Abstract
During an epidemiological study on rotaviruses among diarrheic children in the northeastern and middle belt regions of Nigeria, the distribution of G and P types was investigated in 127 stool specimens. By PCR G typing, the G type of rotaviruses in 97 samples was identified. Interestingly, an unusual G8 type, as well as common G1, G2, and G3 types, was detected more frequently (31 of 112; 27.7%). Eleven samples contained multiple G types, and a G9 strain (Bulumkutu) was identified for one of the probable mixed infections. In PCR P typing, P[6] was detected most frequently, P[8] being the second most common type, while the P type of 73 samples could not be identified. One rotavirus strain with a G8 type specificity could be cultivated in cell culture, and the P type of this strain was found to be P[1], which is usually carried by bovine strains. When the combinations of G and P types were examined, the unusual strains G2P[6] and G8P[1] were often identified. Sequence analysis was performed for the VP7 gene of the G9 strain Bulumkutu and the VP4 and VP7 genes of G8P[1] strain HMG035. The VP7 sequence of the Nigerian serotype G9 was more closely related to that of a Brazilian strain than to those of other African strains. The VP7 and VP4 genes of G8P[1] strain HMG035 were found to be very similar to that of a Thai bovine strain A5, suggesting that bovine strains may have been transmitted directly to humans. These results highlight an unexpected diversity among rotavirus strains in Nigeria and emphasize the need for further serological and genetic surveys on more rotavirus strains in African countries, including Nigeria.
Rotavirus gastroenteritis among infants and young children remains a major cause of mortality in developing countries and a significant cause of morbidity in the developed countries (22). Rotaviruses are classified into groups A to E, group A rotavirus being known to exhibit the highest prevalence and pathogenicity. The virus has an inner capsid and an outer one containing a genome of 11 segments of double-stranded RNA. The two outer capsid proteins of the virus, VP7 (encoded by gene segment 7, 8, or 9, depending on the strain) and VP4 (encoded by gene segment 4), independently specify the G and P types, respectively. Consequently, rotaviruses exhibit diverse and complex serotypic specificities (20).
To date, 14 G serotypes have been defined by neutralization assays and 10 of them have been identified in humans (11). Extensive G serotyping surveys across the globe have shown that serotypes G1 to G4 are the most prevalent worldwide (2, 10, 12, 13, 16, 29, 31). Thus, the first licensed rotavirus vaccine, RotaShield, formulated to cover the epidemiologically important serotypes G1 to G4, was used in the United States (18). However, the vaccine was later withdrawn because of a possible association with intussusception reported with its use (6, 7). There now appears to be a resurgence of enthusiasm for this vaccine, at least in randomized, controlled trials in developing countries (17, 38). In contrast, there has been an increasing number of reports on the detection of rotavirus strains with unusual G serotypes among infants recently (5, 10, 14, 24, 26–32). They include G5, G6, G8, G9, G10, and G12. Some of these G serotypes were detected exclusively in animals in the past; G5 in pigs and G6, G8, and G10 in cattle.
A total of 20 P types has been reported, of which 7 types have been detected in humans, P[8] and P[4] being the most common. Compared to surveys on the distribution of G types, surveys on the distribution of P types have been done less widely because of a lack of rapid and simple serological methods. Precise identification of these VP4 and VP7 antigenic determinants and global epidemiological surveys on the serotype distribution of rotaviruses will provide the basic information necessary to develop effective vaccines. In particular, more complete surveillance of the serotype distribution is required in the African continent, where a higher frequency of unusual serotypes (2–4, 5, 9, 10, 14, 19, 31) appears to be associated with low protection efficacy in vaccine trials (23).
In previous studies (2–4), rotavirus strains bearing G2 specificity have never been detected in Nigeria, and only one strain each bearing G8 and G9 specificities has been identified in addition to a number of untypeable specimens. In this study, however, two unusual strains, exhibiting G2P[6] and G8 type specificity, respectively, were detected in a high proportion. The results of sequence analyses of the VP7 gene of a Nigerian G9 strain and the VP4 and VP7 genes of a Nigerian G8P[1] strain are also presented.
MATERIALS AND METHODS
Stool specimens.
A total of 127 stool specimens was collected from children with diarrhea under 7 years of age in the outpatient pediatric department of the State Specialist Hospital, Maiduguri, Nigeria, and the inpatient pediatric department of the Federal Medical Centre, Makurdi, Nigeria, between November 1999 and April 2000, as previously described (1). The hospitals in which samples were obtained serve patients of different educational and socioeconomic backgrounds living in neighborhoods with distinctly different levels of sanitation. They were stored at −20°C until being transported on ice to Japan, where they were analyzed.
Virus isolation in MA-104 cells in roller tube cultures was attempted from 11 stool specimens of a sufficient amount for cell culture as described previously (37). Briefly, each stool extract was pretreated with 10 to 30 μg of acetylated trypsin (Sigma, St. Louis, Mo.) per ml, inoculated onto MA-104 cells, maintained in the presence of trypsin (3 μg/ml), and then harvested 3 to 5 days after infection. At least three cycles of passage in roller tube cultures were performed.
ELISA.
Enzyme-linked immunosorbent assay (ELISA) with a group-A-common monoclonal antibody (YO-156) directed to VP6 was carried out as described previously (35).
Polyacrylamide gel electrophoresis (PAGE).
Viral RNA was extracted from a fecal suspension or culture fluid with a one-fifth volume of a 6× disruption solution comprising 6% sodium dodecyl sulfate, 0.6% 2-mercaptoethanol, and 300 mM EDTA and then with phenol-chloroform. The RNA was electrophoresed in 10% acrylamide gels (2 mm thick) for 16 h at 20 mA at room temperature. RNA segments were visualized by silver staining.
RT-PCR.
Rotavirus double-stranded RNA was extracted by the guanidine-silica method with an RNAid kit (Bio 101, La Jolla, Calif.), and the extracts were used as templates for reverse transcription-PCR (RT-PCR).
For G typing, a full-length VP7 gene (1,062 bp) was amplified with a pair of primers, T31 and T32, corresponding to the common 5′ and 3′ ends of the gene, respectively. In the second and multiplex seminested PCR, G-serotype-specific primers were used to identify G types (36). Similarly, PCR for P typing was carried out in two steps (first and second amplifications), as described previously (39). Briefly, a pair of primers (T5′END and -3′END) corresponding to the common sequences of nucleotide 11 to 32 and 1072 to 1094 was used for the first amplification, and a mixture of primers specific to each of the variable regions of P1A[8], P1B[4], P2[6], and P3[9] and a primer (T5′END) corresponding to nucleotides 11 to 32 were employed for the second amplification. PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and then visualized with a UV transilluminator.
Nucleotide sequencing.
The purified RT-PCR products from the stool specimen containing rotavirus of the G9 type and from the culture fluid infected with a rotavirus strain, HMG035, with G8P[1] specificity were sequenced directly by the method previously described (1). The 5′ and 3′ sequences comprising 20 to 22 nucleotides were derived from the primer sequences used for the RT-PCR. Gel read lengths of 400 to 500 nucleotides were routinely used, and the sequences corresponding to the primers were sequenced using different PCR products. The sequencing primers and their positions as used individually in the sequence extension reactions are listed in Table 1. Sequence data were analyzed with the Genetyx-Mac software package for sequence alignment and for the construction of a phylogenetic tree using the unweighted-pair group method with arithmetic means.
TABLE 1.
Primers used for RT-PCR and sequence determination of the VP4 and VP7 genes of two Nigerian human rotavirus strains, Bulumkutu and HMG035
| Strain (gene) | Primera | Primer sequence | Position (5′–3′) |
|---|---|---|---|
| HMG035 (VP4) | T5′ENDb | TGGCTTCGTTCATTTATAGACA | 11–32 (F)c |
| T804b | GGCTTTAAAATGGCTTCACTC | 1–21 (F) | |
| T805b | GGTCACATCCTCTGTCAGTTGC | 2341–2362 (R) | |
| T803 | AACACAAACAGATGGTTAGC | 300–320 (F) | |
| T3′ENDb | CTAAATGCTTTTGAATCATCCCA | 1072–1094 (R) | |
| T806b | GACGGTGAAGAAGTGACAGC | 931–951 (F) | |
| T807b | CTCTTCATTACATTCGTCGCC | 1641–1661 (R) | |
| T809 | TGGAGCGTAATTTGTTTGCGC | 130–150 (R) | |
| T810b | CAGTTACGGTTAGACAAGAC | 1470–1489 (F) | |
| T810a | CTACTCTAACTGACTCCCTGT | 1694–1714 (F) | |
| T811 | AGGAGGTGCCATTTGACGTAC | 2107–2127 (F) | |
| HMG035 (VP7) | T31b | GGCTTTAAAAGAGAGAATTTCCGTCTGG | 1–28 (F) |
| T32b | GGTCACATCATACAATTCTAATCTAAG | 1039–1062 (R) | |
| T1 | TTGGCCATCCTTTAGT | 369–385 (R) | |
| T3 | CCATTGGATTACACAACCATTC | 532–553 (R) | |
| T4 | GCTACGTTTTCTCTTGGTCC | 805–824 (R) | |
| T43 | GGAAAAAATGGTGGCAAGT | 914–932 (F) | |
| Bulumkutu (VP7) | T31b | GGCTTTAAAAGAGAGAATTTC CGTCTGG | 1–28 (F) |
| T32b | GGTCACATCATACAATTCTAATCTAAG | 1039–1062 (R) | |
| T730 | ATTAATTTACCAATCACTGG | 211–230 (F) | |
| T1 | TTGGCCATCCTTTAGT | 369–385 (R) | |
| T3 | CCATTGGATTACACAACCATTC | 532–553 (R) | |
| T731 | TTGAATACGCAGACTTTAGG | 640–659 (F) | |
| T738 | CCGATGTTGTTGATGGTGTG | 728–747 (F) |
All the primers, including those used for RT-PCR, were used individually for sequence determinations. For the VP4 gene, three partially overlapping RT-PCR products were amplified by employing T804 and T3′ END, T806 and T807, and T805 and T810 primer pairs, respectively. The sequencing results were unequivocal; hence, one strand of each of the genes was sequenced in both strains.
These primers were used in RT-PCR.
F, forward; R, reverse.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper for the VP4 and VP7 genes of strains HMG035 and Bulumkutu have been submitted to the GenBank database and have been assigned the accession numbers AF361438 (HMG035 VP4), AF359359 (HMG035 VP7), and AF359358 (Bulumkutu VP7).
RESULTS
Rotavirus detection.
A total of 127 stool specimens was analyzed by RNA-PAGE, ELISA, and RT-PCR. Fifteen (11.8%), 29 (22.8%), and 112 (88.2%) samples were found to be positive for rotavirus on ELISA, RNA-PAGE, and RT-PCR, respectively. Two of them contained group C rotavirus strains, whose genome characterization was described elsewhere (1).
G type distribution.
RT-PCR for G typing showed that G1 was the most prevalent type, being found in 44 (39.3%) of the 112 rotavirus-positive specimens. An unusual G8 type was also detected at high frequency (31 of 112; 27.7%). Types G2 and G3 were found in 20 (17.8%) and 2 (1.8%) specimens, respectively, while no G4 types were detected (Table 2). Thirteen specimens (11.6%) contained multiple G types, such as G1+G2, G8+G9, G1+G3, G1+G8, G2+G8, and G1+G2+G8.
TABLE 2.
Distributions of G types, P types, and mixed infections of human rotaviruses in the northeastern and middle belt regions of Nigeria
| P type | Total (%) | Distribution for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G type
|
Mixed infection
|
||||||||||
| G1 | G2 | G3 | G8 | G1G2 | G8G9 | G1G3 | G1G8 | G2G8 | G1G2G8 | ||
| P[6] | 24 (21.4) | 10 | 12 | 2 | |||||||
| P[8] | 10 (8.9) | 10 | |||||||||
| P[6]+P[8] | 2 (1.8) | 2 | |||||||||
| P[1]b | 1 (0.9) | 1 | |||||||||
| NDd | 73 (65.2) | 22 | 8 | 2 | 30 | 2 | 1 | 1 | 5 | 1 | 1 |
| Total (%)a | 112c | 44 (39.3) | 20 (17.8) | 2 (1.8) | 31 (27.7) | 4 (3.6) | 1 (0.9) | 1 (0.9) | 5 (4.4) | 1 (0.9) | 1 (0.9) |
Percentage of number typeable.
The P type could only be determined after successful adaptation to growth in cell culture.
The total number of samples here includes two in which human group C rotaviruses were identified and reported elsewhere (1). Fifteen of the stool specimens were negative for rotavirus with all the methods employed for their analysis.
ND, P type could not be determined.
P type distribution.
The P types could be assigned for only 36 (32.1%) of the positive specimens (Table 2). Of these, P[6] predominated, accounting for 24 (66.7%), followed by P[8] in 10 (27.8%), while two specimens (5.6%) contained a mixture of P[6]+P[8] specificities. P[4] was not detected in this study. None of the specimens with the G8 type were typeable as to the P type, except for one cultivable strain, HMG035, which had P[1] specificity.
G and P type combinations.
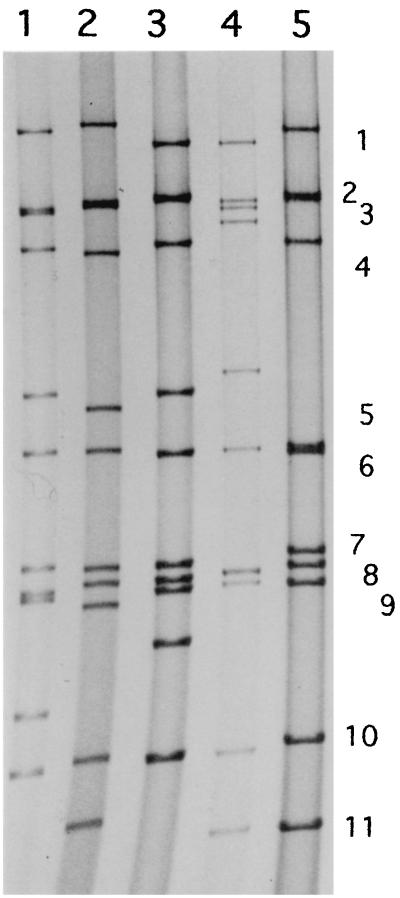
Four distinct G and P type combinations were identified among the 36 stool specimens in which there were 10 (27.8%) of G1P[8] and G1P[6] specificities (Table 2). Two cases each of mixed infections, G1P[6]+P[8] and G1+G2P[6] specificity, respectively, were observed. As mentioned above, a rotavirus strain (HMG035) with G8 specificity was successfully grown in MA-104 cells. RNA extracted from the culture fluid was subjected to RT-PCR for G and P typing, and the G specificity was confirmed. The P type of this cultivable strain was found to be P[1], and the RNA profile is shown in Fig. 1. Its fifth RNA band migrated faster than usual.
FIG. 1.
RNA migration patterns of some Nigerian human group A rotaviruses and laboratory reference strains belonging to group A. Lanes: 1, S2 (G2P1B, human); 2, KU (G1P1A, human); 3, 69M (G8P4, human); 4, A5-10 (G8P[1], bovine); 5, HMG035 (G8P[1], human). The numbers on the right refer to the corresponding gene segment number. The migration is from top to bottom.
Nucleotide sequence analysis.
G8 or G9 human rotaviruses have been highlighted as emerging strains worldwide. Sequence analysis was carried out on the G9 strain Bulumkutu and the G8 strain HMG035. The VP7 gene of strain Bulumkutu in a mixed infection was similar in primary structure to that of reported G9 strains worldwide. It is 1,061 nucleotides long with one base deletion at position 1030, compared to rotaviruses with non-G9 specificity. This gene has an open reading frame encoding a protein of 326 amino acids. Two in-phase initiation codons located at positions 49 to 51 and 136 to 138 were observed, with no potential N-glycosylation site at residues 238 to 240 as has been found in other G9 strains.
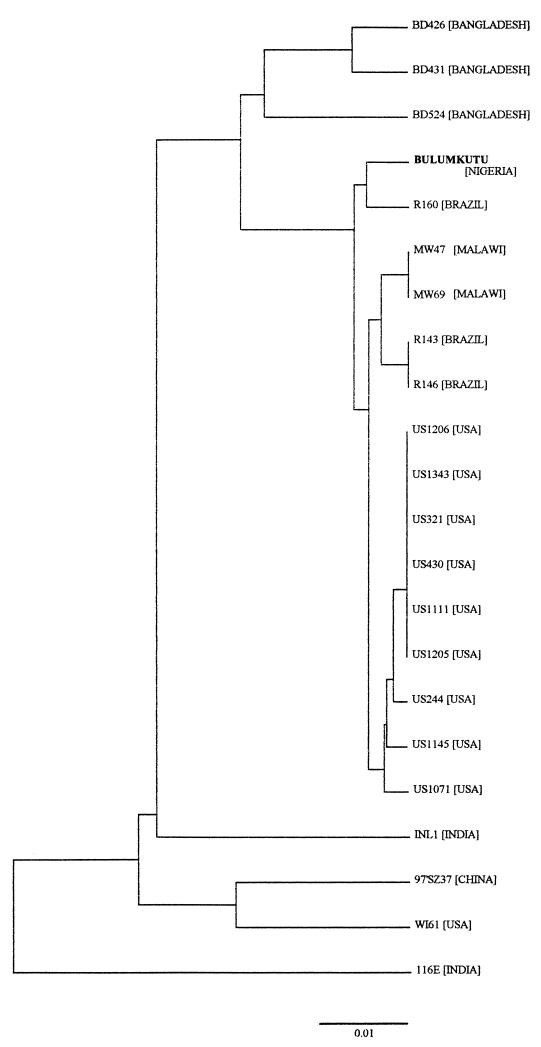
When the sequence of the VP7 gene of Nigerian G9 strain Bulumkutu was compared with other published G9 VP7 gene sequences, closer identity (99.1% at amino acid level) was found with a Brazilian strain, R160 (Table 3). The Nigerian strain is also more similar (98.8%) to other Brazilian, Malawi, and U.S. strains than to Indian, Bangladeshi, and Chinese strains. Strain Bulumkutu exhibited the least homology (91.1%) to Indian strain 116E and only 95.7% identity with prototype G9 strain WI61. On a phylogenetic tree, the Nigerian strain Bulumkutu forms a separate cluster with Brazilian strain R160, to which it is most closely related, but not with African strains MW47 and MW69 from Malawi (Fig. 2).
TABLE 3.
VP7 nucleotide and amino acid sequence homologies of Nigerian G8 rotavirus strain HMG035 with other published strains
| Straina | Origin | G type | Homology (%)
|
|
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| HMG89 | Human | 8 | 97.2 | 97.2 |
| MW23 | Human | 8 | 96.0 | 96.6 |
| MW333 | Human | 8 | 96.0 | 96.9 |
| EGY1850 | Human | 8 | 89.2 | 96.9 |
| HAL1166 | Human | 8 | 84.7 | 96.0 |
| QEH14262 | Human | 8 | 88.2 | 95.1 |
| GR570/85 | Human | 8 | 91.8 | 95.4 |
| DG8 | Human | 8 | 85.0 | 94.8 |
| 69M | Human | 8 | 84.7 | 94.8 |
| B37 | Human | 8 | 83.7 | 92.9 |
| A5 | Bovine | 8 | 87.3 | 95.4 |
| Cody-1801 | Bovine | 8 | 83.1 | 94.2 |
| Wa | Human | 1 | 73.2 | 77.1 |
| S2 | Human | 2 | 72.8 | 74.8 |
| HCR3 | Human | 3 | 77.0 | 82.2 |
| HOCHI | Human | 4 | 73.4 | 72.7 |
| OSU | Porcine | 5 | 76.5 | 80.7 |
| UK | Bovine | 6 | 75.9 | 81.9 |
| 116E | Human | 9 | 76.5 | 79.4 |
| I321 | Human | 10 | 74.9 | 79.1 |
| YM | Porcine | 11 | 76.9 | 82.2 |
| L26 | Human | 12 | 75.0 | 77.3 |
| L338 | Equine | 13 | 74.0 | 75.8 |
| F123 | Equine | 14 | 74.1 | 78.8 |
The accession numbers of the reference sequences used in this analysis are HMG89, X98918; MW23, AJ278254; MW333, AJ278257; EGY1850, AF104102; HAL1166, L20882; QEH14262, AF143689; GR570/85, AF143688; DG8, AF034852; B37, J04334; A5, D01054; Cody-1801, U14999; Wa, K02033; S2, M11164; HCR3, L21666; HOCHI, AB012078; OSU, X04613; UK, X00896; 116E, L14072; I321, L07658; YM, M23194; L26, M58290; L338, D13549; and F123, M61876.
FIG. 2.
Phylogenetic tree of the VP7 proteins of G9 human rotaviruses, including Nigerian strain Bulumkutu. The bar indicates the variation scale.
The complete nucleotide sequence of the VP7 gene of the Nigerian human G8 strain HMG035 was determined and compared with corresponding sequences of strains representing G1 to -14 from human and animal species (Table 3). The Nigerian G8 strain HMG035 was similar to the other strains in primary structure, but the VP7 gene was most closely related to that of another Nigerian G8 strain, HMG89 (97.2%), which was detected in a previous study (4). It also showed close identity with African strains from Malawi and Egypt (96.6 to 96.9%) and with a Thai bovine G8 strain, A5 (95.4%).
The VP4 gene sequence was also determined for Nigerian strain HMG035. It was 2,362 nucleotides in length with an open reading frame extending from nucleotides 10 to 2335 and encoded a protein of 776 amino acids. On comparison with the published VP4 gene sequences of other strains, it was found to be most closely related to bovine strains A5 and NCDV, the amino acid identities being 92.5 and 92.4%, respectively (Table 4). This sequence analysis thus further confirmed the PCR results; i.e., the strain is of a P[1] genotype.
TABLE 4.
VP4 nucleotide and amino acid sequence homologies of Nigerian rotavirus strain HMG035 with representative human and animal strains
| Straina | Origin | P type | Homology (%)
|
|
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| A5 | Bovine | 1 | 81.7 | 92.5 |
| NCDV | Bovine | 1 | 81.8 | 92.4 |
| SA11 | Simian | 2 | 76.8 | 84.3 |
| RRV | Simian | 3 | 77.0 | 85.3 |
| MW333 | Human | 4 | 69.6 | 71.7 |
| L26 | Human | 4 | 69.9 | 71.5 |
| RV-5 | Human | 4 | 69.4 | 71.0 |
| MW23 | Human | 6 | 70.9 | 73.0 |
| US1205 | Human | 6 | 71.0 | 73.8 |
| RV-3 | Human | 6 | 70.6 | 73.4 |
| M37 | Human | 6 | 70.6 | 73.7 |
| ST3 | Human | 6 | 70.1 | 72.8 |
| 1076 | Human | 6 | 70.2 | 71.9 |
| Gottfried | Porcine | 6 | 69.8 | 73.8 |
| OSU | Porcine | 7 | 74.9 | 83.0 |
| Wa | Human | 8 | 70.7 | 71.7 |
| AU-1 | Human | 9 | 68.2 | 70.3 |
| 69M | Human | 10 | 74.9 | 84.0 |
| 116E | Human | 11 | 62.7 | 57.5 |
| H2 | Equine | 12 | 73.2 | 79.0 |
| MDR-13 | Porcine | 13 | 73.0 | 77.7 |
| HAL1166 | Human | 14 | 68.6 | 71.6 |
| Lp14 | Ovine | 15 | 74.5 | 81.7 |
| Eb | Murine | 16 | 68.4 | 73.3 |
| 993/83 | Bovine | 17 | 64.2 | 61.6 |
| L338 | Equine | 18 | 77.5 | 84.5 |
| 4F | Porcine | 19 | 71.8 | 74.6 |
| EHP | Murine | 20 | 71.1 | 79.5 |
The accession numbers of the reference sequences used in this analysis are A5, D13395; SA11, D16346; RRV, M18736; MW333, AJ278256; L26, M36397; RV-5, M32559; MW23, AJ278253; US1205, AF079356; RV-3, U16299; M37, L20877; ST3, L33895; 1076, M88480; Gottfried, M33516; OSU, X13190; Wa, L34161; AU-1, D10970; 69M, M60600; 116E, L07934; H2, D13397; MDR-13, L07886; HAL1166, L20875; Lp14, L11599; Eb, L18992; 993/83, D16352; L338, D13399; 4F, L10359; EHP, U08424.
DISCUSSION
Recent studies (10, 27, 31) have indicated that uncommon human rotavirus strains are emerging as global strains, which has important implications for effective vaccine development. The present study adds to this pool of information and further confirms the emergence of these unusual strains.
Previous studies (3) in Nigeria identified strains of G1P[8], G3P[6], G1P[6], and G3P[8] as the commonly prevalent ones. In the present small study, however, a novel strain, G2P[6], hitherto unidentified in this country, was observed to be the most predominant. Recently, a report (5) from a neighboring West African country, Ghana, indicated that this type accounted for the majority (50%) of the isolates and this may be an emerging West African one. It would be interesting to determine the prevalence of this “putative” neonatal genotype strain in other African countries.
Of interest too in this study is the emergence of strains with serotype G8 whose VP4 genotype is P[1] or remains untypeable. Human G8 strains were first detected in Indonesia (25). However, this serotype now appears to have a worldwide distribution, as evidenced by its detection in other countries of the world. While in some studies the P[1], P[4], or P[6] VP4 genotype has been assigned to these G8 serotype strains (8, 21), in others characterization of the VP4 genotypic specificity was unsuccessful. The G8 serotype identified in Nigeria in 1994 to 1995 had P[6] specificity (4).
In this study, one cultivable G8 strain, HMG035, was found to exhibit P[1] specificity. A bovine rotavirus with the same G8P[1] specificity has been reported in Thailand (33, 34). Furthermore, a G8P[1] bovine rotavirus was recently detected in Nigeria, and we are now characterizing this strain by Northern blotting and sequence analysis in order to elucidate the relationship with the human G8 strain. The stool specimen from which strain HMG035 was isolated did not reveal any nucleic acid upon direct PAGE analysis. This strain might have recently crossed the species boundary from animals to humans and might not have yet fully adapted to humans. If this is correct, it could explain why none of the specimens exhibiting G8 specificity showed any nucleic acid on direct PAGE analysis. The northeastern region of Nigeria where strain HMG035 was isolated is a predominantly rural livestock-producing area, there being close contact between humans and animals. It will be interesting to determine whether the G8P[1] strain detected in a human stool was due to the close contact of the patient with calves which excreted the strains with the same G8P[1] specificity. The direct transmission of animal strains to humans should be a subject for further consideration as to the ecology of rotavirus infection. A number of mixed infections with rotaviruses was also observed in this study. Such an event could facilitate the emergence of rotavirus reassortants, including ones with animal rotaviruses (15). This may reflect the frequent rotavirus infections in heavily contaminated environments in the area surveyed in this study.
As in previous studies in Nigeria (2), serotype G4 was never detected and only one G9 serotype, in a mixed infection with a G8 serotype, was identified in this study. Interestingly, sequence analysis of this G9 strain revealed that it is most closely related to a South American strain from Brazil and not to African strains from Malawi. A similar observation was made about the VP6 sequence of human group C rotavirus strains from Nigeria in our other recent study (1): the VP6 gene of a Nigerian strain was more closely related to that of a Brazilian strain than to those of other Nigerian strains. Thus, it was confirmed that there is an unusual diversity among rotavirus strains in Nigeria. Of importance, however, is that sequence data for Nigerian serotype G9 are now known. This sequence information can now be used to design more efficient PCR primers for detecting strains of the G9 serotype more effectively and could be used to determine the true distribution of this emerging global strain in Nigeria. One study (4) has revealed that mismatches at the primer binding site of a G8 serotype resulted in erroneous typing as a G3 serotype. The sequence data presented here could be used to prevent a similar error in the future typing of G9 serotypes from Nigeria.
A number of samples remained untypeable as to the P type specificity. Such strains may be of types other than P[8], P[4], P[9], or P[6], since only primers specific to these P types were routinely used in this study. Expecting that the G8 strains might have P[1] specificity similar to that of strain HMG035, we also used bovine-specific primers for PCR P typing but failed to obtain conclusive results. Although they may have new P type(s) not yet recognized or nonhuman, nonbovine P types, further studies are needed to identify their P types. Alternatively, the unsatisfactory storage conditions for the stool samples may be related to the low P type identification, since the detection rates of rotavirus on ELISA and RNA-PAGE were also low. Because the sensitivity of our PCR P typing was less than that of PCR G typing, such circumstances might have affected the efficiency of PCR P typing.
The sample size and the distribution of sampling sites are limitations of this study. It has, however, demonstrated some fundamental features of rotavirus epidemiology in Nigeria: the emergence of a novel strain, G2P[6], hitherto unidentified; the emergence of G8P[1] serotypes and a high proportion of mixed infections providing a favorable environment for reassortment to occur; the consistent absence of G4 serotypes in the country; and the presentation of the sequence data of the VP7 genes of Nigerian serotypes G9 and G8. In particular, the G8 serotype is now established as the second most predominant after G1 in Nigeria. These findings highlight the need for continuous monitoring of the G and P type distributions of rotaviruses in Africa to provide information essential for rotavirus vaccine development.
ACKNOWLEDGMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan. M. I. Adah received an award, “The Long Term FY2000 JSPS Invitation Fellowship Program for Research in Japan,” from the Japan Society for the Promotion of Science (JSPS).
We are grateful to the staff and nurses, especially Terna Yalwe, Samuel Ville, Zira Gambo, and Ada Alechenu of the State Specialist Hospital, Maiduguri, Nigeria, and the Federal Medical Center, Makurdi, Nigeria, for their cooperation in the stool sample collection.
REFERENCES
- 1.Adah, M. I., A. Wade, M. Oseto, M. Kuzuya, and K. Taniguchi. First detection of human group C rotaviruses in Nigeria and sequence analysis of their genes encoding VP4, VP6 and VP7 proteins. J. Med. Virol., in press. [DOI] [PubMed]
- 2.Adah M I, Rohwedder A, Olaleye O D, Durojaiye O A, Werchau H. Serotype of Nigerian rotavirus strains. Trop Med Int Health. 1997;2:363–370. doi: 10.1111/j.1365-3156.1997.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 3.Adah M I, Rohwedder A, Olaleye O D, Durojaiye O A, Werchau H. Further characterization of field strains of rotavirus from Nigeria VP4 genotype P6 most frequently identified among symptomatically infected children. J Trop Pediatr. 1997;43:267–274. doi: 10.1093/tropej/43.5.267. [DOI] [PubMed] [Google Scholar]
- 4.Adah M I, Rohwedder A, Olaleye O D, Werchau H. Nigerian rotavirus serotype G8 could not be typed by PCR due to nucleotide mutation at the 3′ end of the primer binding site. Arch Virol. 1997;142:1881–1887. doi: 10.1007/s007050050206. [DOI] [PubMed] [Google Scholar]
- 5.Armah G E, Pager C T, Asma R H, Anto F R, Oduro A B, Binka F, Steele D. Prevalence of unusual human rotavirus strains in Ghanaian children. J Med Virol. 2001;63:67–71. [PubMed] [Google Scholar]
- 6.Bass D M. Rotavirus vaccinology: good news and bad news. J Pediatr Gastroenterol Nutr. 2000;30:10–11. doi: 10.1097/00005176-200001000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Withdrawal of rotavirus vaccine recommendation. Morb Mortal Wkly Rep. 1999;48:1007. [PubMed] [Google Scholar]
- 8.Cunliffe N A, Gentsch J R, Kirkwood C D, Gondwe J S, Dove W, Nakagomi O, Nakagomi T, Hoshino Y, Bresee J S, Glass R I, Molyneux M E, Hart C A. Molecular and serologic characterization of novel serotype G8 human rotavirus strains detected in Blantyre, Malawi. Virology. 2000;274:309–320. doi: 10.1006/viro.2000.0456. [DOI] [PubMed] [Google Scholar]
- 9.Cunliffe N A, Gondwe J S, Broadhead R L, Molyneux M E, Woods P A, Bresee J S, Glass R I, Gentsch J R, Hart C A. Rotavirus G and P types in children with acute diarrhoea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57:308–312. [PubMed] [Google Scholar]
- 10.Cunliffe N A, Kilgore P E, Bresee J S, Steele A D, Luo N, Hart C A, Glass R I. Epidemiology of rotavirus diarrhoea in Africa: a review to assess the need for rotavirus immunization. Bull W H O. 1998;76:525–537. [PMC free article] [PubMed] [Google Scholar]
- 11.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1656. [Google Scholar]
- 12.Gentsch J R, Woods P A, Ramachandran M, Das B K, Leite J P, Alfieri A, Kumar R, Bhan M K, Glass R I. Review of G and P typing results from a global collection of strains: implications for vaccine development. J Infect Dis. 1996;174(suppl. 1):S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 13.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brussow H, Hoshino Y, Flores J. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol. 1992;30:9–16. doi: 10.1128/jcm.30.1.9-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerna G A, Scraisisi A, Zenhlin L, DiMatteo A, Mirando P, Parea A, Battaglia M, Milanese G. Isolation in Europe of 69M-like (serotype 8) human rotavirus strains with either subgroup I or II specificity and a long RNA electropherotype. Arch Virol. 1990;112:27–40. doi: 10.1007/BF01348983. [DOI] [PubMed] [Google Scholar]
- 15.Gouvea V, Brantly M. Is rotavirus a population of reassortants? Trends Microbiol. 1995;3:150–162. doi: 10.1016/s0966-842x(00)88908-8. [DOI] [PubMed] [Google Scholar]
- 16.Gusmao R H, Mascarenhas J D P, Gabbay Y B, Iins-Lainson Z, Ramos F L P, Monteiro T A F, Valente S A, Fagundes-Neto U, Linhares A C. Rotavirus subgroups, G serotypes and electropherotypes in cases of nosocomial infantile diarrhoea in Belem, Brazil. J Trop Pediatr. 1999;45:81–86. doi: 10.1093/tropej/45.2.81. [DOI] [PubMed] [Google Scholar]
- 17.Hall A, Clemens J. Adverse reactions to vaccines in the tropics. Trop Med Int Health. 2000;5:229–230. doi: 10.1046/j.1365-3156.2000.00552.x. [DOI] [PubMed] [Google Scholar]
- 18.Hochwald C, Kivela L. Rotavirus vaccine, live, oral, tetravalent (RotaShield®) Pediatr Nurs. 1999;25:203–207. [PubMed] [Google Scholar]
- 19.Holmes J L, Kirkwood C D, Gerna G, Clemens J D, Rao M R, Naficy A B, Abu-Elyazeed R, Savarino S J, Glass R I, Gentsch J R. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch Virol. 1999;144:1381–1396. doi: 10.1007/s007050050594. [DOI] [PubMed] [Google Scholar]
- 20.Hoshino Y, Kapikian A Z. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J Health Popul Nutr. 2000;18:5–14. [PubMed] [Google Scholar]
- 21.Jagannath M R, Vethanayagam R R, Reddy B S, Raman S, Rao C D. Characterization of human symptomatic rotavirus isolates MP409 and MP480 having “long” RNA electropherotype and subgroup I specificity, highly related to the P6[1], G8 type bovine rotavirus A5, from Mysore, India. Arch Virol. 2000;145:1339–1357. doi: 10.1007/s007050070094. [DOI] [PubMed] [Google Scholar]
- 22.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1657–1708. [Google Scholar]
- 23.Lanata C F, Midthun K, Black R E, Lazo F, Butron B, Linhares A, Huapaya A, Ventura G, Gil A, Kapikian A Z. Safety, immunogenicity, and protective efficacy of one and three doses of the tetravalent rhesus rotavirus vaccine in infants in Lima, Peru. J Infect Dis. 1996;174:268–275. doi: 10.1093/infdis/174.2.268. [DOI] [PubMed] [Google Scholar]
- 24.Leite J P, Alfieri A A, Woods P, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 25.Matsuno S, Hasegawa A, Mukoyama A, Inouye S. A candidate for a new serotype of human rotavirus. J Virol. 1985;54:623–624. doi: 10.1128/jvi.54.2.623-624.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palombo E A, Clark R, Bishop R F. Characterization of a “European-like” serotype G8 human rotavirus isolated in Australia. J Med Virol. 2000;60:56–62. [PubMed] [Google Scholar]
- 27.Palombo E A, Masendycz P J, Bugg H C, Bogdanovic-Sakran N, Branes G L, Bishop R F. Emergence of G9 in Australia. J Clin Microbiol. 2000;38:1305–1306. doi: 10.1128/jcm.38.3.1305-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thaku S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Bresee J S, Glass R I the National Rotavirus Strain Surveillance System (NRSSS) Collaborating Laboratories. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos N, Lima R C C, Pereira C F A, Gouvea V. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J Clin Microbiol. 1998;36:2727–2729. doi: 10.1128/jcm.36.9.2727-2729.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele A D, Parker S P, Peenze I, Pager C T, Taylor M B, Cubitt W D. Comparative studies of human rotavirus serotype G8 strains recovered in South Africa and the United Kingdom. J Gen Virol. 1999;80:3029–3034. doi: 10.1099/0022-1317-80-11-3029. [DOI] [PubMed] [Google Scholar]
- 32.Taniguchi K, Urasawa T, Kobayashi N, Gorziglia M, Urasawa S. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J Virol. 1990;64:5640–5644. doi: 10.1128/jvi.64.11.5640-5644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi K, Urasawa T, Pongsuwanna Y, Choothanom M C, Jayavasu, Urasawa S. Molecular and antigenic analyses of serotypes 8 and 10 of bovine rotaviruses in Thailand. J Gen Virol. 1991;72:2929–2937. doi: 10.1099/0022-1317-72-12-2929. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi K, Urasawa T, Urasawa S. Independent segregation of the VP4 and VP7 genes in bovine rotaviruses as confirmed by VP4 sequence analysis of G8 and G10 bovine rotaivus strains. J Gen Virol. 1993;74:1215–1221. doi: 10.1099/0022-1317-74-6-1215. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi K, Urasawa T, Urasawa S, Yasuhara T. Production of subgroup-specific monoclonal antibodies against human rotaviruses and their application to an enzyme-linked immunosorbent assay for subgroup determination. J Med Virol. 1984;14:115–125. doi: 10.1002/jmv.1890140205. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi K, Wakasugi F, Pongsuwanna Y, Urasawa T, Ukae S, Chiba S, Urasawa S. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol Infect. 1992;109:303–312. doi: 10.1017/s0950268800050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urasawa T, Urasawa S, Taniguchi K. Sequential passages of human rotavirus in MA-104 cells. Microbiol Immunol. 1981;25:1025–1035. doi: 10.1111/j.1348-0421.1981.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 38.Weijer C. The future of research into rotavirus vaccine: benefits of vaccine may outweigh risks for children in developing countries. BMJ. 2000;321:525–526. doi: 10.1136/bmj.321.7260.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, Taniguchi K, Wakasugi F, Ukae S, Chiba S, Ohseto M, Hasegawa A, Urasawa T, Urasawa S. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol Infect. 1994;112:615–622. doi: 10.1017/s0950268800051311. [DOI] [PMC free article] [PubMed] [Google Scholar]