estimation of 2-NBDG fluorescence intensity and lactate estimation (Fig. 1B–C). Additionally, the enhanced glucose uptake and lactate production in infected cells was confirmed by significantly increased levels of key glucose transporter proteins GLUT1, GLUT3 and GLUT4, corroborating the infection mediated upregulation of glucose influx in virus-infected cells (Fig. 1 D). Besides, significantly enhanced levels of key glycolytic enzymes viz. Hexokinase-II (HK-II), Phsopho-fructokinase-I (PFK-1), Pyruvate Kinase (PKM-2), and Lactate dehydrogenase (LDH-A) were observed in virus-infected cells (Fig. 1 D). Analysis of glucose uptake and glycolytic enzymes revealed that virus-infected cells accelerate the glycolysis to meet the high energy demand, supported by the significant increase in ATP production, compared to uninfected cells (Fig. 1C). Moreover, the higher protein levels of glycolytic enzymes are responsible for the regulation of diverting the glucose through glycolysis and conversion of pyruvate to lactate after glycolysis, substantiating the evidence of reprogrammed metabolism towards increased glycolysis in SARS-CoV-2 infected Vero E6 cells.
Fig. 1.
SARS-CoV-2 infection induces glycolysis in Vero E6 cells.
A. Photomicrographs show the merged image of both 2-NBDG (200 μM) and Hoechst-3342 nuclear staining In Vero E6 cells at 48 h post-infection captured under 10 × 10× magnification (see also supplementary Fig. 1). B. An intracellular fluorescence uptake was quantified (derived information of Fig. 1A) using software toolbox CELLSEGM on MATLAB, and the intracellular 2-NBDG uptake was examined in >1000 cells. The violin plot is the derived information of Fig. 1 A, indicating the median signal intensity of 2-NBDG in the indicated treatment groups (from two independent observation). C. In a similar experimental condition, spectrofluorimetric estimation of 2-NBDG (n = 3; from four independent observations), whereas lactate production and luminescence-based ATP determination (n = 3; from three independent observations) were carried out followed by normalization with respective cell number and protein content, respectively. Bar plot presented as relative fold change (RFC) with respect to uninfected (control) in the indicated groups. D. Immunoblot and densitometric evaluation indicating the glucose influx and glycolytic profile of SARS-CoV-2 infected Vero E6 cells and presented as RFC with respective control (additional blot images are also provided in supplementary Fig. 2). All barplots show the mean ± standard deviation (SD), and individual dot represents the mean value of three independent sample observations (n = 3). P-values were calculated with a two-tailed Student's t-test with respect to uninfected control.
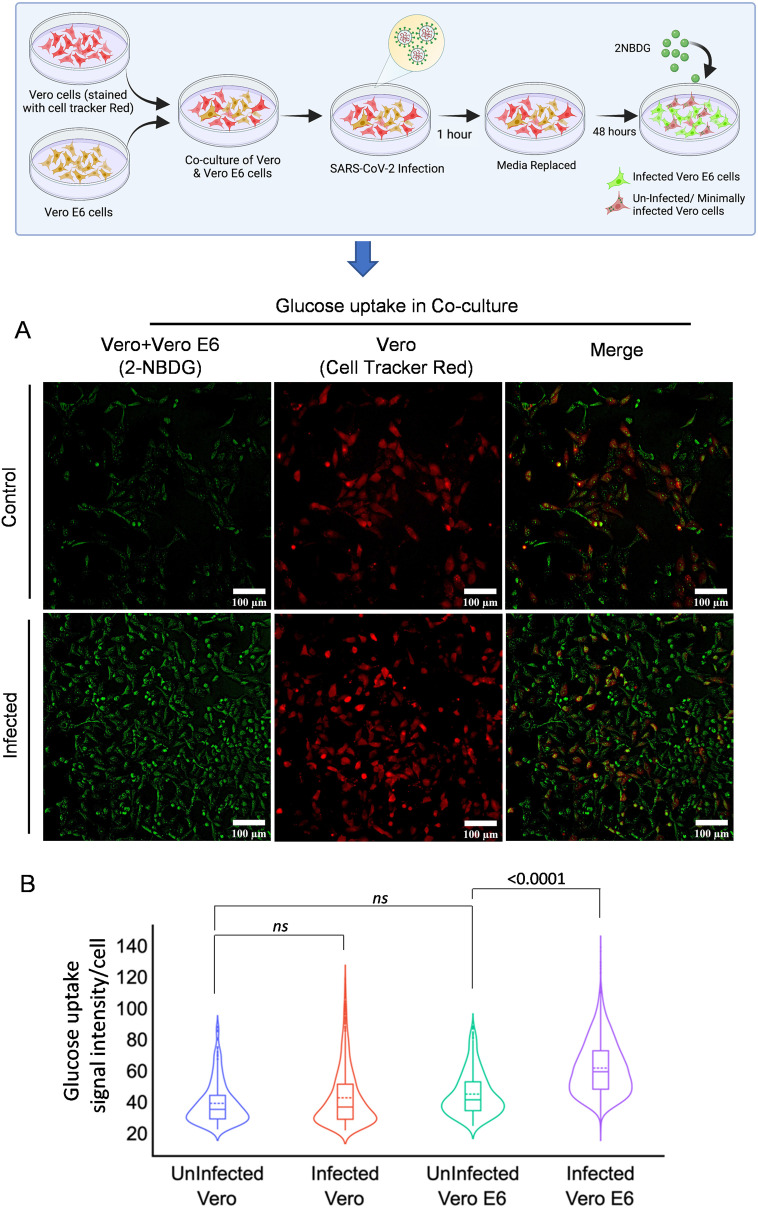
2.2. SARS-CoV-2 infection-mediated enhanced glucose uptake is selective to infected cells
To further deepen our understanding about the differential glucose uptake in high and low or minimally SARS-CoV-2 infected cells, co-culture of Vero and Vero E6 cells was performed using fluorescent analog (2-NBDG) of 2-DG. To rule out the phenotypic differences in basal glucose uptake, cells with similar origins (Vero and Vero-E6) were selected. Vero cells showed compromised infection and poor multiplication efficiency of SARS-CoV-2 virus upon infection with similar MOI (0.3) with respect to Vero E6 (data provided in supplementary Fig. 3) [16]. To differentiate between the cells, Vero cells were stained with CTR (cell tracker red) and co-cultured with Vero E6 cells; the co-culture was infected 24 h later with SARS-CoV-2 (Fig. 2A). Interestingly, the basal level uptake of 2-NBDG in infected Vero cells showed a minimal increase compared to respective control visualized as merged green (2-NBDG) and red (CTR) emission of Vero cells (Fig. 2A). Whereas the mostly acquired intracellular green fluorescence in co-culture corresponds to a significant increase of 2-NBDG uptake by Vero E6 cells following infection, compared to uninfected cells (Fig. 2B). Quantitative estimation of fluorescence intensity from multiple images of these samples confirmed that 2-NBDG uptake is significantly favored by SARS-CoV-2 infection, selectively in highly infected Vero E6 cells (Fig. 2B). These findings suggest that SARS-CoV-2 infection favors increased glucose as well as a 2-DG influx in cells.
Fig. 2.
Glucose uptake is selectively more in highly infected cells.
A. The graphical presentation depicts the methodology of co-culture of Vero and Vero E6 cells. Vero cells with relatively poor ability of infection uptake were stained with CellTracker™ Red and co-cultured with Vero E6 cells to test if 2-NBDG accumulation is preferably favored by SARS-CoV-2 infected cells. Photomicrographs in the left panel show the 2-NBDG (green) uptake in both Vero and Vero E6 cells of indicated treatment groups whereas, the middle panel are representative of CTR (Red) stained only Vero cells in the same field and right panel micrographs are the merged representation of left and right panel for the better appreciation of differential 2-NBDG accumulation in both the cell lines (additional image panels are also provided in supplementary Fig. 4). B. The violin plot is the derived information of Fig. 2 A, indicating the median signal intensity of 2-NBDG in the indicated treatment groups (from two independent observation). The non-significant (ns) and significant change in the indicated groups were calculated with a two-tailed Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
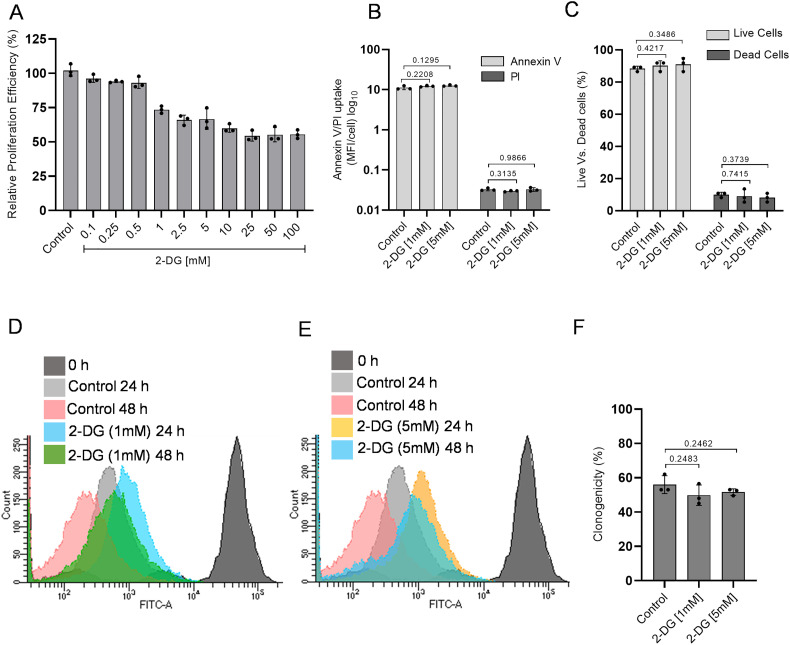
2.3. Effect of glucose antimetabolite 2-DG on cell proliferation and viability
Since SARS-CoV-2 infected cells predominantly showed increased glucose metabolism. Therefore, we hypothesized that selective inhibition of glucose metabolism using 2-DG might reduce the SARS-CoV-2 infection in these cells. To validate our hypothesis first, we examined the safe concentration range of 2-DG in Vero E6 cells using sulforhodamine B (SRB) assay to understand the effect of 2-DG on cell proliferation. The decrease in cell growth was observed in 2-DG treated cells and notified as inhibition range of 1–10% at 0.1–0.5 mM, 10–30% at 1-5 mM, and reached maximum inhibition ~40–50% at 10-100 mM (Fig. 3 A) in Vero E6 cells. It was interesting to observe that 5–100 mM showed a near saturation range of growth inhibition (Fig. 3 A), indicating that the observed growth inhibition is not due to the toxicity of 2-DG. Moreover, no significant change was observed either in Annexin V/PI uptake (Fig. 3B) or live vs. dead cell ratio (Fig. 3C) of control and 2-DG (up to 5 mM) treated cells, which indicates that 2-DG induced cell growth inhibition is not linked with cellular sensitization to death by 2-DG (Fig. 3B&C). As the glycolysis inhibitor 2-DG is known to inhibit the proliferation of rapidly multiplying cells and exert a cytostatic effect. Therefore, we analyzed the effect of 2-DG on cell proliferation using the CFSE probe. Relatively more CFSE fluorescence in 1 mM and 5 mM treated cells as compared to their respective time control indicates the slower proliferation in 2-DG treated Vero E6 cells (Fig. 3 D&E). Further, to understand deeply about the 2-DG induced growth inhibition, we analyzed the reproductive potential/clonogenicity following 2-DG treatment by performing a macrocolony assay (Fig. 3F). The non-significant change in the clonogenicity between control and 2-DG (1 and 5 mM) treated cells showed that 2-DG induced growth inhibition is due to reduced proliferation and cytostatic effect and not due to cytotoxic effect. Therefore, the highest used concentrations of 2-DG (up to 5 mM, in most of the experiments) were found to be safe for Vero E6 cells.
Fig. 3.
2-DG treatment reduces the proliferation efficiency of Vero E6 cells.
A. SRB assay was performed to examine the effect of 2-DG on cell proliferation. The graph shows the relative proliferation efficiency (%) of Vero E6 cells at different indicated concentrations of 2-DG with respect to control. B–C. Bar plots indicate the relative uptake in Annexin V/PI (B) and percent of live vs. dead cell population (C) obtained from trypan blue exclusion assay in the indicated treatment groups. D–E. CFSE proliferation assay was performed, and a differential overlay graph obtained in the indicated treatment groups [1 mM] (D) and [5 mM] (E) shows the proliferation-dependent fluorescence shift in Vero E6 cells. F. The barplot represents the reproductive potential/clonogenicity of 2-DG treated cells at 1 and 5 [mM], with respect to control. All barplots show the mean ± standard deviation (SD) obtained from three independent observations (n = 3) and, P-values were calculated with a two-tailed Student's t-test with respect to uninfected control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. 2-DG inhibits SARS-CoV-2 replication and ATP production
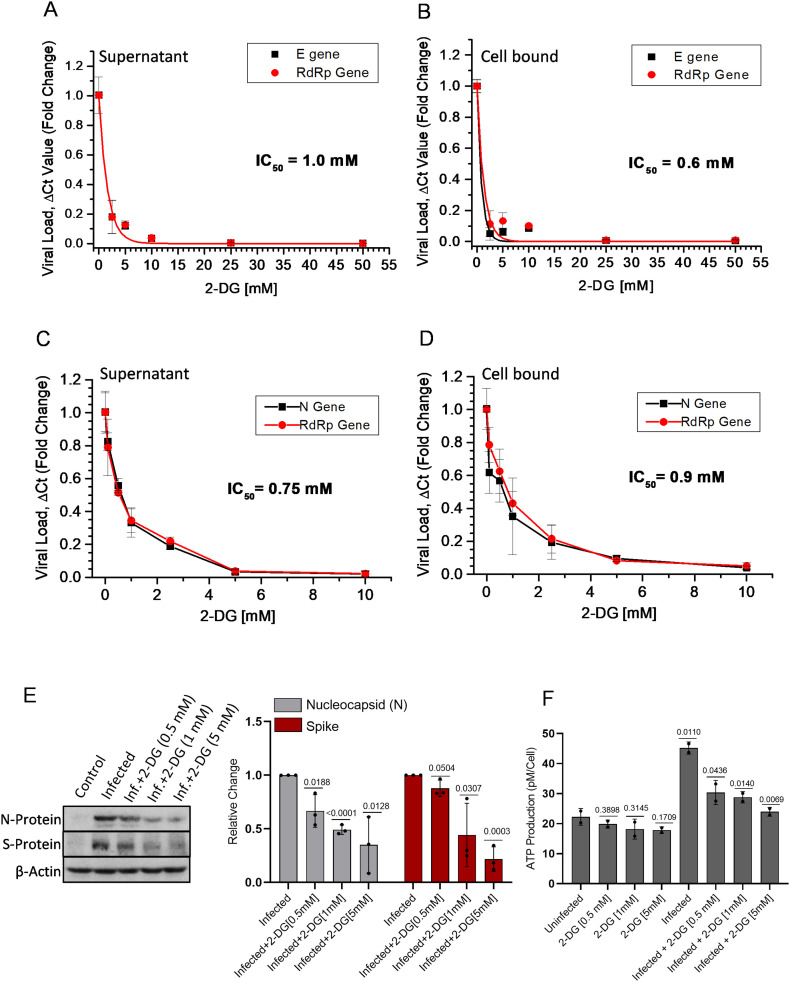
Initially, we assessed the concentration-dependent growth inhibition effect of 2-DG on SARS-CoV-2 (B.6 variant; earlier in May 2020) multiplication (GISAID ID: EPI_ISL_458075; virus ID- hCoV-19/India/TG-CCMB-O2-P1/2020) in Vero cells [17]. The value of half-maximal inhibitory concentration (IC50) on viral E and RdRp gene multiplication was estimated at 1 and 0.6 mM in the supernatant and cell-bound fractions, respectively (Fig. 4A & B). Next, we examined the effect of 2-DG on virus multiplication of B.1.1 variant of SARS-CoV-2 (HV69/70 mutation in S gene, isolated from a patient sample ID INMAS/nCoV/8415) by analyzing the RdRp gene and N gene from secreted virions in cell medium (Fig. 4C) and cell-bound virions in cellular fraction (Fig. 4D). 2-DG strongly inhibited SARS-CoV-2 growth in Vero E6 cells (Fig. 4 C&D). The estimated IC50 value was found to be 0.75 mM and nearly 0.9 mM in released and cell-bound fractions, respectively, which was nearly similar to B.6 variant. It was also observed that 2-DG reduces the level of virus proteins, namely nucleocapsid (N) and spike (S) protein, in a dose-dependent manner (Fig. 4E). Further, in 2-DG treated cells, reduced viral load and protein expression was correlated with the significant reduction in cellular ATP content following SARS-CoV-2 infection (Fig. 4F). It is also pertinent to note here that 2-DG treatment alone did not show a considerable reduction in ATP production (Fig. 4F). These results indicate that 2-DG inhibits glycolysis and thereby faster ATP production, eventually preventing virus multiplication in host cells.
Fig. 4.
2-DG treatment inhibits SARS-CoV-2 replication in Vero and Vero E6 cells.
A-D. RT-PCR of viral E and RdRp genes (A&B) in SARS-CoV-2 infected (B.6 variant) Vero cells; N and RdRP genes (C&D) in infected (variant B.1.1) Vero E6 cells were performed in the supernatant of the infected culture and its cellular fraction. The concentration-dependent inhibitory effect of 2-DG is presented as relative fold change (RFC) in reduced virus load (ΔCt) with respect to the infection control group (n = 3). E. Immunoblotting of virus N and S protein was performed with the whole cell lysate of indicated treatment groups at 24 h post-infection, and the densitometry graph shows the RFC in 2-DG treated [0.5, 1 & 5 mM] fraction with respect to infection alone (from three independent observations; see also supplementary Fig. 5A). F. ATP was estimated at 48 h post-infection in the indicated treatment groups followed by normalization with respective cell numbers and graph presented as ATP production pM/Cell (n = 3) from two independent observations. Data represent the mean ± SD and P-values were calculated using two-tailed Student's t-test.
2.5. 2-DG inhibits SARS-CoV-2 mediated cytopathic effect and cell death
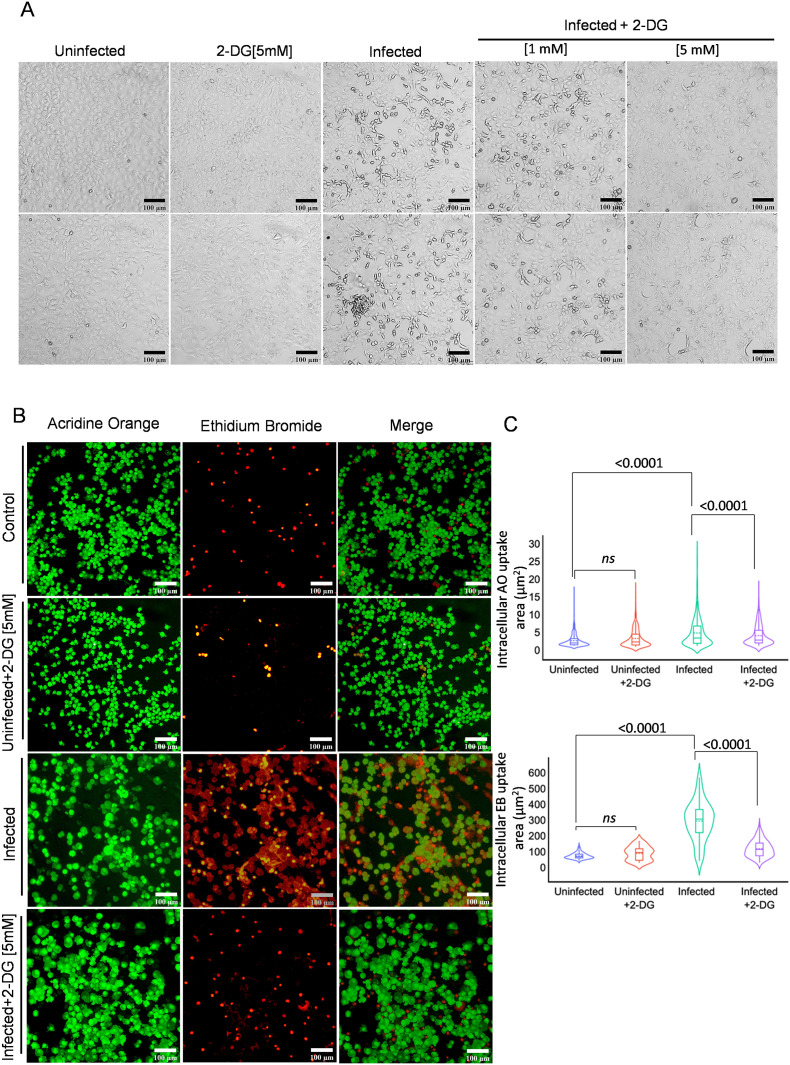
The virus infection-induced cellular deformity, cell lysis, and cell death of host cells are very well characterized phenomenon and commonly known as cytopathic effect (CPE). After analyzing the effect of 2-DG on virus multiplication, next, we analyzed the effect of 2-DG on SARS-CoV-2 infection mediated cytopathic effect (CPE) and induction of cell death. Upon microscopic examination, a profound change in cell morphology was observed in infected cells at 48 h post-infection, whereas this effect tends to be low at 1 mM and visibly absent at 5 mM 2-DG, suggesting considerable low CPE following 2-DG treatment (Fig. 5A). The SARS-CoV-2 infection-induced change in cell death index and integrity of cell membrane was further analyzed by fluorescence imaging of dual DNA binding stain acridine orange (AO: Green) and ethidium bromide (EB: Red), viewed and analyzed in terms of the rate of selective dye ingress and accumulation depending upon the physiological state of the cell. An increased EB ingress in large population of infected cells indicates substantial loss to membrane integrity and cell death. In contrast, 2-DG treated cells showed a significant reduction in cell death confirmed by the quantitative evaluation of cellular fluorescence intensity of both AO/EB stained cells (Fig. 5B–C). Together these results suggest that 2-DG treatment enables cells to overcome the cellular stress caused due to SARS-CoV-2 multiplication and thereby reduces cell death.
Fig. 5.
2-DG treatment reduces SARS-CoV-2 induced cell death.
A. Photomicrographs show the differential change in cellular morphology upon virus infection in the indicated treatment groups performed at 48 h post-infection in Vero E6 cells (n = 4). B. In the similar experimental condition, fluorescence images of acridine orange (AO) and ethidium bromide (EB) (AO/EB; 100 μg/ml each; 1:1) were presented using red and green emission channel respectively in untreated, and 2-DG treated [5 mM] Vero E6 cells (suspension form) captured under 10 × 10 X magnification (additional image panels are also provided in supplementary Fig. 6). C. Photo-morphometric analysis was carried out using algorithms from CELLSEGM and Image Processing toolbox in Matlab 2020a and violin plot (derived information of Fig. 5 B), indicating the the median of AO/EB uptake (obtained from two independent observation) in the indicated treatment groups. The non-significant (ns) and significant change in the indicated groups were calculated with a two-tailed Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.6. Effect of glucose and mannose on 2-DG induced inhibition of virus multiplication
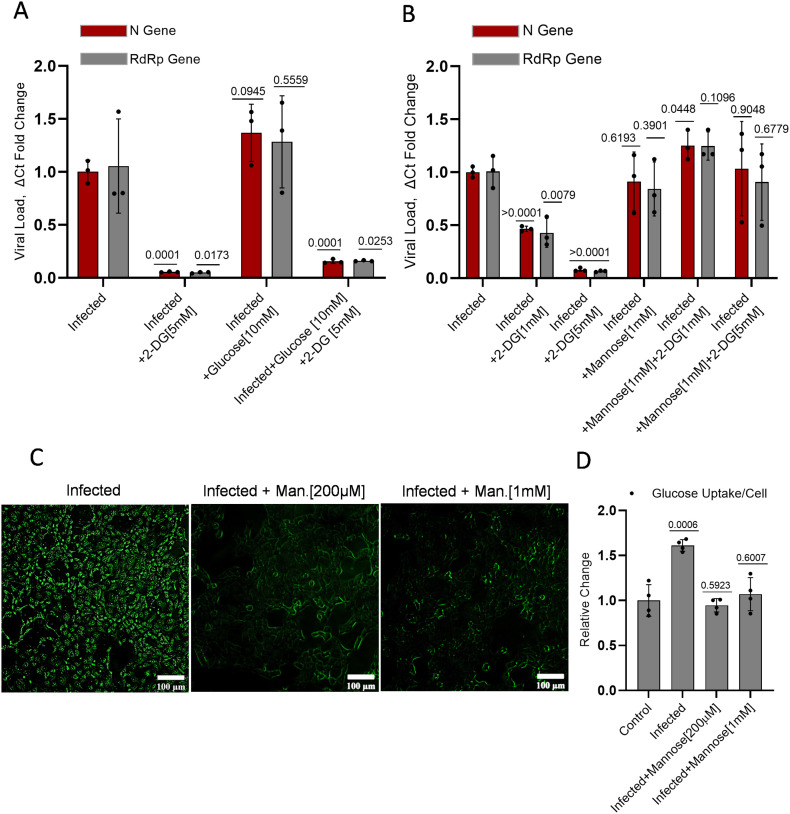
As shown in the previous section, glycolysis is an intrinsic requirement of virus multiplication in host cells, and its inhibition by 2-DG attenuated the virus growth. In a recent study, it was reported that enhanced glucose level favors SARS-CoV-2 growth [6], [7], [8]. To test the effect of increased glucose level on the growth inhibition efficacy of 2-DG, we treated the cells with additional 10 mM Glucose. The 3-fold increased glucose level (5.5 mM in media +10 mM) enhanced the virus multiplication by 32% (Fig 6A) in infected cells. The 2-DG treatment (5 mM) inhibited the virus multiplication by 95% at equimolar glucose concentration in media; however, on the addition of 10 mM glucose, this inhibition was significantly decreased to 88% as compared to its respective control and 84% with respect to infection control (Fig 6A). It is interesting to note that 3-fold increased glucose (~15 mM) could reduce the effect of 2-DG (5 mM) by 7%, only. This finding suggests that glucose is not the primary and efficient competitor of 2-DG uptake in virus-infected cells. Since 2-DG also shares structural similarity with mannose, an epimer of glucose, we tested if mannose can attenuate the effect of 2-DG on SARS-CoV-2 multiplication. Interestingly, we found that mannose is able to reduce the effect of 2-DG on virus multiplication at equimolar and 1/5th of mannose (1 mM) to 2-DG (5 mM) ratio (Fig 6B). We further checked the effect of mannose on glucose uptake. In the qualitative and quantitative examination, it was found that mannose inhibited the uptake of 2-NBDG (a fluorescent analog of glucose/2-DG) in infected cells (Fig. 6C–D). These results suggest that the uptake of 2-DG is affected mainly by mannose and not much by glucose, which reduces the effect of 2-DG on SARS-CoV-2 multiplication.
Fig. 6.
2-DG competes with mannose for intracellular uptake in SARS-CoV-2 infected cells.
A-B The RFC graph presented with respect to the infected group alone indicating the 2-DG induced inhibition of N and RdRP gene and the magnitude of alteration in 2-DG efficacy showed by the separate combination of (A) glucose (glu) and (B) mannose (Man.) at the indicated concentrations. C. Photomicrographs shows the uptake of 2-NBDG [200 μM] affected by equimolar concentration 200 μM and 1 mM of mannose at 48 h post-infection (additional images are also provided in supplementary Fig. 7). D. In the similar experimental condition 2-NBDG uptake was measured on Spectrofluorometer using 465/540 (ex/em.) and the graph represents the RFC of 2-NBDG in the indicated groups. Data represent the mean ± SD and P-values in barplots (A&B) were calculated with respect to infected N and RdRp genes of the infected group (from three independent observations), whereas significance value in barplot (D) is generated as compared to an uninfected control group from (four independent observations) using two-tailed Student's t-test.
2.7. 2-DG weakens the infectivity potential of progeny virions
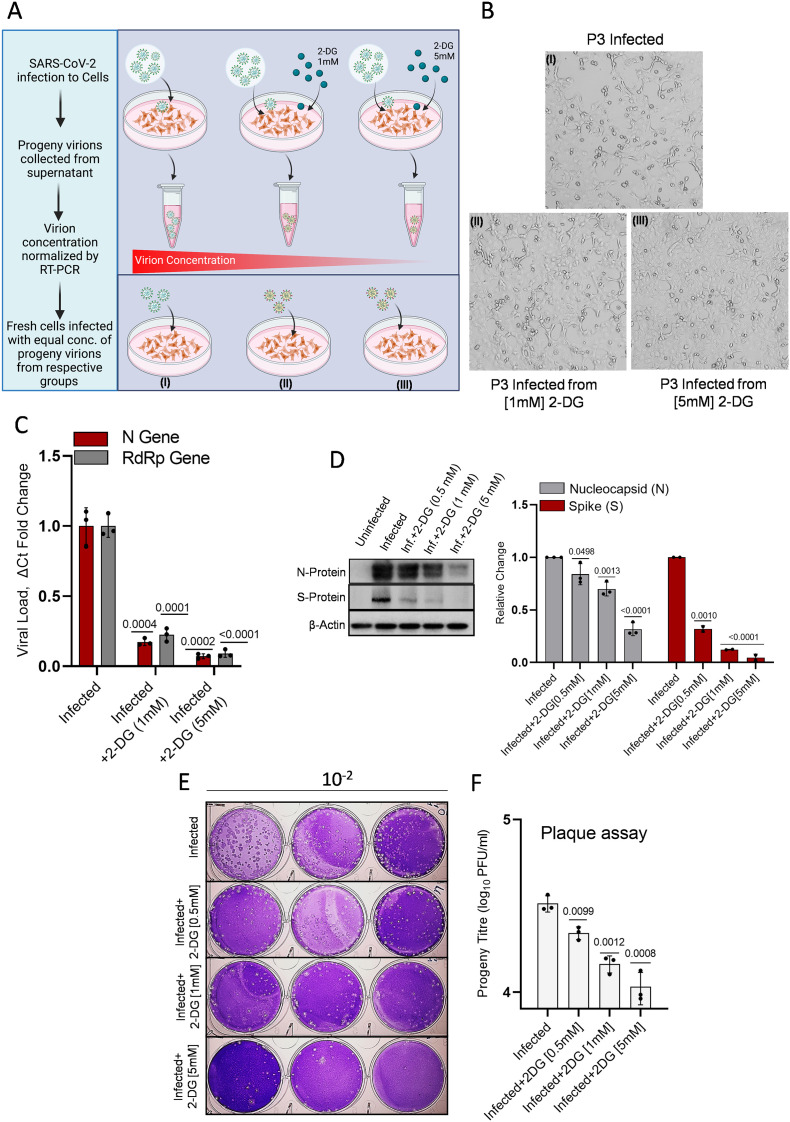
Protein glycosylation of receptor-binding domain (RBD) in spike protein is considered crucial for virus interaction, and internalization to human cells, and 2-DG is known to interfere with the protein glycosylation of newly formed virus proteins in host cells [5]. Therefore, we hypothesized that progeny virions produced from infected and 2-DG treated cells should have compromised infectivity. To test this hypothesis as an alternate mechanism of 2-DG for SARS-CoV-2 attenuation, we collected the secreted virions from media of untreated and 2-DG (1 & 5 mM) treated cells and measured their infectivity in fresh Vero E6 culture (opted method shown by schematic representation in Fig. 7 A) After normalizing the viral concentration based on Ct values, we infected fresh cells with different virions samples (P3, refer to methodology section) without 2-DG treatment and analyzed the virus growth, CPE and plaque-forming ability of progeny virions (Fig 7B). The progeny virions from 2-DG treated cells showed visibly reduced CPE at 48 h post-infection (Fig. 7B). This result was further confirmed by nearly 80%, and 90% reduced virus growth estimated by RT-PCR in cells infected with progeny virions from 1 and 5 mM 2-DG treated cells, respectively (Fig. 7C).
Fig. 7.
2-DG treatment produces defective progeny virions.
A. The graphical depiction indicates the methodology followed for the experiment of progeny virion (P3) infection. The cells were infected with the normalized concentration of progeny virions, and no further 2-DG treatment was given following infection. B. The photomicrographs show the progeny virions induced CPE by bright field imaging in the indicated treatment groups at 48 h post-infection. C. The estimation of N and RdRP genes was performed in the P3 infected samples by RT-PCR assay (n = 3; from three independent observations). Initially, the P3 virions in the supernatant were quantified by RT-PCR from infected, and 2-DG treated cells, and equal virion titer was obtained by volume-dependent concentration normalization depending upon the respective Ct values. Finally, Vero E6 cells were infected with these normalized concentrations of virions from infected and 2-DG treated (1 & 5 mM) groups. D. Immunoblotting of virus N and S protein was performed in cell lysate infected with P3 progeny virion of indicated treatment groups at 12 h post-infection (additional blot images are also provided in supplementary Fig. 5B); The densitometry graph shows the RFC in N and S proteins (from three and two independent observations respectively) among indicated progeny fractions. E. Crystal-violet stained plaques are presented as the plaque-forming unit (PFU)/ml of progeny titer in the indicated treatment groups where Vero E6 cells were infected with the equal concentration of P3 progeny virions from the infected and 2-DG treated (0.5, 1 & 5 mM) groups using 10-fold serial dilution (n = 3). F. The barplot indicates the quantitative estimation of plaques (derived from Fig. 7 E; from two independent observations) in the indicated treatment groups. Data represent the mean ± SD and P-values in all the barplots calculated with respect to the infected group using the two-tailed Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Additionally, these results were also substantiated by the protein expression analysis of N and S proteins in P3 progeny (from 2-DG treated (0.5, 1 & 5 mM) and untreated cells) infected cell lysates. The cells infected with 2-DG treated progeny virions showed a dose-dependent decrease in the levels of N and S proteins, indicating that virus protein synthesis is significantly compromised linked with the low infectivity of progeny virions compared to the untreated cells (Fig. 7D). In line with these observations, progeny virions produced from 2-DG treated cells at different concentrations showed significantly compromised infectivity, estimated in terms of plaque-forming units/ml (PFU/ml; Fig. 7E&F). Interestingly, the reduction in infectivity potential showed a dose-dependent response of 2DG treatment to progeny forming host cells. Reduced levels of virus proteins and infectivity in cells infected with normalized equal particles of progeny virions produced by 2-DG treated cells indicates that the progeny virions produced from 2-DG cells have compromised infective/internalization potential.
2.8. Effect of 2-DG on Virus protein glycosylation and ER stress
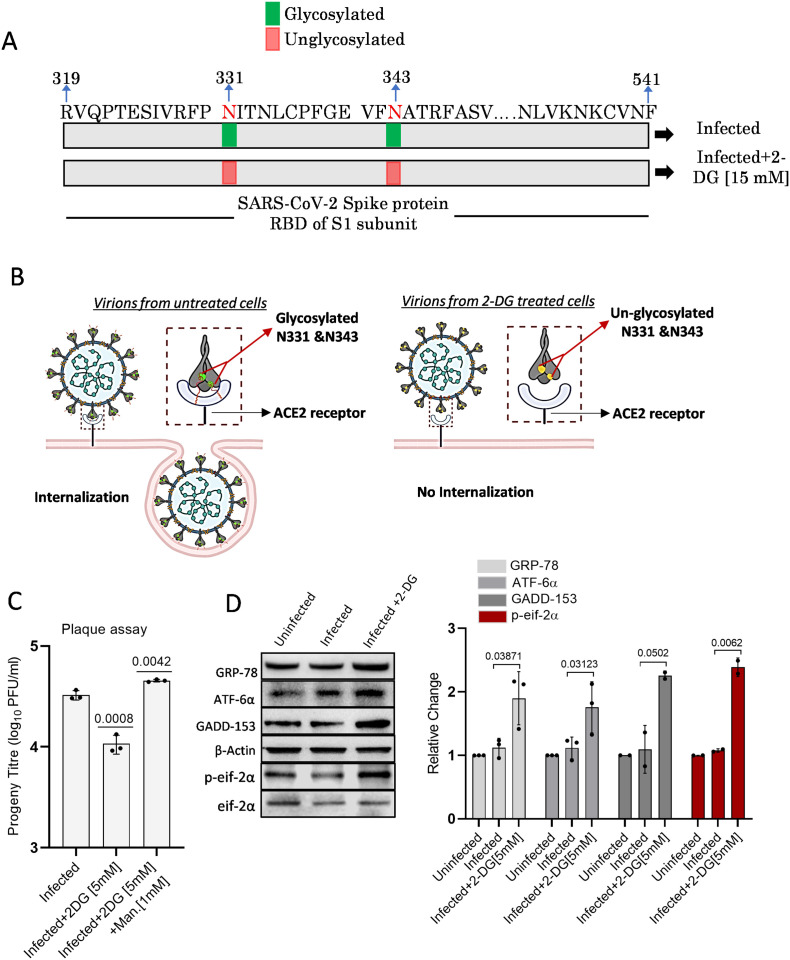
Various amino acids in the spike protein of SARS-CoV-2 were found extensively glycosylated, and certain N-glycosylation sites of the receptor-binding domain (RBD) were found rich in both high mannose and complex type glycans [18], [19]. It is well evident from previous studies that 2-DG as an analog of glucose and mannose interferes with protein glycosylation, resulting in misglycosylated or unglycosylated envelop proteins [5]. The mass spectrometry (MS) analysis (supplementary Fig. 8) of SARS-CoV-2 proteins from secreted virions revealed that in contrast to infected cells, progeny virions from 2-DG treated cells showed 23.6% (at 1 mM) and 38.76% (at 5 mM) increased unglycosylation of the complete retrieved N-glycosites (Source file 1). In addition, further increased unglycosylation (1.29 and 2.66 times at 1 and 5 mM, respectively) was observed on two important N-glycosites (N331 and N343) of RBD in secreted virions from 2-DG treated cells as compared to virions from untreated cells (Source file 1). Interestingly, highly mannosylated N331 and N343 glycosites of RBD are reported to be critical for the interaction of the virus at ACE2 receptor and entry to human cells [20]. Therefore, these results indicates that, by interfering with N-glycan biosynthesis at crucial N-glycosites of RBD (N331 and N343), 2-DG may possibly intervene in the SARS-CoV-2 binding at ACE-2 receptor and subsequent internalization of the virus at the host cell surface as shown by schematic representation (Fig. 8A &B). Mannose is known to reverse the unglycosylation effect of 2-DG; hence we tested the infective potential of progeny virions produced from 2-DG and mannose +2-DG treated cells using PFU assay. Interestingly, virions from co-treated (mannose and 2-DG) cells showed significantly higher infectivity than 2-DG alone treated cells and even higher than infection control (Fig. 8C). Besides, 2-DG is known to induce Endoplasmic Reticulum stress (ER stress) by inhibiting glycolysis and glycosylation [21]. Further to investigate on 2-DG induced unglycosylation of crucial residues in RBD, we analyzed if 2-DG treatment induces ER stress in infected cells. Comparatively, a significant 1.7 to 2.4 fold increase in the levels of ER stress markers like GRP-78, ATF-6α, GADD-153 was observed in infected cells treated with 2-DG (Fig. 8D). The activation of p-eif-2α was also observed in the 2DG treated cells (Fig. 8D). These results indicate that 2-DG induced unglycosylation is causing ER stress in infected cells, which may interfere in virion packaging and production.
Fig. 8.
2-DG induces unglycosylation and ER stress.
A. Schematic representation of N-glycosites (N331 and N343) on RBD (319–541) of SARS-CoV-2 S protein (S1 subunit) shows the glycosylation status in the 2-DG treated and untreated cells following SARS-CoV-2 infection (please refer to source file 1 and supplementary fig. 8). B. Graphical explanation of glycosylated and unglycosylated N-glycosites (N-331 & N-343) mediated virus-host interaction and internalization in untreated and 2-DG treated cells. C. Plaque assay was performed to examine if mannose treatment reverts the effect of 2-DG. Barplot indicates the progeny titer as PFU/ml in the indicated treatment groups. D. Immunoblot and densitometric evaluation indicating the differential expression pattern of mentioned ER stress marker proteins in the untreated and 2-DG (5 mM) treated cells following SARS-CoV-2 infection at 24 h post-infection in Vero E6 cells (additional blot images are also provided in supplementary Fig. 9). Individual dot in barplot indicates the mean value of respective independent observations. Data represent the mean ± SD and P-values in barplots calculated with respect to the infected group using a two-tailed Student's t-test.
3. Discussion
Today whole world is struggling with the serious problem of the COVID-19 pandemic and looking for an effective therapeutic solution. The identification of mutations in the SARS-CoV-2 genome at a rapid rate and the possibility of mutations in prone genes is a huge concern for the efficacy of the vaccine as well as therapeutic resistance [7], [22]. Whenever new virus-induced pathogenesis is identified, it takes years to identify, characterize and develop a specific prophylactic and therapeutic approach against it. Therefore, to counter any virus outbreak at its commencement, there is a need to develop an effective broad-spectrum anti-viral strategy to prevent it from becoming an epidemic and/or pandemic.
Being an obligate parasite, virus hijacks host cell machinery and reprograms it to favor its own multiplication [5]. A compromised mitochondrial metabolism and an enhanced glycolytic pathway upon SARS-CoV-2 infection is one of the examples of virus infection-induced metabolic reprogramming [6], [7]. Normal/uninfected cells show metabolic plasticity due to the presence of both glycolytic and mitochondrial metabolism. However, predominantly glycolytic metabolism in SARS-CoV-2 infected host cells makes glycolysis a lucrative target for developing therapeutic strategies. Therefore, we used a well-known glycolytic inhibitor, 2-DG, to counter the SARS-CoV-2 multiplication in host cells. This molecule has been tested extensively in lab and clinical trials.
We found that glucose uptake, lactate and ATP production was significantly upregulated in infected cells. This observation was also supported by enhanced protein levels of three major glucose transporters, GLUT1, GLUT3, and GLUT4 in infected cells (Fig. 1). While GLUT1 and GLUT4 both have a medium-range affinity for glucose, GLUT3 exhibits a very high affinity for glucose with a very low Km value in the range of 1 mM. Hence, GLUT3 plays a crucial role in the glucose transport of cell types with a high demand for energy in glucose-hungry cells, even at a very low surrounding glucose concentration [23]. Therefore, in addition to the virus-induced levels of GLUT1/4, an upregulation of GLUT3 together ensure the high influx of glucose to infected cells even at the low concentration of extracellular glucose. In addition, increased levels of key glycolytic regulatory enzymes HK-II, PFK-1, and PKM-2 ensure the metabolism of glucose through glycolysis and other anabolic pathways. Further, an increased level of LDH-A confirms the Warburg phenotype of infected host cells (Fig. 1D). Since there is significantly higher glucose demand in SARS-CoV-2 infected cells, the intake of 2-DG (glucose mimic) will also be selectively high in infected cells at low 2-DG concentration, which was validated by increased uptake of 2-NBDG (at 0.2 mM, a fluorescent analog of glucose/2-DG). 2-DG caused 30% to 40% inhibition of cell proliferation in the range of 1 to 5 mM with no significant effect on cell death, and clonogenicity indicates that 2-DG is not cytotoxic/genotoxic upto 5 mM (Fig 3B, C&F). Reduced 30% to 40% proliferation observed in 2-DG treated cells is due to the cytostatic effect of this molecule. Moreover, it is pertinent to note here that the inhibition of proliferation by 2-DG is not a matter of concern as the SARS-CoV-2 infected host cells in humans are primarily non-proliferating in nature, and the cytostatic effect exerted by 2-DG is transient and can be reversed after removal of the drug [24].
Considering the selective higher uptake of 2-DG in SARS-CoV-2 infected cells and its potential to inhibit the virus multiplication, we tested its efficacy on B.6 variant at CCMB, Hyderabad in May 2020 and later on B.1.1 at INMAS, Delhi. 2-DG is able to significantly inhibit the virus multiplication at concentrations ranging from 0.5 mM to 5 mM, which was further verified by significantly reduced protein levels of SARS-CoV-2 nucleo-capsid protein (Fig. 4A-E) and reduced CPE (Fig. 5A). We observed profoundly high cytopathic effect and cell death in SARS-CoV-2 infected cells, which was significantly reversed by 2-DG treatment (Fig. 5A-C). Taken together, these observations suggest that 2-DG inhibits the virus multiplication in infected host cells and thereby reduces the infection-induced CPE and cell death. However, 2-DG on its own did not contribute to cell death in this concentration range. This was further substantiated by the observation that 2-DG significantly reduced the ATP production in infected cells; however, it could not induce a meaningful reduction in ATP levels in uninfected cells (Fig. 4F). It is also pertinent to mention here that the cellular debris of dying cells is one of the major causes of inflammation therefore reduced cell death in the 2-DG treated sample will result in reduced inflammation [25]. Supporting this, an earlier study on rhinovirus infection reported that 2-DG treatment reduces rhino virus-induced lung inflammation in a murine model [11].
A recently published study reported that enhanced glucose levels favor SARS-CoV-2 growth, and thereby diabetes is turning to be one of the primary comorbid conditions responsible for the poor outcome in the treatment of COVID patients [6]. Therefore, we tested the effect of increased glucose on the efficacy of 2-DG. Irrespective of a 300% increase in glucose, the efficacy of 2-DG induced inhibition of virus multiplication was reduced by 7% (from 95% to 88%) at a given concentration of 2-DG (Fig. 6A). This result shows that the high relative glucose concentration could only minimally compromise the 2-DG efficacy, suggesting that the inhibition potential of 2-DG can be maintained at a low 2-DG to glucose ratio, also. The reversal of the inhibitory effect of virus multiplication by 2-DG and glucose uptake at an equimolar concentration of mannose (structurally similar to 2-DG), indicates that mannose can inhibit the 2-DG uptake and compromise the efficacy of 2-DG. It is noteworthy to mention here that the uptake of mannose can take place at very low plasma concentrations (30 to 50 micromolar), and cells use high-affinity glucose transporters like GLUT3 or dedicated mannose transporter which are insensitive to glucose [23], [26]. The important point to note here is that similar to mannose, 2-DG uptake can also take place even at its lower concentrations, and a higher concentration of glucose may not be able to compromise the effectiveness of the 2-DG uptake and efficacy profoundly. Our study provides a kind of evidence that 2-DG uptake is also mediated through either high-affinity glucose transporters like GLUT3 or selective mannose transporter in infected cells, which warrants further investigation.
N-glycosylation is a common feature of the viral envelope proteins [27]. SARS envelope proteins are reported to be highly glycosylated and responsible for the virus to host cell interaction, infection, and immune evasion by glycan shielding. Being the obligate parasites, viruses are dependent on host-cell machinery to glycosylate their own proteins in the process of replication/multiplication [28]. In line with earlier observations on 2-DG induced de/mis-glycosylation of viral envelop proteins, we also found that two crucial residues N331 and N343 in RBD of the spike protein of the secreted SARS-CoV-2 virus from 2-DG treated cells were un-glycosylated (Fig. 8 A&B and Source file 1). It is reported that glycosylation of N331 and N343 is critical for virus binding and infection in host cells [20]. This is further supported by the concentration-dependent reduction in infectivity of progeny virions collected from media of 2-DG treated cells (Fig. 7) suggest the defective and compromised infectivity potential of these virions, supporting the proposition that 2-DG induced un-glycosylation is leading to the production of defective/weaker virions probably resulting in reduced cell binding and internalization (Fig. 7&8). Altered glycosylation of proteins leads to accumulation of misfolded proteins in ER causing ER stress [21]. Increased level of ER stress markers (Fig. 8D) also indicates that 2-DG induced un-glycosylation may be leading to misfolding of viral proteins resulting in ER stress. Moreover, it was observed that the inhibition of viral multiplication by 2-DG was reversed upon addition of mannose (Fig. 6B), suggesting that mannose may inhibit 2-DG uptake (Fig. 6D) and reverse the glycosylation status of both glycosites (N331 and N343), leading to normal virus multiplication in host cells, even in the presence of 2-DG. These results strengthen the observation that 2-DG also inhibits the virus multiplication by interfering with glycosylation of spike protein of SARS-CoV-2. However, more information is required to establish the role of 2-DG induced un-glycosylation in the formation of defective virions. The doses at which 2-DG has been used in clinical trials, its maximum plasma concentration varies in the range of 1 mM [29]. Therefore, it is also pertinent to note here that 1 mM of 2-DG inhibits the virus multiplication by approximately 65% (0.35 fold remaining), and progeny virions produced from 1 mM treated cells lose infectivity and result in 80% (0.2 fold remaining) reduced multiplication and secretion of the virus. Thus, the effective inhibition of SARS-CoV-2 multiplication obtained at 1 mM 2-DG was 93% (0.35 × 0.2 = 0.07 fold remaining).
Although the anti-SARS-CoV-2 activity of 2DG has been demonstrated in various human cell lines from different tissue origins; however, the anti-viral mechanistic action of 2-DG outlined in this study is limited to African green monkey kidney cells (Vero E6), which is one of the limitations of this study. Additionally, the detailed mechanistic investigation on the intervention of 2-DG in metabolic profile, glycosylation status of different N glycosites of spike protein, and ER stress in infected cells remains to be elucidated, which warrants further investigation. These studies are currently in progress in our lab to decipher the detailed mechanism of anti-viral effects of 2-DG following SARS-CoV-2 infection in human cells.
4. Conclusion
The findings presented in this manuscript highlight the SARS-CoV-2 infection mediated enhanced glucose metabolism in host cells, which can be targeted for therapeutic application. 2-DG exploits the inherent and natural mechanism of infected host cells for selective, high accumulation of the drug without compromising uninfected/normal cell functioning significantly. By inhibiting both catabolic and anabolic pathways, 2-DG reduces the virus replication and infectivity of the progeny virions, which has a compromised potential of infection in neighboring cells (Graphical Abstract). Although the effect of 2-DG has been analyzed on only 2 different SARS-CoV-2 variants (B.6 and B.1.1), its anti-viral property is suggested to be universal on all the variants of SARS-CoV-2, as 2-DG interferes with the metabolic requirement of virus-infected host cells. In summary, we demonstrate that glycolytic inhibitor 2-DG exhibits significant potential to be developed as a therapeutic to combat the COVID. These experimental evidence and previous clinical trial experience of 2-DG made way for this molecule to reach clinical trials in COVID-19 patients in India.
5. Material and methods
5.1. Cell culture, virus propagation, and virus quantification
Vero E6 cell line was maintained at 37 °C with 5% CO2 in an incubator; the culture was propagated in minimum essential medium (MEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, Life Technologies, Paisley, UK). The SARS-CoV-2 virus was isolated from a positively tested nasopharyngeal swab at BSL-3 facility INMAS-DRDO, Delhi. Briefly, Vero E6 cells were infected with filtered positive viral transport medium (filtered through 0.2 μm filter) and mixed in the ratio 1:1 with MEM supplemented with 2% FBS, followed by incubation at 37 °C in 5% CO2 with repeated mild agitation for 1 h. The inoculum was removed post-incubation, and the culture was washed with PBS, and further, the resulting positive culture was re-supplemented with MEM having 10% FBS and maintained at 37 °C & 5% CO2 up until cytopathic effects were apparent in the cells. The resulting SARS-CoV-2 viral stock (P1) was then collected and confirmed by RT-PCR. The viral stock was used for one more passage to obtain a working stock (P2) which was then used for all the experiments. Plaque assay was performed for the quantification of viral titrations. Briefly, Vero E6 cells were seeded in 6-well plates at a density of 0.75 × 106 cells/well, and crystal violet staining was performed at 4–5 days post-infection on the appearance of proper plaques followed by plaque enumeration. Finally, Plaque forming units (PFU) were calculated using the formula: PFU/ml = Number of plaques/(Dilution factor×volume of diluted virus per well). The multiplicity of infection (MOI) was derived from the formula: MOI = PFU of virus used for infection/ No. of cells, and in the present study, the MOI was kept low (0.3), which was standardized to ensure the experimental study and optimal analysis of SARS-CoV-2 induced intracellular stress behavior in Vero E6 cells. A similar protocol was followed for establishing B.6 variant in Vero cells at the BSL-3 facility, CSIR-CCMB, Hyderabad.
5.2. Virus growth kinetics and Bright-field imaging
For SARS-CoV-2 infection, cells were seeded into 24 well-plates with the density of 0.05 × 106 cells/well and were infected with MOI of 0.3. For extracellular virus RNA quantification, media was collected from each well at 24 h, and virus RNA was isolated using MagMAX Virus/Pathogen II Nucleic Acid Isolation Kit (Thermo Fisher Scientific) and automated extraction was carried out using KingFisher™ Flex Purification System, KingFisher with 96 Deep-well Head (Thermo Fisher Scientific). For intracellular virus RNA quantification, TRIzol® Reagent (Ambion, Life Technologies) was used, further isolation and extraction were performed as mentioned earlier. Multiplex rRT-PCR kit (Allplex™ 2019-nCoV assay, Seegene) based identification of the three target genes associated with SARS CoV-2, i.e., Nucleocapsid (N) gene, RNA dependent RNA polymerase (RdRP) gene, and Envelop (E) gene were performed following the manufacturer's protocol in CFX96 touch real-time PCR detection system (Biorad). Moreover, morphological changes associated with cytopathic effects (CPE) caused by SARS-CoV-2 infection in Vero E6 cells were observed at 24 h and 48 h post-infection under Bright Field (10× objective) on an inverted microscope (Nikon ECLIPSE Ts2).
5.3. Analysis of infectivity
The comparative analysis of infection potential of progeny virions (P3) from SARS-CoV-2 infected untreated and 2-DG treated cells was examined using RT-PCR estimation of N and RdRp genes from the supernatant of respective samples of Vero E6 cells. The P3 virions in the supernatant were quantified by RT-PCR from infected untreated and 2-DG treated cells, and obtained values were used to normalize the concentration of virions in each group. Then freshly cultured Vero E6 cells were infected with a normalized equal concentration of virions (P3) from each group. Further N and RdRP gene analysis was performed to examine the infectivity potential of P3 virions. PFU/ml of P3 progeny virions were calculated using the method as described above.
5.4. Estimation of glucose uptake, lactate, and ATP determination
The qualitative and quantitative estimation of glucose uptake was performed using 2-NBDG (200 μM) in Vero E6 cells. Briefly, cells were seeded with density 0.05 × 106 cells/well in 24 well plates. Whereas glucose uptake in Vero and Vero E6 cells was analyzed using co-culture of both the cell lines. To differentiate Vero from Vero E6 cell population, initially Vero cells (0.025 × 106) were stained with CellTracker™ Red CMPTX (Invitrogen) using (5 μM; 30mins; 37 °C) followed by normal growth medium replacement and co-culture with the same number of unstained Vero E6 cells (0.025 × 106). The next day similar treatment infection protocol opted for 2-NBDG uptake either in Vero E6 or co-cultured Vero-Vero E6 cells carried out as described in the previous section. Additionally, 2-DG and mannose (man.) treatment were given as mentioned in the respective figure legend. Finally, glucose uptake was monitored at 48 h post-infection by incubating infected and respective uninfected cells with 2-NBDG (200 μM; 20mins; 37 °C) probe buffer solution containing MgCl2 and CaCl2 (1 mM each) in PBS. Further cells were washed twice with PBS, and fluorescence images were captured under a fluorescence microscope (Nikon ECLIPSE Ts2-FL) using 10 × 10 X magnification. The quantitative estimation of 2-NBDG fluorescence was carried out at 465/540 (ex/em.), and lactate estimation was performed using Lactate Enzymatic UV kit (DIALAB; Austria) as per the manufacturer protocol on Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek Instruments, USA). Estimated fluorescence values of both 2-NBDG uptake and lactate production were further normalized with cell numbers from respective treatment groups and graph plotted as relative fold change with respect to control. In a similar experimental condition, Luminescence based ATP production was measured using ATP determination kit (Molecular Probes™; Invitrogen) as per the manufacturer protocol, and estimated values were normalized with the protein content of respective treatment groups. Subsequently, all the 2-NBDG fluorescence images were subjected to morphometric analysis to quantify the intracellular uptake of the fluorophore. For this purpose, the study employed algorithms from CELLSEGM and Image Processing toolbox in Matlab 2020a (Mathworks). It initially segmented the foreground (nucleus, cell) from background pixel intensity and segmented more than 1000 cell objects from multiple fields of view, and the segmented cell object's median pixel intensity was calculated [30].
5.5. Cell proliferation assay, Annexin V/PI uptake, trypan blue exclusion assay, and determination of clonogenicity
Sulforhodamine B (SRB) assay was employed to derive the relative proliferation efficiency of Vero E6 cells following 2-Deoxy-d-Glucose (2-DG) treatment. Vero E6 cells were seeded in a 96 well plate at a density of 0.003 × 106 cells/well and left overnight in a CO2 incubator at 37 °C. The next day cells were treated with various concentrations of 2-DG ranging from 0.1 mM to 100 mM, and the assay was performed as previously described method [31]. To examine the 2-DG induced cell toxicity, AnnexinV/PI uptake and trypan blue exclusion assay was performed in Vero E6 cells. Cells were seeded in 35 mm petri dishes (PD) with the density of 0.075 × 106 Cells/PD and kept in CO2 incubator followed by 2-DG [1&5 mM] treatment at 24 h post-incubation. For Annexin V/PI uptake, cells were stained in binding buffer (1×) with Annexin V (Alexa Fluor™ 488 conjugate; Invitrogen) and propidium iodide (BD Pharmingen™) followed by fluorimetric estimation using 495/519 (ex/em.) and 535/617 (ex/em.) respectively. Estimated values were further normalized with the cell number of respective treatment groups and graph plotted as mean fluorescence intensity/cell (MFI/cell). Whereas trypan blue exclusion was performed using trypan blue (0.4%) in a 1:1 dilution with cell suspension in PBS and cell numbers were enumerated with Neubauer improved counting chamber (Paul Marienfeld GmbH & Co. KG, Germany) under 10 X objective, and 10× eyepiece magnification with a compound light microscope (Olympus CH30, Japan). Trypan blue stained dead cells were excluded from the live cell population, and the percentage of Live vs. Dead cells was calculated in treatment groups. CFSE (Sigma) staining for cellular proliferation was performed in accordance with the manufacturer's protocol. Vero E6 cells were stained with CFSE (5 μM; 20 min, at room temp.) in MEM with 2% FBS. After incubation, cells were washed with a normal growth medium and reseeded 35 mm petri dishes (PD) with the density of 0.25 × 106 Cells/PD and kept overnight in the dark in a CO2 incubator. 2-DG treatments were given the following day, and the cells were terminated and processed for flow cytometry (BD LSR II) based analysis at 0 h, 24 h, and 48 h. Macrocolony assay was performed for determining the plating efficiency of 2-DG treated cells at 1 mM and 5 mM in Vero E6 cells. Cells were seeded in 60 mm petri dishes and were treated with 2-DG for 24 h. Further, these cells were reseeded with 200 cells/PD with the control group and kept for 7–8 days in a humidified incubator to allow the formation of macroscopic colonies. After that, cells were washed once with PBS and stained with crystal violet, followed by an enumeration of colonies consisting of >50 cells. Finally, the plating efficiency was calculated as the ratio of (Number of colonies formed/ Number of cell-seeded) × 100 and presented as % clonogenicity.
5.6. Apoptosis assay
Apoptosis assay was performed using ethidium bromide (EB)/acridine orange (AO) staining as described previously with brief modifications [32] in Vero E6 cell line at 48 h post-infection in the treatment groups (as mentioned in the respective figure legend). Vero E6 cells were seeded in 24 well plates (0.05 × 106cells/well) and kept at 37 °C in a CO2 incubator. The next day cells were infected with SARS-CoV-2, and 2-DG treatment was carried out (as described in the previous section). At the required time point, cells were dislodged by trypsinization and floater was collected followed by centrifugation (100 g; 10mins), and each group's cells were transferred to a 96 well plate. Further cells were stained with AO/EB dye in the ratio of 1:1 (100 μg/ml each) to each well. Finally, cell images were captured using a fluorescence microscope (Nikon ECLIPSE Ts2-FL) using 10 × 10 X (objective and eyepiece) magnification. Finally, the acquired fluorescence images of AO/EB cells were subjected to the morphometric analysis to estimate the median area of AO/EB uptake in fluorescence images of uninfected, 2-DG treated, infected, and infected +2-DG treated cells. For this purpose, the study employed the algorithms from CELLSEGM and Image Processing toolbox in Matlab 2020a (Mathworks). At first, fluorescence images were smoothened and subjected to the segmentation process to isolate cells having little overlap with other cells. This cell segmentation was carried out at many fields of view images, and more than 1000 non-overlapping cell objects were isolated, and their median area was calculated for each uninfected, 2-DG treated, infected, and infected with 2-DG treated category.
5.7. Immunoblotting
Immunoblotting was performed to examine the differential expression of glucose transporters, glycolytic proteins, and drug-induced change in virus nucleocapsid proteins as mentioned in the respective figure legend. Briefly, cells were seeded at 0.5 × 106 density in 60 mm tissue culture dishes and kept in a CO2 incubator at 37 °C. The following day, virus and drug treatments were performed (as described previously). At the time of analysis, whole-cell lysate was processed in RIPA buffer containing Tris/HCl: (50 mM; pH 7.4), Na3VO4 (1 mM), EDTA (1 mM), NaCl (150 mM), NaF (1 mM) PMSF (2 mM), NP-40 1%; supplemented with protease inhibitor cocktail (1×), and protein was estimated by BCA method. Equal quantities of lysates (40 μg and 60 μg were used for detection of nucleocapsid protein and glycolytic proteins, respectively) were resolved on 10% or 12% SDS-PAGE gel (depending on the molecular weight of the respective proteins). Further protein samples were electro-blotted onto PVDF membrane (MDI) followed by membrane blocking with 5% BSA for 1 h (according to the manufacturer protocol). Incubation of the primary antibody was performed in the dilution of (1:1000; according to the manufacturer instructions) for 14 h at 4 °C then washed in Tris-buffered saline supplemented with 0.1% Tween-20 (TBST) followed by HRP conjugated secondary antibody incubation (1:2500) for 2 h. Finally, blots were washed again with TBST and developed using ECL chemiluminescence detection reagent on Luminescent image analyzer (Image Quant LAS 500, Japan). Densitometry was performed for each blot using Image J software and obtained values were normalized with respective loading control β-actin, and the graph presented as relative fold change among the treatment groups.
5.8. Virus harvesting, inactivation, and glycosylation study
Vero E6 cells were infected with SARS-CoV-2, as mentioned in the previous section, followed by 2-DG treatment (1 and 5 mM). Virus harvesting, purification, and inactivation were performed using the experimental protocol as described elsewhere [33] with some modifications. Briefly, the cell culture supernatant (25 ml) of infected and 2-DG treated cells were collected at 72 h post-infection and stored at −80 °C overnight followed by centrifugation (4500 g; 10mins.). Further supernatant was overlayed on 20% sucrose (w/v), HEPES (25 mM), in PBS and ultracentrifugation was performed at 103,745 g for 5 h using Beckman Coulter Euro Center SA. Finally, the virus pellet was dissolved in TNE buffer (10 mM Tris, 0.2 M NaCl, 10 mM EDTA in PBS; pH 7.4) and inactivated by incubating samples at 56 °C for 10 min. in Tris–HCL (100 mM) supplemented with SDS (5%). For the glycosylation study, proteolytic digestion of 30 μg protein was performed and the obtained peptides were separated using liquid chromatography and eluates were directly infused into Electrospray ionization mass spectroscopy setup followed by the acquisition of MS and MS/MS data. Raw data was mapped with the SARS-CoV-2 (S) protein FASTA sequence, and the analysis of peptide mapping and glycosylation status was carried out using BioPharma Finder software.
5.9. Statistical analysis
Unless otherwise stated, bar plots are the representation of the arithmetic mean obtained from three independent experiments (n = 3), with error bars indicating standard deviations. Statistical analysis of the data was performed as a two-tailed Student t-test using GraphPad Prism8 software, and values of P < 0.05 were considered statistically significant.
CRediT authorship contribution statement
Anant Narayan Bhatt: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing, Data curation. Abhishek Kumar: Investigation, Writing – original draft, Writing – review & editing, Data curation. Yogesh Rai: Investigation, Writing – original draft, Writing – review & editing, Data curation. Neeraj Kumari: Method and data illustration. Dhiviya Vedagiri: Investigation. Krishnan H. Harshan: Investigation, Writing – review & editing. Vijayakumar Chinnadurai: Formal analysis. Sudhir Chandna: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are thankful to Dr. Rakesh Mishra, Director CSIR-CCMB, for facilitating the 2-DG testing against SARS-CoV-2 at the BSL-3 facility of CCMB during the nationwide lockdown. We are thankful to Dr. Sankar Bhattacharya from THSTI and Dr. Chandru from RCB, Faridabad, for their help in providing Vero and Vero E6 cell lines for virus culture. We are also thankful to Dr. Jubilee Purkaystha, in charge of BSL-3 facility INMAS, for helping us in conducting the experiments at this facility. We also want to express our gratitude to Dr. Viney Jain and Dr. B S Dwarakanath for useful discussions and advice. AK is a recipient of a fellowship from CSIR. The study was funded by DRDO, Ministry of Defence, Govt. of India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.lfs.2022.120411.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2: Source file 1
Data availability
Data will be made available on request.
References
- 1.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datta C., Bhattacharjee A. Cytokine storm and its implication in coronavirus disease 2019 (COVID-19) J.Immunol.Sci. 2020;4(3) [Google Scholar]
- 3.Wang C., Wang Z., Wang G., JYN Lau, Zhang K., Li W. Signal Transduction And Targeted Therapy. Vol. 6. Springer Nature; 2021. COVID-19 in early 2021: current status and looking forward. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer K.A., Stöckl J., Zlabinger G.J., Gualdoni G.A. Hijacking the supplies: metabolism as a novel facet of virus-host interaction. Front. Immunol. 2019;10(JULY):1–12. doi: 10.3389/fimmu.2019.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thaker S.K., Ch'ng J., Christofk H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019;17(1):59. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Codo A.C., Davanzo G.G., de Brito Monteiro L., de Souza G.F., Muraro S.P., Virgilio-da-Silva J.V., et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab. 2020;32(3):437–446. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Guo R., Kim S.H., Shah H., Zhang S., Liang J.H., et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat. Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-21903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt A.N., Chauhan A., Khanna S., Rai Y., Singh S., Soni R., et al. Transient elevation of glycolysis confers radio-resistance by facilitating DNA repair in cells. BMC Cancer. 2015;15(1):1–12. doi: 10.1186/s12885-015-1368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H.T., Hwang E.S. 2-Deoxyglucose: an anticancer and antiviral therapeutic, but not any more a low glucose mimetic. Life Sci. 2006;78(12):1392–1399. doi: 10.1016/j.lfs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Gualdoni G.A., Mayer K.A., Kapsch A.M., Kreuzberg K., Puck A., Kienzl P., et al. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc. Natl. Acad. Sci. U. S. A. 2018;115(30):E7158–E7165. doi: 10.1073/pnas.1800525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passalacqua K.D., Lu J., Goodfellow I., Kolawole A.O., Arche J.R., Maddox R.J., et al. Glycolysis is an intrinsic factor for optimal replication of a norovirus. MBio. 2019;10(2) doi: 10.1128/mBio.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontaine K.A., Sanchez E.L., Camarda R., Lagunoff M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015;89(4):2358. doi: 10.1128/JVI.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich A., Uhl S., Ioannidis K., Hofree M., tenOever B.R., Nahmias Y. 2020. The SARS-CoV-2 transcriptional metabolic signature in lung epithelium. Available at SSRN 3650499. [Google Scholar]
- 15.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., et al. SARS-coronavirus-2 replication in vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J.Gen.Virol. 2020;101(9):925. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davda J.N., Frank K., Prakash S., Purohit G., Vijayashankar D.P., Vedagiri D. 2020. An inexpensive RT-PCR endpoint diagnostic assay for SARS-CoV-2 using nested PCR: direct assessment of detection efficiency of RT-qPCR tests and suitability for surveillance. bioRxiv. [Google Scholar]
- 18.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q., Hughes T.A., Kelkar A., Yu X., Cheng K., Park S., et al. Inhibition of SARS-CoV-2 viral entry upon blocking N-and O-glycan elaboration. eLife. 2020;9 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S.-M., Kim S.-J. Endoplasmic reticulum stress (ER-stress) by 2-deoxy-D-glucose (2DG) reduces cyclooxygenase-2 (COX-2) expression and N-glycosylation and induces a loss of COX-2 activity via a Src kinase-dependent pathway in rabbit articular chondrocytes. Exp. Mol. Med. 2010;42(11):777–786. doi: 10.3858/emm.2010.42.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. Iscience. 2021;24(1) doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cura A.J., Carruthers A. Role of monosaccharide transport proteins in carbohydrate assimilation, distribution, metabolism, and homeostasis. Compr.Physiol. 2011;2(2):863–914. doi: 10.1002/cphy.c110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X.D., Deslandes E., Villedieu M., Poulain L., Duval M., Gauduchon P., et al. Effect of 2-deoxy-D-glucose on various malignant cell lines in vitro. Anticancer Res. 2006;26(5A):3561–3566. [PubMed] [Google Scholar]
- 25.Rock K.L., Kono H. The inflammatory response to cell death. Annu. Rev. Pathol. Mech. Dis. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panneerselvam K., Freeze H.H. Mannose enters mammalian cells using a specific transporter that is insensitive to glucose. J. Biol. Chem. 1996;271(16):9417–9421. doi: 10.1074/jbc.271.16.9417. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhao W., Mao Y., Chen Y., Wang S., Zhong Y., et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol.Cell.Proteomics. 2020 Oct;19(20) doi: 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim.Biophys.ActaGen.Subj. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein M., Lin H., Jeyamohan C., Dvorzhinski D., Gounder M., Bray K., et al. Targeting tumor metabolism with 2-deoxyglucose in patients with castrate-resistant prostate cancer and advanced malignancies. Prostate. 2010;70(13):1388–1394. doi: 10.1002/pros.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodneland E., Kögel T., Frei D.M., Gerdes H.-H., Lundervold A. CellSegm-a MATLAB toolbox for high-throughput 3D cell segmentation. Source CodeBiol.Med. 2013;8(1):1–24. doi: 10.1186/1751-0473-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vichai V., Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006;1(3):1112. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 32.Ribble D., Goldstein N.B., Norris D.A., Shellman Y.G. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005;5(1):1–7. doi: 10.1186/1472-6750-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pralow A., Nikolay A., Leon A., Genzel Y., Rapp E., Reichl U. Site-specific N-glycosylation analysis of animal cell culture-derived zika virus proteins. Sci. Rep. 2021;11(1):5147. doi: 10.1038/s41598-021-84682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2: Source file 1
Data Availability Statement
Data will be made available on request.