Abstract
Recent advancements in chemotaxonomic and molecular biology-based identification methods have clarified the taxonomy of the genus Actinomyces and have led to the recognition of several new Actinomyces and related species. Actinomyces-like gram-positive rods have increasingly been isolated from various clinical specimens. Thus, an easily accessible scheme for reliable differentiation at the species level is needed in clinical and oral microbiology laboratories, where bacterial identification is mainly based on conventional biochemical methods. In the present study we designed a two-step protocol that consists of a flowchart that describes rapid, cost-efficient tests for preliminary identification of Actinomyces and closely related species and an updated more comprehensive scheme that also uses fermentation reactions for accurate differentiation of Actinomyces and closely related species.
The genus Actinomyces consists of a heterogeneous group of gram-positive, mainly facultatively anaerobic or microaerophilic rods with various degrees of branching (22). Actinomyces species are frequently found as members of the normal microflora, especially in the mouth; but they are also found to be etiologic agents in infections, such as in classical actinomycosis, human bite wounds and abscesses at different body sites, eye infections, and oral, genital, and urinary tract infections (20, 23). Detection of Actinomyces species in clinical specimens is important, as it may affect the prognosis and patient management, but identification by conventional biochemical methods can be difficult.
At present, 15 different Actinomyces species are found in humans, with 9 found in the oral cavity. Actinomyces israelii is known as the key species responsible for classical actinomycosis (23), but it is often isolated in connection with other oral infections, such as peri-implantitis (N. Sarkonen, E. Könönen, E. Tarkka, P. Laine, M. Könönen, and H. Jousimies-Somer, J. Dent. Res. 79(special Issue):620, abstr. 3813, 2000). Actinomyces odontolyticus, Actinomyces naeslundii, and Actinomyces viscosus are the primary Actinomyces species in infants' mouths (21) as well as in early dental plaque (13, 17). Actinomyces georgiae, Actinomyces gerensceriae, and Actinomyces meyeri have been isolated from gingival crevices of periodontally healthy individuals (3, 10). Two new Actinomyces species of oral origin have been described recently: Actinomyces radicidentis from infected root canals (4) and Actinomyces graevenitzii from respiratory tract secretions (19) and infants' saliva (21). During the past few years, several other new species from nonoral sources have been included in the genus Actinomyces (6, 7, 12, 16, 27) and some former Actinomyces species have been moved to the closely related genera Arcanobacterium and Actinobaculum (11, 18). The natural habitats of these species have remained obscure, and their clinical relevance as a part of a polymicrobial infection is not fully established (8, 20). The recent changes in nomenclature among the Actinomyces species and closely related genera are presented in Table 1.
TABLE 1.
Recent taxonomic changes among Actinomyces and closely related genera from human sources
| Year | Current name | Previous nomenclature or taxonomic position | Source | Reference |
|---|---|---|---|---|
| 1994 | Actinomyces neuii subsp. anitratus | CDC group 1 coryneform | Abscess, blood | 7 |
| 1994 | Actinomyces neuii subsp. neuii | CDC group 1-like coryneform | Abscess, blood | 7 |
| 1995 | Actinomyces radingae | A. pyogenes-like (APL1) | Polymicrobial infection | 27 |
| 1995 | Actinomyces turicensis | A. pyogenes-like (APL10) | Polymicrobial infection | 27 |
| 1995 | Actinomyces europaeus | New species | Abscess | 6 |
| 1997 | Actinomyces graevenitzii | New species | Respiratory tract | 19 |
| 2000 | Actinomyces radicidentis | New species | Oral cavity | 4 |
| 2000 | Actinomyces urogenitalis | New species | Urogenital tract | 16 |
| 2001 | Actinomyces funkei | New species | Blood | 12 |
| 1997 | Actinobaculum schalii | New species | Blood | 11 |
| 1997 | Arcanobacterium bernardiae | Actinomyces bernardiae | Abscess, blood | 18 |
| 1997 | Arcanobacterium pyogenes | Actinomyces pyogenes | Polymicrobial infection | 18 |
The identification and differentiation of the gram-positive rods that belong to the genus Actinomyces may pose major problems for clinical and oral microbiology laboratories in terms of labor, time, and cost when conventional biochemical methods are used. Furthermore, currently available commercial identification kits do not include most of newer species in their databases. Sophisticated novel methods such as pyrolysis mass spectrometry, amplified 16S ribosomal DNA restriction analysis (8, 14), and 16S rRNA sequencing will greatly help in the identification of the most problematic Actinomyces species. Unfortunately, these methods are still available only in research and reference laboratories. The aim of the present study was to create an easily accessible flowchart that describes rapid, cost-efficient tests for the preliminary identification of Actinomyces and closely related species and an updated biochemical scheme for the more definite differentiation of Actinomyces species and closely related species in routine clinical and oral microbiology laboratories.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study consisted of 19 reference strains from international culture collections (see Table 2), including 15 Actinomyces spp., 3 Arcanobacterium spp., and 1 Actinobaculum sp., and 70 clinical Actinomyces isolates from oral and nonoral sources. The clinical isolates, which originated from infants' saliva (n = 29), peri-implantitis samples (n = 20), submandibular abscesses (n = 6), and nonoral sites (n = 15, of which 13 were a kind gift from V. Hall, University Hospital of Wales) in adults, were presumptively assigned as members of the genus Actinomyces on the basis of the fact that they were gram-positive branching rods and produced succinic acid as the major end product of glucose metabolism, as determined by gas-liquid chromatography. All strains were revived from frozen (−70°C) stocks, subcultured twice, on brucella blood agar, and incubated anaerobically at 37°C for 3 to 4 days before testing.
TABLE 2.
Identification scheme for Actinomyces and closely related speciesa
| Species and strain(s) | Pigmentation | Catalase production | Nitrate reduction | CAMP test | Hydrolysis of:
|
Production of:
|
Fermentation of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urea | Esculin | α-Fucosidase | α-Glucosidase | β- NAG | β-Galalactosidase | Arabinose | Maltose | Mannitol | Raffinose | Rhamnose | Sucrose | Xylose | Trehalose | |||||
| A. europaeus CCUG 32789AT | − | − | +b | − | − | +b | − | + | − | + | − | + | − | − | − | − | +b | − |
| A. funkei CCUG 42773T | − | − | + | +c | − | − | +d | + | + | + | +e | + | − | − | − | + | + | − |
| A. georgiae | ||||||||||||||||||
| ATCC 49285T | − | − | + | − | − | + | − | + | − | + | − | w | − | − | w | + | + | + |
| 1 clinical strain | − | − | − | − | − | + | − | + | − | + | − | + | − | − | + | + | + | + |
| A. gerencseriae | ||||||||||||||||||
| ATCC 23860T | − | − | + | − | − | + | − | + | − | + | − | + | + | −f | +f | + | +f | + |
| 12 clinical strains | − | − | v | − | − | + | − | + | − | + | − | + | − | + | − | + | w | v |
| A. graevenitzii | ||||||||||||||||||
| CCUG 27294T | +g | − | − | − | − | − | − | − | + | + | − | + | − | + | − | + | − | − |
| 9 clinical strains | + | − | v | − | − | − | − | v | + | + | − | + | − | − | − | + | w | v |
| A. israelii | ||||||||||||||||||
| ATCC 10049 | − | − | + | − | − | + | − | + | − | + | + | + | − | + | + | + | + | + |
| 10 clinical strains | − | − | + | − | − | + | − | + | − | + | + | + | w | + | v | + | + | + |
| A. meyeri | ||||||||||||||||||
| ATCC 35568T | − | − | − | +h | − | − | − | + | +i | +i | − | + | − | − | − | + | + | − |
| 4 clinical strains | − | − | v | + | − | − | − | + | + | + | + | + | − | − | − | + | + | − |
| A. naeslundii | ||||||||||||||||||
| ATCC 12104T | − | − | + | − | + | + | − | + | − | + | − | + | − | + | − | + | − | + |
| 8 clinical strains | − | − | v | − | + | v | − | + | − | + | − | + | − | + | − | + | − | + |
| A. neuii subsp. neuii CCUG 32252T | − | + | + | + | − | − | − | + | − | + | + | + | + | −j | wj | + | + | −j |
| A. neuii subsp. anitratus CCUG 32253T | − | + | − | + | − | +k | − | + | − | + | − | + | + | + | − | + | + | + |
| A. odontolyticus | ||||||||||||||||||
| ATCC 17929T | + | − | + | − | − | w | + | +l | − | +l | − | +m | − | − | +m | +m | w | − |
| 10 clinical strains | + | − | + | − | − | v | v | v | − | + | − | + | − | − | + | + | + | − |
| A. radicidentis CCUG 36733T | +n | + | + | − | − | w | − | + | − | + | − | + | + | + | − | + | − | + |
| A. radingae | ||||||||||||||||||
| CCUG 32394T | − | − | − | +o | − | + | + | + | + | + | + | + | − | − | − | + | + | − |
| 5 clinical strains | − | − | + | + | − | + | + | + | v | + | + | + | − | w | − | + | v | v |
| A. turicensis | ||||||||||||||||||
| CCUG 34269T | − | − | − | − | − | − | + | + | − | − | − | + | − | − | − | + | + | −p |
| 5 clinical strains | − | − | − | − | − | −+ | v | + | − | − | − | + | − | − | − | + | + | v |
| A. urogenitalis CCUG 38702T | +q | − | + | − | − | + | − | + | + | + | w | w | − | −r | w | + | + | w |
| A. viscous | ||||||||||||||||||
| ATCC 15987T | − | + | + | − | + | − | − | + | − | + | − | + | − | + | − | + | − | − |
| 6 clinical strains | − | + | + | − | − | − | − | + | − | v | − | + | − | + | − | + | v | − |
| Arcanobacterium bernardiae CCUG 33419T | − | − | − | − | − | − | +s | + | +t | − | w | + | − | − | w | − | − | − |
| Arcanobacterium haemolyticum ATCC 9345T | − | − | − | +Rev | − | − | + | + | + | + | − | + | − | − | − | + | − | − |
| Arcanobacterium pyogenes CCUG 13230T | − | − | − | − | − | − | − | +u | −v | + | − | + | w | − | − | w | + | + |
| Actinobaculum schalii CCUG 27420T | − | − | − | w | − | − | − | + | − | − | + | + | − | − | + | + | + | − |
All enzyme reactions in this table are based on results obtained with Rosco diagnostic tablets; fermentation reactions are based on tests with PRAS biochemicals. The other footnotes describe the reactions that are discrepant compared with previously published data. Abbreviations ATCC, American Type Culture Collection, Manassas, Va.; CCUG, Culture Collection, University of Gothenborg, Gothenborg, Sweden; +, positive reaction or result; −, negative reaction or result; w, weak reaction; v, variable reaction; Rev, reverse.
In the original description (6), the type strain is nitrate, esculin, and xylose negative with the API CORYNE kit.
In the original description (12), the strain is not reported to be CAMP test positive.
In the original description (12), the strain is negative with API systems.
In the original description (12), acid is not produced from l-arabinose with API systems.
In the original description (10), the type strain does not ferment rhamnose or xylose and ferments raffinose.
In the original description (19), the strain is nonpigmented.
Wüst et al. (27) reported a negative reaction by the CAMP test.
Schaal (22), in Bergey's manual, reports a negative reaction with the API ZYM system.
In the original description (7), acid is produced from raffinose and trehalose but acid is not produced from l-rhamnose with the API 50CH system.
In the original description (7), the strain is reported to be esculin negative with the API CORYNE kit.
Schaal (22), in Bergey's manual, reports a negative reaction with the API ZYM kit.
Johnson et al. (10) report a negative reaction with PRAS biochemicals.
In the original description (4), the strain is not reported to be a pigment producer.
Vandamme et al. (25) reported a negative CAMP test reaction.
Vandamme et al. (25) reported a positive reaction with a tryptone soy broth plus horse serum (Oxoid) and a variable reaction with the API CORYNE kit.
In the original description (16), the strain was not reported to be a pigment producer.
Nikolaitchouk et al. (16) reported a positive reaction for d-raffinose with the API RAPID ID 32 STREP kit.
In the original description (18), the strain was reported to have a negative reaction with the API ZYM kit.
Funke et al. (6) and Lawson et al. (11) reported negative reactions (the test system was not traceable).
Lawson et al. (11) reported a negative reaction.
Lawson et al. (11) reported a positive reaction.
Morphological and biochemical characteristics.
The identification of the isolates was performed by established biochemical methods. Briefly, colony morphology was examined under a dissecting microscope, pigmentation was assessed on brucella and rabbit laked blood agar media after incubation for 5 days, and cell morphology was assessed with Gram-stained preparations. Growth patterns in ambient air, in 5% CO2, and under anaerobic conditions were recorded after prolonged incubation (5 to 10 days). Production of catalase was tested with 15% H2O2, and reduction of nitrate was tested by a disk test (24). Staphylococcus aureus ATCC 25923 was used as an indicator strain for the CAMP test (synergistic hemolysis) on brucella blood agar. The enzyme tests described in Fig. 1 and 2 and in Table 2 were performed, and incubation was at 36°C for 4 h in air, according to the manufacturer's instructions, with individual diagnostic tablets (Rosco, Taastrup, Denmark). The tests were for hydrolysis of urea and esculin and production of α-fucosidase, α-glucosidase, β-galactosidase (o-nitrophenyl-β-d-galactopyranoside [ONPG]), β-N-acetyl-glucosaminidase (β-NAG), α-mannosidase, and arginine dihydrolase (the last two tests were conducted only for A. israelii and A. gerencseriae), l-arabinose, and β-xylosidase (for differentiation of Arcanobacterium bernardiae and Actinomyces turicensis, see Fig. 2). To assess the uniformity of reactivity by different test systems, the reference strains were additionally tested in parallel with the API ZYM kit (bioMerieux, Marcy l'Etoile, France) by incubation at 36°C for 4 h and a test based on substrates linked to 4-methylumbelliferyl [4-MU; Sigma, St. Louis, Mo.; 20 μl of substrate in N-tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid buffer plus a loopful of bacterial cells from colonies on a blank paper disk (Oxoid, Unipath, Basingstow, England)], with incubation at 36°C for 15 to 30 min (5, 15). Inocula for the testing of enzyme activities were from 3 to 4 days of growth on brucella plates and were adjusted to a cell turbidity equal or greater than a McFarland no. 4 standard in saline for Rosco diagnostic tablets and a McFarland 5 to 6 standard in sterile water for the API-ZYM kit. Tests for the fermentation of arabinose, glucose, maltose, mannitol, raffinose, rhamnose, sucrose, trehalose, and xylose used prereduced, anaerobically sterilized (PRAS) biochemical media incubated at 36°C for a minimum of 5 days (24). If no or scanty growth (<2+) was obtained, 50 μl of 10% Tween 80 was added to 5 ml to promote growth.
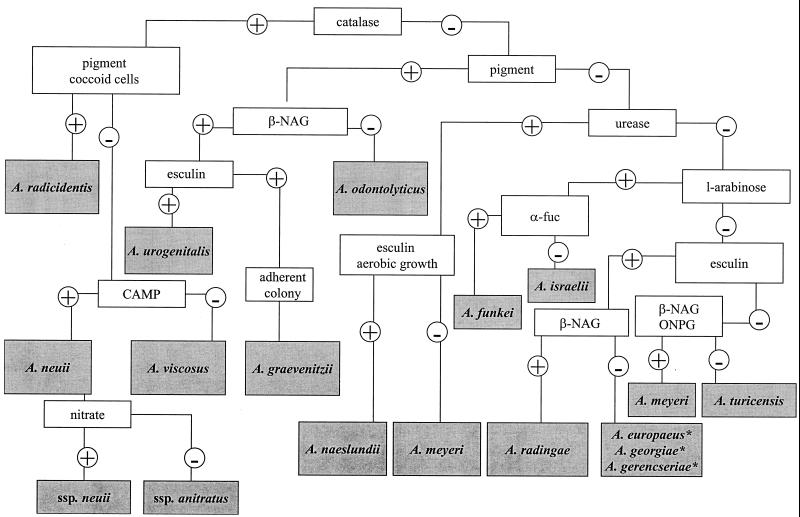
FIG. 1.
Flowchart for preliminary identification of Actinomyces species. All enzyme reactions were performed with Rosco diagnostic tablets. ∗, see Table 2; ssp., subsp.; α-fuc, α-fucosidase.
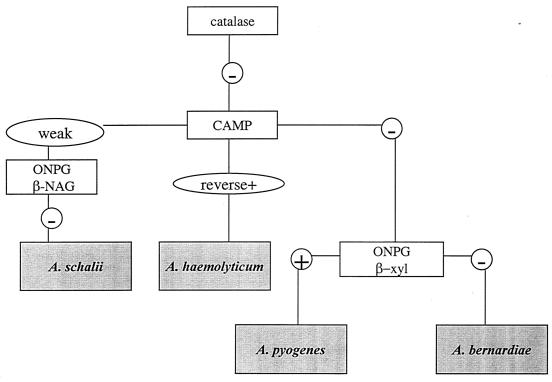
FIG. 2.
Flowchart for preliminary identification of Arcanobacterium species and A. schalii. All enzyme reactions were performed with Rosco diagnostic tablets.
RESULTS AND DISCUSSION
The flowchart for the preliminary identification of Actinomyces species by a limited number of rapid, cost-effective tests is depicted in Fig. 1. The flowchart for the preliminary identification of Arcanobacterium species and Actinobaculum schalii is depicted in Fig 2. The more comprehensive identification scheme is presented in Table 2.
The group of gram-positive, non-spore-forming bacilli consisting of several genera can reliably be differentiated from each other only by their metabolic end products. Gas-liquid chromatography should be used for differentiation of these genera. The separation of Arcanobacterium, Actinobaculum, and Actinomyces from other genera can be very difficult without the demonstration of succinic acid as a metabolic end product. In addition to succinic acid, the first two genera produce acetic acid and Actinomyces produces considerable amounts of lactic acid. Furthermore, the CAMP test reaction, catalase production nitrate reduction, and the production of β-galactosidase, β-NAG, and β-xylosidase are important tests for discrimination of these three genera from each other (Table 2; Fig. 2). Classically, Actinomyces species have been described as branching rods, but many of the recently described species are seldom branching.
In a deviation from the information in the current literature, we noticed that not only A. odontolyticus but also three other Actinomyces species, namely, A. graevenitzii, A. radicidentis, and A. urogenitalis, produced pigment. All colonies of A. odontolyticus showed brown or purple red pigmentation, A. graevenitzii showed a dark, almost black, pigmentation, A. radicidentis showed brown pigmentation and Actinomyces urogenitalis showed a reddish pigmentation on rabbit laked blood agar after incubation for 5 days. However, on brucella agar A. graevenitzii colonies were nonpigmented, confirming the original description by Pascual Ramos et al. (19). Colonies of the type strain of A. radicidentis were brownish, whereas those of A. urogenitalis were pinkish beige on brucella agar (after 5 days) and resembled colonies of A. odontolyticus (pinkish, “old rosa”). It is noteworthy that many other Actinomyces strains may exhibit some brownish color after prolonged incubation (6 to 11 days) (2); however, this is not usually regarded as real pigment production but, rather, is a result of medium decomposition.
In contrast to smooth and nonadherent colonies of A. odontolyticus, A. radicidentis, and A. urogenitalis, colonies of A. graevenitzii were rough and dry and adhered to blood agar, as described previously (19). In addition to deviating colony characteristics, in our study the definite differentiation of A. odontolyticus, A. graevenitzii, and A. urogenitalis was accomplished by testing for production of β-NAG: A. graevenitzii and A. urogenitalis were positive and A. odontolyticus was negative. Furthermore, esculin hydrolysis discriminates A. graevenitzii (negative) and A. urogenitalis (positive). The strikingly coccoid microscopic morphology of A. radicidentis (4) easily separated it from the other three pigment producers. On the other hand, this atypical morphology may lead one to falsely suspect the presence of gram-positive cocci and thus result in failure to identify the species as a member of the Actinomyces genus.
Catalase production has previously been considered the key characteristic for A. viscosus only. However, two additional catalase-producing Actinomyces species, Actinomyces neuii (two subspecies [7]) and A. radicidentis (4), currently exist in the genus. To confirm the separation of these newly described catalase-positive Actinomyces species from A. viscosus, we recommend testing for pigment production and the CAMP test reaction (Table 2). A. neuii can be further differentiated to the subspecies level by the nitrate reaction (7). We also found the type strain of A. neuii subsp. anitratus to be lipase positive and the type strain of A. neuii subsp. neuii to be lipase negative, characteristics that may be used for the separation of these two subspecies.
Previous published data on enzyme reactions and fermentation tests can be very difficult to interpret because they are often obtained by using different commercial kits and in-house systems that deviate in their substrate specificities, buffering capacities, and hence, sensitivities. Therefore, to allow direct comparisons, it is of utmost importance to carefully describe in publications the system or method by which the reactions were obtained (see the footnotes to Table 2).
In the present study, to compare different systems for testing, of enzyme activity, the reactivities of α-fucosidase, α-glucosidase, β-galactosidase, and β-NAG were tested for the reference strains in parallel by using individual Rosco tablets, 4-MU-linked substrates as a rapid filter paper spot test, and API ZYM kits. Table 3 presents the reactions obtained by these three test methods. Variation was seen mainly with α-glucosidase reactivities (three negative reactions with 4-MU-linked substrates and two negative reactions and one positive reaction with the API ZYM kit) and β-galactosidase reactivities two negative reactions with the API ZYM kit). In addition to reference strains, the α-fucosidase reactivities of 33 clinical strains of A. odontolyticus were tested in parallel by using Rosco tablets and 4-MU-linked substrates. Thirteen (33%) of these A. odontolyticus strains were α-fucosidase positive by using Rosco tablets, whereas only one (3%) isolate was positive by the method with 4-MU-linked substrates. The discrepancy may be explained by the substrate avidities or the specificities of the different test systems (1).
TABLE 3.
Enzyme reactions of three different test methodsa
| Strain | Rosco diagnostic tabletsb
|
4-MU-linked substratesb
|
APIZYM kitc
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α-Fucosidase | α-Glucosidase | β- NAG | β-Galactosidase | α-Fucosidase | α-Glucosidase | β- NAG | β-Galactosidase | α-Fucosidase | α-Glucosidase | β- NAG | β-Galactosidase | |
| A. europaeus | − | + | − | + | − | − | − | + | 0 | 3 | 0 | 5 |
| A. funkei | + | + | + | + | + | + | + | + | 0 | 4 | 5 | 3 |
| A. georgiae | − | + | − | + | − | + | − | + | 0 | 3 | 0 | 0 |
| A. gerencseriae | − | + | − | + | − | + | − | + | 0 | 5 | 0 | 5 |
| A. graevenitzii | − | − | + | + | − | − | + | + | 0 | 2 | 5 | 5 |
| A. israelii | − | + | − | + | − | + | − | + | 0 | 5 | 0 | 5 |
| A. meyeri | − | + | + | + | − | + | − | w | 0 | 3 | 0 | 3 |
| A. naeslundii | − | + | − | + | − | w | − | w | 0 | 0 | 0 | 3 |
| A. neuii subsp. anitratus | − | + | − | + | − | + | − | + | 0 | 4 | 0 | 5 |
| A. neuii subsp. neuii | − | + | − | + | − | − | − | + | 0 | 5 | 0 | 5 |
| A. odontolyticus | + | + | − | + | w | w | − | + | 1 | 1 | 0 | 2 |
| A. radicidentis | − | + | − | + | − | + | − | + | 0 | 4 | 0 | 5 |
| A. radingae | + | + | + | + | + | + | + | w | 5 | 5 | 5 | 5 |
| A. turicensis | + | + | − | − | + | + | − | − | 5 | 1 | 0 | 0 |
| A. urogenitalis | − | + | + | + | − | + | + | + | 0 | 5 | 4 | 5 |
| A. viscosus | − | + | − | + | − | − | − | w | 0 | 1 | 0 | 5 |
| A. bernardiae | + | + | + | − | − | + | + | − | 0 | 5 | 5 | 0 |
| A. haemolyticum | + | + | + | + | w | + | + | + | 0 | 3 | 4 | 3 |
| A. pyogenes | − | + | − | + | − | + | − | + | 0 | 0 | 0 | 0 |
| A. schalii | − | + | − | − | − | + | − | − | 0 | 5 | 0 | 0 |
The results with major discrepancies are indicated in boldface. β-galactosidase substrates were ONPG for Rosco diagnostic tablets, β-d-galactopyranoside for the 4-MU-linked substrates, and 2-naphthyl-β-d-galactopyranoside for the API ZYM kit.
−, negative reaction; +, positive reaction; w, weak reaction.
Color intensities: 0, negative; 1 to 2, weakly positive; 3 to 5, positive.
The phenotypic differentiation of A. israelii and A. gerencseriae (previously A. israelii serotype II) may pose problems due to a lack of discriminatory tests. Their biochemical reactions are very similar; however, the capability of A. israelii to ferment arabinose seems to separate it from A. gerencseriae (Table 2). According to the original description by Johnson et al. (10), the majority (89%) of A. israelii strains ferment arabinose. In contrast, in a recent study in which species-specific oligonucleotide probes were used for identification of A. gerencseriae and A. israelii, Jauh-Shun et al. (9) reported that only the reference strain of A. israelii fermented arabinose but that none of the clinical strains fermented arabinose. The result may be due to different substrate specificities and buffering conditions in their commercial biochemical test kit (Microbact 24AN system; Pacific Diagnostics) compared to those for PRAS biochemicals. In the present study, the arabinose-fermenting strains were identified as A. israelii and arabinose-nonfermenting strains were identified as A. gerencseriae. The separation was supported by the finding that all clinical A. israelii strains tested were positive for mannitol fermentation and arginine dihydrolase, whereas all strains of arabinose-negative A. gerencseriae were negative for these reactions. By using the 4-MU-linked fluorogenic substrates, Maiden et al. (15) reported negative α-mannosidase reactivity for A. israelii but positive α-mannosidase reactivity for A. gerencseriae. This reactivity pattern is listed in the user's guide for Rosco diagnostic tablets as well. Therefore, using Rosco diagnostic tablets, we tested both the type strains and seven clinical isolates representing each species for α-mannosidase reactivity. The type strain and six clinical strains of A. israelii were negative, as described previously (15), whereas only the type strain and one clinical strain of A. gerencseriae were positive.
In our flowchart (Fig. 1), esculin hydrolysis was used to separate A. meyeri and A. turicensis (negative) from Actinomyces radingae (positive). Furthermore, tests for production of β-NAG glucosaminidase and β-galactosidase were positive for A. radingae and negative for A. turicensis. Although we found that both type strains were α-fucosidase positive (with Rosco tablets, 4-MU-linked substrates, and the API ZYM kit), our previous experience shows that the production of α-fucosidase is a variable feature of A. turicensis among clinical strains. This probably reflects the vast heterogeneity of the former Actinomyces pyogenes-like (26) and A. meyeri-like (2) organisms that are included in A. turicensis (25). The separation of A. meyeri from A. turicensis is difficult. However, the type strain and four clinical isolates of A. meyeri (confirmed by molecular biology-based methods) were positive for both β-galactosidase and β-NAG by the test with Rosco tablets, whereas the type strain and five clinical isolates of A. turicensis were negative (see Table 2). Classically, A. meyeri has been described as an obligatory anaerobic organism (3). As A. turicensis grows both anerobically and aerobically (25), aerotolerance may also be a phenotypic test that can be used to discriminate between these two phenotypically close species. A. turicensis may be differentiated from Arcanobacterium bernardiae by positivity for xylose fermentation or a rapid β-xylosidase reaction (Fig. 2; Table 2). An unexpected finding was that Actinomyces funkei, A. meyeri, and A. radingae isolates, including the type strains, were positive for the CAMP test reaction.
The rapid enzyme tests that were used in our flowchart (Fig. 1) failed to separate Actinomyces europaeus, A. georgiae, and A. gerencseriae from each other. Instead, the fermentation of raffinose, rhamnose, sucrose, and trehalose could be used for identification (Table 2). According to the original descriptions (6, 10), A. europaeus does not ferment any of these carbohydrates, whereas A. georgiae ferments rhamnose, sucrose, and trehalose and A. gerencseriae ferments raffinose, sucrose, and trehalose. Surprisingly, in our tests, in which we also used the PRAS biochemicals, the type strain of A. gerencseriae did not ferment raffinose but was positive for rhamnose fermentation. However, the results for 12 clinical strains tested confirmed the original description of A. gerencseriae (10).
Although the identification of these gram-positive rods to the species level possesses major problems, it is important to clarify their roles in both oral and nonoral ecologies and infections. The phenotypic scheme presented here can help to identify the current members of the genera Actinomyces, Arcanobacterium, and Actinobaculum to the species level. In cases of unresolved results with the current scheme for potential actinomycete isolates from invasive sites, such as blood, and from clinically significant infections, the strains should be sent to a reference laboratory for definite confirmation of their identities. Commercial identification kits are widely used in clinical laboratories; however, the lack of data on the novel species interferes with successful precise identification. Therefore, evaluation of the applicability and accuracy of commercial kits for the rapid identification of Actinomyces species is in progress in our laboratory with the intent to further facilitate the task of clinical and oral microbiology laboratories.
ACKNOWLEDGMENT
This work was partially funded by the Finnish Dental Society and Research Foundation of Orion Corporation, Espoo, Finland.
REFERENCES
- 1.Bascomb S, Manafi M. Use of enzyme tests in characterization and identification of aerobic and facultatively anaerobic gram-positive cocci. Clin Microbiol Rev. 1998;11:318–340. doi: 10.1128/cmr.11.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brander M, Jousimies-Somer H. Evaluation of the RapID ANA II and API ZYM systems for identification of Actinomyces species from clinical specimens. J Clin Microbiol. 1992;30:3112–3116. doi: 10.1128/jcm.30.12.3112-3116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cato E, Moore W, Nygaard G, Holdeman L. Actinomyces meyeri sp. nov., specific epithet rev. Int J Syst Bacteriol. 1984;34:487–489. [Google Scholar]
- 4.Collins M D, Hoyles L, Kalfas S, Sundquist G, Monsen T, Nikolaitchouk N, Falsen E. Characterization of Actinomyces isolates from infected root canals of teeth: description of Actinomyces radicidentis sp. nov. J Clin Microbiol. 2000;38:3399–3403. doi: 10.1128/jcm.38.9.3399-3403.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durmaz B, Jousimies-Somer H R, Finegold S M. Enzymatic profiles of Prevotella, Porphyromonas, and Bacteroides species obtained with the API ZYM system and Rosco diagnostic tablets. Clin Infect Dis. 1995;20(Suppl. 2):192–194. doi: 10.1093/clinids/20.supplement_2.s192. [DOI] [PubMed] [Google Scholar]
- 6.Funke G, Alvarez N, Pascual C, Falsen E, Akervall E, Sabbe L, Schouls L, Weiss N, Collins M D. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:687–692. doi: 10.1099/00207713-47-3-687. [DOI] [PubMed] [Google Scholar]
- 7.Funke G, Stubbs S, von Graevenitz A, Collins M D. Assignment of human-derived CDC group 1 coryneform bacteria and CDC group 1-like coryneform bacteria to the genus Actinomyces as Actinomyces neuii subsp. neuii sp. nov., subsp. nov., and Actinomyces neuii subsp. anitratus subsp. nov. Int J Syst Bacteriol. 1994;44:167–171. doi: 10.1099/00207713-44-1-167. [DOI] [PubMed] [Google Scholar]
- 8.Hall V, O'Neill G L, Magee J T, Duerden B I. Development of amplified 16S ribosomal DNA restriction analysis for identification of Actinomyces species and comparison with pyrolysis-mass spectrometry and conventional biochemical tests. J Clin Microbiol. 1999;37:2255–2261. doi: 10.1128/jcm.37.7.2255-2261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jauh-Shun C, Vinh T, Davies J K, Figdor D. Molecular approaches to the differentiation of Actinomyces species. Oral Microbiol Immunol. 1999;14:250–256. doi: 10.1034/j.1399-302x.1999.140409.x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson J L, Moore L V H, Kaneko B, Moore W E C. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int J Syst Bacteriol. 1990;40:273–286. doi: 10.1099/00207713-40-3-273. [DOI] [PubMed] [Google Scholar]
- 11.Lawson P, Falsen E, Åkervall E, Vandamme P, Collins M D. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schalii sp. nov. Int J Syst Bacteriol. 1997;47:899–903. doi: 10.1099/00207713-47-3-899. [DOI] [PubMed] [Google Scholar]
- 12.Lawson P, Nikolaitchouk N, Falsen E, Westling K, Collins M D. Actinomyces funkei sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 2001;51:853–855. doi: 10.1099/00207713-51-3-853. [DOI] [PubMed] [Google Scholar]
- 13.Liljemark W F, Bloomquist C G, Bandt C L, Pihlström B L, Hinrichs J E, Wolff L F. Comparison of the distribution of Actinomyces in dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease. Oral Microbiol Immunol. 1993;8:5–15. doi: 10.1111/j.1399-302x.1993.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 14.Magee J. Whole-organism fingerprinting. In: Goodfellow M, O'Donnell A G, editors. Handbook of bacterial systematics. London, United Kingdom: Academic Press; 1993. pp. 383–427. [Google Scholar]
- 15.Maiden M, Tanner A, Macuch P. Rapid characterization of periodontal bacteria isolates using fluorogenic substrate tests. J Clin Microbiol. 1996;34:376–384. doi: 10.1128/jcm.34.2.376-384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolaitchouk N, Hoyles L, Falsen E, Grainger J M, Collins M D. Characterization of Actinomyces isolates from samples from the human urogenital tract: description of Actinomyces urogenitalis sp. nov. Int J Syst Evol Microbiol. 2000;50:1649–1654. doi: 10.1099/00207713-50-4-1649. [DOI] [PubMed] [Google Scholar]
- 17.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 18.Pascual Ramos C, Foster G, Collins M D. Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. Int J Syst Bacteriol. 1997;47:46–53. doi: 10.1099/00207713-47-1-46. [DOI] [PubMed] [Google Scholar]
- 19.Pascual Ramos C, Falsen E, Alvarez N, Åkervaii E, Sjöden B, Collins M D. Actinomyces graevenitzii sp. nov., isolated from human clinical specimens. J Clin Microbiol. 1997;61:2011–2014. doi: 10.1099/00207713-47-3-885. [DOI] [PubMed] [Google Scholar]
- 20.Sabbe L, van de Merwe D, Schouls L, Bergmans A, Vaneechoutte M, Vandamme P. Clinical spectrum of infections due to the newly described Actinomyces species A. turicensis, A. radingae, and A. europaeus. J Clin Microbiol. 1999;37:8–13. doi: 10.1128/jcm.37.1.8-13.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkonen N, Könönen E, Summanen P, Kanervo A, Takala A, Jousimies-Somer H. Oral colonization with Actinomyces species in infants by two years of age. J Dent Res. 2000;79:864–867. doi: 10.1177/00220345000790031301. [DOI] [PubMed] [Google Scholar]
- 22.Schaal K P. Genus Actinomyces. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1383–1418. [Google Scholar]
- 23.Schaal K P, Lee H J. Actinomycete infections in humans—a review. Gene. 1992;115:201–211. doi: 10.1016/0378-1119(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 24.Summanen P, Baron E J, Citron D M, Strong C A, Wexler H M, Finegold S M. Wadsworth anaerobic bacteriology manual. 5th ed. Belmont, Calif: Star Publishing; 1993. [Google Scholar]
- 25.Vandamme P, Falsen E, Vancanneyt M, Van Esbroeck M, Van de Merwe D, Bergmans A, Schouls L, Sabbe L. Characterization of Actinomyces turicensis and Actinomyces radingae strains from human clinical samples. Int J Syst Bacteriol. 1998;48:503–510. doi: 10.1099/00207713-48-2-503. [DOI] [PubMed] [Google Scholar]
- 26.Wüst J, Martinetti Lucchini G, Lüthy-Hottenstein J, Brun F, Altwegg M. Isolation of gram-positive rods that resemble but are clearly distinct from Actinomyces pyogenes from mixed wound infections. J Clin Microbiol. 1993;31:1127–1135. doi: 10.1128/jcm.31.5.1127-1135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wüst J, Stubbs S, Weiss N, Funke G, Collins M D. Assignment of Actinomyces pyogenes-like (CDC coryneform group E) bacteria to the genus Actinomyces as Actinomyces radingae sp. nov. and Actinomyces turicensis sp. nov. Lett Appl Microbiol. 1995;20:76–81. doi: 10.1111/j.1472-765x.1995.tb01290.x. [DOI] [PubMed] [Google Scholar]