Abstract
In the summer of 1999, the incidence of Salmonella enterica serotype Infantis infections in Alberta rose dramatically. Subsequent laboratory and epidemiological investigations established that an outbreak of human disease caused by this organism was occurring across Canada and was associated with pet treats for dogs produced from processed pig ears. Laboratory investigations using phage typing and pulsed-field gel electrophoresis (PFGE) established that isolates of Salmonella serotype Infantis from pig ear pet treats and humans exposed to pig ear pet treats comprised a well-defined subset of all isolates analyzed. Of the 53 subtypes of Salmonella serotype Infantis obtained around the time of the outbreak as defined by PFGE and phage typing, only 6 subtypes were associated with both human infection and isolation from pig ears. Together with information from epidemiological studies, these investigations established pig ear pet treats as the cause of the Salmonella serotype Infantis outbreak. The results are consistent with a model in which contaminated pig ear pet treats constitute a long-term, continuing vehicle for infection of the human population rather than causing temporally delimited point-source outbreaks. During the course of this outbreak, several other Salmonella serotypes were also isolated from pet treats, suggesting these products may be an important source of enteric infection in both humans and dogs. Though isolates of Salmonella serotypes other than Salmonella serotype Infantis from pet treats were also subjected to PFGE and phage typing, no link with human disease could be definitively established, and the contribution of pig ear pet treats to human disease remains unclear. Elimination of bacterial contamination from pet treats is required to reduce the risk of infection from these products.
Infections with nontyphoidal Salmonella enterica serovars are most often the result of ingestion of contaminated foods and are only rarely attributed to other causes. Salmonella is the most frequently detected cause of bacterial illness in parts of Europe and Korea and is the second most frequently detected cause of bacterial illness in the United States, England, and Australia (20, 28, 30). The estimated annual cost of food-borne salmonellosis in the United States is $0.9 to 3.5 billion (3). Around the world, Salmonella is the most important agent causing food-borne illness, with Salmonella enterica serovar Enteritidis and Salmonella serovar Typhimurium predominating (28). In Canada, Salmonella serotype Typhimurium and Salmonella serotype Enteritidis have also caused a majority of cases of sporadic diseases and are responsible for most outbreaks due to Salmonella since 1983 (5, 6, 8, 14–18). While other serotypes, including Salmonella serotypes Heidelberg, Hadar, Agona, Infantis, Thompson, and Newport, are important causes of human disease in Canada, for the most part these organisms are thought to be acquired through ingestion of contaminated food (11, 18). The characterization of a Salmonella serotype Infantis outbreak in Canada in 1999 (P. N. Sockett et al., unpublished data) led to the identification of pet treats produced from pig ears as a common source of human illness for many of the outbreak cases, suggesting that there are alternate ways for these bacteria to infect the human population.
Salmonella serotype Infantis is a pathogen of both humans and animals. This organism has caused outbreaks of human disease in England (2), Finland (12), the United States (21), and Canada (23), and isolates of Salmonella serotype Infantis have been recovered from pigs in Africa (22). A Danish surveillance program for Salmonella in fresh meat, instituted after an epidemic of Salmonella serotype Infantis in 1993 in that country, found 3.1% of pork cuts were contaminated with Salmonella serotype Infantis (27). In Finland, Salmonella serotype Infantis has been isolated frequently from chickens and cattle and is the third most common cause of human salmonellosis (24, 29). Isolates from a cattle outbreak of Salmonella serotype Infantis associated with contaminated feed in Finland were analyzed by pulsed-field gel electrophoresis (PFGE), plasmid analysis, ribotyping, and IS200 typing (19).
Salmonella serotype Infantis has been among the top 10 Salmonella serovars from humans in Canada characterized by the National Laboratory for Enteric Pathogens (NLEP) in recent years (5, 6,14–17), ranking second as a cause of human illness in Canada in 1983 and then decreasing to consistently rank sixth or seventh by the early 1990s (18). This was accompanied by a corresponding decrease in prevalence in nonhuman isolates during this time. Illness due to this organism has been seen in all provinces, although more than half of the human cases identified from 1993 through 1996 were from Ontario, Canada. In the absence of identified outbreaks, it has been difficult to trace the source of human infection by this organism.
In this study, we describe the typing and fingerprinting methods used to characterize isolates from pet treats associated with the Salmonella serotype Infantis outbreak in Canada in 1999. The results indicate that contact with pet treats made from pig ears or with dogs who had eaten these pet treats was the predominant cause of recent human Salmonella serotype Infantis illness. A variety of other pet treats were also contaminated with additional Salmonella serotypes. Although none of these isolates appeared to be associated with specific outbreaks, the disease burden in terms of sporadic cases due to serotypes other than Salmonella serotype Infantis to human disease is not known. Possible reasons for the predominance of Salmonella serotype Infantis in the present outbreak remain to be determined.
MATERIALS AND METHODS
Bacterial isolates.
Bacterial isolates were obtained from a number of provincial government and private laboratories across Canada. Isolates were forwarded either to the NLEP in Winnipeg, Manitoba, Canada, or to the Laboratory for Foodborne Zoonoses in Guelph, Ontario, Canada. After subculture, strains were prepared for long-term storage by inoculation onto slants of Institut Pasteur maintenance medium or were frozen at −80°C in brain heart infusion broth containing 15% (vol/vol) glycerol.
Bacterial strains used for analysis included Salmonella serotype Infantis isolates from humans gathered during the outbreak period (December 1998 to September 1999) as well as two animal isolates collected in 1998. These strains included 99 isolates from humans collected from all Canadian provinces except Newfoundland. They also included 48 pig or pig ear isolates from Alberta, Saskatchewan, Ontario, Québec, Newfoundland, and Nova Scotia; 6 poultry isolates from Alberta; and animal isolates from bovine stool (Alberta), an iguana (New Brunswick), and two unspecified animal sources. All but 2 of the 157 specimens analyzed were isolated in 1999.
Isolation and characterization of Salmonella.
Salmonella was isolated from human stool and urine samples in hospital laboratories by standard methods. During the outbreak investigation, screening of pet treats and animal products for Salmonella was conducted at Provincial Laboratories of Public Health, the Canadian Food Inspection Agency laboratories, and private laboratories by methods established by the Canadian Health Protection Branch (7, 26).
Salmonella isolates were characterized biochemically (9) and were initially confirmed with commercially available somatic polyvalent and monovalent antisera. Confirmation of the serotyping reactions (13, 25) was performed using a full set of polyclonal absorbed rabbit antiserum prepared and standardized by the NLEP.
Phage typing.
All Salmonella strains isolated were phage typed when appropriate using schemes developed at the Central Public Health Laboratory, Colindale, United Kingdom, and at the NLEP, Winnipeg, Manitoba, Canada (1; R. Khakhria, D. Duck, and H. Lior, Conjoint Meet. Inf. Dis., abstr. BC = 26, 1982).
PFGE.
PFGE was done according to previously described protocols (4, 10), summarized as follows. Bacteria grown overnight in brain heart infusion broth were collected by centrifugation, washed in TE wash buffer (100 mM Tris [pH 8.0], 100 mM EDTA), and suspended in TE wash buffer to give a density reading of 0.50 with a Dade turbidity meter. Proteinase K was added to washed cells, which were then directly embedded in 1.2% SeaKem Gold agarose (Mandel Scientific Co., Guelph, Ontario, Canada) prepared with TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) containing 1% sodium dodecyl sulfate. Solidified plugs were transferred to 1.5 ml of lysis buffer (50 mM Tris [pH 8.0], 50 mM EDTA, 1% sarcosine, 0.5 mg of proteinase K/ml) and incubated at 54°C in a water bath with shaking at 150 rpm for 2 h. Plugs were washed twice at 50°C with distilled water (18 MΩ quality) and then washed three times for 15 min with TE buffer with shaking. Plug slices were equilibrated in 100 μl of buffer H (Roche Diagnostics, Laval, Quebec, Canada) at 37°C for 15 min and digested at 37°C for 2 h with 80 U of XbaI in 100 μl of fresh buffer H. Selected isolates were also restricted with SpeI. After digestion, plugs were equilibrated in 0.5× TBE containing 0.89 M Tris, 0.89 M boric acid, and 0.02 M disodium EDTA (pH 8.4) (Roche Diagnostics) for 5 min. Electrophoresis was performed with the CHEF DR III unit (Bio-Rad Laboratories Canada, Ltd., Mississauga, Ontario, Canada) in 1% PFGE agarose at 14°C. Initial and final switch times were 2.2 and 63.8 s, respectively, and the total run time was 22 h. Following electrophoresis the gels were stained with ethidium bromide (0.5 μg/ml) and imaged with the Alpha Imager 2000 (Canberra Packard Canada, Mississauga, Ontario, Canada). The interpretation of the PFGE patterns was aided by use of Molecular Analyst Software Fingerprinting PLUS, version 1.6 (Applied Maths, Kortrijk, Belgium). All associations obtained using these software packages were checked visually by at least two people. For the isolates analyzed in this study, each unique pattern was given its own number. Dendrograms were created using the unweighted pair group method with arithmetric means within the Molecular Analyst software.
RESULTS
Isolation of Salmonella spp. from humans and dog treats.
Of 94 pig ear samples obtained from retail outlets at the time of the investigation, 48 (51%) were positive for Salmonella. Other retail products tested at this time included treats containing lamb, turkey, or beef products and those labeled as beef chew, rawhide, chew stick, beef hoof, braided chew, lamb chunks, hoof delight, and roulé rôti, or just “pet treats.” Several (15 of 39, or 38%) of these treats also contained Salmonella. In addition to retail products, pig ears and other pet treats or animal parts were obtained directly from production plants during the investigation. Salmonella was found in products from 5 of the 12 plants investigated in Alberta, Manitoba, Ontario, Québec, and New Brunswick and was found in plants from three of the five provinces. Overall, 49 of 171 (29%) of pig ear pet treats from these plants contained Salmonella; 7 other pet treat products (as listed above) were also contaminated with Salmonella. Some products contained as many as four different serotypes of Salmonella, although the relative levels of each serotype in these products were not determined.
Molecular typing of Salmonella serotype Infantis: evidence for transmission of bacteria from pig ear dog treats to humans.
Phage typing and PFGE using XbaI and SpeI were used to discriminate among isolates confirmed as Salmonella serotype Infantis by biochemical testing and serological identification (Table 1). Nine phage types (PTs) were found among the strains tested, as well as one isolate that gave an atypical phage typing pattern and another that was untypeable (Table 2). Of the 156 isolates tested by phage typing, 96 (62%) were PT 4, and 20 (13%) were PT 26. Together, these two PTs accounted for 116 (74%) of all isolates tested. PT 4 isolates comprised 53% of isolates from human patients, 85% of isolates from pig ears, and approximately one-third of the isolates from chickens. Other PTs were found in isolates from a more restricted number of hosts: PT 3 and PT 7 in isolates from humans and pigs, PT 8 in isolates from humans and an iguana, PT 9 in isolates from humans and chickens, and PTs 10, 13, and 29 in humans only.
TABLE 1.
Characteristics of serotype Infantis present in humans and pigs only
| Typea | PFGE type (XbaI) | PT | No. of isolates
|
|
|---|---|---|---|---|
| Human | Pig | |||
| 1 | 1 | 4 | 14 | 9 |
| 2 | 2 | 4 | 3 | 8 |
| 3 | 3 | 4 | 26 | 14 |
| 4 | 4 | 26 | 2 | 1 |
| 5 | 6 | 4 | 1 | 7 |
| 6 | 7 | 7 | 1 | 2 |
Combined PFGE type and PT.
TABLE 2.
Characteristics of all serotype Infantis characterized in the NLEP during the period of study
| Source(s) | No. of types | No. of isolates | Type characteristic(s) (PFGE:PT)a |
|---|---|---|---|
| Humans and pigs | 6 | 88 | 1:4, 2:4, 3:4, 4:26, 6:4, 7:7 |
| Humans only | 36 | 51 | 3:26, 6:26, 9:26, 10:3, 10:10, 11:10, 12:8, 12:26, 13:7, 14:13, 15:26, 16:4, 16:10, 19:8, 19:29, 20:4, 21:7, 22:7, 22:10, 24:UT, 25:26, 26:26, 27:26, 28:26, 29:4, 30:4, 30:7, 31:7, 32:7, 34:13, 35:9, 36:4, 37:13, 39:26, 40:13, 41:4 |
| Pigs only | 6 | 7 | 1:26, 5:4, 7:4, 8:4, 15:atypical, 19:3 |
| Humans and poultry | 1 | 5 | 13:9 |
| Poultry only | 2 | 2 | 17:4, 18:4 |
| Cows only | 1 | 1 | 38:26 |
| Other sources | 3 | 3 | 14:4, 23:ND, 33:8 |
UT, untypeable; ND, not determined.
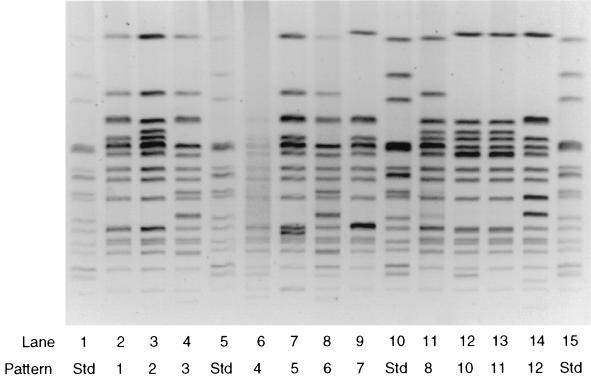
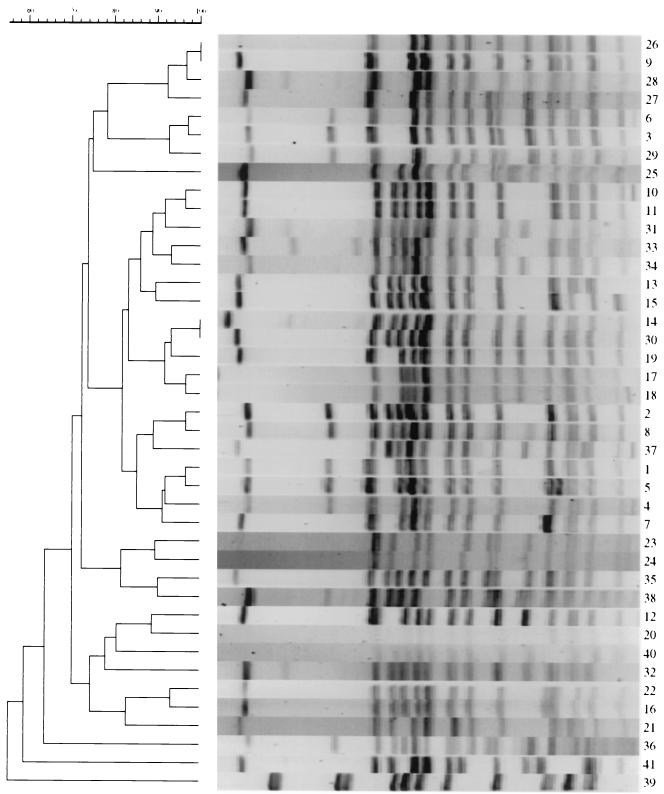
PFGE of Salmonella serotype Infantis isolates using XbaI demonstrated 41 types with patterns differing by one or more bands (Table 2). Forty isolates were tested using SpeI restriction digestion for PFGE, but no additional discrimination was obtained. As the distribution of patterns obtained using SpeI correlated well with the XbaI patterns, XbaI was used for all further analysis. PFGE patterns 1 and 3 comprised 41 of 99 (41%) of human isolates and 25 of 48 (52%) of isolates obtained from pigs or pig ear dog treats. These patterns differed by three bands (Fig. 1) and were widely separated in the dendrogram shown in Fig. 2, exhibiting relatedness of less than 80%. PFGE pattern 1 was more closely related to patterns 2, 3, 4, and 7, while patterns 3 and 6 were closely related (Fig. 2).
FIG. 1.
PFGE profiles of Salmonella serotype Infantis strains restriction enzyme digested with XbaI showing representative PFGE patterns. Lanes 1, 5, 10, and 15 contained the Salmonella serotype Newport standard strain AM01144.
FIG. 2.
Dendrogram of Salmonella serotype Infantis PFGE types.
Most (four of six) of the poultry isolates had PFGE pattern 13, which was also found in isolates from humans who did not have exposure to pig ears (Table 2). Unique PFGE XbaI patterns were found in the remaining two poultry isolates (patterns 17 and 18) and a bovine isolate (pattern 38). Relationships among PFGE XbaI patterns are summarized in Fig. 2.
A combination of PT and XbaI PFGE types was used to further assess relationships among the isolates tested. Though other methods, such as randomly amplified polymorphic DNA analysis, were used to investigate strain variability, they were much less discriminatory than PFGE or phage typing and were not used to assign specific types. There were 53 types that differed by PT and at least one band in PFGE XbaI macrorestriction patterns. A limited number of types were found in both humans and pigs (Table 1). Type 1 and 3 isolates constituted the two most prevalent types found in human disease. Most human strains with these characteristic types were found in Alberta (19 isolates), although lower numbers were also found from human cases in British Columbia (6), Saskatchewan (1), Manitoba (2), Ontario (4), Québec (7), and Prince Edward Island (2). Type 1 and 4 strains were found in animals or pig ear dog treats from Alberta (15 isolates), Saskatchewan (2), and Nova Scotia (2). Further investigations are being carried out to determine whether isolates having the same PTs and PFGE types were also present in other provinces.
Strains isolated from human cases with PFGE pattern 19 and PT 8 were very similar to a human strain with PFGE pattern 19 and PT 29 and a pig ear dog treat-associated strain with PFGE pattern 19 and PT 3, differing only by PT but having an identical PFGE type. All three types were found only in Québec.
Most types found were from strains isolated only from pig ear dog treats or only from humans. Human isolates with types unrelated to those found in pig ear dog treats yielded 31 PFGE types (Table 2). PFGE analysis subdivided most PTs into several groups, and several PFGE patterns were shared by two or more PTs (Table 3).
TABLE 3.
PFGE patterns in specific PTs
| PT | PFGE pattern no. |
|---|---|
| 3 | 10, 19 |
| 4 | 1–8, 14, 16–18, 20, 29, 30, 36, 41 |
| 7 | 7, 13, 21, 22, 30, 31, 32 |
| 8 | 12, 19, 33 |
| 9 | 13, 35 |
| 10 | 10, 11, 16, 22 |
| 13 | 14, 34, 37, 40 |
| 26 | 1, 3, 4, 6, 9, 12, 15, 25–28, 38, 39 |
| 29 | 19 |
| Atypical | 15 |
| Untypeable | 24 |
| Not tested | 23 |
Salmonella serotype Infantis was the serotype most often isolated from pig ear pet treats, comprising 18% of all isolates obtained (Table 4). Salmonella serotype Infantis was also isolated more frequently than other serotypes from pet treats other than pig ear pet treats, comprising 28% of the total isolates obtained. The two serotypes with similar but slightly reduced frequencies were Salmonella serotype Typhimurium (11%) and Salmonella serotype Derby (10%); all other serotypes were isolated much less frequently from pig ear pet treats. Salmonella serotype Banana and Salmonella serotype Typhimurium var. copenhagen were found in pet treats during the course of this investigation but were not isolated from humans during either 1998 or 1999.
TABLE 4.
Incidence of Salmonella serotypes in humans and in pet treats
| Salmonella serotype | Incidence in humans ina:
|
Frequency of isolation inb:
|
||
|---|---|---|---|---|
| 1998 | 1999 | Pig ear | Other | |
| Agona | 164 | 151 | 2/122 | 1/29 |
| Agoueve | 1 | 1 | 0 | 1/29 |
| Anatum | 52 | 26 | 5/122 | 0 |
| Banana | 0 | 0 | 0 | 1/29 |
| Bovis-morbificans | 59 | 23 | 3/122 | 1/29 |
| Brandenberg | 110 | 45 | 4/122 | 1/29 |
| California | 6 | 0 | 4/122 | 0 |
| Derby | 57 | 29 | 12/122 | 1/29 |
| Havana | 17 | 5 | 0 | 2/29 |
| Heidelberg | 1,241 | 821 | 5/122 | 0 |
| Infantis | 155 | 193 | 22/122 | 8/29 |
| Livingstone | 2 | 1 | 1/122 | 0 |
| Mbandaka | 100 | 62 | 2/122 | 1/29 |
| Meleagridis | 19 | 5 | 0 | 1/29 |
| Montevideo | 43 | 54 | 1/122 | 2/29 |
| Muenster | 16 | 8 | 1/122 | 0 |
| Ohio | 13 | 13 | 5/122 | 0 |
| Panama | 27 | 34 | 1/122 | 0 |
| Schwarzengrund | 29 | 49 | 2/122 | 0 |
| Typhimurium | 1,590 | 1,401 | 13/122 | 2/29 |
| Typhimurium var. copenhagen | 0 | 0 | 1/122 | 1/29 |
| Uganda | 15 | 8 | 1/122 | 0 |
| Worthington | 5 | 23 | 5/122 | 0 |
Information regarding the incidence of human disease in Canada in 1999 was taken from the National Enteric Surveillance Report, a tool for collecting and collating weekly data on bacterial isolations provided by provincial laboratories. Though the information is validated weekly, it is regarded as provisional until a final validation is completed after the year's end. Data for 1999 should be regarded as provisional.
The frequency of isolation refers to isolates obtained as part of this study. The number of each Salmonella serotype isolated is expressed as the ratio of all Salmonella isolates obtained from either pig ears or from other sources. “Other” indicates different kinds of pet treats (braided chew, hoof, roulé rôti) or pig parts. This does not include other animal isolates summarized in Table 1.
Molecular typing of other Salmonella serotypes.
Phage and PFGE typing information are available only for some of the other serotypes of Salmonella isolated from pig ear pet treats (Table 5). Relatively low numbers of some serotypes were isolated from humans in Canada in 1998 and 1999. These isolates included the Salmonella serotypes Agoueve, Banana, Bovis-morbificans, Brandenberg, California, Derby, Havana, Livingstone, Mbandaka, Meleagridis, Montevideo, Muenster, Ohio, Panama, Schwarzengrund, and Uganda. PFGE was not done with human isolates of these serotypes unless they were involved in outbreaks of human disease during the course of the year. Pig ear pet treat isolates of a few serotypes were analyzed by phage typing and by PFGE to give baseline data for future comparisons with human isolates (see Table 5).
TABLE 5.
PT and PFGE type found in Salmonella serotypes other than serotype Infantis from pig ears and humansa
| Salmonella serotype | PT
|
PFGE type
|
||
|---|---|---|---|---|
| Pig ear | Human | Pig ear | Human | |
| Agona | Pattern G | Patterns C and E | 16 | 1–3, 7–8, 9–15 |
| Anatum | Pattern A | Pattern A | 2 | 1 |
| Heidelberg | 19 | Many | 1 | 1–24 |
| Typhimurium | 12, 104, 104a | Many | 1–4 | 1–5 |
| Worthington | NT | NT | 2–5 | 1, 2 |
There are 49 PTs in the serotype Heidelberg phage typing scheme and 262 PTs in the serotype Typhimurium scheme. Many of these have been found in Salmonella isolates in Canada, including those found in pig ears. NT, not tested (isolates were not sent to the NLEP for testing).
The E1 PFGE pattern of an environmental Salmonella serotype Muenchen strain obtained from a packing plant that produced pig ear pet treats was quite different from the A1, A2, B1, B2, C1, and D1 patterns previously found among human isolates of this organism. Both the PT and PFGE type of Salmonella serotype Agona from pig ear pet treats were distinct from types characteristic of human strains (Table 5). Similarly, while B1, B2, B3, and C3 PFGE patterns were found in four pig ear isolates of Salmonella serotype Worthington, only one human strain isolated in British Columbia in 1999 carried the B1 pattern. Seven other human isolates associated with a previous outbreak in British Columbia had the A1 PFGE pattern. There is no convincing evidence that pig ear pet treats are associated with human disease caused by these organisms.
Salmonella serotype Typhimurium PFGE types 1, 2, 3, and 4 have been isolated from pig ear dog treats obtained from Toronto, Ontario (types 1 and 2), Halifax, Nova Scotia (types 3 and 4), and Calgary, Alberta (type 3). Strains with PFGE type 3 have been isolated recently from a number of patients in Mississauga, Ontario, while those with PFGE type 1 have been isolated from patients in British Columbia. Isolates with the PFGE type 1 pattern were also obtained recently from samples of cheese found in Toronto. Interestingly, some of the Salmonella serotype Typhimurium isolated from pig ear dog treats were definitive type (DT) 104 strains exhibiting the ACSSuT multiple antibiotic resistance phenotype.
A single strain of Salmonella serotype Heidelberg PT 19 and PFGE type 1 was isolated in Calgary from pig ear pet treats. This combination of types was found in a large number of Salmonella serotype Heidelberg isolates from British Columbia and from other sources. PFGE type 1 is the predominant pattern detected by the NLEP and may be carried by the majority of Salmonella serotype Heidelberg strains. Strains with both PT 19 and PFGE type 1 are less prevalent.
DISCUSSION
The evidence presented here indicates that pig ears and other pig products are a potential source of human infection by many serotypes of Salmonella. Though pet treats made from pig ears and other porcine products were contaminated with many different kinds of bacteria, only Salmonella serotype Infantis was identified as causing an increased number of human infections. While the reasons for this are not clear, it may be that this organism was more virulent or was present in higher numbers in the pet products; however, quantitative assessments of Salmonella serotype Infantis in these products were not made. A second possibility is that Salmonella serotype Infantis grows better in dogs than other serotypes, thus serving to amplify the organism, and that dogs are an intermediate source of infection for many humans. During the outbreak, infection occurred in patients whose sole exposure was to pig ears. Few isolations from dogs were attempted, though case reports suggested that dogs become sick as a result of exposure to pet treats. It is not clear that methods currently available would be adequate for this kind of analysis. Identifying the absolute numbers of bacteria contaminating these treats may be of limited value until the role of dogs in amplifying pathogen numbers and causing infection is understood.
Salmonella serotype Infantis characterized in this study constituted a diverse group of organisms. Isolates from humans showed considerable variation in their PFGE patterns and PTs, and in some cases particular types may be associated with specific sources or routes of infection. The presence of identical PFGE and PTs in strains of Salmonella serotype Infantis associated with both humans and pig ear pet treats supports the idea that pig ear pet treats were the original source of these strains. Pigs and pig ear pet treats appeared to be contaminated with a limited variety of Salmonella serotype Infantis PFGE and PTs, suggesting that limited type diversity is not a likely reason for the isolation of identical PTs and PFGE types from both pig ear dog treats and humans. The prolonged outbreak of Salmonella serotype Infantis appeared to result from multiple introductions of the organism into pigs or pig ears and from there into the human population, rather than introduction of bacteria from a point source. Support for this premise comes from the fact that the PFGE patterns 1 and 3 responsible for most of the human cases are widely separated in the dendrogram of Salmonella serotype Infantis patterns and have less than 80% pattern similarity.
A single pig ear isolate with a PFGE type 19 pattern was found in Québec, where there were also a number of isolates from human patients with identical PFGE patterns. The finding of these and closely related types only in Québec suggests a possible route of transmission from pet treats to humans specific for this area and indicates that there may be regional differences in the Salmonella serotype Infantis types contaminating these treats. It is possible that the prevalence of the type 1 and 3 patterns among Salmonella serotype Infantis isolates from humans largely reflects the distribution of pet treats from suppliers in western Canada throughout the rest of the country (Sockett et al., unpublished).
Salmonella serotype Infantis is present in pigs, poultry, chicken eggs, cattle, and animal feeds. In 1996, Salmonella serotype Infantis of animal origin was predominantly recovered from pork or pork products, especially in Québec and Prince Edward Island (14). Very few isolates of this organism were found in pork in 1995 (15). Poultry, especially in Ontario, was the predominant source of nonhuman Salmonella serotype Infantis in 1993 and 1994 (16, 17). Interestingly, a single isolate of Salmonella serotype Infantis was obtained from dog snacks in 1994, although not in 1995 or 1996. It is not clear how this pathogen emerged in pork, though its present prevalence seems to be relatively recent.
Both PFGE and phage typing were required for an adequate description of the strain characteristics of all Salmonella serotypes analyzed in this study. Phage typing discriminated among most of the Salmonella serotype Infantis PFGE types and less frequently among PFGE types of other Salmonella serotypes. PFGE similarly subdivided PTs of most Salmonella serotypes tested. It is important to note that pentaresistant Salmonella serotype Typhimurium DT 104 was isolated from pig ears, suggesting that this may constitute a source of human infection with this organism as well.
The sharp increase in the numbers of Salmonella serotype Infantis infections is a recent trend in human infection, and dog treats may have contributed to this increase. Clearly, the causal relationship between human cases and pig ear dog treats was clarified by molecular typing, which also served as the trigger for an epidemiological investigation of this outbreak. Comprehensive and detailed descriptions of the phenotypic, genetic, and molecular fingerprint characteristics of the Salmonella isolates associated with pig ear pet treats are necessary to evaluate the effects of interventions designed to eliminate these treats as a source of human infection. For instance, the fact that phage typing and PFGE appear to measure biological traits that vary independently of each other suggests that the use of both methods is necessary for adequate typing of related isolates. The laboratory and epidemiological components of the National Enteric Surveillance Program for enteric bacteria currently track trends in Salmonella infections throughout Canada to evaluate the effect of such interventions and to determine the contribution of pig ear pet treats to the disease burden.
Pig ears and other dog treats from similar sources, produced under similar conditions, may have been a continuing source of human disease for quite some time and may have contributed to the background of sporadic cases seen in Canada and elsewhere. Continued epidemiological and laboratory monitoring of changes in the background of sporadic human cases, especially of uncommon serotypes, will further define the scope of the problem.
ACKNOWLEDGMENTS
We thank the staff of the provincial Public Health Laboratories, the Canadian Food Inspection Agency (CFIA), and MAPAQ, who provided initially characterized isolates to the NLEP along with information from plant inspections and analysis of retail samples. We also thank Karen Grimsrud, Linda Chui, Shelley Johnson, Walter Demczuk, and Jennifer Campbell for their valuable contributions to the epidemiological and laboratory investigations.
REFERENCES
- 1.Anderson E S, Williams R E O. Bacteriophage typing of enteric pathogens and Staphylococci and its use in epidemiology. J Clin Pathol. 1956;9:94–127. doi: 10.1136/jcp.9.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrell R A E. Isolation of Salmonella from human foods in the Manchester area. Epidemiol Infect. 1987;98:277–284. doi: 10.1017/s0950268800062038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzby J C, Roberts T. Economic costs and trade impacts of microbial foodborne illness. World Health Stat Q. 1997;50:57–66. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Atlanta, Ga: The National Molecular Subtyping Network for Foodborne Disease Surveillance. Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 5.Cuff W, Khakhria R, Woodward D L, Ahmed R, Clark C G, Rodgers F G. Enteric pathogens identified in Canada: annual summary 1997. Winnipeg, Manitoba, Canada: Laboratory Centre for Disease Control; 2000. [Google Scholar]
- 6.Cuff W, Khakhria R, Woodward D L, Ahmed R, Clark C G, Rodgers F G. Enteric pathogens identified in Canada: annual summary 1998. Winnipeg, Manitoba, Canada: Laboratory Centre for Disease Control; 2001. [Google Scholar]
- 7.D'Aoust J-Y, Purvis U. Isolation and identification of Salmonella from foods. MFHPB-20. In: Warburton D, editor. Compendium of analytical methods. Laval, Quebec, Canada: Health Protection Branch, Health Canada. Polyscience Publications; 1998. [Google Scholar]
- 8.Ellis A. National foodborne, waterborne, and enteric outbreak summary report 1997–1998. LCDC report. Can J Infect Dis. 1999;10:201–206. [Google Scholar]
- 9.Ewing W H. Edwards and Ewings's identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. The genus Salmonella; pp. 181–340. [Google Scholar]
- 10.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez T M, Motarjemi Y, Miyagawa S, Käferstein F K, Stöhr K. Foodborne salmonellosis. World Health Stat Q. 1997;50:81–89. [PubMed] [Google Scholar]
- 12.Hatakka M. Salmonella outbreak among railway and airline passengers. Acta Vet Scand. 1992;33:253–260. doi: 10.1186/BF03547291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kauffman F. The bacteriology of Enterobacteriaceae. Baltimore, Md: The Williams & Wilkins Co.; 1966. [Google Scholar]
- 14.Khakhria R, Woodward D, Cuff W, Rodgers F, Johnson W. Enteric pathogens identified in Canada: annual summary 1996. Winnipeg, Manitoba, Canada: Laboratory Centre for Disease Control; 1999. [Google Scholar]
- 15.Khakhria R, Woodward D, Johnson W. Salmonellae, shigellae, pathogenic E. coli, campylobacters and Aeromonas identified in Canada: annual summary 1995. Ottawa, Ontario, Canada: Laboratory Centre for Disease Control; 1997. [Google Scholar]
- 16.Khakhria R, Woodward D, Johnson W. Salmonellae, shigellae, pathogenic E. coli, campylobacters and Aeromonas identified in Canada: annual summary 1994. Ottawa, Ontario, Canada: Laboratory Centre for Disease Control; 1996. [Google Scholar]
- 17.Khakhria R, Woodward D, Johnson W. Salmonellae, shigellae, pathogenic E. coli, campylobacters and Aeromonas identified in Canada: annual summary 1993. Ottawa, Ontario, Canada: Laboratory Centre for Disease Control; 1995. [Google Scholar]
- 18.Khakhria R, Woodward D, Johnson W M, Poppe C. Salmonella isolated from humans, animals, and other sources in Canada, 1982–92. Epidemiol Infect. 1997;119:15–23. doi: 10.1017/s0950268897007577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindqvist N, Heinikainen S, Toivonen A-M, Pelkonen S. Discrimination between endemic and feedborne Salmonella Infantis infection in cattle by molecular typing. Epidemiol Infect. 1999;122:497–504. doi: 10.1017/s095026889900237x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meehan P J, Atkeson T, Kepner D E, Melton M. A foodborne outbreak of gastroenteritis involving two different pathogens. Am J Epidemiol. 1992;136:611–616. doi: 10.1093/oxfordjournals.aje.a116539. [DOI] [PubMed] [Google Scholar]
- 22.Ngoma M, Pandey G S, Suzuki A, Sato G, Chimana H. Prevalence of Salmonella in apparently healthy slaughtered cattle and pigs in Zambia. Indian J Anim Health. 1996;35:197–200. [Google Scholar]
- 23.Page W E, Shimes V, Vair L. Salmonella infantis in a hospital obstetrical department—Ontario. Canada Dis Wkly Rep. 1988;14:42–43. [PubMed] [Google Scholar]
- 24.Pelkonen S, Romppanen E-L, Siitonen A, Pelkonen J. Differentiation of Salmonella serovar Infantis isolates from human and animal sources by fingerprinting IS200 and 16S rrn loci. J Clin Microbiol. 1994;32:2128–2133. doi: 10.1128/jcm.32.9.2128-2133.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popoff M Y, LeMinor L. W. H. O. Collaborating Centre for Reference and Research on Salmonella. 6th ed. Paris, France: Institut Pasteur; 1992. Antigenic formulas of the Salmonella serovars. [Google Scholar]
- 26.Poppe C, Mann E D. Procedure for the isolation of Salmonella species by the modified semi-solid Rappaport Vassiliadis (MSRV) method. MFLP-75. In: Warburton D, editor. Compendium of analytical methods. Health Protection Branch, Health Canada. Laval, Québec, Canada: Polyscience Publications; 1998. [Google Scholar]
- 27.Sørensen L L, Bager F. Salmonella—forekomst i svinekød 1993–1994. Dansk Veterinærtidsskr. 1995;78:159–162. [Google Scholar]
- 28.Todd E C. Epidemiology of foodborne diseases: a worldwide review. World Health Stat Q. 1997;50:30–50. [PubMed] [Google Scholar]
- 29.Vasa M. Epidemiological aspects of Salmonella infantis and Salmonella typhi infections in Finnish broiler chickens. Nord Vet Med. 1984;36:317–323. [PubMed] [Google Scholar]
- 30.Wheeler J G, Sethi D, Cowden J M, Wall P G, Rodrigues L C, Tomkins D S, Hudson M J, Roderick P J. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. The Infectious Intestinal Disease Study Executive. Br Med J. 1999;318:1046–1050. doi: 10.1136/bmj.318.7190.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]