Abstract
Tumor-associated macrophages (TAM) are an important component of the tumor microenvironment (TME) that can promote tumor progression, metastasis, and resistance to therapies. Although TAMs represent a promising target for therapeutic intervention, the complexity of the TME has made the study of TAMs challenging. Here, we established a physiologically relevant in vitro TAM polarization system that recapitulates TAM pro-tumoral activities. This system was used to characterize dynamic changes in gene expression and protein phosphorylation during TAM polarization and to screen phenotypic kinase inhibitors that impact TAM programming. BMS-794833, a multi-targeted compound, was identified as a potent inhibitor of TAM polarization. BMS-794833 decreased pro-tumoral properties of TAMs in vitro and suppressed tumor growth in mouse triple-negative breast cancer models. The effect of BMS-794833 was independent of its primary targets (MET and VEGFR2) but was dependent on its effect on multiple signaling pathways, including focal adhesion kinases, SRC family kinases, STAT3, and p38 MAP kinases. Collectively, these findings underline the efficacy of polypharmacological strategies in reprogramming complex signaling cascades activated during TAM polarization.
Keywords: Tumor-associated macrophage, Breast cancer, Tumor microenvironment, Kinase inhibitor screening, Cell-cell interaction
Introduction
Solid tumors comprise heterogeneous populations of cancer and non-cancerous cells that interact through direct contact and secreted factors, thus establishing the tumor microenvironment (TME). TME alters the behavior of non-cancerous cells, resulting in phenotypes with tumor-supportive properties. For example, tumor cells and immune cells that reside within TME release a number of anti-inflammatory and pro-angiogenic factors that result in pro-tumoral polarization of tumor-associated macrophages (TAMs), one of the abundant types of immune cells found in TME. Thus, polarized TAMs can further promote tumor progression by stimulating proliferation, invasion, angiogenesis, immunosuppression, metastasis, endowing resistance against chemo- and radiotherapy, and decreased efficacy of immunotherapy (1,2). Further, clinical evidence supports that patients with higher TAM infiltration have poor prognoses in several cancers(3). Given their role in tumor progression and the clinical evidence, TAMs have been proposed as a promising therapeutic target (4–6). However, physiological TAMs are scarce and difficult to study using existing experimental strategies. Therefore, mechanisms that drive TAM pro-tumor polarization, as well as potential targeting opportunities for drug development remain incompletely understood.
Accumulated evidence suggests that TAMs are highly plastic and dynamic cells with complex signaling networks optimized to rapidly adjust the phenotype in response to external stimuli(7). Therefore, TAMs exhibit heterogenous(8) and continuum phenotypes between inflammatory and anti-inflammatory(9). One strategy to model TME in vitro is to employ tumor conditioned medium (CM) of cancer cells, which has been shown to contain a near-complete set of secreted factors and metabolites found in TME and has been used in several TAM studies aimed at improving our understanding of TAM biology (10–14). However, quantitative, phenotypic drug screening assays aimed at TAMs have not been described before, thus limiting the ability to discover compounds with potential therapeutic benefits in this context. To address this need, we established an in vitro quantitative TAM polarization model using human monocyte cell line THP-1 and CM from multiple cancer models. Using cellular elongation measurement as a quantitative measure of TAM polarization, we screened a library of 85 kinase inhibitors targeting most of the human kinome. We identified BMS-7948933, a dual-targeting inhibitor of c-MET and VEGFR2, as the most potent blocker of cellular elongation and pro-tumoral function of TAMs. BMS-794833 suppressed tumor growth on mouse triple-negative breast tumor models. Surprisingly, we found the TAM inhibitory function of BMS-794833 does not involve c-MET or VEGFR2 but a range of other targets that include focal adhesion kinases (FAK) and cytoskeletal-related proteins, SRC family kinases, STAT3, and p38 MAP kinases. We also observed that targeting a single pathway exhibited little to no effect on TAM polarization, whereas BMS-794833-mediated targeting of multiple signaling pathways exhibited the most potent effect on TAM polarization. Our study underlines the complex regulation of signaling pathways during TAM polarization and the necessity of concomitant blocking of multiple signaling pathways to re-program pro-tumoral TAM function.
Material and Methods
Cell lines and culture
Cell lines including 4T1, CT26, Py8119, ACHN, THP-1, and Jurkat cells were purchased from American Type Culture Collection (ATCC). HCC70, HCC1419, and HCC1937 were gifts from Dr. Peggy Porter, Fred Hutchinson Cancer Research Center (FHCRC). 4T1, CT26, HCC70, HCC1419, HCC1937 cells were maintained in RPMI1640 media (Gibco by Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% Penicillin-Streptomycin (P/S, final concentration: 100 units/ml of penicillin, 100 μg/ml of streptomycin) (Gibco). Py8119 cells were maintained in F-12K medium (ATCC) with 5% FBS. THP-1 cells were cultured in RPMI1640 media supplemented with 10% FBS, 1% P/S, 1 mM sodium pyruvate (Lonza, Basel, Switzerland), and 55 nM β-mercaptoethanol (Gibco). Jurkat cells were maintained in RPMI1640 media with 10% FBS, 1% P/S, 10 mM HEPES (Santa Cruz Biotechnology, Dallas, TX, USA), and 1 mM sodium pyruvate. All cells were cultured in humidified 37°C incubators with 5% CO2 atmosphere.
Collection of conditioned medium (CM)
For the collection of CM from tumor tissue and cancer cells, serum-free Tumor slice culture (TSC) medium (15,16) was used. Cancer cells grown to subconfluent were washed with PBS twice and incubated in TSC medium for 1–2 days. For the collection of CM from tumor tissues, tumors were cut into 4–5 mm cubes, washed with PBS twice, and incubated with 10 ml of TSC medium per 1 g tumor for one day. After collecting the spent medium, fresh TSC medium was added to the tumor pieces to collect the second pool. The collected medium was centrifuged at 2,000 ×g for 10 min, and the supernatant was used as CM. TSC medium without conditioning was used as an experimental control. For the collection of TAM CM, TAMs polarized for 3 days were washed with PBS twice, then incubated for 24 h in RPMI1640 medium for THP-1 maintenance. The supernatant was centrifuged at 2,000 ×g for 10 min, to remove cell debris. RPMI1640 medium for THP-1 culture without conditioning was used for experimental control.
TAM polarization model with cancer CM
THP-1 cells were differentiated into macrophages by inducing with phorbol 12-myristate 13-acetate (PMA, LC Laboratories, Woburn, MA, USA) at 25 ng/ml for one day. The cell densities used were; 1.3–1.5×104/well for 96-well plate, 4×105 cells/well for 6-well plate, 2.4×106 cells for 10 cm dish. The medium was replaced with medium containing cancer CM at 25–50% (v/v) with volume adjustment to 50% with TSC medium, and 50% (v/v) RPMI1640 for THP-1 cell culture. Cellular elongation was measured with NeuroTrack analysis software accompanied with IncuCyte Zoom Live Imaging system (Sartorius, Goettingen, Germany).
Kinase inhibitor screening
THP-1 cells were seeded at 1.5×104 cells per well of 96-well plates with PMA at 25 ng/ml. After a day, the medium was replaced to CM mix (25% 4T1 cell CM, 25% TSC medium, with 50% RPMI1640 for THP-1 culture, YOYO-3 at 1:10,000 dilution) together with kinase inhibitors at 8 serial concentrations of 10, 3.3, 1.1, 0.37, 0.12, 0.04, 0.01, and 0 μM with triplicates. The 85 inhibitors tested are listed in Table S1. All small molecules were constituted in DMSO for the stock solution, and DMSO (0.1%, up to 1% based on the volume of inhibitor solution) was supplemented as vehicle control. A red fluorescent viability dye, YOYO-3 (Thermo Fisher Scientific) was supplemented to the culture to detect cellular death. The phase contrast and red fluorescent images were taken every 2 hours over 3–5 days using IncuCyte Zoom instrument. The target kinase profiles of BMS-794833 in the acellular system were described in Rata, et al.(17).
Co-culture of TAM and cancer cells
THP-1 cells at 1.5×104 cells per well of 96-well plate were induced with PMA at 25 ng/ml and polarized for 3 days with the indicated tumor CM with or without inhibitors. The TAMs were washed with PBS twice, and 4T1-nucGFP cells were seeded at 3×103 cells per well density with RPMI1640 supplemented with 0.5% FBS. The cell numbers of 4T1 were counted based on nuclear GFP using the software accompanied with IncuCyte Zoom.
Tube formation assay
Tube formation assay of endothelial cells was performed using IncuCyte Angiogenesis 96-well PrimeKit Assay (Sartorius) with modifications. For the angiogenesis assay of TAM CM, the cells were seeded following the manufacturer’s protocol. Three days post seeding, TAM CM was supplemented at 50% concentration diluted in the assay medium included in the kit. The cells were incubated for 6 days with a medium replacement on day 2 and 5 during the culture. The plate was scanned and analyzed with IncuCyte Zoom. For assay of BMS-794833-treated TAM CM, NHDF and GFP-expressing human umbilical vein endothelial cells (GFP-HUVECs) were minimally expanded. NHDF were seeded at 1×104 cells per well of 96-well plate with DMEM with 10% FBS and 1% P/S and incubated for 2 days till confluent. GFP-HUVECs were seeded at 2×103 cells per well in EGM-2 on top of the NHDF layer. On the next day, the medium was replaced with 50% TAM CM diluted in EGM-2, and cells were incubated for 6 days. A half volume of medium was replated every 2 days. The plate was scanned and analyzed with IncuCyte S3.
Jurkat cell chemotaxis assay
Jurkat cells were washed with PBS and seeded onto a 24-well transwell culture insert with 3 μm pores (Celltreat Scientific Products, Pepperell, MA, USA) at 5×105 cells/well with RPMI1640 supplemented with 2.5% FBS. The inserts were placed onto wells containing TAM CM or control RPMI1640 medium for THP-1, and incubated for 19 h. The relative number of cells migrated to the bottom chamber was quantified using Celltiter Glo (Promega, Madison, WI, USA). The remaining cells in the inserts were analyzed with Caspase-Glo 3/7 assay (Promega) to confirm that Jurkat cells incubated with TAM CM did not induce apoptosis compared to the control medium.
Mouse experiment
All animal studies were approved by the IACUC committee of FHCRC. BALB/c and C57BL/6J females were sourced from The Jackson Laboratory. NSG female mice were inbred in-house by Comparative Medicine of FHCRC. 4T1 cells or Py8119 cells suspended at 1–2 million/150 μl of 33% (v/v) matrigel (Fisher Scientific) were transplanted subcutaneously to the right flank of 6–8 week-old BALB/c and NSG, or C57BL/6J female mice, respectively. After tumors reached 50–100 mm3, mice were randomized into treatment and untreated groups. The mice received an intratumoral injection of BMS-794833 at 25 mg/kg dose, BMS-5 at 30 mg/kg dose, or vehicle control twice weekly. BMS-794833 (Selleckchem, Houston, TX, USA) was dissolved into 4% (v/v) DMSO (Sigma-Aldrich), 45% (v/v) PEG300 (Sigma-Aldrich), and 5% (v/v) Tween 80 (Fisher Scientific). BMS-5 (Medchem Express, Monmouth, NJ, USA) was dissolved into 5% DMSO, 30% PEG400, 5% propylene glycol, and 0.5% Tween 80. The tumor size was measured by caliper twice weekly and by weight at the experimental endpoint. The tumor volumes were calculated with the following equation: V= 4/3π×(L/2)×(W/2)×(H/2), (L=length, W=width, H=height). HCI010 model was kindly gifted from Dr. Alana Welm (University of Utah) and expanded orthotopically in NSG female mice as described before (18). Py8119 tumors used for CM collection used in THP-1-derived TAM polarization were prepared by injecting Py8119 cells subcutaneously to NSG female mice.
Histology
At the end of the animal study, tumors and lungs were harvested from mice and fixed in 10% neutral-buffered formalin (MilliporeSigma, Burlington, MA, USA) for 7 days. The tissues were embedded into paraffin blocks and cut into sections. Hematoxylin-Eosin (H&E) staining of lung sections was performed by the Histopathology core at the FHCRC. The entire tissue was scanned with TissueFAXS (TissueGnostics, Vienna, Austria). The number and area of metastatic foci were evaluated using QuPath software(19). For detection and characterization of macrophages, the staining and analyses were performed by the Histopathology core at the FHCRC using ARG1, F4/80, and iNOS as primary antibodies.
Statistics
All the statistical analyses were performed using Graphpad Prism 8.0.1 software. The statistical analyses used for each data are indicated in Figure Legends. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Heatmaps were prepared using pheatmap package in R Studio.
For more detailed methods, see Supplementary Material and Methods.
Results
Development and characterization of an in vitro TAM polarization model
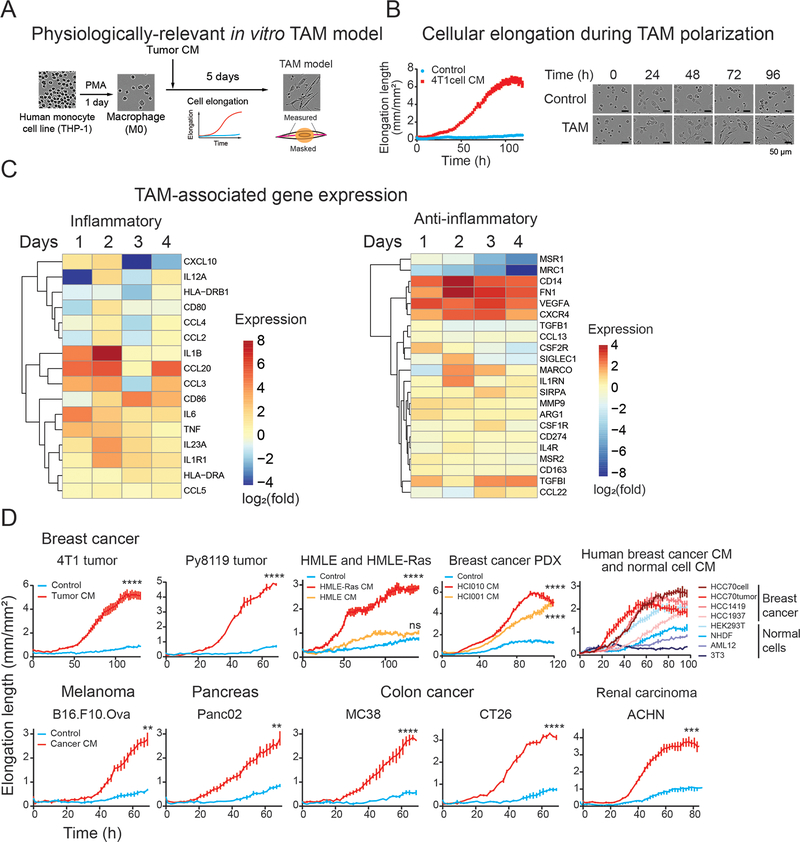
To develop a physiologically relevant in vitro TAM polarization model, we exposed macrophages differentiated from THP-1 cells, a human monocyte cell line, to tumor CM collected from multiple tumor models (Figure 1A). CM from the mouse triple-negative breast cancer (TNBC) cell line 4T1 induced protrusions in the THP-1-derived macrophages, causing them to exhibit drastically elongated cellular morphology starting from day 2 with peaking at around day 4 (Figure 1B). Given that previous studies have shown that TAM cellular elongation correlates strongly with pro-tumor phenotypes (10,12–14,20–23), we adopted cellular elongation quantified by live-cell imaging as an indicator of TAM polarization (Figure 1A). We then assessed whether the TAM polarization model reproduces the molecular characteristics of TAMs in vivo using a gene set panel adapted from (8). Gene expression profiles of macrophages induced by the CM from 4T1 cells upregulated TAM-associated genes and temporally induced inflammatory marker genes compared to macrophages cultured without 4T1 CM (Figure 1C). The TAM-related secretory proteins were also detected from the 4T1 CM-induced TAMs (Figure S1A). Consistent with previous reports (24), the TAM model induced both inflammatory (CCL1, Groα, IL-1β, IL-6, IL-8, IL-12, MIP-1α, MIP-1β, CCL5, TNFα, sICAM) and anti-inflammatory cytokines (IL-1RA, IL-4 IL-10, SerpinE1) to 1.4–13.2 and 1.2–1.8 folds, respectively, over control macrophages (Figure S1A). Similar cellular elongation during TAM polarization was observed with CM collected from other breast cancer models (4T1 tumor, Py8119 tumor, a Ras-expressing human mammary epithelial cells HMLE-Ras, breast cancer PDX models, human breast cancer cells: HCC70, HCC1419, HCC1937, HCC70 tumor) (Figure 1D, top), melanoma (B16.F10.Ova), pancreas cancer (Panc02), colon cancer (MC38 and CT26), and human renal carcinoma (ACHN) (Figure 1D, bottom). In contrast, CM from untransformed HMLE cells, human primary fibroblast cells (NHDF), mouse hepatocytes (AML12), and mouse fibroblast (3T3) did not polarize TAMs as effective as cancer CM (Figure 1D, top), except for human embryonic kidney cells (HEK293T) that partially elongated cells. These results provide validation that our tumor CM-induced TAM polarization system is reliable in vitro model for studying TAM function.
Figure 1. Establishment and characterization of an in vitro TAM polarization model.
(A) A schematic showing establishment of an in vitro TAM polarization model. Human monocyte THP-1 cells were induced by phorbol 12-myristate 13-acetate (PMA) to differentiate into macrophages, followed by culturing in the presence of tumor conditioned medium (CM). Cellular polarization was assessed by cellular elongation measurement via live-cell imaging, as depicted in the illustration.
(B) Morphological alteration during TAM polarization in culture. THP-1-derived macrophages were incubated with CM collected from 4T1 cells. Live-cell images were captured every 2 hours under a live cell imaging microscope. The cellular protrusion length per image was measured using image analysis software.
(C) Gene expression of in vitro TAM model. THP-1 cells were polarized using CM from 4T1 tumor and collected at the indicated time. The expression of TAM-related genes was analyzed by qPCR. Data are presented as the log2 fold change of expression of CM-treated cells over control cells for each day.
(D) Validation of CM from multiple tumor models for cellular elongation of THP-1-derived macrophages. (Top) Validation in other breast cancer models. THP-1-derived macrophages were cultured with CM collected from 4T1 tumors, Py8119 tumors, human mammary epithelial cell line and its derivative cancerous cells generated by introducing Ras oncogene, (HMLE and HMLE-Ras), TNBC PDX tumors (HCI010 and HCI001), human breast cancer cell lines (HCC70, HCC1419, HCC1937), HCC70 tumors, and non-cancerous cell lines; human embryonic kidney cells (HEK293T), human primary fibroblasts (NHDF), mouse hepatocytes (AML12), and mouse fibroblasts (3T3). (Bottom) TAM polarization by CM from other cancer types. CMs from mouse melanoma (B16.F10.Ova), pancreatic cancer (Panc02), colon carcinoma (MC38 and CT26), and human renal carcinoma (ACHN) were used to induce THP-1-derived macrophages. Cellular elongation was measured under live-cell imaging system. The graphs indicate mean ± SEM of measurement at each time point. **p<0.01, ***p<0.001, **** p<0.0001, Student’s t-test (2 groups) or One-way ANOVA with Dunnett’s multiple comparison test (3 groups) at the endpoint.
In vitro-derived TAMs promote cancer proliferation and angiogenesis and inhibits T cell chemotaxis
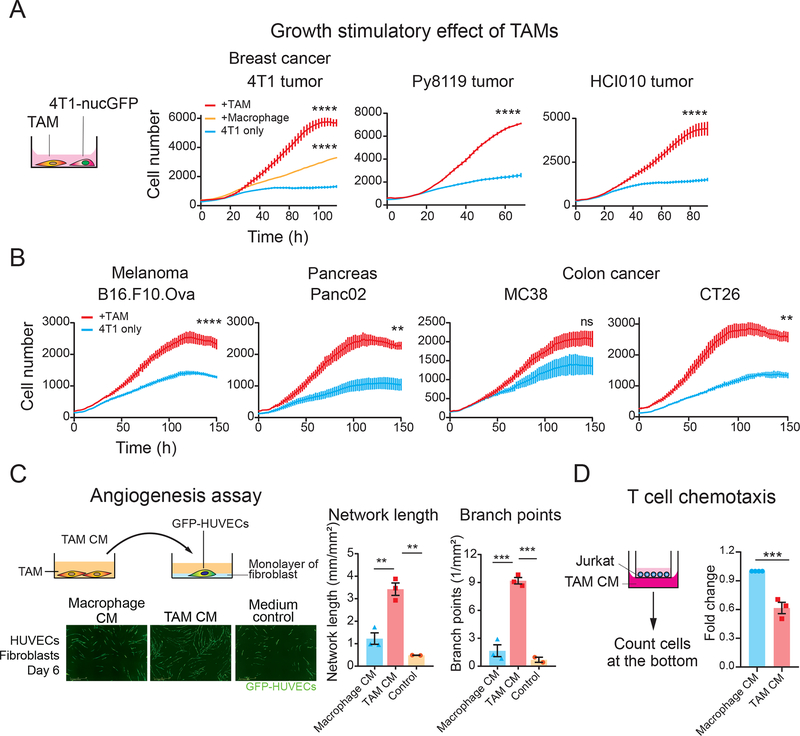
To further investigate the pro-tumoral behavior of our TAM model and further validate that this model recapitulates in vivo physiology of TAMs, we performed a series of co-culture experiments. Co-culture of 4T1 breast cancer cells with breast cancer CM-induced TAMs stimulated cancer cell proliferation, recapitulating previously-reported growth stimulus effects of TAMs (Figure 2A). Similar cancer cell growth stimulation by TAM was observed with TAMs induced by CM from other cancer models (melanoma, pancreas, and colon cancer) (Figure 2B). To assess pro-angiogenesis ability of TAM, we treated HUVECs with TAM CM that was enriched with secretion factors from polarized TAMs, and monitored the ability of HUVECs forming tube-like structures. Our data show that TAM CM exposure stimulates both length and network branching of endothelial cells (Figure 2C). It is known that TAMs impede T cell infiltration into the tumor by secretory factors (25–27). Consistently, TAM CM suppressed Jurkat cell migration in a trans-well migration assay compared to macrophage CM (Figure 2D). Together, these results further validate our in vitro TAM polarization model as the system recapitulates in vivo ability of TAMs to promote cell proliferation and angiogenesis and suppress T cell chemotaxis. Therefore, this model offers an opportunity to conduct screening campaigns to identify compounds that re-program TAMs with pro-tumoral traits into anti-tumoral traits.
Figure 2. Tumor CM-induced TAMs exhibit pro-tumoral phenotypes.
(A) Growth stimulation effect of TAMs on cancer cells. 4T1 cells labeled with nuclear GFP (4T1-nucGFP) were co-cultured with breast tumor CM-induced TAMs. THP-1-derived macrophages were polarized with CM collected from indicated breast cancer models (TAMs) or control medium (Macrophages) for 3 days. After TAMs were washed, 4T1-nucGFP cells were seeded on top of TAMs with the serum-reduced medium. For control, 4T1-nucGFP was cultured without macrophages (4T1 only). The number of 4T1 cells was counted based on nuclear GFP by the image analysis software. The bars indicate the mean ± SEM of measurement at each time point.
(B) Growth stimulation effect of TAMs induced by CM from melanoma (B16.F10.Ova), pancreatic cancer (Panc02), and colon cancer cell lines (MC38 and CT26). The experimental details are the same as (A).
(C) Stimulation of tube formation of HUVECs by TAMs. GFP-expressing HUVECs were seeded on a monolayer of fibroblasts. HUVECs were cultured with TAM CM collected from TAM induced by 4T1 tumor CM. (Left) Wide-field image obtained after 6 days of incubation. (Right) Network length and branch points of GFP-HUVEC networks on day 6 by live cell imaging analysis. The bar graphs show mean ± SEM of n=3 (macrophage CM, TAM CM), n=2 (medium control).
(D) T cell chemotaxis assay by TAM model. Jurkat cells seeded in culture inserts were incubated with TAM CM collected from TAM induced with HCI010 CM. The relative number of Jurkat cells migrated to the bottom was quantified by luminescent-based cell viability assay. Individual experimental values are shown as dots (n=3 for TAM, n=4 for the rest). *p<0.05, **p<0.01, *** p<0.001, **** p<0.0001, Student’s t-test (2 groups) or One-way ANOVA with Dunnett’s multiple comparison test (3 groups) at the endpoint for (A), One-way ANOVA with Tukey’s multiple comparison tests for (C).
Kinase inhibitor screening on the in vitro TAM model identified BMS-794833 as a potent inhibitor of TAM polarization
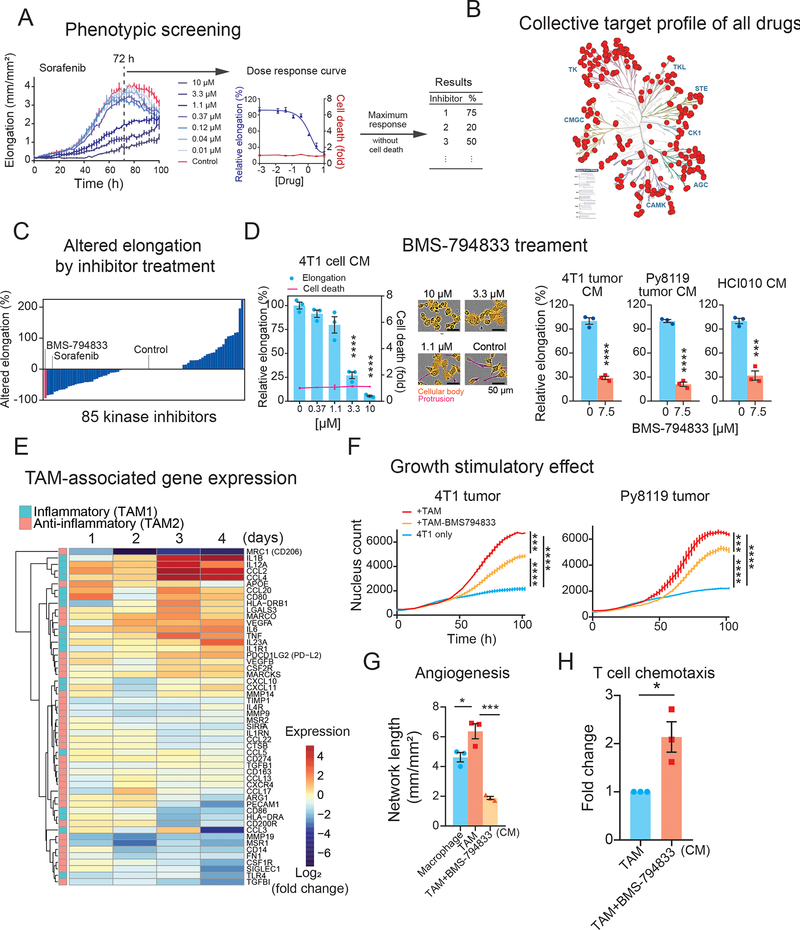
Protein kinases (kinases) are critical components of cellular signaling networks that transmit extra/intracellular stimuli to cellular responses. Given that TAM polarization is an external stimuli-regulated process, we hypothesized that kinases will play a major role in this process, and therefore, kinase inhibitors may represent useful pharmacological agents to inhibit pro-tumoral TAM polarization. To examine this further, we used our model system and cellular elongation as an indicator of TAM polarization levels to screen a curated collection of 85 kinase inhibitors (Figure 3A) that achieves broad kinome coverage (289 kinases out of 298 kinases measured), with lower than 50% residual activities at 0.5 μM (Figure 3B) (28). We tested this inhibitor collection at eight serially-diluted concentrations (0 – 10 μM) on our TAM model using THP-1 cells induced with CM from 4T1 cultured cells (Figure 3A). The efficacy of each inhibitor on TAM polarization was quantified as inhibition of cellular elongation compared to vehicle control. To omit inhibitors causing cellular death, we supplemented the assay with YOYO-3, a red fluorescent dye that stains membrane compromised cells. The response to each inhibitor was evaluated at the highest concentration ranging from 0 to 10 μM that did not exhibit cellular death. As a result, of the 85 tested inhibitors, 33 inhibitors caused a reduction in elongation by 3–93%, 26 inhibitors showed no change and/or induced cellular death, and surprisingly, 26 caused an increase in elongation by 14–225% (Figure 3C, Table S1). Sorafenib, a multikinase inhibitor of Raf, VEGFR, and PDGFR, has been previously shown to inhibit M2 (anti-inflammatory) macrophage phenotype and restore inflammatory cytokine expression (29–31). In our assay, sorafenib efficiently suppressed cellular elongation at 3.3 μM or higher, with 81.5% inhibition at 10 μM (Figure 3A) and was the fourth-most effective inhibitor (Figure 3C), validating the reliability of the screening system.
Figure 3. Kinase inhibitor screening in the in vitro TAM polarization model identified BMS-794833 as a potent TAM polarization inhibitor.
(A) A schematic showing the screening procedure for 85 kinase inhibitors in an in vitro TAM polarization model. Response to sorafenib is shown as a representative. TAMs were polarized under eight serial doses of kinase inhibitors ranging from 0 to 10 μM. Relative cellular elongation levels of inhibitor-treated TAMs compared to control were measured as the effect of inhibitors. A red fluorescent cell death indicator dye, YOYO-3, which detects disrupted cellular membranes, was supplemented to detect cellular deaths caused by kinase inhibitors.
(B) Collective target kinase profiles of the 85 screened kinase inhibitors. The selected kinase inhibitors cover 289 kinases from various kinase families, with lower than 50% residual activities at 0.5 μM. The kinase tree was prepared using KinMap (28).
(C) Changes in cellular elongation in response to inhibitor treatment. Inhibitors are plotted based on the maximum altered elongation levels at up to 10 μM dose. Inhibitors causing cellular death were regarded as causing no change in elongation.
(D) Inhibitory effect of BMS-794833 on cellular elongation of THP-1-derived TAM polarization. (Left) Elongation of THP-1-derived TAMs treated with a serial dose of BMS-794833 in the presence of 4T1 cell CM. The relative cellular death of inhibitor-treated TAMs was overlayed on the graph. The bars and plots are the means with ± SEM of n=3. (Middle) Representative images of THP-1-derived TAM with BMS-794833 at the indicated concentration at 72 h from polarization. Cell protrusions (measured as cellular elongation) and cellular body masked are labeled with magenta and orange, respectively. (Right) TAM elongation with CM from 4T1 tumor, Py8119 tumor, and breast PDX HCI010 with or without BMS-794833 at 7.5 μM.
(E) Time-course analyses of TAM-associated gene expression of BMS-794833-treated TAMs. Log2- transformed fold changes of gene expression of BMS-794833-treated TAMs at 7.5 μM compared to control TAMs were shown in heatmaps. Genes with inflammatory or anti-inflammatory properties are shown in the left color panels.
(F) Growth stimulation effect of TAM treated with BMS-794833. 4T1-nucGFP were co-cultured with BMS-794833-treated TAMs at 7.5 μM induced by 4T1 tumor CM or Py8119 tumor CM under a reduced serum condition. The growth of 4T1 was evaluated by nuclear GFP counts. The bars indicate mean ± SEM of measurement at each timepoint.
(G) Tube formation ability of TAM treated with BMS-794833. CM was collected from TAM with or without BMS-794833 treatment at 7.5 μM induced by 3 independent batches of HCI010 CM. The TAM CM was supplemented to tube formation assay of GFP-expressing HUVECs grown on fibroblast layer.
(H) T cell chemotaxis assay. Jurkat cells migrated through the bottom of the chamber with CM from TAMs polarized with BMS-794833 at 7.5 μM. The relative number of migrated cells was evaluated by luminescent-based cell assay. CM from TAM induced by three independent batches of HCI010 CM were used for this study.
*p<0.05, *** p<0.001, **** p<0.0001, unpaired two-tailed Student’s t-test ((D, right), (H)), One-way ANOVA followed by Dunnett’s multiple comparison test ((D, left)) or Tukey’s multiple comparisons ((F) at endpoint, (G)).
Among the tested inhibitors, BMS-7948333, a dual inhibitor of c-MET and VEGFR2(32), was the most potent inhibitor of cellular elongation during TAM polarization, with 93% inhibition at 10 μM with no evidence of cell death (Figure 3D, Figure S1B). We confirmed the inhibitory effect of BMS-794833 on cellular elongation in our other TAM polarization models using breast cancer CM (4T1 tumor, Py8119 tumor, and PDX) as well as CM from other cancer models (Figure 3D, Figure S1C). Time-course transcriptional analyses of BMS-794833-treated cells showed a marked decrease in expression of the anti-inflammatory macrophage marker (MRC1/CD206), and enhanced expressions of inflammatory cytokine-coding genes, such as IL1B, CCL2, CCL4, IL12A, IL6, and TNFA (Figure 3E). Cytokines secreted from BMS-794833-treated TAMs were also measured using protein arrays and absolute quantification using Luminex (Figure S1D). BMS-794833 treatment suppressed secretion of anti-inflammatory cytokines IL-4, IL-13, and IL-10, and enhanced inflammatory cytokines such as IL-6, IL-8, IL-27, MIP-1α/β, CCL2, TNFα, and IL-1β. These results demonstrate that BMS-794833 could abrogate TAM polarization and re-program TAMs to proinflammatory phenotypes.
BMS-794833 inhibits TAM polarization and pro-tumoral phenotypes
We next investigated the effect of BMS-794833 on the protumoral activities of TAMs. BMS-7949833 treatment decreased the growth stimulatory effect on 4T1 cells following co-culture with 4T1 or Py8119 tumor CM-induced TAMs (Figure 3F), as well as other cancer models (Figure S1E). Additionally, CM collected from BMS-794833-treated TAMs reduced angiogenesis stimulation potential and increased T cell chemotaxis ability compared to untreated TAMs (Figure 3G, H). Coculturing with BMS-794833-treated TAMs increased expression of PDCD1 (PD-1-coding gene) and Jurkat cell adhesion, indicating stimulation of inflammatory response (Figure S1F). We further validated the inhibitory effect of BMS-794833 on TAM polarization models prepared using bone marrow-derived cells (BMDCs) from BALB/c and C57BL/6J mice induced with CM from 4T1 and Py8119 tumors, respectively. In agreement with our previous results, BMDC-derived TAMs exhibit elongated morphology and induced expression of TAM-related anti-inflammatory genes, and BMS-794833 treatment suppressed both cellular elongation and expression of these genes (Figures S1G, H). Taken together, these results demonstrate that BMS-794833 identified through the phenotypic screen can impede TAMs from enacting their protumoral functions in vitro and re-program them into more pro-inflammatory phenotypes.
BMS-794833 treatment suppressed breast tumor growth
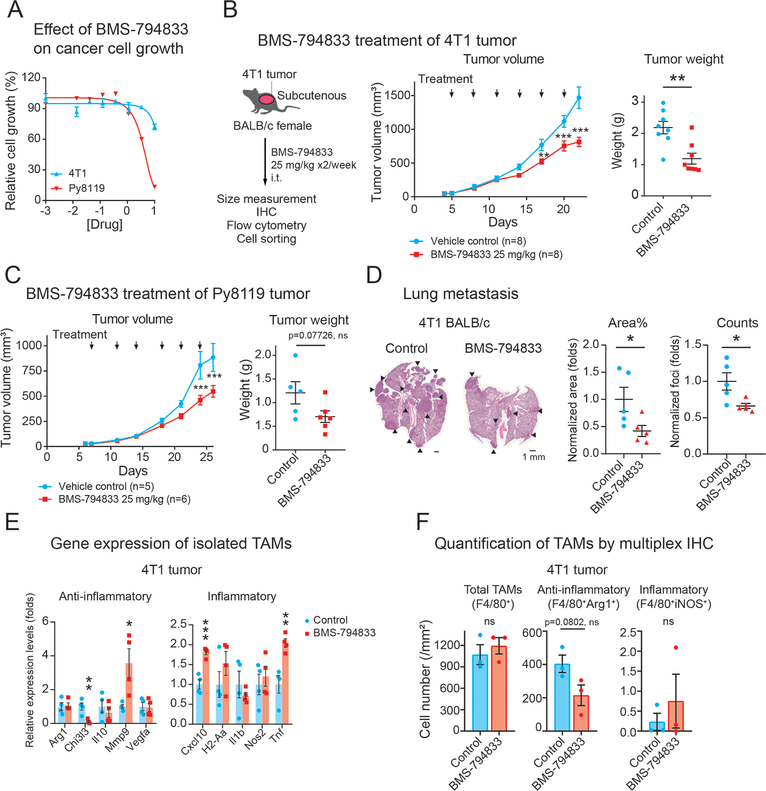
We next investigated the effect of BMS-794833 on primary breast tumor growth in mice. First, we evaluated the effect of BMS-794833 on the proliferation of breast cancer cells in monoculture. BMS-794833 inhibited cellular growth of 4T1 breast cancer cells by 28% at 10 μM (Figure 4A). Py8119 cells were slightly more sensitive, with IC50 at 4.1 μM and inhibition by 87% at 10 μM. These results showed that BMS-794833 has a modest inhibitory effect on breast cancer cell growth. Next, we administered BMS-794833 to 4T1 tumor transplanted subcutaneously in BALB/c mice. Biweekly treatment of BMS-794833 at 25 mg/kg dose intratumorally significantly reduced tumor weight by 52% compared to vehicle control without a decrease of body weight (Figure 4B, Figure S2A). The tumor-suppressive effect of BMS-794833 on 4T1 was similar in immunodeficient NSG mice (Figure S2B). Reduction of tumor growth was also observed in Py8119 tumors grown subcutaneously in C57BL/6J mice with no obvious systemic toxicity (Figure 4C, Figure S2A). Immunohistochemistry of cleaved caspase-3 in BMS-794833-treated tumors revealed that BMS-794833-treatment caused an increased trend of apoptosis (Figure S2C). In addition, BMS-794833-treated 4T1 tumors developed fewer microvessels (Figure S2D). The BMS-794833 treatment also decreased lung metastasis in the 4T1 tumor model (Figure 4D). Py8119-bearing mice did not develop visible lung metastatic foci in either control or treated tumors within the experimental period. These results demonstrate that BMS-794833 suppresses tumor growth and metastasis, and tumor suppression is likely due to its effect on cancer and stromal cells.
Figure 4. BMS-794833 treatment suppressed breast tumor growth.
(A) Effect of BMS-794833 on the proliferation of TNBC cell lines, 4T1 and Py8119 cultured in a plate. 4T1-nucGFP or Py8119 were cultured under the presence of a serial dose of BMS-794833. Cell proliferation of 4T1 was evaluated by nuclear GFP count, and of Py8119 by cell confluency. N=3, with mean ± SEM.
(B) Tumor growth of 4T1 tumors treated with BMS-794833. (Left) A schematic of experimental design. BALB/c female mice with subcutaneous 4T1 tumor were treated biweekly with either BMS-794833 (25 mg/kg) or vehicle control with intratumoral injections. (Middle) Growth curves of 4T1 tumors. The timing of drug administration is indicated with black arrows. n=8. (Right) Tumor weight at the experimental endpoint. Mean ± SEM.
(C) Tumor growth of Py8119 tumors treated with BMS-794833. (Left) Growth curves of Py8119 tumors. CB57BL/6J female mice with subcutaneous Py8119 tumor were treated biweekly with either BMS-794833 (25 mg/kg) or vehicle control with intratumoral injections. The timing of drug administrations is indicated with black arrows. n=5 (control), n=6 (treatment). (Right) Tumor weight at the experimental endpoint. Mean ± SEM.
(D) (Left) Representative images of H&E staining of lung sections from BMS-794833-treated or vehicle-treated 4T1 tumor-bearing mice. Black arrows indicate metastatic tumors. (Middle) Normalized percentage of the tumor area and (right) the normalized number of metastatic foci in the lung sections. The lines indicate the mean ± SEM.
(E) Gene expression of TAMs (CD45+CD11b+Ly6C−Ly6G−F4/80+) isolated from 4T1 tumor treated with BMS-794833. Expression of inflammatory and anti-inflammatory genes in the sorted TAM population (the gate is shown in Figures S2E, F) was measured by qPCR.
(F) Quantification of inflammatory and anti-inflammatory TAMs by fluorescent multiplex IHC. Total and subtypes of TAMs in the representative 4T1 tumors with or without BMS-794833 treatment from Figure 4B were quantified by multiplex IHC using antibodies against F4/80, Arg1, and iNOS. Cells were counted based on nuclear segmentation and were normalized to the analyzed surface area. The graph represents mean ± SEM with individual data points. *p<0.05, **p<0.01, **p<0.001, multiple t-test at each timepoint with Holm-Sidak correction (α<0.05) for tumor growth of (B) and (C), unpaired two-tailed Student’s t-test ((B, right), (C, right), (D, right), (E), (F)).
BMS-794833 abrogates TAM polarization to retain inflammatory phenotypes
To investigate whether BMS-794833 treatment impacts the profiles and characteristics of TAMs and other immune cells, we analyzed 4T1 and Py8119 tumors treated or untreated with BMS-794833 by flow cytometry (Figure S3A). BMS-794833 caused a significant decrease in live cells (Figure S3B), consistent with cleaved caspase-3 staining observed in tumor sections (Figure S2C). In both tumor models, BMS-794833 treatment did not change the percentage of overall immune populations (CD45+ cells) (Figure S3C), myeloid populations (CD45+CD11b+ cells) and their most marker expressions (Figures S3D–H), and most lymphoid populations (Figure S3I). TAM population (CD45+CD11b+Ly6G−Ly6C−F4/80+) exhibited a slight decrease under treatment but was not statistically significant (Figure S3E). The most prominent change in population was in monocytes, defined by Ly6C immature marker, which was significantly increased in 4T1 and less significantly in the Py8119 model (Figure S3E). Blood-derived monocytes are a major source of TAMs, continuously replenishing TAM in tumor tissues (33). An increase in monocytes may be due to enhanced monocyte recruitment by tissue-regenerative responses induced by cellular death, increased chemoattractants in cancer and stromal cells (such as CCL2 and MIP-1α/β increased in THP-1-derived TAM by BMS-794833 treatment (Figures 3E, S1D)), or inhibition of polarization into TAMs. To investigate whether BMS-794833 causes incomplete TAM polarization, surface markers of THP-1-derived TAMs treated or untreated with BMS-7948333 were analyzed. Untreated TAMs exhibited increased expression of CD11b (a myeloid cell marker), CD68 (a pan-macrophage marker), and CD14 (monocytes/macrophage marker) compared to the parental THP-1 cells (Figure S2E). BMS-794833 treated TAMs retained a similar level of CD11b+ cells, but decreased CD68 and CD14 expression at the cell surface. These results suggest that the BMS-794833 has inhibitory roles on complete differentiation/polarization of monocytes to TAM.
Next, we investigated gene expression of TAMs and their precursor cells, monocytes, in BMS-794833-treated tumors. The TAM and Ly6C+ monocyte populations of 4T1 and Py8119 tumors were isolated through FACS (Figures S2F, G). Gene expression analyses of TAMs isolated from BMS-794833-treated 4T1 tumors significantly upregulated inflammatory marker genes (Cxcl10, Tnf), and suppressed protumoral gene expression (Chi3l3) (Figure 4E). Ly6C+ monocytes from BMS-794833-treated 4T1 tumors exhibited a similar decrease in the anti-inflammatory Chi3l3 and increase in inflammatory markers, but to a lesser extent (Figure S2H). In the Py8119 tumors, the gene expression of TAM and Ly6C+ monocytes were less significant; neither TAM nor monocytes of Py8119 tumor induce TNFα, but notably, TAM increased expression of Nos gene, which encodes inducible NO synthase causing cytotoxicity (Figure S2H). These results demonstrate that BMS-794833 induced inflammatory phenotypes in intratumoral TAMs and Ly6C+ monocytes. We further analyzed the distribution and activation status of TAMs by multiplex immunohistochemistry (Figure 4F). TAMs in both treated and untreated tumors were distributed throughout the tissues as clusters. Total TAMs (F4/80+), anti-inflammatory (F4/80+Arg1+) and inflammatory (F4/80+iNOS+) TAMs were quantified within the intact tissue area. The total TAM distribution remained at the same level, and inflammatory TAMs were scarce, but anti-inflammatory TAMs showed a decreasing trend in BMS-794833 treated tumors (Figure 4F). These results further support that BMS-794833 suppressed anti-inflammatory phenotypes of TAMs in vivo.
BMS-794833 suppresses TAM polarization through polypharmacological effect
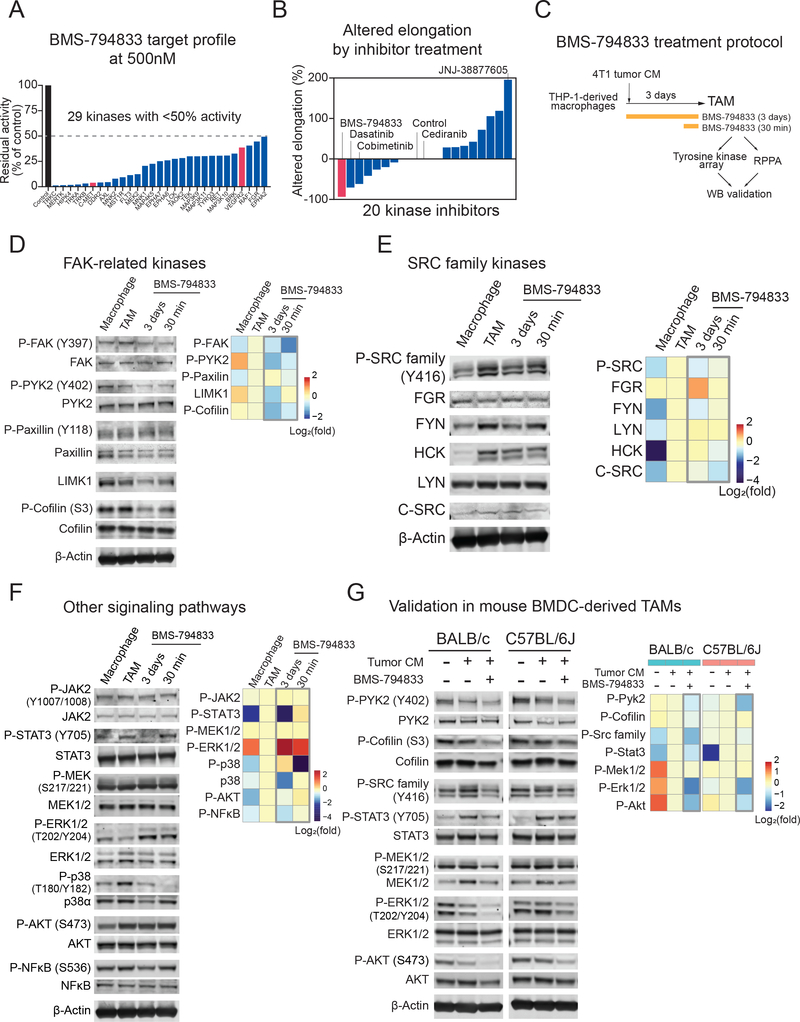
We sought to investigate the mechanism by which BMS-794833 impairs TAM polarization. Although c-MET and VEGFR2 are considered to be two main targets of BMS-794833, in vitro target profiling revealed that BMS-794833 inhibited 29 kinases to below 50% residual activity at 0.5 μM (Figure 5A, Figure S4A, Table S2). To examine whether c-MET and VEGFR2 are responsible for inhibition in TAM polarization, we first tested selective inhibitors for c-MET and VEGFR2, JNJ-38877605 and cediranib, respectively, on the TAM model. Neither compound inhibited TAM elongation (Figure S4B), suggesting that the inhibitory effect of BMS-794833 results from inhibition of either another single target or a polypharmacological effect on multiple targets. To test this further, we also inhibited the top 18 kinases targeted by BMS-794833 using selective inhibitors, but a majority of these inhibitors were not as potent as BMS-794833 (Figure 5B). Only inhibitors chemically-analogous to BMS-794833 showed a similar level (cabozantinib) or less (BMS-777607) inhibition on cellular elongation during TAM polarization (Figure S4C). Cabozantinib is newly recommended as standard-of-care combined with nivolumab for renal cell carcinoma (34). We validated that CM from human renal carcinoma, ACHN cell line, polarized THP-1-derived TAMs (Figure 1D), and cabozantinib inhibited cellular elongation but was less potent than BMS-794833 (Figure S4D). These results suggest that targeting a single kinase is insufficient to suppress TAM elongation, and that BMS-794833 was the most potent inhibitor to inhibit multiple signaling pathways to effectively block TAM polarization.
Figure 5. BMS-794833 targets multiple signaling pathways in TAMs.
(A) Target kinase profile of BMS-794833. Residual activities of kinases under the presence of BMS-794833 at 0.5 μM were evaluated in in vitro acellular system with synthetic substrates (17). Also, see Figure S4A and Table S2.
(B) Changes in cellular elongation caused by inhibitors targeting the top BMS-794833-targeted 18 kinases to below 30% at 0.5 μM. A total of 20 inhibitors, including BMS-794833 as a comparison and a VEGFR2 inhibitor cediranib are shown. Inhibitors are plotted based on the maximum altered elongation observed at below 10 μM dose. Inhibitors that caused cell death were regarded as no change in elongation.
(C) Experimental design for protein analysis of BMS-794833-treated TAM. Macrophages prepared from THP-1 were polarized with 4T1 tumor CM for 3 days. BMS-794833 was supplemented at 7.5 μM final concentration simultaneously with CM addition, or 30 min before sample collection on day 3. The resulting TAMs were analyzed with protein arrays followed by validation by western blotting.
(D-G) Detection of phosphorylated and total protein amount by immunoblot. (Left) Representative images of the blot. (Right) Heatmap showing quantification of signal intensities. The log2 fold change of normalized signal intensity is shown.
(D) Phosphorylation of FAK and cytoskeleton-related proteins. Signal intensity was normalized to either total protein or β-actin (LIMK1).
(E) Phosphorylation of SRC family kinases. The antibody for phosphorylated SRC family kinases detects all SRC family kinases. The signal intensity was normalized signal intensity to β-actin.
(F) Phosphorylation of STAT3, MAPKs, AKT, and NFκB. The signal intensity was normalized to either total protein or β-actin.
(G) Kinase phosphorylation of of mouse BMDC-derived in vitro TAM model cells. BALB/c or C57BL/6J-derived BMDC were induced with M-CSF for 2 days, followed by 4T1 (BALB/c) or Py8119 (C57BL/6J) tumor CM supplemented with M-CSF for 5 days, with or without BMS-794833 at 7.5 μM. The signal intensity was normalized to total protein.
Inhibitory effect of BMS-794833 on time-dependent signal activation during TAM polarization
To characterize the effect of BMS-794833 further, we performed a phosphorylated tyrosine kinase array (Figure S4E, F) and reverse-phase protein array (RPPA) screening followed by WB validation (Figure 5C). TAMs polarized with 4T1 CM were treated with BMS-794833 for full-time (3 days) or the last 30 min of the polarization period. BMS-794833 treatment decreased phosphorylation of proteins related to focal adhesion-cytoskeleton regulation, SRC family kinases, JAK-STAT3, p38 MAP kinase, AKT, and NF-κB (Figures 5D, E, F). The results also revealed differential response at the time of inhibition by BMS-794833; phosphorylation of FAK, PYK, cofilin, and p38 MAPK can be inhibited at the last 30 minutes of the polarization period, whereas total LIMK1, phosphorylated STAT3, AKT, and NFκB were downregulated only upon full-time treatment. In detail, phosphorylated FAK, PYK2, and cofilin decreased within 30 min and remained low after 3 days of BMS-794833 treatment, while the total protein level of LIMK1, an upstream kinase that phosphorylates cofilin (35), decreased after 3 days (Figure 5D). Among the 9 SRC family kinases (36), THP-1-derived TAMs expressed HCK, FGR, LYN, FYN, and a low amount of c-SRC (Figure 5E). Phosphorylation of total SRC family kinases was induced in TAM polarization and decreased by 3-days BMS-794833 treatment (Figure 5E). Phosphorylated STAT3, a transcription regulator of immunosuppressive cytokines, MMPs, and angiogenesis factors (7) was decreased significantly after long-term BMS-794833 treatment (Figure 5F). The phosphorylation level of p38 MAPK, a conditional regulator of pro- or anti-inflammatory response (37), dropped within 30 min and also decreased total protein amount after 3 days. Phosphorylated ERK1/2 was increased by BMS-794833 treatment within 30 min and remain high on 3 days (Figure 5F). However, in the validation experiment with mouse BMDC-derived TAMs, phosphorylation of Erk1/2 and Mek1/2 decreased, indicating that the increase of phosphorylated ERK is a THP-1-specific observation (Figure 5G). Downregulation of phosphorylated Src family, Pyk2, Cofilin, Stat3, and Akt by BMS-794833 were validated in the mouse BMDC-derived TAM model (Figure 5G).
As TAMs dynamically regulate signaling activation over time, we further investigated time-dependent activation of signaling pathways during TAM polarization, leveraging the advantage of our in vitro system (Figure S4G). We observed an increase in the phosphorylation of AKT, ERK1/2, and p38 MAPK within one hour in response to 4T1 tumor CM, followed by an increase in the phosphorylation of NF-κB, which peaked at 24 hours, indicating activation of these proteins is an early event in TAM polarization. In contrast, phosphorylation of FAK, cofilin, SRC family, and STAT3 was increased from day 2, indicating that these proteins are activated in a late event. Importantly, BMS-794833 treatment suppressed induction of p38 MAPK, NF-κB, FAK, Cofilin, SRC family, and STAT3, suggesting that BMS-794833 targets both early and late-stage protein signaling during TAM polarization. Together, these data highlight temporal activation of multiple pathways during TAM polarization, and the effect of BMS-794833 on these signal cascades.
Systems-based analysis of TAM polarization and BMS-794833 treatment
TAMs exhibit dynamic changes in gene expression and protein phosphorylation during the polarization period (Figures 1, 3, 5). To evaluate relative contributions of each change to TAM polarization and to the response to BMS-794833, we performed partial least square regression (PLS-R) analysis of time-course gene expression and protein phosphorylation of TAMs treated or untreated with BMS-794833, with cellular elongation as a response (Figure S5). Component 1 (40% explained variance) delineated time-dependent TAM polarization, and component 2 (19% explained variance) discriminated BMS-794833 treated and untreated groups (Figure S5A). The top 30 standardized coefficient of component 2 includes positive coefficients (towards BMS-794833 treated group) of inflammatory cytokine-coding genes and negative coefficients (towards untreated group) of phosphorylated STAT3, cofilin, p38 MAPK, and total LIMK1 (Figure S5B, C). These results indicate that the effect of BMS-794833 can be explained by both increases in inflammatory cytokine expression and a decrease in phosphorylated/total proteins involve in TAM polarization.
Targeting multiple signaling pathways is necessary for abrogating TAM polarization
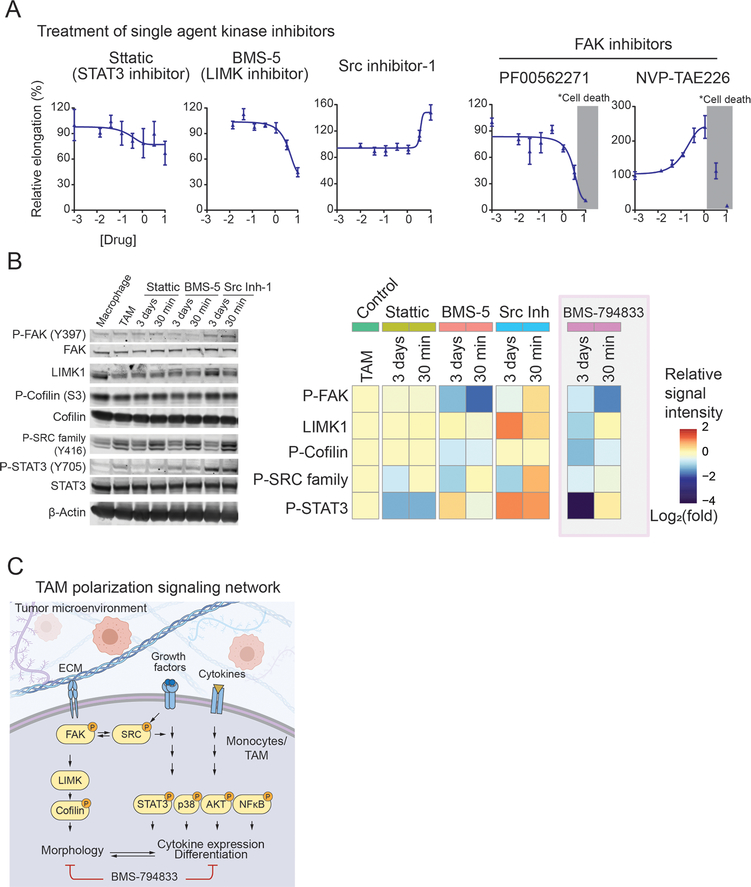
To investigate the contribution of the individual signaling pathway in TAM polarization, we tested selective inhibitors for FAK, LIMK1, SRC family, and STAT3 (Figure 6A). Two potent FAK inhibitors (PF00562271 and NVP-TAE226) showed an inconsistent effect on TAM elongation and did not provide conclusive evidence for the importance of FAK. A highly-selective LIMK inhibitor BMS-5 inhibited cell elongation, consistent with the role of LIMK1 in actin polymerization. LIMK1 is predominantly expressed in the TAM population of breast cancer patient tumors (Figure S6A). BMS-5 treatment upregulated secretion of inflammatory cytokines (Figure S6B) and lowered the growth stimulation effect of TAMs (Figure S6C). However, BMS-5 did not show any tumor-suppressive effect on 4T1 tumor-bearing mice (Figure S6D). Western blot analyses revealed that BMS-5 suppresses phosphorylation of FAK, cofilin, and SRC family kinases, but phosphorylation of STAT3 remained unchanged (Figure 6B). A STAT3 inhibitor, Stattic (38), had a modest effect on TAM elongation at 10 μM (Figure 6A). The immunoblot confirmed suppressed phosphorylation of STAT3 (Figure 6B), and also showed decreased phosphorylated SRC. However, phosphorylation of FAK, cofilin, and total LIMK remained unchanged. Src inhibitor-1, which inhibits most SRC family kinase, stimulated cellular elongation (Figure 6A). Immunoblot showed that Src inhibitor-1 induces phosphorylated SRC, FAK, and STAT3 within 30 min, and increased LIMK1 expression after 3 days (Figure 6B). The results indicate that SRC signal blockade results in compensatory induction of SRC and other TAM-related signals.
Figure 6. Targeting multiple signaling cascades is required for impeding TAM polarization.
(A) Dose-response curve of selective inhibitors for STAT3 (Stattic), LIMK1 (BMS-5), SRC family kinases (Src inhibitor-1), and FAK (PF-00562271 and NVP-TAE226) on TAM polarization using 4T1 tumor CM. Inhibitors were tested within 0 to 10 μM range and dose response was plotted in log10 scale. The concentration causing cellular death is shaded with gray. The error bars depict the mean ± SEM of 3 replicates.
(B) Immunoblots of TAM polarized in the presence of Stattic, BMS-5, and Src Inhibitor-1 (Src Inh-1). Inhibitors were administered to TAM polarization culture at the time of 4T1 tumor CM induction starts (3 days), or the last 30 min before sample collection at day 3 (30 min) at 10 μM concentration. (Right) Heatmap showing quantification of signal intensities. The log2 fold change of normalized signal intensity to either total protein or β-actin is shown. Also, the heatmap of BMS-794833 WB signal intensities from Figure 5 are included for comparison.
(C) A schematic showing signaling pathways activated during TAM polarization. Pathways inhibited by BMS-794833 are shown.
As both BMS-5 and Stattic could inhibit TAM elongation with complementary inhibition of phosphorylated cofilin and STAT3, we tested whether the combination treatment of BMS-5 and Stattic potentiate inhibitor effect on TAM polarization. A combination of BMS-5 and Stattic suppressed TAM elongation more potently than either BMS-5 or Stattic alone at the same dose (5 μM, Figure S6E). The Bliss combination index (39) was 1.02, suggesting that combination treatment resulted in an additive effect. These results indicate that targeting both LIMK1actin-related signal and STAT3 better suppress TAM polarization.
Overall, the results above suggest that in vitro TAM polarization assay system enables investigations of the state of signaling pathways during polarization, and our findings indicate targeting a single kinase/pathway is not sufficient to block TAM polarization. The results reiterate that the potency of BMS-794833 is due to its multi-targeting of SRC, actin regulation, and STAT3, inactivating surrogate pathways that support the plastic nature of TAMs.
Discussion
TAMs receive aggregated signals in the form of growth factors, cytokines, chemokines, extracellular vesicles, and metabolites accumulated in TME. How TAMs process this multitude of signals to alter their morphology and function to support the tumor growth and spread remains poorly understood. Current in vitro model systems that rely on cytokine-supplemented cultures, such as IL-4 and IL-13, do not adequately recapitulate the complex nature of TME to study TAM function (40). To overcome this limitation, we used media collected from tumor tissues, tumor conditioned medium (CM), to induce a complex signaling and phenotypic response through a mixture of secreted factors contained in the CM. Although our in vitro system does not model specific cellular interactions that may impact TAM polarization, we confirmed that tumor CM was sufficient to induce pro-tumoral phenotypes known in TAMs. The monocyte cell line and cancer CM-based system provided a relatively uniform and accessible source for TAMs that are compatible with phenotypic drug screening and functional characterization. Using this model system, we could demonstrate interactions between in vitro-generated TAMs and (i) cancer cells that led to increased cancer cell growth and motility, (ii) endothelial cells leading to the promotion of microvessel formation, and (iii) T cells causing inhibition of T cell migration. Collectively, we present a physiologically-relevant model system to study TAMs.
To document the signaling cascades that regulate complex TAM function, we performed molecular characterization of our in vitro-derived TAMs. We observed alterations of both gene expression and phosphorylation of proteins previously known to be involved in TAM function, including VEGFA, FN1, CXCR4, IL1B(8), CCL20(41), phosphorylated STAT3(42), AKT (43), and NF-κB (44). The in vitro systems traced the TAM polarization process and allowed us to study time-dependent changes in multiple signaling pathways during the polarization (Figure S4G). Pro-inflammatory transcription factor NF-κB was phosphorylated on day 1, which was then downregulated at later time points, suggesting that tumor CM first induces inflammatory responses in TAMs then shifts towards anti-inflammatory phenotypes. In contrast, STAT3 was unchanged in earlier timepoints but phosphorylated at later timepoints. These results describe serially programmed signaling cascades during TAM polarization and suggest that re-programming this process may offer an opportunity for inhibiting the pro-tumor activity of TAMs.
A combination of rich and uniform sources of in vitro-generated TAMs and robust and quantitative cellular elongation phenotype provides a reliable assay for high-throughput screening. To identify compounds that could compromise TAM function, we carried out a chemical inhibitor screen using a set of 85 kinase inhibitors covering most of the kinome. We identified BMS-794833 as a potent suppressor of cellular elongation in multiple cancer models, and suppresses protumoral activities of TAMs in vitro, underscoring the potential utility of our model system for phenotypic-based screening in drug discovery. The mechanism of action of BMS-794833 involves inhibition of phosphorylation status of multiple proteins, including FAK, SRC, p38 MAPK, and STAT3 (Figure 5, 6C). When administered to breast tumor-bearing mice, BMS-794833 could suppress tumor growth and metastasis (Figures 4B–D). Although BMS-794833 programs TAMs towards more anti-tumoral phenotypes in vivo (Figures 4E, F S3E), the tumor-suppressive effect of BMS-794833 seems primarily derived from tumoricidal activity and effects on non-immune stromal cells, including endothelial cells (Figures 4B, D, S2B–D). Thus, a single agent, like BMS-794833, that targets multiple cell types in the tumor microenvironment could be desirable in clinical settings.
Several TAM-targeting therapies are currently in clinical studies, including CSF1-CSF1R, either as monotherapy or in combination with conventional treatment to augment therapeutic effects (45,46). However, clinical studies have shown that monotherapies exhibit moderate effects (47), indicating the limited efficacy of targeted therapies in a complex environment. Consistently, our data showed that targeting single kinases was ineffective in inhibiting cellular elongation-based TAM polarization (Figure 6A) while BMS-794833, which exhibits broad polypharmacology, effectively inhibits TAM polarization (Figure 3D). Collectively, these data suggest simultaneous targeting of multiple signaling pathways in TAM polarization yields more effective treatment options than the use of more selective agents.
Overall, our study highlights the complex interplay of macrophage polarization with components of the TME. Broadly, our study provides the cancer community with a platform that allows analyses of TAMs under near-physiological conditions. As we enter a new era of data-rich cancer biology, one of the primary challenges is integrating the knowledge of cellular and molecular information into a holistic understanding of cancer as a biological system. Quantitative assays and technologies that enable pharmacological assessment with cellular and molecular phenotypes in the physiologically relevant environment, like the one described in this study, will form an essential component of system-based investigations.
Supplementary Material
Statement of Significance.
A physiologically relevant in vitro system of tumor-associated macrophage polarization uncovers signaling pathways that regulate polarization and identifies strategies to target macrophage reprogramming to suppress cancer growth.
Acknowledgments
This work was supported by grants from the Breast Cancer Research Foundation [BCRF 17-035] and the American Cancer Society (133870-RSG-19-197-01-CDD). NNA is a recipient of the Fred Hutch Interdisciplinary Training Grant in Cancer Research and the Japan Society for the Promotion of Science Overseas Research Fellowship. This research was supported by the Comparative Medicine, Scientific Imaging, and Flow Cytometry, and Experimental Histopathology Shared Resources of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704). We thank Drs. Milka Kostic, Thomas Bello, and Aleena Arakaki, and members of the Gujral lab for helpful comments and suggestions for the manuscript.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 2018;17:887–904 [DOI] [PubMed] [Google Scholar]
- 2.Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduction and Targeted Therapy 2021;6:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung KY, Cho SW, Kim YA, Kim D, Oh B-C, Park DJ, et al. Cancers with higher density of tumor-associated macrophages were associated with poor survival rates. Journal of pathology and translational medicine 2015;49:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol 2002;196:254–65 [DOI] [PubMed] [Google Scholar]
- 5.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34 [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Qu J, Sun Y, Wang J, Liu X, Wang F, et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget 2017;8:30576–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irey EA, Lassiter CM, Brady NJ, Chuntova P, Wang Y, Knutson TP, et al. JAK/STAT inhibition in macrophages promotes therapeutic resistance by inducing expression of protumorigenic factors. Proc Natl Acad Sci U S A 2019;116:12442–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018;174:1293–308 e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res 2010;70:5728–39 [DOI] [PubMed] [Google Scholar]
- 10.Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell 2014;25:605–20 [DOI] [PubMed] [Google Scholar]
- 11.Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, et al. Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J Immunol 2010;185:642–52 [DOI] [PubMed] [Google Scholar]
- 12.Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, et al. Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc Natl Acad Sci U S A 2017;114:580–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabanel M, Brand C, Oliveira-Nunes MC, Cabral-Piccin MP, Lopes MF, Brito JM, et al. Epigenetic Control of Macrophage Shape Transition towards an Atypical Elongated Phenotype by Histone Deacetylase Activity. PLoS One 2015;10:e0132984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benner B, Scarberry L, Suarez-Kelly LP, Duggan MC, Campbell AR, Smith E, et al. Generation of monocyte-derived tumor-associated macrophages using tumor-conditioned media provides a novel method to study tumor-associated macrophages in vitro. J Immunother Cancer 2019;7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida-Aoki N, Bondesson AJ, Gujral TS. Measuring Real-time Drug Response in Organotypic Tumor Tissue Slices. J Vis Exp 2020 [DOI] [PubMed] [Google Scholar]
- 16.Sivakumar R, Chan M, Shin JS, Nishida-Aoki N, Kenerson HL, Elemento O, et al. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. Oncoimmunology 2019;8:e1670019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rata S, Gruver JS, Trikoz N, Lukyanov A, Vultaggio J, Ceribelli M, et al. An optimal set of inhibitors for Reverse Engineering via Kinase Regularization. bioRxiv 2020:2020.09.26.312348 [Google Scholar]
- 18.DeRose YS, Gligorich KM, Wang G, Georgelas A, Bowman P, Courdy SJ, et al. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol 2013;Chapter 14:Unit14 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vereyken EJ, Heijnen PD, Baron W, de Vries EH, Dijkstra CD, Teunissen CE. Classically and alternatively activated bone marrow derived macrophages differ in cytoskeletal functions and migration towards specific CNS cell types. J Neuroinflammation 2011;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porcheray F, Viaud S, Rimaniol AC, Leone C, Samah B, Dereuddre-Bosquet N, et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol 2005;142:481–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu G, Su Y, Kang BH, Fan Z, Dong T, Brown DR, et al. High-throughput phenotypic screen and transcriptional analysis identify new compounds and targets for macrophage reprogramming. Nat Commun 2021;12:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollmen M, Roudnicky F, Karaman S, Detmar M. Characterization of macrophage--cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Sci Rep 2015;5:9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grugan KD, McCabe FL, Kinder M, Greenplate AR, Harman BC, Ekert JE, et al. Tumor-associated macrophages promote invasion while retaining Fc-dependent anti-tumor function. J Immunol 2012;189:5457–66 [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Qiu X, Li J, Zheng S, Li L, Zhao H. TGF-beta secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle 2018;17:2766–78 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zhang S, Che D, Yang F, Chi C, Meng H, Shen J, et al. Tumor-associated macrophages promote tumor metastasis via the TGF-beta/SOX9 axis in non-small cell lung cancer. Oncotarget 2017;8:99801–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunderson AJ, Yamazaki T, McCarty K, Fox N, Phillips M, Alice A, et al. TGFbeta suppresses CD8(+) T cell expression of CXCR3 and tumor trafficking. Nat Commun 2020;11:1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eid S, Turk S, Volkamer A, Rippmann F, Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics 2017;18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards JP, Emens LA. The multikinase inhibitor sorafenib reverses the suppression of IL-12 and enhancement of IL-10 by PGE(2) in murine macrophages. Int Immunopharmacol 2010;10:1220–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprinzl MF, Puschnik A, Schlitter AM, Schad A, Ackermann K, Esposito I, et al. Sorafenib inhibits macrophage-induced growth of hepatoma cells by interference with insulin-like growth factor-1 secretion. J Hepatol 2015;62:863–70 [DOI] [PubMed] [Google Scholar]
- 31.Deng YR, Liu WB, Lian ZX, Li X, Hou X. Sorafenib inhibits macrophage-mediated epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget 2016;7:38292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fargnoli J, Henley BJ, Wautlet BS, Borzilleri R. 106 Preclinical studies and characterization of BMS-794833, a small molecule inhibitor of Met and VEGFR-2 kinases. Eur J Cancer Suppl 2010;8:41 [Google Scholar]
- 33.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011;475:222–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choueiri TK, Powles T, Burotto M, Escudier B, Bourlon MT, Zurawski B, et al. Nivolumab plus Cabozantinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2021;384:829–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 1998;393:809–12 [DOI] [PubMed] [Google Scholar]
- 36.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol 2009;6:587–95 [DOI] [PubMed] [Google Scholar]
- 37.Raza A, Crothers JW, McGill MM, Mawe GM, Teuscher C, Krementsov DN. Anti-inflammatory roles of p38alpha MAPK in macrophages are context dependent and require IL-10. J Leukoc Biol 2017;102:1219–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006;13:1235–42 [DOI] [PubMed] [Google Scholar]
- 39.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 2015;3:e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu K, Lin K, Li X, Yuan X, Xu P, Ni P, et al. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front Immunol 2020;11:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samaniego R, Gutierrez-Gonzalez A, Gutierrez-Seijo A, Sanchez-Gregorio S, Garcia-Gimenez J, Mercader E, et al. CCL20 Expression by Tumor-Associated Macrophages Predicts Progression of Human Primary Cutaneous Melanoma. Cancer Immunol Res 2018;6:267–75 [DOI] [PubMed] [Google Scholar]
- 42.Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat Rev Cancer 2019;19:82–96 [DOI] [PubMed] [Google Scholar]
- 43.Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol 2017;198:1006–14 [DOI] [PubMed] [Google Scholar]
- 44.Mancino A, Lawrence T. Nuclear factor-kappaB and tumor-associated macrophages. Clin Cancer Res 2010;16:784–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowal J, Kornete M, Joyce JA. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 2019;11:677–89 [DOI] [PubMed] [Google Scholar]
- 47.Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, Ruttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 2017;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.